Abstract
To date, no polymerase chain reaction diagnostic technique exists to directly identify mammalian blood meals from mosquitoes by sized DNA fragments following agarose gel electrophoresis. We have developed a vertebrate-specific multiplexed primer set based on mitochondrial cytochrome b to identify the mammalian blood hosts of field-collected mosquitoes. Although designed for the study of African malaria vectors, the application of this tool is not restricted to this disease system. Validation of this diagnostic technique on dried anopheline and culicine field specimens collected in Zambia and Mali demonstrated that blood meals could be identified 2–7 months after collection. Time course experiments showed that host DNA was detectable in frozen mosquito abdomens 24–30 hours post-feeding. Additionally, multiple blood meals from different mammals could be detected in a single mosquito. This diagnostic assay will be a valuable tool for identifying the blood meals of field-collected mosquitoes where people and alternative mammal hosts are present.
INTRODUCTION
Blood meals of hematophagous arthropods have traditionally been identified by serologic techniques such as the precipitin test, latex agglutination, and the enzyme-linked immunosorbent assay (ELISA).1–4 These methods have provided countless valuable data over the years, especially in mosquito blood meal identification. However, an alternative method may be desired in laboratories not set up to perform immunologic assays, or if samples are already in the form of extracted DNA. The polymerase chain reaction (PCR) has been successfully used in forensic applications, detection of animal tissues in ruminant feeds, DNA profiling of mosquito blood meals, and blood host identification in hematophagous arthropods.5–11 The PCR-based identification of arthropod blood meals provides a convenient alternative for laboratories using primarily DNA-based techniques, and may be necessary when the study design already requires the use of individual DNA extractions for multiple purposes such as species confirmation, determination of infection status for various pathogens, and vector population genetic studies. Furthermore, engorged specimens collected in the field may be preserved dry, stored for long periods of time, and tested at facilities that may be physically distant from the point of collection.
Cytochrome b is a well-characterized protein from complex III of the mitochondrial oxidative phosphorylation system, and the only protein in this complex encoded by the mitochondrial genome.12,13 This gene has been used to resolve vertebrate evolutionary questions as well as served as a target for molecular diagnostics.13–17 Cytochrome b has a proven utility for identifying arthropod blood meals due to high copy number as a mitochondrial gene and sufficient genetic variation at the primary sequence level among vertebrate taxa for reliable identification. Kirstein and Gray designed degenerate primers to amplify homologous DNA fragments from 11 different animals commonly fed on by the tick Ixodes ricinus L.14 The mammalian host was then identified based on unique restriction patterns when the amplified DNA fragments were digested with Hae III and Dde I restriction enzymes. Boake and others and Lee and others developed primers to PCR amplify a universal cytochrome b fragment from mammalian and avian hosts that were then differentiated by heteroduplex analysis.15,16 Similarly, Ngo and Kramer developed an order-specific PCR diagnostic assay based on cytochrome b to identify avian blood meals from mosquitoes.17 Other markers that have been used to identify blood meals from arthropods include vertebrate 18S ribosomal DNA from I. ricinus ticks,18 the hypervariable region 2 from human mitochondrial DNA in bloodfed crab lice (Pthirus pubis L.),6 and TC-11 and VWA (HUMVWFA31/A, a repeat polymorphism in the von Willebrand factor gene) human short tandem repeat loci from blooded Anopheles gambiae Giles.19 Of the currently available diagnostic tests for identifying mammalian blood from arthropods by cytochrome b, each requires the added time and expense of a restriction enzyme digestion or heteroduplex analysis following the PCR. A PCR diagnostic test that directly identifies the mammalian host source would greatly increase the speed and cost-effectiveness at which blood meal identifications could be performed.
We have developed a vertebrate-specific multiplexed primer set based on cytochrome b to identify the mammalian blood hosts of engorged mosquitoes caught during field collections. The advantage of this novel PCR diagnostic test is that mammalian blood hosts can be identified directly by sizespecific fragments following agarose gel electrophoresis. This diagnostic test was designed for use in differentiating between blood meals of mosquitoes involved in malaria transmission caught in African villages where the potential hosts are primarily humans and domestic animals. However, the assay can be applied to any environment where people, cattle, dogs, goats, and pigs are present as potential blood hosts.
MATERIALS AND METHODS
Mosquito collection and handling
Engorged field specimens of An. arabiensis Patton, An. funestus s.s. Giles, An. leesoni Evans, Mansonia uniformis Theobald, and Culex pipiens quinquefasciatus Say used for method validation were collected by pyrethrum spray catch in Macha, Zambia (16.39292S, 26.79061E) in March, May, and November 2004. Field samples of An. gambiae s.s. Giles were collected indoors by aspiration in Pimperena, Mali (11.467N, 5.700E) in August 2004. Field specimens were individually placed in tubes containing silica gel desiccant (J. T. Baker, Phillipsburg, NJ) and cotton immediately following collection and stored at room temperature for 2–7 months until processing. Prior to homogenization, dry field specimens were rehydrated at room temperature in 20 µL of double-distilled water for 20 minutes. Colony mosquitoes used as positive controls were killed by freezing at designated times post-feeding and processed immediately. Because these samples were fresh, no rehydration step was necessary. For all mosquitoes, heads/thoraces were separated from abdomens and only the DNA isolated from engorged abdomens were used in the blood meal diagnostic analysis. Unfed An. gambiae s.s., An. funestus, An. stephensi (Liston), and Cx. p. quinquefasciatus were also tested simultaneously for nonspecific amplification by the mammalian primer set.
