Abstract
Objective
To specify the problem of patient-level temporal aggregation from clinical text and introduce several probabilistic methods for addressing that problem. The patient-level perspective differs from the prevailing natural language processing (NLP) practice of evaluating at the term, event, sentence, document, or visit level.
Methods
We utilized an existing pediatric asthma cohort with manual annotations. After generating a basic feature set via standard clinical NLP methods, we introduce six methods of aggregating time-distributed features from the document level to the patient level. These aggregation methods are used to classify patients according to their asthma status in two hypothetical settings: retrospective epidemiology and clinical decision support.
Results
In both settings, solid patient classification performance was obtained with machine learning algorithms on a number of evidence aggregation methods, with Sum aggregation obtaining the highest F1 score of 85.71% on the retrospective epidemiological setting, and a probability density function-based method obtaining the highest F1 score of 74.63% on the clinical decision support setting. Multiple techniques also estimated the diagnosis date (index date) of asthma with promising accuracy.
Discussion
The clinical decision support setting is a more difficult problem. We rule out some aggregation methods rather than determining the best overall aggregation method, since our preliminary data set represented a practical setting in which manually annotated data were limited.
Conclusion
Results contrasted the strengths of several aggregation algorithms in different settings. Multiple approaches exhibited good patient classification performance, and also predicted the timing of estimates with reasonable accuracy.
Keywords: Patient classification, Asthma epidemiology, Natural language processing, Information extraction
Background and significance
With the rapid adoption of electronic medical records (EMRs) and the increase of clinical text in electronic form, clinical natural language processing (NLP) is a pivotal research concern in the medical informatics community. Extensive innovation has been applied to core language-related problems like negation detection,1 named entity recognition (NER),2 and coreference resolution.3 4 Successful applications of NLP techniques have ranged from specific clinical problems like peripheral arterial disease5 to ancillary information like smoking status.6 Many of these applications are interested in NLP-based inferences about whole patients and their status at the present time. However, NLP techniques have overwhelmingly been evaluated at the term,7 event,8 sentence,9 document,6 or visit,10 11 level.
In this article, we develop and evaluate techniques for patient-level temporal aggregation. We do so through the lens of text-based ascertainment of a pediatric asthma status for clinical care and research.12 This is medically significant because asthma is the most common chronic illness in childhood, has significant comorbidities,13 14 and is often not promptly diagnosed (or treated).15 16
The use case of pediatric asthma reveals some interesting aspects of patient-level analysis. First, patient-level analysis by its nature faces the challenge of time-distributed evidence—relevant information can be distributed across multiple documents at different time points. This is particularly important for studying chronic diseases such as asthma. Second, a patient's status may evolve over time, and estimates of status change can be made. Third, clinical text is an excellent (but not exhaustive) source of clinical information, and can be harnessed with information extraction techniques. Finally, patient-level evaluations require a substantial amount of infrastructure and resources: longitudinal electronic records must be both available and annotated consistently.
Note that our purpose is not to design the most accurate system for pediatric asthma status ascertainment, as in other work,12 but to provide a preliminary testbed for the novel problem of patient-level temporal aggregation. Our evaluations explore the temporal evolution of a patient's health while preserving the common task of patient classification. Our previous work on the same data set12 used only the most basic of temporal aggregation techniques (Sum aggregation) and a different machine learning classifier. While temporal expressions and temporal relation extraction have been explored in clinical NLP,17 18 they have not typically been approached from the standpoint of patient-level temporal aggregation. In contrast, structured data have been visualized by systems such as LifeLines,19 KNAVE,20 and TimeLine,21 but the data have been infrequently integrated with NLP processing or with secondary use problems. Furthermore, the visualization of time-stamped observations does not solve the problem of how disparate observations should be weighed against each other for a given task. Another active area of research is reasoning and inference over the temporal events, often using ontologies or standards such as CNTRO.8 This event-level reasoning is important, but different from the patient-level status ascertainment we discuss here.
Below, we describe the problem setting, motivate and introduce six methods for patient-level temporal aggregation, present evaluation results, and discuss comparisons, limitations, and the conclusions of our work.
Materials and methods
Problem setting: pediatric asthma
As context for the issue of temporal aggregation, figure 1 shows our overall study design. We evaluated NLP-based systems (bottom branch) against manual medical record reviews (top branch).
