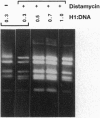
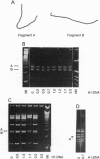
Abstract
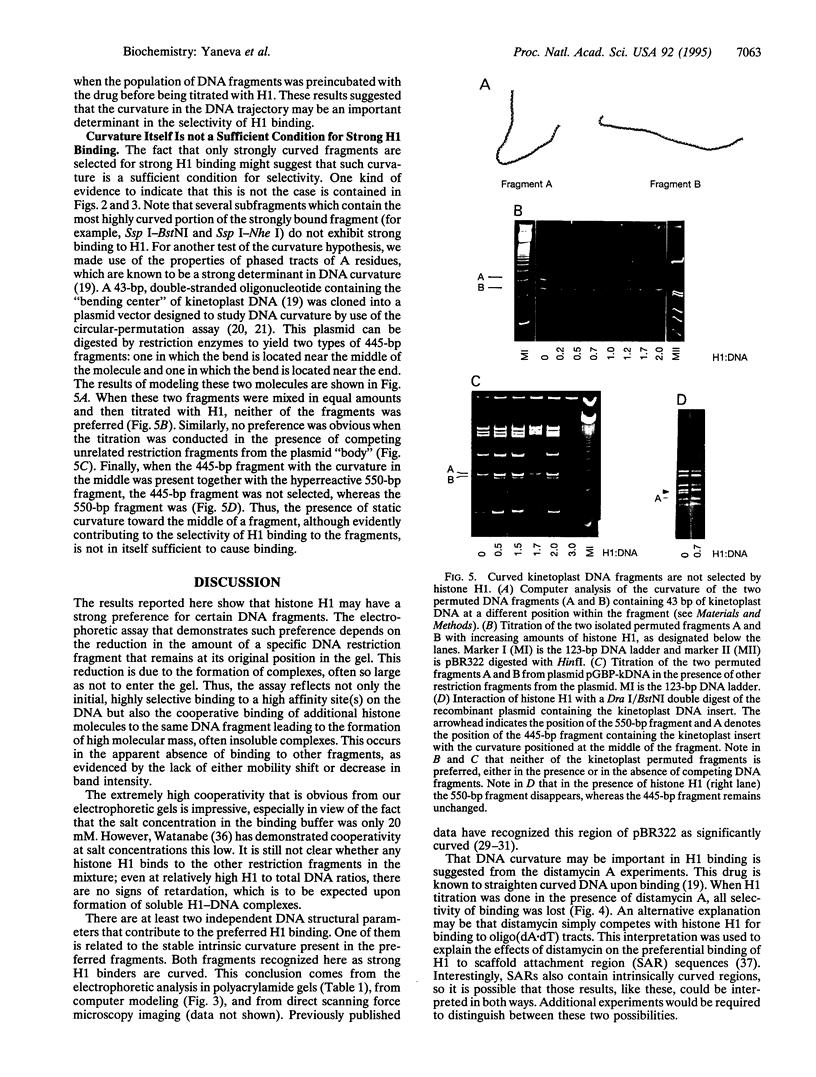
The interaction of histone H1 isolated from chicken erythrocytes with restriction fragments from plasmids pBR322 and pUC19 was studied by gel electrophoresis. Certain restriction fragments exhibited unusually high affinity for the histone, forming high molecular mass complexes at protein to DNA ratios at which the other fragments did not show evidence for binding. The highly preferred fragments are intrinsically curved, as judged by their electrophoretic mobility in polyacrylamide gels, by computer modeling, and by imaging with scanning force microscopy. However, control experiments with either curved portions of the same fragments or highly curved kinetoplast DNA fragments showed that the presence of curvature alone was not sufficient for preferential binding. By using various restriction fragments centered around the highly preferred sequence, it was found that the high-affinity binding required in addition the presence of specific sequences on both sides of the region of curvature. Thus, both curvature and the presence of specific sites seem to be required to generate high affinity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. N. Detection, sequence patterns and function of unusual DNA structures. Nucleic Acids Res. 1986 Nov 11;14(21):8513–8533. doi: 10.1093/nar/14.21.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchev T., Srebreva L., Zlatanova J. Purification of histone H10 and its subfractions under non-denaturing conditions. Biochim Biophys Acta. 1991 Jan 23;1073(1):230–232. doi: 10.1016/0304-4165(91)90208-x. [DOI] [PubMed] [Google Scholar]
- Bolshoy A., McNamara P., Harrington R. E., Trifonov E. N. Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2312–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Harrington R. E. Studies of DNA bending and flexibility using gel electrophoresis. Electrophoresis. 1993 Aug;14(8):732–746. doi: 10.1002/elps.11501401116. [DOI] [PubMed] [Google Scholar]
- Krylov D., Leuba S., van Holde K., Zlatanova J. Histones H1 and H5 interact preferentially with crossovers of double-helical DNA. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5052–5056. doi: 10.1073/pnas.90.11.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käs E., Izaurralde E., Laemmli U. K. Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA).oligo(dT) tracts. J Mol Biol. 1989 Dec 5;210(3):587–599. doi: 10.1016/0022-2836(89)90134-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leuba S. H., Yang G., Robert C., Samori B., van Holde K., Zlatanova J., Bustamante C. Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11621–11625. doi: 10.1073/pnas.91.24.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S., Travers A. A. DNA sequence organization in chromatosomes. J Mol Biol. 1994 Jan 21;235(3):855–870. doi: 10.1006/jmbi.1994.1044. [DOI] [PubMed] [Google Scholar]
- Muzard G., Théveny B., Révet B. Electron microscopy mapping of pBR322 DNA curvature. Comparison with theoretical models. EMBO J. 1990 Apr;9(4):1289–1298. doi: 10.1002/j.1460-2075.1990.tb08238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T., Nagumo M., Hirota Y., Sakuma S. Alteration of the curved helical structure located in the upstream region of the beta-lactamase promoter of plasmid pUC19 and its effect on transcription. Nucleic Acids Res. 1992 Apr 11;20(7):1617–1622. doi: 10.1093/nar/20.7.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radic M. Z., Lundgren K., Hamkalo B. A. Curvature of mouse satellite DNA and condensation of heterochromatin. Cell. 1987 Sep 25;50(7):1101–1108. doi: 10.1016/0092-8674(87)90176-0. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V., Finch J. T., Graziano V., Lee P. L., Sweet R. M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993 Mar 18;362(6417):219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Schroth G. P., Siino J. S., Cooney C. A., Th'ng J. P., Ho P. S., Bradbury E. M. Intrinsically bent DNA flanks both sides of an RNA polymerase I transcription start site. Both regions display novel electrophoretic mobility. J Biol Chem. 1992 May 15;267(14):9958–9964. [PubMed] [Google Scholar]
- Stellwagen N. C. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983 Dec 20;22(26):6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- Thoma F. Nucleosome positioning. Biochim Biophys Acta. 1992 Feb 28;1130(1):1–19. doi: 10.1016/0167-4781(92)90455-9. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. DNA in profile. Trends Biochem Sci. 1991 Dec;16(12):467–470. doi: 10.1016/0968-0004(91)90181-t. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P., Zlatanova J., Leuba S. H., Schroth G. P., van Holde K. Binding of histones H1 and H5 and their globular domains to four-way junction DNA. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3525–3529. doi: 10.1073/pnas.91.9.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P., van Holde K., Zlatanova J. Preferential binding of histone H1 to four-way helical junction DNA. J Biol Chem. 1993 Oct 5;268(28):20699–20700. [PubMed] [Google Scholar]
- Vogel T., Singer M. F. Interaction of f1 histone with superhelical DNA. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2597–2600. doi: 10.1073/pnas.72.7.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T., Singer M. F. The effect of superhelicity on the interaction of histone f1 with closed circular duplex DNA. J Biol Chem. 1976 Apr 25;251(8):2334–2338. [PubMed] [Google Scholar]
- Watanabe F. Cooperative interaction of histone H1 with DNA. Nucleic Acids Res. 1986 Apr 25;14(8):3573–3585. doi: 10.1093/nar/14.8.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellman S. E., Sittman D. B., Chaires J. B. Preferential binding of H1e histone to GC-rich DNA. Biochemistry. 1994 Jan 11;33(1):384–388. doi: 10.1021/bi00167a049. [DOI] [PubMed] [Google Scholar]
- Woodcock C. L., Grigoryev S. A., Horowitz R. A., Whitaker N. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9021–9025. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Yaneva J., Zlatanova J. Histone H1 interacts specifically with certain regions of the mouse alpha-globin gene. DNA Cell Biol. 1992 Mar;11(2):91–99. doi: 10.1089/dna.1992.11.91. [DOI] [PubMed] [Google Scholar]
- Yang G., Leuba S. H., Bustamante C., Zlatanova J., van Holde K. Role of linker histones in extended chromatin fibre structure. Nat Struct Biol. 1994 Nov;1(11):761–763. doi: 10.1038/nsb1194-761. [DOI] [PubMed] [Google Scholar]
- Zlatanova J. Histone H1 and the regulation of transcription of eukaryotic genes. Trends Biochem Sci. 1990 Jul;15(7):273–276. doi: 10.1016/0968-0004(90)90053-e. [DOI] [PubMed] [Google Scholar]
- Zlatanova J., Van Holde K. Histone H1 and transcription: still an enigma? J Cell Sci. 1992 Dec;103(Pt 4):889–895. doi: 10.1242/jcs.103.4.889. [DOI] [PubMed] [Google Scholar]
- Zlatanova J., Yaneva J. DNA sequence specific interactions of histone H1. Mol Biol Rep. 1991 Feb;15(1):53–56. doi: 10.1007/BF00369901. [DOI] [PubMed] [Google Scholar]
- Zlatanova J., Yaneva J. Histone H1-DNA interactions and their relation to chromatin structure and function. DNA Cell Biol. 1991 May;10(4):239–248. doi: 10.1089/dna.1991.10.239. [DOI] [PubMed] [Google Scholar]