Isolation of DNA
Whole blood was extracted with a QiaAmp DNA Mini Kit (Qiagen Inc.,Valencia, CA) according to manufacturer’s instructions and eluted in 200 µL of double-distilled water. DNA was extracted from mosquito abdomens by a modified salt procedure as described previously.20 Briefly, mosquitoes were homogenized in 100 µL of extraction buffer containing 0.1 M NaCl, 0.2 M sucrose, 0.1 M Tris-HCl, 0.05 M EDTA, pH 9.1, and 0.5% sodium dodecyl sulfate (SDS) and incubated at for one hour at 65°C. To precipitate the SDS, 15 µL of cold 8 M potassium acetate was added to each homogenate and samples were incubated on ice for 45 minutes. Samples were then centrifuged for 10 minutes to remove cellular debris, and DNA was precipitated by adding 250 µL of 100% ethanol to the transferred supernatant. Following a 5-minute incubation at room temperature, DNA was pelleted by a 15-minute centrifugation. Finally, the supernatant was discarded and dried pellets were resuspended in 10 µL of 0.1× SSC (15 mM NaCl, 1.5 mM sodium citrate) plus 40 µL of double-distilled water, The relative quality of all DNA extractions was evaluated by PCR amplification of a fragment of the mitochondrial NADPH dehydrogenase subunit 4 (ND4) using arthropod-specific primers.21,22
Blood sample sources and blood feeding
Blood was obtained from Pel-Freez Biologicals (Rogers, AR) and Pocono Rabbit Farm and Laboratory (Canadensis, PA). Cow (Bos tarsus), pig (Sus scrofa), and goat (Nupian strain) (Capra hircus) blood contained one part sodium citrate to nine parts blood. Dog (Canis familiaris) and human (Homo sapiens) blood was preserved in K3 EDTA anticoagulant. Positive control mosquitoes were generated at the Johns Hopkins Bloomberg School of Public Health (Baltimore, MD) by feeding colonized An. stephensi on fresh, non-sterile, whole blood warmed to 37°C through a Parafilm® (American Can Co., Greenwich, CT) membrane for two hours. Four hours post-feeding, blooded mosquitoes were collected and frozen at −20°C until processed. For the time course experiments, mosquitoes were held for various time points post-feeding to determine how long host DNA was detectable in extracted abdomens. Four to six mosquitoes were removed from each host group at 24-, 48-, 72-, 96-, and 120-hours post-feeding and frozen at −20°C until analyzed.
Primer design and PCR
One universal reverse primer and five animal-specific forward primers were manually selected from a multiple alignment of cytochrome b sequences obtained from GenBank: An. gambiae (NC002084), cow (B. tarsus AB090987; AB090986), human (H. sapiens AY495285), pig (S. scrofa AY237531; AY237528), goat (C. hircus D84201; AB110597), and dog (C. familiaris NC002008). To prevent cross-annealing with non-target templates, animal-specific primers were designed to polymorphic regions where the target sequence had at least 5, but up to 13, nucleotide differences from homologous regions in the other animal sequences. Primer sequences were checked for melting temperature (Tm) compatibility and self-complementarity with Primer3 software.23 Expected products were designed to differ in size by at least 100 bases for unmistakable identification on agarose gels. Two alternative primers, UNFOR403 and COWREV371, were also designed to substitute for Cow121F in the multiplexed reaction mixture (Table 1).
Table 1.
Primer sequences for the cytochrome b-based polymerase chain reaction blood meal identification assay
| Primer | 5′-3′ sequence | Product size with UNREV1025 |
|---|---|---|
| Pig573F | cctcgcagccgtacatctc | 453 |
| Human741F | ggcttacttctcttcattctctcct | 334 |
| Goat894F | cctaatcttagtacttgtacccttcctc | 132 |
| Dog368F | ggaattgtactattattcgcaaccat | 680 |
| Cow121F | catcggcacaaatttagtcg | 561 |
| Cow371R | gagctagaattagtaagagggcc | – |
| UNFOR403 | tgaggacaaatatcattctgagg | 623 |
| UNREV1025 | ggttgtcctccaattcatgtta | – |
Sequences from additional domestic and wild animals were compared by multiple alignment with the sequences of animals included in the diagnostic assay to predict the compatibility of the primer set with other hosts within each taxon: sheep (Ovis aries NC001941), wart hog (Phacochoerus aethiopicus AJ314551, Phacochoerus africanus AJ314548), water buffalo (Bubalus bubalis D88637), Cape buffalo (Synceros caffer D82888), African hunting dog (Lycaon pictus S69130), baboon (Papio hamadryas Y16590), African green monkey (Cercopithecus aethiops U63128), and chimpanzee (Pan troglodytes AY585837). Avian hosts were not considered in this diagnostic assay due to the availability of alternative PCR diagnostic assays designed specifically to identify avian blood meals in mosquitoes.16,17
The multiplexed PCR was optimized for both single and mixed templates. An initial denaturation of 5 minutes at 95°C was followed by 35 cycles at 95°C for 1 minute, 58°C for 1 minute, and 72°C for 1 minute. The final extension step was at 72° for 7 minutes. Each 25-µL PCR consisted of 10 mM Tris, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 1.0 mM dNTPs, 0.5 units of Taq polymerase, and 50 pmol of Pig573F, Human741F, Goat894F, Dog368F, Cow121F, and UNREV1025 primers. It was found that 15 ng (0.5 µL) of extracted DNA from whole blood was sufficient template for use in positive control reactions. DNA amplifications were completed on a MJ Research® PTC-200 thermal cycler (Bio- Rad Laboratories, Inc., Hercules, CA) and visualized after electrophoresis on ethidium bromide–stained 2% agarose gels. Electrophoresis was conducted with GeneRuler 100-basepair (bp) molecular mass marker (Fermentas Life Science, Hanover, MD).
Multiple host feeding analysis
Additional experiments were conducted to evaluate the sensitivity of the diagnostic assay on disproportionate ratios of mixed animal bloods. These experiments were intended to simulate the potential situation in the field where a mosquito may take multiple blood meals from different mammalian hosts during one gonotrophic cycle. Human and cow blood samples were mixed at ratios of 50:50, 30:70, and 70:30 and fed to starved mosquitoes by methods described above. Two to five engorged mosquitoes were removed from each group and frozen at 4 hours and 12 hours post-feeding. DNA was extracted from mosquito abdomens as described above and tested for the presence of both host templates by PCR. Excess mixed whole blood samples used to feed the mosquitoes were simultaneously extracted and used as positive controls in the PCRs.
RESULTS
We have developed an animal-specific PCR assay based on the mitochondrial cytochrome b gene to identify the mammalian blood meals of field-collected mosquitoes. The diagnostic assay correctly identified the blood meal of control mosquitoes fed on different mammalian bloods 100% of the time and yielded no amplification of unfed mosquito template DNA from various mosquito vectors (Figure 1). An alternative primer combination using primers UNFOR403, UNREV1025, COWREV371, Human741F, Cow121F, Goat894F, Dog 368F, and Pig573F also gave an array of diagnostic fragments distinguishable by size by agarose electrophoresis (Figure 2). This alternative multiplexed PCR generated a 623-bp control product with UNFOR403 and UNREV1025 for all five species, and a 313-bp cow-specific product produced by UNFOR403 and COWREV371.