Figure 1.
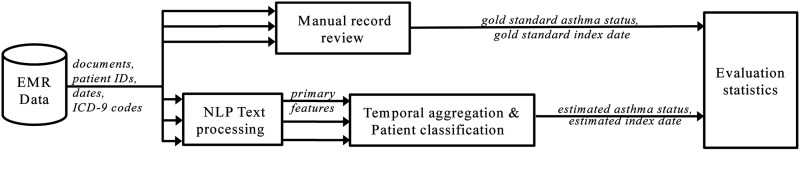
Overall study design, comparing automatic methods (NLP+classification) to manual record review. Multiple arrows indicate that each patient may have more than one document. EMR,electronic medical record; NLP, natural language processing.
The gold standard asthma status was labeled according to manual chart review as a binary 0 or 1 for each patient, following established asthma criteria22 (see figure 2). The criteria were found to have high inter-annotator agreement and have been extensively used in research for asthma epidemiology.22–31 Each system or algorithm produced an estimated asthma status that could be compared to the gold standard asthma status for accuracy.
Figure 2.
Established criteria for retrospective asthma ascertainment. Highlighted terms were named entity (NE) concepts that were eventually used as primary features in classification.
Multiple arrows in figure 1 illustrate the setting of time-distributed evidence: namely, that each patient may have multiple documents associated with his or her medical record, which may or may not suggest an asthma status. To capture the temporal progression of asthma status, manual record reviews tracked an index date, that is, the first date (document) for which the patient should have been considered to have asthma. Thus, non-asthmatic patients had no index date associated with their records. To determine whether there is a delay in diagnosis for patients who have asthma, the temporal aggregation and patient classification step (bottom branch of figure 1) must also produce an estimated index date—an inference as to when a diagnosis should have occurred. Direct comparison of these dates yields an evaluation metric of time-estimation accuracy.
We used days as the unit of time in all our analyses unless otherwise specified. Also, the abstraction date was recorded, indicating when the manual chart review was performed; for all analyses, records beyond the abstraction date were excluded for fair comparison.
NLP as feature extraction
In this work, automatic classification of patients’ asthma statuses was done using both machine learning methods and a logic-based method, using a limited set of features that we will describe here.12 A filtered set of custom named entities (NEs), reflecting the criteria of figure 2, were considered to be primary features in the data set; NEs found in historical sections or in negated contexts were excluded. Thus, for a document at time t, a valid NE was considered to be a positive binary feature f(t)=1; if an NE was filtered out or not present at all, a binary feature f(t)=0 was assigned to the document.
Basic processing of the clinical text was done with cTAKES V.1.3.2. This clinical NLP tool analyzes text at the document level, sentence level, and word (token) level; discovers grammatical structure; and most importantly, finds identifiable medical concepts in text (the task of NER). We modified cTAKES V.1.3.2 to discover the criteria-based primary features. Variants of the highlighted words in figure 2 were included as NEs in the custom dictionary; we also crafted additional high-precision long-range negation detection rules.
Note that finding some of the features using other EMR data sources (eg, structured data from laboratories) is completely consistent with the temporal aggregation approach to be introduced. We have constrained ourselves to an NLP-based feature extraction because of the need to resolve instances when both structured and unstructured data are present. For example, if structured data describe the results of a test, but clinical text describes one the day before, should the patient get ‘credit’ for two events? Although this problem of coreference also exists in the text itself, we have sought to reduce the variability of timing estimates by excluding structured data sources in this exploratory methodological work.
