Abstract
Background
Re-emergence of pertussis has been reported in Iran despite a high rate of vaccination coverage. Low efficacy of the vaccine might be due to the genetic divergence between clinical versus vaccine strains. In the current study, the genetic profiles of clinical isolates and vaccine strains of Bordetella pertussis (B. pertussis) were assessed by using Pulsed Field Gel Electrophoresis (PFGE).
Methods
Following phenotypic and molecular identification of isolates, XbaI-digested genomic DNA of 5 clinical isolates, 2 vaccine strains and a Tohama I strain were analyzed by PFGE along with B. parapertussis as a control.
Results
Seven distinct PFGE profiles were found among all examined isolates/strains. In 5 clinical isolates, 4 profiles were identified whereas the vaccine strains displayed 2 distinct profiles. The reference strain, Tohama I had a distinct profile. Vaccine and clinical profiles had low similarity, with relatedness of approximately 40%.
Conclusion
The genetic profiles of B. pertussis were different between circulating isolates and vaccine strains used in the national vaccination programs. Since new genetic profiles of B. pertussis can be disseminated periodically, the profiles of isolates circulating in the population should be monitored over the course of the re-emergence.
Keywords: Bordetella pertussis, PFGE profile, Vaccination, Whooping cough
Introduction
Bordetella pertussis is a strict human pathogen causing severe contagious respiratory infection, whooping cough or pertussis (1). Pertussis remains one of the ten leading causes of death from infectious diseases among children (2). The development of whole-cell pertussis (wP) vaccines since 1950s, and the broad use of this vaccine has greatly reduced the incidence of pertussis in developed and developing countries. Despite the Expanded Program of Immunization (EPI), pertussis has not been eradicated and the re-emergence of the disease is reported worldwide (3). Re-emergence of pertussis is attributed to several factors such as waning of vaccine-induced immunity, pathogen adaptation, antigenic divergence between vaccine strains and circulating clinical isolates, and the resurgence of new variants of B. pertussis (4–6).
According to the World Health Organization (WHO) reports, the diphtheria toxoid, tetanus toxoid and pertussis (DTP) vaccine coverage is estimated to be greater than 96% in Iran since the year 2000 (7). However, the pertussis cases increased from 464 in 2010 to 650 in 2011 and the national vaccination programs had a limited impact on the incidence of pertussis in various districts of the country in recent years (8). One study in assessment of the immune response against B. pertussis in Iranian children showed that half of the vaccinated children aged 8 months to 6 years did not display protective antibody levels to pertussis antigens (9). Evaluation of the immunogenicity of Diphtheria, Tetanus and whole-cell Pertussis (DTwP) vaccine used in Iran also demonstrated lower immunogenicity against pertussis compared to diphtheria and tetanus (10). Moreover, the majority of reported pertussis cases in 2009-2010 were children vaccinated by DTwP (11).
Since one reason for low efficacy of the vaccine might be the genetic divergence between vaccine and clinical strains, studies should be designed to characterize the genomic pattern of the circulating isolates and vaccine strains of B. pertussis. The aim of this study was to determine the genetic profiles of clinical isolates and vaccine strains of B. pertussis using a golden standard typing method with high discriminatory power, PFGE.
Materials and Methods
Isolates/strains and growth condition
A total of 11 suspected clinical isolates of B. pertussis named Bp1-11 (Pertussis Reference Laboratory, Pasteur Institute of Iran, Tehran, Iran), Tohama I (ATCC® BAA-589™) as reference strain, and 2 vaccine strains (Department of Aerobic Bacterial Vaccines, Razi Vaccine and Serum Research Institute) were studied. B. parapertussis (ATCC:15311) was included as a control. Two vaccine strains used in this study were Bp509, and Bp134. These two strains have been used for production of DTwP vaccine in Iran. Two subcultures of Bp509 strain, including Bp509/1997 and Bp509/2000 were available. These isolates were prepared and frozen in 1997 and 2000 from the original stock of Bp509. Frozen bacteria were cultured onto Bordet-Gengou Agar (BGA) (Becton Dickinson, USA) supplemented with 1% glycerol, peptone, and 20% defibrinated sheep blood and were incubated at 35°C for 72 hr.