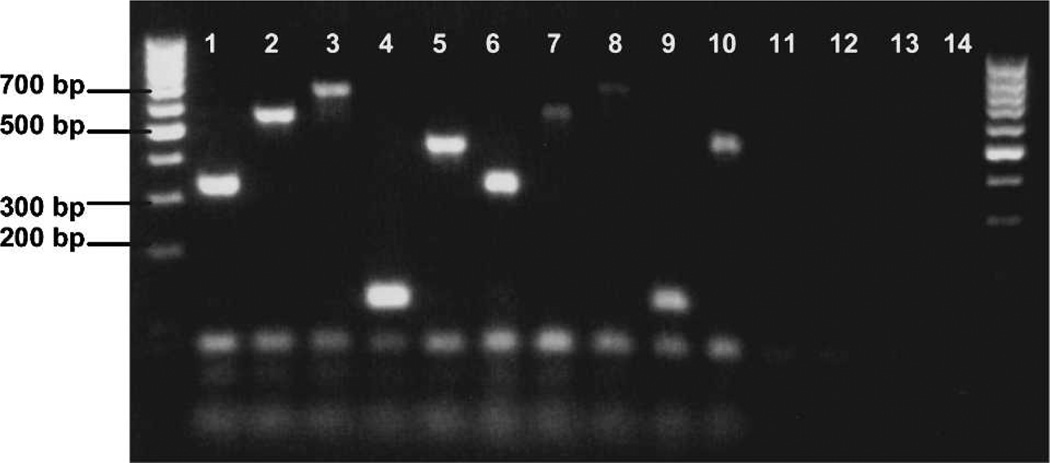
Figure 1.
Ethidium bromide–stained agarose gel showing hostspecific cytochrome b polymerase chain reaction products amplified from whole blood DNA extractions and Anopheles stephensi mosquitoes fed on each blood source. Control products amplified from whole blood are shown in lanes 1 (human, 334 bp), 2 (cow, 561 bp), 3 (dog, 680 bp), 4 (goat, 132 bp), and 5 (pig, 453 bp). Control products from engorged mosquitoes are shown in lanes 6 (human), 7 (cow), 8 (dog), 9 (goat), and 10 (pig). Lanes 11–13, unfed An. stephensi, An. gambiae, and Culex pipiens quinquefasciatus, respectively. Lane 14, negative control. Outside lanes are 100-basepair DNA ladders. bp = basepairs.
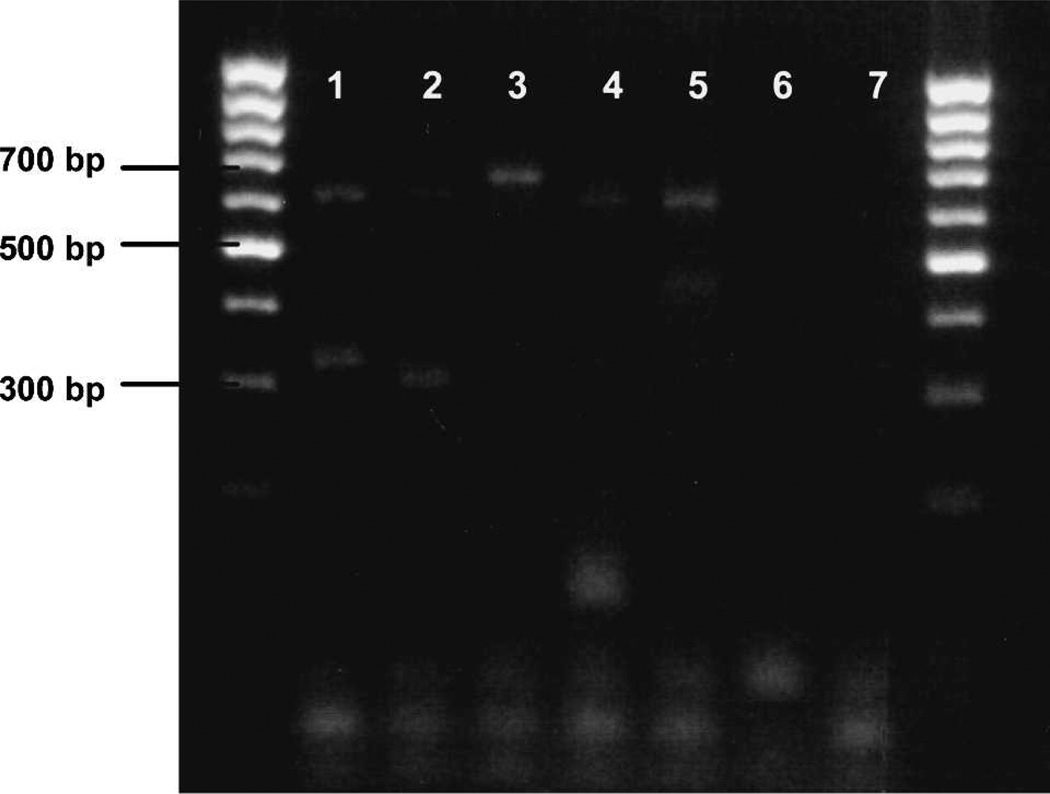
Figure 2.
Alternative primers UNFOR403 and COWREV 371 used in place of Cow371F, showing a 623-bp control band for all vertebrate species and a 313-bp cow-specific band. Lanes 1–5, Anopheles stephensi mosquitoes fed on animal blood of known origin. Lane 1, human (623 bp, 334 bp); lane 2, cow (623 bp, 313 bp); lane 3, dog (680 bp); lane 4, goat (623 bp, 132 bp); lane 5, pig (623 bp, 453 bp); lane 6, unfed An. stephensi control; lane 7, negative control. Outside lanes are 100-basepair DNA ladders. bp =basepairs.
Only the five most common domestic animals in the Zambian study region were included in the development of this PCR diagnostic assay. The probability of obtaining amplification from other species within each animal group that may be encountered in other study areas was evaluated by multiple sequence alignment. Comparison of goat and sheep sequences showed five nucleotide differences in the Goat894F priming region of the sheep sequence. Therefore, if a more universal amplification of ovid template is desired, Goat894F may need to be redesigned, or an additional sheep forward primer designed for inclusion in the diagnostic assay. Similarly, there are five nucleotide differences between the Dog368F forward primer and homologous wild dog sequence, rendering the primer pair unlikely to amplify a product from wild dog. Comparison of the domestic pig sequence with both P. africanus and P. aethiopicus showed only 2–3 nucleotide differences in the pig forward priming region and 1 nucleotide difference in the UNREV1025 reverse priming region. Therefore, these primers would be expected to work for both warthog and domestic pig templates. Comparison of primate sequences showed between 8 and 10 nucleotide differences between Human741F and baboon, vervet monkey, and chimpanzee sequences; therefore, these primers are not predicted to cross-anneal to non-human primate template. Finally, Cow121F is not predicted to anneal to water buffalo DNA due to six nucleotide differences in the forward priming region. However, these primers may amplify Cape buffalo cytochrome b, with only three nucleotide differences in the forward priming site and a solid 3′ alignment.