Characteristics of time-distributed evidence
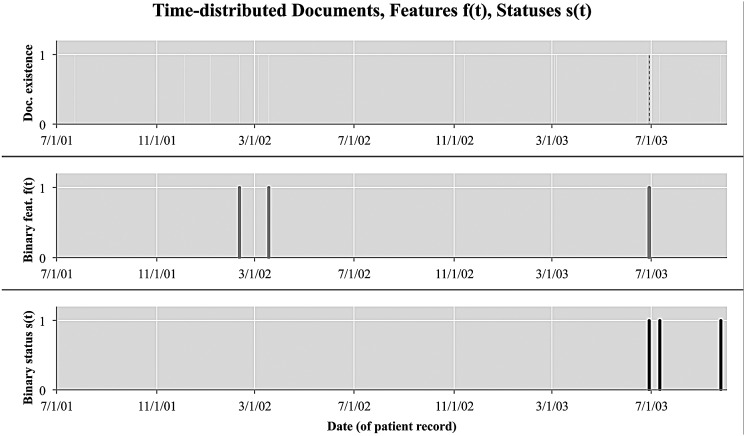
Here, we typify the time-distributed evidence setting, and analyze the temporal characteristics of patient-level data. Consider figure 3, in which a patient will have associated clinical documents distributed over a timeline (top). Some of these documents have some relevant evidence in the form of binary features, that is, f(t)=1, and some do not, that is, f(t)=0 (middle). Furthermore, given a manually annotated index date, here 6/28/03, we can calculate the gold standard asthma status of a patient at each point in time (bottom). In this work, we only consider time points t at which an EMR document was generated about a patient.
Figure 3.
Just as medical documents for a patient are distributed over time, extracted features f(t) and asthma statuses s(t) are also distributed over time. s(t) has been inferred as positive for all documents after the manually annotated index date.
Practically speaking, we can thus infer asthma statuses at any point in the document history (see the final column of table 1 discussed in the next section). If the same patient's history had been examined 2 weeks earlier on 6/13/03, documentation up to that point would still have insufficient evidence to consider the patient asthmatic. On 6/28/03, new information added to the patient history allowed that patient to be classified as asthmatic. On 7/11/03, the patient was still asthmatic, even though there was no further evidence of asthma based on feature f(t).
Table 1.
Features f(t), six different aggregation methods on the 23rd primary feature (bronchodilator use), and statuses s(t) for a set of time-distributed documents from one de-identified patient
| Date | f23(t) | Or | Sum | PDF-IN | CDF | PDF-NC | PDF-ST | s23(t) |
|---|---|---|---|---|---|---|---|---|
| 7/23/01 | 0 | 0 | 0 | 0 | 0 | 0.96×10−3 | 0 | 0 |
| 12/5/01 | 0 | 0 | 0 | 0 | 0 | 1.25×10−3 | 0 | 0 |
| 1/5/02 | 0 | 0 | 0 | 0 | 0 | 1.32×10−3 | 0 | 0 |
| 2/10/02 | 1 | 1 | 1 | 0.58×10−3 | 0.396 | 1.39×10−3 | 0.482 | 0 |
| 3/5/02 | 0 | 1 | 1 | 0.59×10−3 | 0.410 | 1.42×10−3 | 0.500 | 0 |
| 3/18/02 | 1 | 1 | 2 | 1.17×10−3 | 0.813 | 1.43×10−3 | 0.991 | 0 |
| 11/13/02 | 0 | 1 | 2 | 0.97×10−3 | 1.078 | 1.34×10−3 | 1.282 | 0 |
| 3/4/03 | 0 | 1 | 2 | 0.87×10−3 | 1.180 | 1.34×10−3 | 1.354 | 0 |
| 3/6/03 | 0 | 1 | 2 | 0.87×10−3 | 1.181 | 1.34×10−3 | 1.355 | 0 |
| 6/13/03 | 0 | 1 | 2 | 0.79×10−3 | 1.263 | 1.37×10−3 | 1.415 | 0 |
| 6/28/03 | 1 | 1 | 3 | 1.36×10−3 | 1.671 | 1.36×10−3 | 1.904 | 1 |
| 7/11/03 | 0 | 1 | 3 | 1.36×10−3 | 1.688 | 1.36×10−3 | 1.921 | 1 |
| 9/24/03 | 0 | 1 | 3 | 1.31×10−3 | 1.789 | 1.31×10−3 | 2.009 | 1 |
CDF, cumulative index date.