Phenotypic identification
Colonies were identified as B. pertussis by a combination of colony morphology, growth rate, Gram stain, and biochemical tests such as oxidase, catalase, urease, nitrate reductase, and lack of growth on the MacConkey and blood agar (12).
Molecular identification
Genomic DNA extraction was performed using the standard phenol-chloroform extraction method (13). Specific primers targeting insertion sequence 481 (IS481) and pertussis toxin promoter (ptxP) were used to amplify 181 base pair (bp) and 573 bp products (14, 15), respectively (Table 1). Amplification of IS481 region was carried out in a total reaction volume of 20 µl containing 2 µl 10× PCR buffer (CinnaGen co., Iran), 1.5 mM MgCl2 (CinnaGen co., Iran), 0.2 mM deoxynucleotide (CinnaGen co., Iran), 0.25 mM of each primer (Bioneer, Seoul, South Korea), 0.5 U Taq polymerase (CinnaGen co., Iran), and 10 ng DNA. The Thermocycler (Peqlab, Germany) was set with the following conditions: Initial denaturation for 5 min at 95 °C and 30 cycles including denaturation for 30 s at 95°C, annealing for 30 s at 54°C, extension for 2 min at 72°C, and final extension for 10 min at 72°C. For amplification of ptxP, in a total volume of 25 µl, the same reaction mix was used as described above, with the exceptions that Taq polymerase was increased to 1 U, 10% of dimethyl sulfoxide (CinnaGen co., Iran) was added, and annealing temperature was increased to 58°C. Electrophoresis was performed in a 1% agarose (Invitrogen, USA) gel and was stained with 0.5 µg/ml ethidium bromide (Sigma, USA).
Table 1.
Specific primers sequences targeting IS481 and ptxP
| Target region | Primer sequence (5′-3′) | Amplicon size (bp) | Reference |
|---|---|---|---|
| IS481 | 5′-GATTCAATAGGTTGTATGCATGGTT-3′ 5′-TTCAGGCACACAAACTTGATG-3′ | 181 | 14 |
| PtxP | 5′-AATCGTCCTGCTCAACCGCC-3′ 5′-GGTATACGGTGGCGGGAGGA-3′ | 573 | 15 |
Pulsed field gel electrophoresis (PFGE)
A single colony of each clinical isolate and vaccine strain confirmed as B. pertussis were subcultured onto BGA. The bacterial suspension was prepared in Cell Suspension Buffer (CSB) containing 100 mM Tris-HCl (Merck, Germany) and 100 mM EDTA (Promega, UK) with an optical density of 0.5 at 650 nm. Each plug was prepared by adding 120 µl of bacterial suspension and 20 µl of 50 mg/ml proteinase K (Sigma, USA) to 120 µl of 1% Seakem Gold agarose (FMC BioProducts, Rock land, ME) and was incubated in lysis buffer [100 mM Tris-HCl, 100 mM EDTA, 1% lauroyl sarcosine (Sigma, USA), and 0.5 mg/ml solution of proteinase K (Sigma, USA)] at 55°C overnight. The plugs were washed with deionized water and 1× TE (10 mM Tris-HCl, 1 mM EDTA). The plug slices were incubated with 30 U of XbaI (Fermentas, Lithuania) at 37°C for 16 hr. PFGE was performed with 1% agarose gel in a contour-clamped homogeneous electric field (CHEF-DRIII) (Bio-Rad, USA) apparatus at 6 V/cm with a 120°C angle for 23 hr at 12°C with initial switch time of 2.2 s and the final switch of 55 s. The genomic DNA of Salmonella enterica serotype Braenderup H9812 was used as the size marker. The obtained XbaI PFGE profiles were analyzed using GelComparII version 4 (Applied Maths, Sint-Martens-Latem, Belgium). The Unweighted Pair Group Method with arithmetic mean Algorithm (UPGMA) was used as the clustering method, with a 1% band tolerance, 1% optimization, and the Dice's coefficient (16). The band pattern of each strain was confirmed visually. Isolates with a DNA band pattern differing by ≥1 band were defined as single type or distinct PFGE profile.