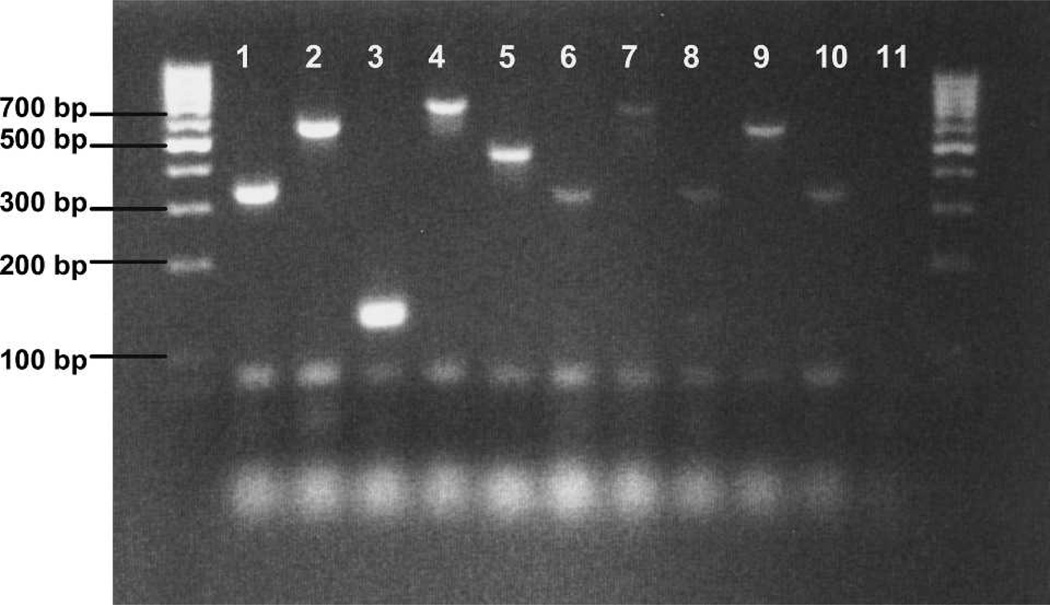
Figure 3 illustrates the validation of the diagnostic assay on blood-fed An. arabiensis, An. funestus s.s., An. leesoni, and Cx. p. quinquefasciatus collected by pyrethrum spray catch in Macha, Zambia. Positive blood meal identifications were obtained from 100 (81%) of 123 field-collected mosquitoes: 61 of 74 An. gambiae s.s., 19 of 20 An. arabiensis, 8 of 15 An. funestus s.s., 8 of 8 An. leesoni, 1 of 1 Ma. uniformis, and 3 of 5 Cx. p. quinquefasciatus. Reamplification using 1 µL of template from the initial PCRs produced visible product from 16% of individuals that did not appear to work using a single round of amplification. The considerably smaller body size and therefore smaller blood meal size of An. funestus s.l. compared with An. arabiensis, An. gambiae s.s., and Ma. uniformis was problematic in obtaining visible amplification from these specimens, especially if these blood meals were either incomplete or partially digested. Human blood was successfully detected from An. gambiae s.s., An. arabiensis, An. funestus s.s., Ma. uniformis, and Cx. p. quinquefasciatus. Cow blood was detected in An. gambiae and An. leesoni, goat blood in An. leesoni, and dog blood in Cx. p. quinquefasciatus (Figure 3). Although pigs are common in rural Zambia, pig blood was not detected in any field samples tested.
Figure 3.
Ethidium bromide–stained agarose gel showing blood meal identifications of engorged field samples. Lanes 1–5 show positive controls amplified from animal whole blood extractions, and lanes 6–10 show products obtained from field samples. Lane 1, human; lane 2, cow; lane 3, goat; lane 4, dog; lane 5, pig; lane 6, human blood meal detected in Culex p. quinquefasciatus; lane 7, dog blood meal detected in Cx. p. quinquefasciatus; lane 8, human blood meal detected in Anopheles arabiensis; lane 9, cow blood meal detected in An. leesoni; lane 10, human blood meal amplified from An. funestus s.s.; lane 11, negative control. Outside lanes are 100-basepair DNA ladders. bp = basepairs.
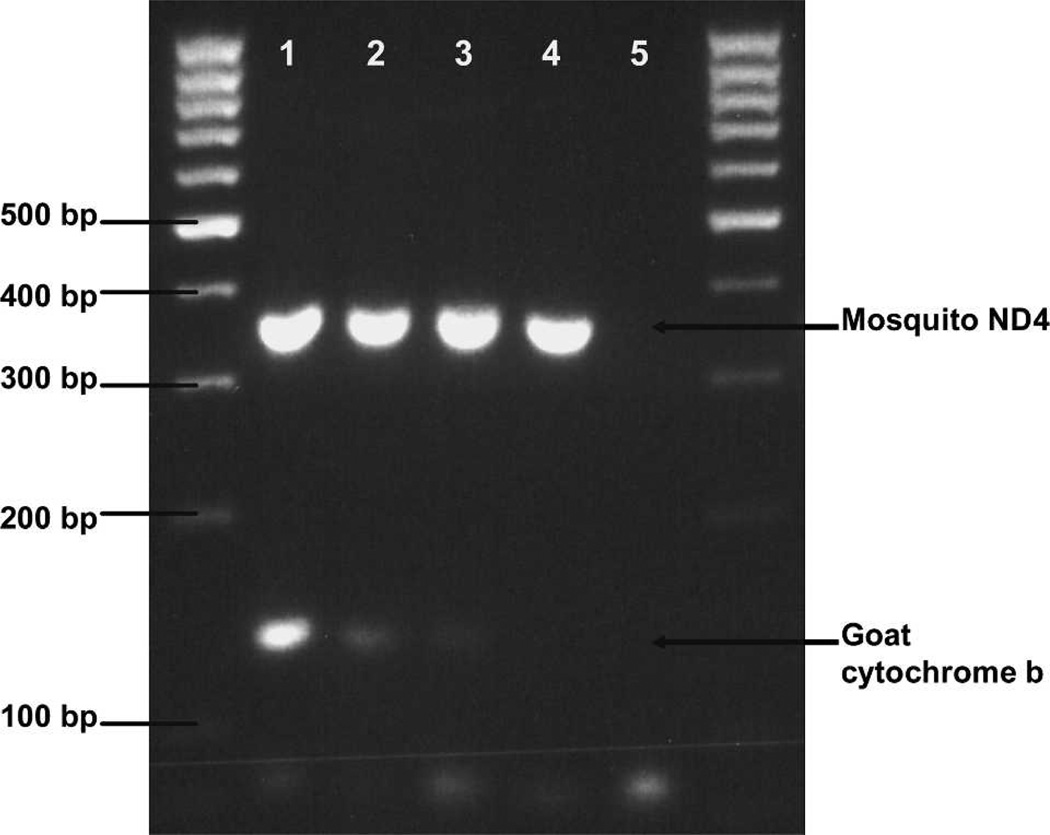
Time course analysis showed that host DNA was detectable in frozen mosquito abdomens 24–30 hours post-feeding on known animal bloods. All animal blood meals tested from mosquitoes frozen on the day of feeding and 24 and 30 hours post-feeding were successfully identified. However, at 48 hours post-feeding, only the blood meals from mosquitoes fed on goat blood could be positively identified (Figure 4). Reamplification of 48-hour PCR products did not produce any product, suggesting that insufficient host DNA remained to be amplified. The ND4 PCR products subjected to electrophoresis simultaneously with cytochrome b PCR products for comparative purposes confirmed that the relative quality of all DNA templates was approximately equal (Figure 4).