Patient-level temporal aggregation methods
Here, we introduce several methods to aggregate features from the document level to the patient level. In each aggregation method, we re-assigned feature values 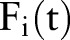 in place of the primary features
in place of the primary features 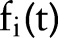 , and the aggregated features were used for training and testing machine learning classifiers. After some preliminary cross-validation tests comparing different classifiers in the Weka machine learning environment, our experiments used a simple logistic regression classifier. However, the re-assigned temporal aggregation values are applicable to any machine learning classifier that can handle real-valued features. For feature i at time t, we denote
, and the aggregated features were used for training and testing machine learning classifiers. After some preliminary cross-validation tests comparing different classifiers in the Weka machine learning environment, our experiments used a simple logistic regression classifier. However, the re-assigned temporal aggregation values are applicable to any machine learning classifier that can handle real-valued features. For feature i at time t, we denote 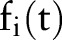 as the binary feature value,
as the binary feature value, 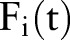 as the aggregated value, and
as the aggregated value, and 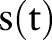 as the manually annotated binary asthma status.
as the manually annotated binary asthma status.
Or aggregation
Possibly the most common (but under-examined) way to do patient-level aggregation is to just consider all documents for a patient at the same time. This reduces the patient-level classification problem into a document classification problem. If any feature is discovered in a document before the current time t, it gets counted as having been discovered for that feature:
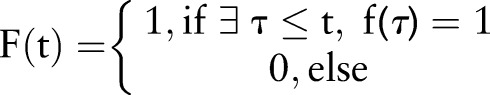 |
This Or operation across documents is extremely simple; however, it is somewhat naive in that it does not consider additional occurrences of a feature. Furthermore, it is sensitive to noise, since it considers any positive feature to be persistent throughout a patient's history, even if that feature was hedged, mistaken, or more or less predictive over time.
Sum aggregation
Another simple strategy that is often used (eg, in our previous work) is to count the frequency of positive features up to time t. All previous occurrences of a feature are considered to be accumulated evidence over the history of a patient. This addresses one weakness of Or aggregation, because additional evidence over time is recorded in the summed frequency count:
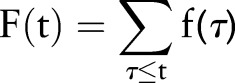 |
Note that, similar to Or aggregation, this is not truly a temporal aggregation method; it does not reflect when the aggregated features happened. For example, if a patient had two bouts of wheezing 5 years before time t, it would have the same effect as wheezing twice in the 5 weeks just before t.
Index date probability (PDF-IN) aggregation
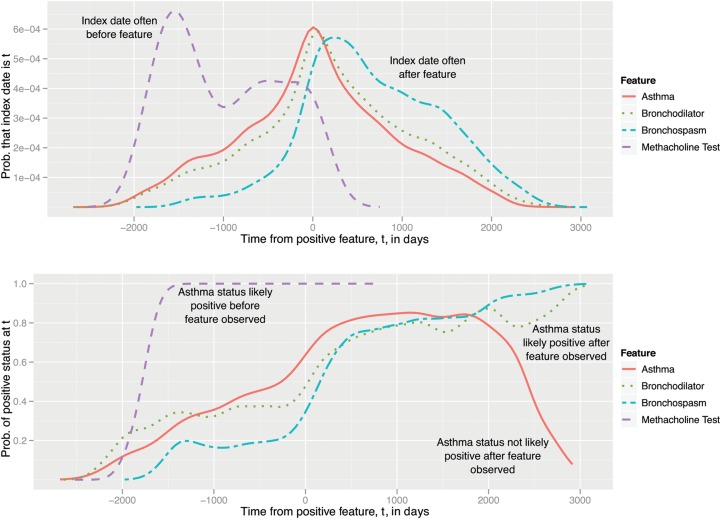
To account for the temporal aspect of evidence, we employed probability density functions (PDFs) using kernel density estimation, as we now describe. With conditional PDFs that specify probabilities at time t, we can examine how the gold standard annotations and features may be related to each other temporally. Two variations on PDF-based plots are shown in figure 4; these are estimated using a kernel density estimator with a Gaussian kernel, which is similar to a continuous version of a histogram.
Figure 4.
Top: Probability density function (PDF) for index date (p(t), used in PDF-IN, CDF, and PDF-NC), as estimated by kernel density estimation on a training set. Bottom: PDF for positive asthma status (a(t), used in PDF-ST) of non-zero primary features. CDF, cumulative index date.