Results
Isolates as small, smooth, gray colonies which appeared at a minimum of 72 hr on BGA were identified as B. pertussis and they were stained as Gram negative coccobacilli. B. pertussis colonies did not grow on common laboratory media such as MacConkey agar and blood agar and were distinguished from other Bordetella species by oxidase- and catalase-positive but urease- and nitrate-negative reactions. As indicated in Table 2, five of 11 suspected clinical isolates and all vaccine strains as well as Tohama I were verified as B. pertussis based on phenotypic and biochemical characteristics.
Table 2.
Differential characteristics of vaccine strains, clinical isolates, and reference strains of B. pertussis and B. parapertussis
| Strain/Isolate Characteristic | Bp509/1997 | Bp509/2000 | Bp 134 | Tohama I | B. parapertussis | Bp1 | Bp2 | Bp3 | Bp4 | Bp5 | Bp6 | Bp7 | Bp8 | Bp9 | Bp10 | Bp11 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Growth rate * (Days) | 4 | 4 | 4 | 4 | 1 | 1 | 1 | 4 | 1 | 5 | 1 | 5 | 5 | 5 | 1 | 1 |
| Catalase | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Oxidase | + | + | + | + | - | + | + | + | + | + | + | + | + | + | + | + |
| Nitrate reductase | - | - | - | - | - | + | - | - | - | - | - | - | - | - | - | - |
| Urease | - | - | - | - | + | - | + | - | + | - | + | - | - | - | + | + |
| Growth on | ||||||||||||||||
| Blood agar | - | - | - | - | + | - | + | - | - | - | - | - | - | - | - | - |
| MacConkey agar | - | - | - | - | V | - | + | - | - | - | - | - | - | - | - | - |
Growth rate represents minimum days of appearance of colonies on BGA; Bp: Bordetella pertussis, V: Variable growth patterns
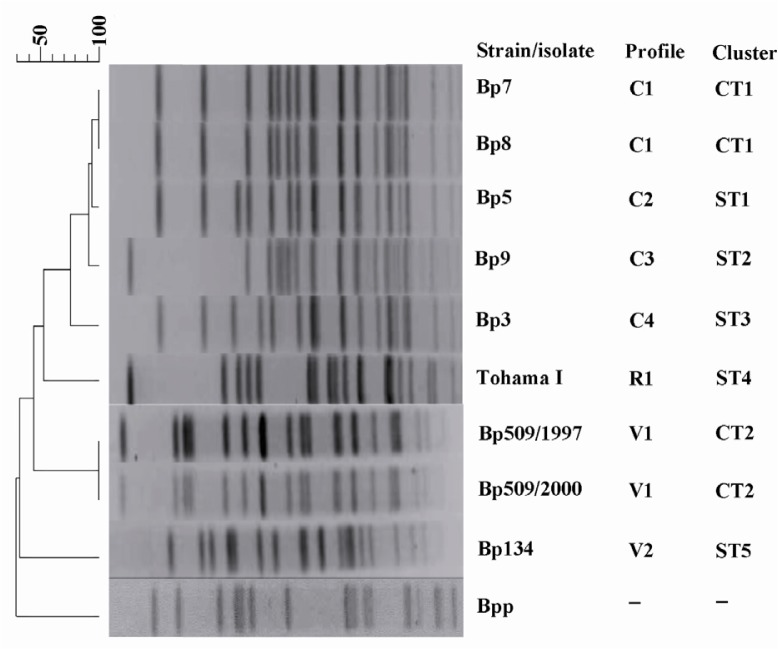
Five of the 11 clinical isolates and all vaccine strains yielded 181 bp (Figure 1) and 573 bp (Figure 2) amplicons through amplification of IS481 and ptxP, respectively, confirming the identity of B. pertussis. A total of 5 clinical isolates, 2 vaccine strains, and Tohama I strain were firmly confirmed via both phenotypic and molecular methods. Chromosomal DNA of these confirmed strains were typed using PFGE. Seven distinct PFGE profiles were found among all examined strains. In the 5 clinical isolates, 4 profiles were identified and named C1 to C4. Two clinical isolates with the same number as well as same profiles for bands size were grouped in common type 1 (CT1). The other clinical isolates (Bp3, Bp5, and Bp9) had 80% of similarity, therefore belonged to three different types, named single type 3, 1, and 2 (ST3, ST1, and ST2), respectively. The 2 vaccine strains