Figure 4.
Time course analysis for mosquitoes fed on goat blood. Lane 1, day 0; lane 2, 24 hours post-feeding; lane 3, 48 hours; lane 4, 72 hours; lane 5, negative control. Each lane also includes NADPH dehydrogenase (ND4) polymerase chain reaction products simultaneously subjected to electrophoresis as a template control. Outside lanes are 100-basepair DNA ladders. bp = basepairs.
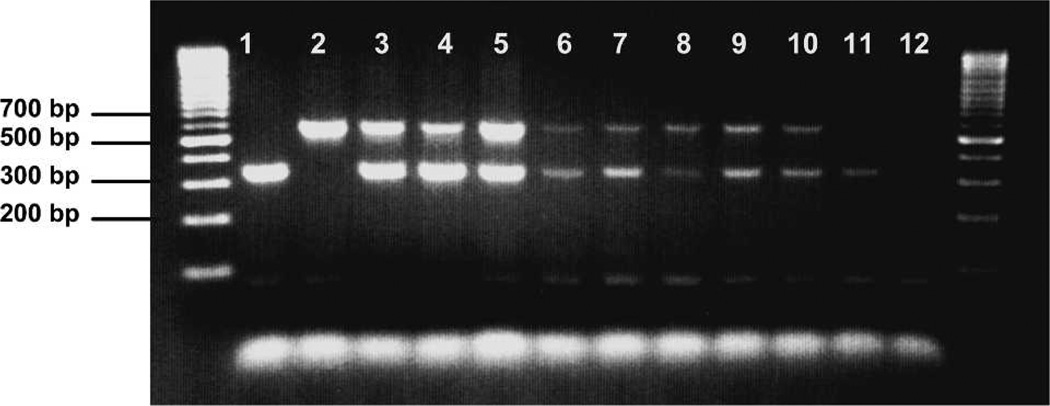
Finally, amplification of template derived from mosquito abdomens containing mixed blood confirmed that multiple blood meals from different mammals could be detected in a single mosquito. Figure 5 shows PCR products obtained from mosquitoes fed on disproportionate ratios of mixed cow and human blood. Both cow and human bloods were clearly detected in all mosquitoes tested for each proportion and timepoint. Multiple blood meals were also detected in three field samples; a blood meal containing human plus cow blood was detected in one An. gambiae s.s., human plus goat blood in one An. funestus s.s., and cow plus goat blood in one An. leesoni.
Figure 5.
Ethidium bromide–stained agarose gel simulating multiple feedings in single mosquitoes. Mosquitoes were fed on disproportionate ratios of human and cow blood and tested 4 and 12 hours post-feeding. Lanes 1–5, whole blood positive controls. Lanes 6–11, DNA extracted from control mosquitoes fed on mixed animal bloods. Lane 1, human blood; lane 2, cow blood; lane 3, 50:50 cow:human blood; lane 4, 30:70 cow:human blood; lane 5, 70:30 cow:human blood; lane 6: 50:50 cow:human, 4 hours; lane 7, 30:70 cow:human, 4 hours; lane 8, 70:30 cow:human, 4 hours; lane 9, 50:50 cow:human, 12 hours; lane 10, 70:30 cow:human, 12 hours; lane 11, 30:70 cow:human, 12 hours; lane 12, negative control. Outside lanes are 100-basepair DNA ladders. bp = basepairs.
DISCUSSION
Species-specific cytochrome b fragments of the predicted size were amplified from cow, human, pig, goat, and dog blood using a novel, multiplexed, vertebrate-specific primer set. Successful amplification was obtained using template DNA from whole blood, mosquitoes fed on blood of known animal origin, and blood-fed field specimens (Figures 1 and 3). For field specimens where blood meal identification failed, host DNA concentrations may be insufficient for detection due to blood meal volume, the processes of digestion may have denatured host DNA, or the mosquito may have fed on an animal not included in the diagnostic assay. In 16% of field specimens from which blood meals failed to amplify initially, reamplification of those apparently negative PCR products resulted in visible products, suggesting that host DNA was at low concentration in the source extraction. To demonstrate that the lack of host cytochrome b PCR product was not due to poor extraction quality, a PCR for arthropod mitochondrial ND4 was conducted for all samples. All ND4 PCRs produced a strong product, which confirmed that the quality of the extractions was sufficient and that the lack of a host cytochrome b PCR product after 48 hours in the time course experiments and in some field samples was most likely due to the process of digestion within the mosquito gut. Multiple sequence alignments with additional domestic and wild animals not included in the diagnostic assay showed that this multiplexed primer set may also amplify cytochrome b from wart hog and Cape buffalo. However, these primers are not predicted to amplify sheep, wild dog, water buffalo, baboon, vervet monkey, or chimpanzee cytochrome b due to at least five nucleotide differences in the forward priming region.