PDF-IN (top plot) is a PDF based on the index date for asthma. At time t, all previous occurrences of a feature are considered, but they are weighted by how likely it is that the feature at time  implies that the index date is at time t:
implies that the index date is at time t:
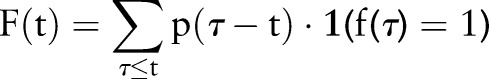 |
where the term 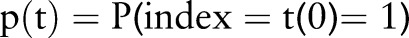 is the PDF and
is the PDF and 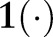 is ‘1’ when the condition is met and ‘0’ when it is not. This differs from Sum aggregation in that the feature counts are weighted by the index date probability.
is ‘1’ when the condition is met and ‘0’ when it is not. This differs from Sum aggregation in that the feature counts are weighted by the index date probability.
In the top plot of figure 4, the x-axis represents time t where a feature such as a mention of ‘Asthma’ would have been observed at time t=0. Each line is an estimated PDF for feature 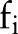 , answering the question: ‘We've observed feature
, answering the question: ‘We've observed feature 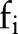 in a patient's record; what is the probability that the index date will occur t days away?’ This estimation only accounts for patients who are, in fact, positive for asthma.
in a patient's record; what is the probability that the index date will occur t days away?’ This estimation only accounts for patients who are, in fact, positive for asthma.
Analysis of this PDF yields some interesting characteristics. For example, the ‘Methacholine test’ feature most likely has its index date before the feature appears at t=0; thus, if we observe that a methacholine test is administered to a patient, the patient is almost sure to have already met the criteria for asthma. In contrast, the ‘Bronchodilator’ feature has a peak and substantial probability mass after the feature at t=0; this implies that bronchodilator use (with f(0)=1) commonly begins before the index date is assessed.
Cumulative distribution function (CDF) aggregation
While PDF-IN treats the temporal aspect of evidence aggregation, it is not clear that the probability of the index date is the ‘correct’ weighting to use. For example, in a clinical decision support setting, it might be more relevant to estimate whether the index date was likely to have occurred at any time before the current time t. Thus, we define a cumulative distribution function (CDF)-based aggregation method:
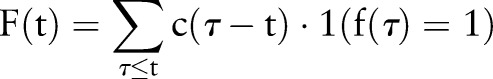 |
where the weighting function 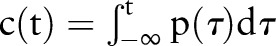 is the cumulative distribution function corresponding to PDF-IN.
is the cumulative distribution function corresponding to PDF-IN.
With a CDF, the weighting for a feature always increases over time, since CDFs are by definition non-decreasing functions. However, a weakness is that there may be features whose evidence becomes less confident over time, rather than more confident. Consider a feature that co-occurs with asthma but does not predict asthma long term. Having a positive feature 5 weeks ago should be better evidence than seeing it 5 years ago, but a CDF will always have higher weights for a feature more distant in the past.
Positive status probability (PDF-ST) aggregation
The bottom of figure 4 is an alternative probability-based aggregation method, which we term the positive status probability (PDF-ST). This distribution compares the probability of a positive binary asthma status to that of a negative one, at a given time t:
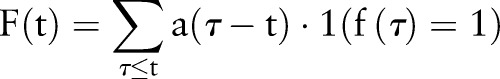 |
where 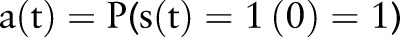 and these conditional probabilities are calculated as:
and these conditional probabilities are calculated as:
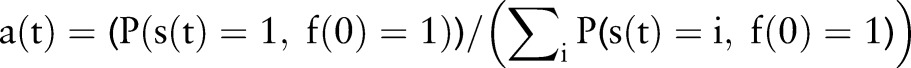 |
Multiple occurrences of a feature are weighted by how likely it is that the feature at time  implies the asthma status is positive at time t.
implies the asthma status is positive at time t.
This differs from PDF-IN in that it directly considers asthma status rather than index date. Examining the bottom of figure 4, if a ‘Methacholine test’ feature is observed, every document in the previous 1500 days would have been considered positive for asthma. Virtually all ‘bronchodilator’ users eventually become asthmatic (3000 days after that feature is observed), which emulates the CDF scenario of temporal feature weighting. However, a mention of ‘asthma’ itself does not necessarily indicate that a patient will eventually be indexed for asthma, and the PDF-ST method accounts for this. It is estimated with all cases of positive asthma status rather than only the index date.