displayed 2 distinct profiles and were named V1 and V2. Two vaccine isolates which were derived from the Netherlands strain Bp509 had exactly the same profile and were grouped in CT2. The vaccine strain, Bp134, had unique PFGE profile and was defined as ST5. As shown in Figure 3, Tohama I strain was included in dendrogram as a reference strain (R1) with unique profile and was considered as ST4. Vaccine profiles and clinical profiles had low similarity, with relatedness of approximately 40%. B. parapertussis which was used as a control had distinct PFGE profile with low relatedness (<40%) to all typed B. pertussis isolates/strains.
Figure 1.

The IS481 amplification using specific primers yielded a product of 181 bp typical to clinical isolates and vaccine strains of B. pertussis. L: 100 bp ladder (Fermentas, Lithuania); 1: Tohama I strain as reference strain; 2-6: clinical isolates of B. pertussis; 7-9: vaccine strains of B. pertussis; 10-15: non-B. pertussis clinical isolates; 16: B. parapertussis; 17: Negative control (distilled water)
Figure 2.
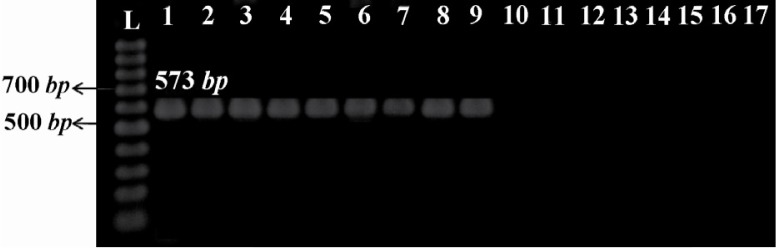
The ptxP amplification using specific primers yielded a product of 573 bp typical to clinical isolates and vaccine strains of B. pertussis. L: 100 bp ladder (Fermentas, Lithuania); 1: Tohama I strain as reference strain; 2-6: clinical isolates of B. pertussis; 7-9: vaccine strains of B. pertussis; 10-15: non-B. pertussis clinical isolates; 16: B. parapertussis; 17: negative control (distilled water)
Figure 3.
Genomic analysis of B. pertussis using PFGE. The dendrogram shows PFGE profiles of clinical isolates (C), vaccine strains (V), and reference strain Tohama I (R1). B. parapertussis (Bpp) was used as a control. The PFGE clusters are indicated as common type (CT) or single type (ST)
Discussion
In this study, the genetic profiles of a limited number of B. pertussis isolates recovered from Iranian patients were analyzed and compared with the two strains which were used for production of the national whole-cell pertussis vaccine. PFGE as a typing method was used and different PFGE patterns among the clinical isolates were found. This finding indicated that different B. pertussis isolates were circulating despite high vaccination coverage in Iran. However, low heterogeneity has been found among the clinical isolates and these isolates belonged to different profiles with an approximately 80% of genetic relatedness. Therefore, the results were consistent with previous studies that classified B. pertussis as a genetically monomorphic pathogen (17, 18).