Abdomens were extracted separately from the head and thorax to maximize the host DNA concentration in each sample; however, detection of miniscule amounts of host DNA in a blood meal remained challenging. Mukabana and others successfully amplified human blood from An. gambiae blood meals estimated to contain between 2 and 82 ng of human DNA.19 Our experimentation suggested that at least 50 ng, but usually 150–200 ng, of template DNA from engorged mosquito abdomens was necessary for strong, visible amplification from blooded mosquito abdomens, particularly dry field specimens. This concentration was readily optimized by using 2–3 µL template from a 50-µL DNA extraction in each PCR. Furthermore, anopheline mosquitoes are known to concentrate erythrocytes during blood feeding by a factor of 1.2–2 times the normal human hematocrit, therefore increasing the concentration of host DNA in the midgut and the probability of obtaining good-quality template for PCR identification.24,25
An alternative multiplexed primer set is also reported that gives a series of species-specific bands distinguishable by size on an agarose gel (Figure 2). All products amplified at the predicted size, except for preferential amplification of the 680-bp band in mosquitoes fed on dog blood. Although either primer set may be used successfully for blood meal identification, the first multiplexed set (Figure 1) is recommended over the alternative primer set due to the increased sensitivity derived from amplification of a single diagnostic band from each host.
Validation of this PCR diagnostic assay used field specimens that were stored dry on silica desiccant for 2–7 months prior to DNA extraction. These conditions further demonstrate the utility of this approach to field-based research when specimens cannot always be processed immediately. Interestingly, these preliminary field data suggest exophagic and endophilic behaviors for An. leesoni in Zambia, since the eight blood meals tested and identified were taken from either cattle or goat, and mosquitoes had been collected inside human sleeping huts. A member of the An. funestus species complex, An. leesoni, has previously been collected resting indoors, but this species is not a known vector of malaria and no data currently exist on its feeding behavior.26–29 The human blood meals detected in An. gambiae s.s., An. arabiensis, and An. funestus s.s. are consistent with their status as primary vectors of Plasmodium falciparum in both Zambian and Malian study regions.26,27 This diagnostic assay will be extremely useful for entomologically based research projects involving blood-feeding behavior and vectorial capacity of mosquito vectors in areas of the world endemic for malaria and other mosquito-transmitted pathogens.
The time course experiments showed that host DNA was detectable in fed control mosquitoes as long as 48 hours. Blood meals from mosquitoes fed on all the different animal bloods could be clearly identified 24–30 hours post-feeding. However, goat blood meals were the only blood meals identifiable as long as 48 hours following blood ingestion. These results could be due to higher priming efficiency of the goatspecific primer, or the relatively small size of the goat-specific product (only 132 bp versus 334–680 bp for the other animals) may have increased its likelihood of detection after 48 hours of exposure to digestive enzymes in the mosquito midgut. These results demonstrate that identification of the blood meals in mosquitoes is possible between 24 and 48 hours post-feeding. Therefore, the blood meals of mosquitoes collected inside houses either by aspiration or pyrethrum spray catch are identifiable by this PCR diagnostic assay for the 24–36-hour duration spent resting inside houses before exiting in search of oviposition sites.
These data fall within the range of expectations based the results of previous studies. Mukabana and others examined how digestion affected the amplification success of human DNA from the blood meals of An. gambiae. They found that digestion between 0 and 8 hours had no significant effect on the ability to obtain PCR products; however, between 8 and 32 hours post-feeding, the proportion of blood meals that could be successfully profiled decreased steadily, dropping below 50% at 15 hours.19 Boake and others detected human blood meals in black flies (Simulium damnosum s.l.) up to 72 hours post-feeding.15 Similarly, Ngo and Kramer were able to detect avian blood meals in Cx. p. pipiens L. up to 72 hours post-feeding.17 Lastly, Lee and others detected Japanese quail blood meals in Cx. tarsalis Coquillett as long as seven days post-feeding.16 Variation in the DNA yields from different extraction procedures, different digestive processes in black flies compared with mosquitoes, or the higher DNA content of nucleated bird blood versus mammalian blood may account for these longer detection periods. These time course results are directly comparable with those obtained with an ELISA, in which human blood meals were accurately detected up to 32 hours post-feeding in dried specimens and 23 hours post-feeding in frozen specimens.2
Gonotrophic discordance, or taking multiple blood meals during the gonotrophic cycle, has been reported in many different mosquito taxa.30–37 In these studies, ≤10% to upwards of 40% of blood-fed field specimens contained multiple blood meals. This host-feeding behavior can influence pathogen transmission through increased frequency of vector-human contact, or possibly reduce vector-human contact if some blood meals are taken from alternative mammalian hosts. To address these possibilities, templates from mosquitoes fed on mixed blood of known animal sources in disproportionate ratios were tested to simulate conditions where multiple blood meals present in a mosquito abdomen will either have undergone differing amounts of digestion at the time of detection or may have been imbibed in differing volumes. It was possible to detect blood meals from multiple sources in both control mosquitoes fed on mixed blood, as well as in field samples using this diagnostic assay, a tool that we anticipate will be extremely useful in studies concerning mosquitofeeding behavior. We detected one mixed cow plus human blood meal in An. gambiae s.s. from 61 individuals in which we obtained an identifiable product. Cattle feeding by this species has also been reported in An. gambiae s.s. from western Kenya and The Gambia.38,39 In addition to our single occurrence of An. funestus s.s. with a mixed human plus goat blood meal, Wanji and others also reported one field specimen of An. funestus s.s. that had fed on both human and goat blood in Cameroon.40 Goat feeding by An. funestus s.s. was also found in Kenya by Mwangangi and others.41
It is our hope that this diagnostic assay will serve as a valuable tool for blood meal identification that may be applied to mosquito vectors with catholic feeding behaviors. This multiplexed primer set successfully identified the blood meals from five mammal species, was validated for use on dry field specimens, and could reliably identify all five animal blood meals up to 24–30 hours post-feeding. Additionally, multiple blood meals from different mammalian hosts were detected from single mosquito abdomens. A final important advantage of this assay is that it directly identifies the mammalian host by size-differentiated DNA fragments on agarose gels in a single step. This diagnostic assay was designed for use in African villages where mosquitoes such as An. arabiensis may feed on a variety of domestic animals in addition to humans. However, the application of this tool would be appropriate in any environment or disease system where vector arthropods exhibit zoophilic feeding behavior for non-human mammals, for example Japanese encephalitis virus where pigs and humans are hosts, and Rift Valley fever virus where cattle and humans are affected hosts for mosquito vectors.42
Acknowledgments
We thank Dr. Sungano Mharakurwa and Dr. Phil Thuma for their time and effort spent coordinating mosquito collections and field team operations in Zambia, as well as Harry Hamapumbu, Patricia Muleya, Petros Moono, Fidelis Chanda, Lushomo Chikobolo, Collence Munsanje, Rodwell Moono, Peter Simakwati, Guide Hansumo, Seen Mudenda, Betham Dubeka, and the Macha Malaria Research Institute staff for their assistance. We also thank Dr. Guimogo Dolo for his assistance with collections in Mali.