Two strains containing the same PFGE pattern were also identified among clinical isolates. They were more likely to be related and to have a common source. Two PFGE patterns among the vaccine strains were observed in this study. These 2 profiles belonged to strains used in vaccine production in Iran. Two isolates included in V1 profile were subcultures or derivatives of a vaccine strain of the Netherlands origin, Bp509, which were cultured at different time intervals. The exactly similar PFGE patterns of our vaccine strains suggest that repeated subculture did not lead to genetic divergence over time, as previously described by Advani et al (19). However, two studies showed that the PFGE profiles changed upon repeated subculture (20, 21). The XbaI digested DNA of the reference strain (Tohama I) had PFGE profile exactly similar to profile obtained by others who clustered Tohama I in PFGE group II (22). The DNA of vaccine strains Bp509 and Bp134 were also digested with XbaI and the profiles were comparable with that of other studies. The strain Bp509 showed exactly similar profile same as several other studies (23–25), but a controversy was found in just one study in which Bp509 and Bp134 were considered the same (23). In this study, strain Bp134 showed the same profile as reported in previous studies (26, 27), but Bottero et al reported similar profiles for Bp509 and Bp134 in their dendrogram (23). This controversy may be due to an error in processing the stocks of bacteria or interpretation of the dendrogram.
Comparison of the genetic profiles of vaccine strains and clinical isolates demonstrated that vaccine strains had different PFGE patterns and were not matched to circulating isolates. One explanation for this variation might be associated to the low efficacy of currently used vaccine which may lead to adaptation of the pathogen and selection of new variants of B. pertussis (5). Another explanation is that the circulating bacteria may evade the host responses and in turn evolve efficient mechanisms to overcome the vaccine-induced immunity (28, 29). It is also possible that such gross genetic variations might not be clinically relevant and reflect random mutations introduced in the genome of Bp isolates obtained from the patients. However, more detailed genetic and structural investigations are required to address this issue.
Several studies have compared the PFGE profiles of clinical isolates and vaccine strains of B. pertussis in different countries. The majority of these studies have indicated that non-vaccine isolates were recovered from patients with pertussis disease and these isolates were genetically different from the vaccine strains (19, 22, 30). However, in the study of Elomaa et al, the profile of one of the two assessed vaccine strains was detected among the circulating isolates (31).
Conclusion
It was demonstrated that non-vaccine isolates of B. pertussis were different from vaccine strains used in the national vaccination program. Since new genetic profiles of B. pertussis can be disseminated periodically, the profiles of isolates circulating in the population should be monitored over the course of the re-emergence. PFGE is used as a golden standard method for determining the gross polymorphisms within restriction enzyme Recognition Sites (RS) in whole genome and for identifying the extensive rearrangements, deletions, and insertions that affect the restriction fragment profile (32). However, some genetic variations such as single nucleotide polymorphism, short tandem repeat polymorphism, non-RS repetitive sequence polymorphisms, and mutations changing the genetic code could not be assessed via PFGE. Therefore, sequencing based methods need to be used to characterize and interpret such minute and small-scale changes. Further monitoring may provide more information about the strains or antigens needed for development of more effective vaccines.
Acknowledgement
This research has been supported in part by Tehran University of Medical Sciences and Health Services grants.
References
- 1.Miyaji Y, Otsuka N, Toyoizumi-Ajisaka H, Shibayama K, Kamachi K. Genetic analysis of Bordetella pertussis isolates from the 2008-2010 Pertussis epidemic in Japan. PLoS One. 2013;8(10):e77165. doi: 10.1371/journal.pone.0077165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McIntyre P, Wood N. Pertussis in early infancy: disease burden and preventive strategies. Curr Opin Infect Dis. 2009;22(3):215–223. doi: 10.1097/QCO.0b013e32832b3540. [DOI] [PubMed] [Google Scholar]
- 3.de Melker HE, Conyn-van Spaendonck MA, Rumke HC, van Wijngaarden JK, Mooi FR, Schellekens JF. Pertussis in The Netherlands: an outbreak despite high levels of immunization with whole-cell vaccine. Emerg Infect Dis. 1997;3(2):175–178. doi: 10.3201/eid0302.970211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preston A. Bordetella pertussis: the intersection of genomics and pathobiology. Can Med Assoc J. 2005;173(1):55–62. doi: 10.1503/cmaj.050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mooi FR, Van Loo IH, King AJ. Adaptation of Bordetella pertussis to vaccination: a cause for its reemergence? Emerg Infect Dis. 2001;7(3 Suppl):526–528. doi: 10.3201/eid0707.017708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mooi FR, Hallander H, Wirsing von Konig CH, Hoet B, Guiso N. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur J Clin Microbiol Infect Dis. 2000;19(3):174–181. doi: 10.1007/s100960050455. [DOI] [PubMed] [Google Scholar]
- 7. http://apps.who.int/immunization_monitoring/globalsummary/timeseries/tscoveragedtp1.html.
- 8. http://www.who.int/gho/publications/world_health_statistics/EN_WHS2013_Full.pdf.
- 9.Eslamifar A, Aghakhani A, Banifazl M, Gachkar L, Khadem Sadegh A, Ramezani A. Seroprevalence of Bordetella pertussis antibodies in different age groups. Iran J Infect Dis Tropic Med. 2010;15(49):43–47. [Google Scholar]
- 10.Zarei S, Jeddi-Tehrani M, Mehdi Akhondi M, Zera-ati H, Ferydonfar AA, Nasernia J, et al. Immunogenicity and reactogenicity of two diphtheria-tetanus-whole cell pertussis vaccines in Iranian pre-school children, a randomized controlled trial. Hum Vaccin Immunother. 2013;9(6):1316–1322. doi: 10.4161/hv.24093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shahcheraghi F, Nakhost Lotfi M, Parzadeh M, Sadat Nikbin V, Shouraj F, Zahraei M. Isolation of Bordetella pertussis and Bordetella parapertussis from clinical specimens at different provinces of Iran in 2009-2010. J Mazand Univ Med Sci. 2012;22(88):2–8. [Google Scholar]
- 12.Loeffelholz MJ. Bordetella. In: Murray PR, editor. Manual of clinical microbiology. America: American Society for Microbiology; 1999. pp. 780–786. [Google Scholar]
- 13.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Reischl U, Lehn N, Sanden GN, Loeffelholz MJ. Real-time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J Clin Microbiol. 2001;39(5):1963–1966. doi: 10.1128/JCM.39.5.1963-1966.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mooi FR, van Loo IH, van Gent M, He Q, Bart MJ, Heuvelman KJ, et al. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis. 2009;15(8):1206–1213. doi: 10.3201/eid1508.081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mooi FR. Bordetella pertussis and vaccination: the persistence of a genetically monomorphic pathogen. Infect Genet Evol. 2010;10(1):36–49. doi: 10.1016/j.meegid.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Nar Otgun S, Durmaz R, Karagoz A, Esen B, Ertek M. Pulsed-field gel electrophoresis characterization of Bordetella pertussis clinical isolates circulating in Turkey in 2001-2009. Eur J Clin Microbiol Infect. 2011;30(10):1229–1236. doi: 10.1007/s10096-011-1217-y. [DOI] [PubMed] [Google Scholar]