Financial support: This research was supported in part by financial assistance to Douglas E. Norris from the Johns Hopkins Malaria Research Institute, an award from the United National Development Program/World Bank/World Health Organization Special Program for Research and Training in Tropical Diseases (TDR) (A10360) to Douglas E. Norris, a Johns Hopkins Malaria Research Institute predoctoral fellowship award to Rebekah J. Kent, and a National Institute of Environmental Health Sciences training award (T32ES07141) to Rebekah J. Kent.
REFERENCES
- 1.Boorman J, Mellor PS, Boreham PFL, Hewett RS. A latex agglutination test for the identification of blood-meals of Culicoides (Diptera: Ceratopogonidae) Bull Entomol Res. 1977;67:305–311. [Google Scholar]
- 2.Beier JC, Perkins PV, Wirtz RA, Koros J, Diggs D, Gargan TP, II, Koech DK. Bloodmeal identification by direct enzyme-linked immunosorbent assay (ELISA), tested on Anopheles (Diptera: Culicidae) in Kenya. J Med Entomol. 1988;25:9–16. doi: 10.1093/jmedent/25.1.9. [DOI] [PubMed] [Google Scholar]
- 3.Washino RK, Tempelis CH. Mosquito host bloodmeal identification: methodology and data analysis. Annu Rev Entomol. 1983;28:179–201. doi: 10.1146/annurev.en.28.010183.001143. [DOI] [PubMed] [Google Scholar]
- 4.Gomes LAM, Duarte R, Lima DC, Diniz BS, Serrão ML, Labarthe N. Comparison between precipitin and ELISA tests in the bloodmeal detection of Aedes aegypti (Linnaeus) and Aedes fluviatilis (Lutz) mosquitoes experimentally fed on feline, canine, and human hosts. Mem Inst Oswaldo Cruz. 2001;96:693–695. doi: 10.1590/s0074-02762001000500020. [DOI] [PubMed] [Google Scholar]
- 5.Gokool S, Curtis CF, Smith DF. Analysis of mosquito bloodmeals by DNA profiling. Med Vet Entomol. 1993;7:208–215. doi: 10.1111/j.1365-2915.1993.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 6.Lord WD, DiZinno JA, Wilson MR, Budowle B, Tapalin D, Meinking TL. Isolation, amplification, and sequencing of human mitochondrial DNA obtained from human crab louse, Pthirus pubis (L.), blood meals. J Forensic Sci. 1998;43:1097–1100. [PubMed] [Google Scholar]
- 7.Bataille M, Crainic K, Leterreux M, Durigon M, deMazancourt P. Multiplex amplification of mitochondrial DNA for human and species identification in forensic evaluation. Forensic Sci Int. 1999;99:165–170. doi: 10.1016/s0379-0738(98)00185-6. [DOI] [PubMed] [Google Scholar]
- 8.Chow-Shaffer E, Sina B, Hawley WA, deBenedictis J, Scott TW. Laboratory and field evaluation of polymerase chain reaction-based forensic DNA profiling for use in identification of human blood meal sources of Aedes aegypti (Diptera: Culicidae) J Med Entomol. 2000;37:492–502. doi: 10.1603/0022-2585-37.4.492. [DOI] [PubMed] [Google Scholar]
- 9.Ansell J, Hu JT, Gilbert SC, Hamilton KA, Hill AV, Lindsay SW. Improved method for distinguishing the human source of mosquito blood meals between close family members. Trans R Soc Trop Med Hyg. 2000;94:572–574. doi: 10.1016/s0035-9203(00)90092-0. [DOI] [PubMed] [Google Scholar]
- 10.Bottero MT, Dalmasso IA, Nucera D, Turi RM, Rosati S, Squadrone S, Goria M, Civera T. Development of a PCR assay for the detection of animal tissues in ruminant feeds. J Food Prot. 2003;66:2307–2321. doi: 10.4315/0362-028x-66.12.2307. [DOI] [PubMed] [Google Scholar]
- 11.deBenedictis J, Chow-Shaffer E, Costero A, Clark GA, Edman JD, Scott TW. Identification of the people from whom engorged Aedes aegypti took blood meals in Florida, Pureto Rico, using polymerase chain reaction-based DNA profiling. Am J Trop Med Hyg. 2003;68:4437–4446. [PubMed] [Google Scholar]
- 12.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 13.Irwin DM, Kocher TD, Wilson AC. Evolution of the cytochrome b gene of mammals. J Mol Evol. 1991;32:128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- 14.Kirstein F, Gray JS. A molecular marker for the identification of the zoonotic reservoirs of Lyme borreliosis by analysis of the blood meal in its European vector Ixodes ricinus. Appl Environ Microbiol. 1996;62:4060–4065. doi: 10.1128/aem.62.11.4060-4065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boake DA, Tang J, Truc P, Merriweather A, Unnasch TR. Identification of bloodmeals in haematophagous Diptera by cytochrome B heteroduplex analysis. Med Vet Entomol. 1999;13:282–287. doi: 10.1046/j.1365-2915.1999.00193.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee JH, Hassan H, Hill G, Cupp EW, Higazi TB, Mitchell CJ, Godsey MS, Jr, Unnasch TR. Identification of mosquito avian-derived blood meals by polymerase chain reaction-heteroduplex analysis. Am J Trop Med Hyg. 2002;66:599–604. doi: 10.4269/ajtmh.2002.66.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ngo KA, Kramer LD. Identification of mosquito bloodmeals using polymerase chain reaction (PCR) with order-specific primers. J Med Entomol. 2003;40:215–222. doi: 10.1603/0022-2585-40.2.215. [DOI] [PubMed] [Google Scholar]
- 18.Pichon B, Egan D, Rogers M, Gray J. Detection and identification of pathogens and host DNA in unfed host-seeking Ixodes ricinus L. (Acari: Ixodidae) J Med Entomol. 2003;40:723–731. doi: 10.1603/0022-2585-40.5.723. [DOI] [PubMed] [Google Scholar]
- 19.Mukabana WR, Takken W, Seda P, Killeen GF, Hawley WA, Knols BGJ. Extent of digestion affects the success of amplifying human DNA from blood meals of Anopheles gambiae (Diptera: Culicidae) Bull Entomol Res. 2002;92:233–239. doi: 10.1079/BER2002164. [DOI] [PubMed] [Google Scholar]