- 19.Advani A, Donnelly D, Hallander H. Reference system for characterization of Bordetella pertussis pulsed-field gel electrophoresis profiles. J Clin Microbiol. 2004;42(7):2890–2897. doi: 10.1128/JCM.42.7.2890-2897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beall B, Cassiday PK, Sanden GN. Analysis of Bordetella pertussis isolates from an epidemic by pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33(12):3083–3086. doi: 10.1128/jcm.33.12.3083-3086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caro V, Njamkepo E, Van Amersfooorth SC, Mooi FR, Advani A, Hallander HO, et al. Pulsed-field gel electrophoresis analysis of Bordetella pertussis populations in various European countries with different vaccine policies. Microbes Infect. 2005;7(7-8):976–982. doi: 10.1016/j.micinf.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Advani A, Hallander HO, Dalby T, Krogfelt KA, Guiso N, Njamkepo E, et al. Pulsed-field gel electrophoresis analysis of Bordetella pertussis isolates circulating in Europe from 1998 to 2009. J Clin Microbiol. 2013;51(2):422–428. doi: 10.1128/JCM.02036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bottero D, Gaillard ME, Fingermann M, Weltman G, Fernandez J, Sisti F, et al. Pulsed-field gel electrophoresis, pertactin, pertussis toxin S1 sub-unit polymorphisms, and surfaceome analysis of vaccine and clinical Bordetella pertussis strains. Clin Vaccine Immunol. 2007;14(11):1490–1498. doi: 10.1128/CVI.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shuel M, Jamieson FB, Tang P, Brown S, Farrell D, Martin I, et al. Genetic analysis of Bordetella pertussis in Ontario, Canada reveals one predominant clone. Int J Infect Dis. 2013;17(6):e413–e417. doi: 10.1016/j.ijid.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Bottero D, Gaillard ME, Basile LA, Fritz M, Hozbor DF. Genotypic and phenotypic characterization of Bordetella pertussis strains used in different vaccine formulations in Latin America. J Appl Microbiol. 2012;112(6):1266–1276. doi: 10.1111/j.1365-2672.2012.05299.x. [DOI] [PubMed] [Google Scholar]
- 26.Hallander H, Advani A, Riffelmann M, von Konig CH, Caro V, Guiso N, et al. Bordetella pertussis strains circulating in Europe in 1999 to 2004 as determined by pulsed-field gel electrophoresis. J Clin Microbiol. 2007;45(10):3257–3262. doi: 10.1128/JCM.00864-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caro V, Bouchez V, Guiso N, Gatti B, Agosti MR, Ayala SE. Pertussis in Argentina and France. Vaccine. 2007;25(22):4335–4339. doi: 10.1016/j.vaccine.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 28.Mooi FR, van Oirschot H, Heuvelman K, van der Heide HG, Gaastra W, Willems RJ. Polymorphism in the Bordetella pertussis virulence factors P.69/ pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine driven evolution. Infect Immun. 1998;66(2):670–675. doi: 10.1128/iai.66.2.670-675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fry NK, Neal S, Harrison TG, Miller E, Matthews R, George RC. Genotypic variation in the Bordetella pertussis virulence factors pertactin and pertussis toxin in historical and recent clinical isolates in the United Kingdom. Infect Immun. 2001;69(9):5520–5528. doi: 10.1128/IAI.69.9.5520-5528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosiej E, Augustynowicz E, Zawadka M, Dabrowski W, Lutynska A. Strain variation among Bordetella pertussis isolates circulating in Poland after 50 years of whole-cell pertussis vaccine use. J Clin Microbiol. 2011;49(4):1452–1457. doi: 10.1128/JCM.01487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elomaa A, Advani A, Donnelly D, Antila M, Mertsola J, He Q, et al. Population dynamics of Bordetella pertussis in Finland and Sweden, neighbouring countries with different vaccination histories. Vaccine. 2007;25(5):918–926. doi: 10.1016/j.vaccine.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Finney M. Pulsed-Field Gel Electrophoresis. In: Ausubel FM, editor. Current protocols in molecular biology. New York: Greene Pub. Associates and Wiley-Interscience; 1988. pp. 2.5B.1–2.5B.9. [Google Scholar]