- 20.Norris DE, Shurtleff AC, Touré YT, Lanzaro GC. Microsatellite DNA polymorphism and heterozygosity among field and laboratory populations of Anopheles gambiae ss (Diptera: Culicidae) J Med Entomol. 2001;38:336–340. doi: 10.1603/0022-2585-38.2.336. [DOI] [PubMed] [Google Scholar]
- 21.Gorrochotegui-Escalante N, DeLourdes Munoz M, Fernandez- Salas I. Genetic isolation by distance among Aedes aegypti populations along the northeastern coast of Mexico. Am J Trop Med Hyg. 2000;62:200–209. doi: 10.4269/ajtmh.2000.62.200. [DOI] [PubMed] [Google Scholar]
- 22.Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701. [Google Scholar]
- 23.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. http:// frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi. [DOI] [PubMed] [Google Scholar]
- 24.Vaughan JA, Noden BH, Beier JC. Concentrations of human erythrocytes by anopheline mosquitos (Diptera: Culicidae) during feeding. J Med Entomol. 1991;28:780–786. doi: 10.1093/jmedent/28.6.780. [DOI] [PubMed] [Google Scholar]
- 25.Briegel H, Rezzonico L. Concentration of host blood protein during feeding by anopheline mosquitoes (Diptera: Culicidae) J Med Entomol. 1985;22:612–618. doi: 10.1093/jmedent/22.6.612. [DOI] [PubMed] [Google Scholar]
- 26.Gillies MT, DeMeillon B. The Anophelinae of Africa South of the Sahara (Ethiopian Zoogeographical Region) Second edition. Johannesburg, South Africa: Publication of the South African Institute for Medical Research No. 54; 1968. [Google Scholar]
- 27.Gillies MT, Coetzee M. A Supplement to the Anophelinae of Africa South of the Sahara. Johannesburg, South Africa: Publication of the South African Institute for Medical Research No. 55; 1987. [Google Scholar]
- 28.Hargreaves K, Koekemoer LL, Brooke BD, Hunt RH, Mthembu J, Coetzee M. Anopheles funestus resistant to pyrethroid insecticides in South Africa. Med Vet Entomol. 2000;14:181–189. doi: 10.1046/j.1365-2915.2000.00234.x. [DOI] [PubMed] [Google Scholar]
- 29.Koekemoer LL, Kamau L, Hunt RH, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am J Trop Med Hyg. 2002;6:804–811. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
- 30.Boreham PF, Jenahan JK, Boulzaguet R, Storey J, Ashkar TS, Nambian, Matsushima T. Studies on multiple feeding by Anopheles gambiae s.l. in a Sudan savanna area of northern Nigeria. Trans R Soc Trop Med Hyg. 1979;73:418–423. doi: 10.1016/0035-9203(79)90167-6. [DOI] [PubMed] [Google Scholar]
- 31.Scott TW, Chow E, Strickman D, Kittayapong P, Wirtz RA, Lorenz LH, Edman JD. Blood-feeding patterns of Aedes aegypti (Diptera: Culicidae) collected in a rural Thai village. J Med Entomol. 1993;30:922–927. doi: 10.1093/jmedent/30.5.922. [DOI] [PubMed] [Google Scholar]
- 32.Klowden MJ, Briegel H. Mosquito gonotrophic cycle and multiple feeding potential: contrasts between Anopheles and Aedes (Diptera: Culicidae) J Med Entomol. 1994;31:618–622. doi: 10.1093/jmedent/31.4.618. [DOI] [PubMed] [Google Scholar]
- 33.Wekesa JW, Yuval B, Washino RK. Multiple blood feeding in Anopheles freeborni (Diptera: Culicidae) Am J Trop Med Hyg. 1995;52:508–511. doi: 10.4269/ajtmh.1995.52.508. [DOI] [PubMed] [Google Scholar]
- 34.Anderson RA, Brust RA. Field evidence for multiple host contacts during blood feeding by Culex tarsalis, Cx. restuans and Cx. nigripalpis (Diptera: Culicidae) J Med Entomol. 1995;32:705–710. doi: 10.1093/jmedent/32.5.705. [DOI] [PubMed] [Google Scholar]
- 35.Koella JC, Sorensen FL, Anderson RA. The malaria parasite, Plasmodium falciparum increases the frequency of multiple feeding of its mosquito vector, Anopheles gambiae. Proc R Soc Lond B Biol Sci. 1998;265:763–768. doi: 10.1098/rspb.1998.0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amerasinghe PH, Amerasinghe FP. Multiple host feeding in field populations of Anopheles culicifacies and An. subpictus in Sri Lanka. Med Vet Entomol. 1999;13:124–131. doi: 10.1046/j.1365-2915.1999.00160.x. [DOI] [PubMed] [Google Scholar]
- 37.Scott TW, Amerasinghe PH, Morrison AC, Lorenz LH, Clark GG, Strickman D, Kittayapong P, Edman JD. Longitudinal studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: blood feeding frequency. J Med Entomol. 2000;37:89–101. doi: 10.1603/0022-2585-37.1.89. [DOI] [PubMed] [Google Scholar]
- 38.Shililu JI, Maier WA, Seitz HM, Orago AS. Seasonal density, sporozoite rates and entomological inoculation rates of Anopheles gambiae and Anopheles funestus in a high-altitude sugarcane growing zone in Western Kenya. Trop Med Int Health. 1998;3:706–710. doi: 10.1046/j.1365-3156.1998.00282.x. [DOI] [PubMed] [Google Scholar]
- 39.Bogh C, Clarke SE, Pinder M, Sanyang F, Lindsay SW. Effect of passive zooprophylaxis on malaria transmission in The Gambia. J Med Entomol. 2001;38:822–828. doi: 10.1603/0022-2585-38.6.822. [DOI] [PubMed] [Google Scholar]
- 40.Wanji S, Tanke T, Atanga SN, Ajonina C, Nicholas T, Fontenille D. Anopheles species of the mount Cameroon region: biting habits, feeding behaviour and entomological inoculation rates. Trop Med Int Health. 2003;8:643–649. doi: 10.1046/j.1365-3156.2003.01070.x. [DOI] [PubMed] [Google Scholar]
- 41.Mwangangi JM, Mbogo CM, Nzovu JG, Githure JI, Guiyun Y, Beier JC. Blood-meal analysis for anopheline mosquitoes sampled along the Kenyan coast. J Am Mosq Control Assoc. 2003;19:371–375. [PubMed] [Google Scholar]
- 42.Eldridge BF, Scott TW, Day JF, Tabachnick TJ. Arbovirus Diseases. In: Eldridge BF, Edman JD, editors. Medical Entomology: A Textbook on Public Health and Veterinary Problems Caused by Arthropods. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 415–461. [Google Scholar]