Because PDF-ST directly estimates the probability of the current status, we can say that it is ‘designed’ for a clinical decision support scenario, in which the status at the current time is all that matters (the index date is less salient). Intuitively, this means that it may be a poorer fit to retrospective epidemiology settings.
Non-causal index date (PDF-NC) aggregation
Here, we define non-causal index date (PDF-NC) aggregation, which is based on the index date probability PDF-IN, but further tailored to a retrospective epidemiology setting:
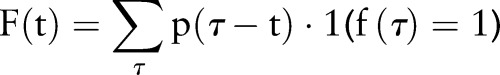 |
where 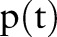 is the index date probability PDF-IN, as above. This differs from PDF-IN in that time points
is the index date probability PDF-IN, as above. This differs from PDF-IN in that time points 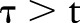 are considered during aggregation. In other words, it is a non-causal aggregation using the PDF (a causal system is one in which the output only depends on time points
are considered during aggregation. In other words, it is a non-causal aggregation using the PDF (a causal system is one in which the output only depends on time points 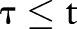 ). This is not a realistic way to ascertain asthma status for clinical decision support, since it uses the contribution of future features. However, it tests the hypothesis that, from a retrospective vantage point, all previous evidence should be used at each point in time.
). This is not a realistic way to ascertain asthma status for clinical decision support, since it uses the contribution of future features. However, it tests the hypothesis that, from a retrospective vantage point, all previous evidence should be used at each point in time.
A worked example of aggregation methods
To clarify the similarities and differences between the aggregation methods, consider the example in table 1 of six evidence aggregation methods. For the 23rd feature, ‘Bronchodilator,’ for each time t that a document is generated about a patient, 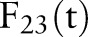 was recalculated to include the aggregated information. Recall that
was recalculated to include the aggregated information. Recall that 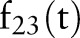 is the feature value and
is the feature value and 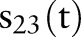 is the asthma status at time t.
is the asthma status at time t.
Note that the scale of these 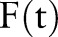 values is a superficial difference that is accounted for in many classifiers, including our implementation's logistic regression classifier. Visually, figure 5 scales these values by their largest value, so that we can compare the effect of each function. Gray bars show the dates on which a ‘bronchodilator’ feature was observed, and the black bar shows the index date.
values is a superficial difference that is accounted for in many classifiers, including our implementation's logistic regression classifier. Visually, figure 5 scales these values by their largest value, so that we can compare the effect of each function. Gray bars show the dates on which a ‘bronchodilator’ feature was observed, and the black bar shows the index date.
Figure 5.
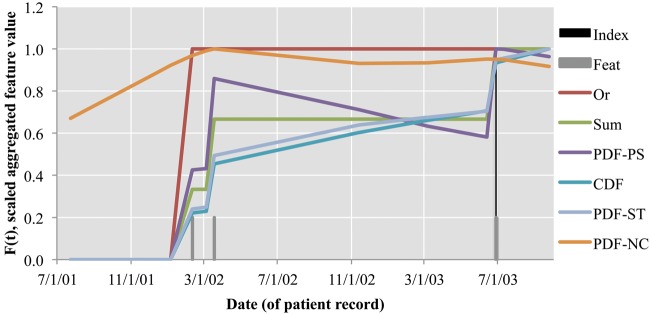
Relative F(t) of aggregation methods on the ‘bronchodilator’ primary feature.
First, note that Or and Sum values are step functions that only change value at the time of an observed feature, whereas the values in PDF-related versions give some ‘partial credit’ based on a probability estimate. In particular, note the span from the second to the third ‘bronchodilator’ features (3/18/02 to 6/28/03). The PDF-IN estimate is initially more likely to assign positive asthma status than the Sum estimate, but this fades over time. The CDF and PDF-ST versions both increase in confidence about a positive diagnosis even when no additional features are being observed. The CDF does this because its probabilities are always increasing, while PDF-ST does this because previous examples have shown that bronchodilator features often result in positive asthma status at a later time point. The PDF-NC is clearly different from the others, with the highest value at 3/18/02 because it gets ‘partial credit’ for both the past (2/10/02) and future (6/28/03) features.
Evaluation
Data source
As in previous work,12 our test set consisted of 112 study subjects, children under 4 years of age who were enrolled in the Mayo Clinic sick-child daycare program,32 33 of whom 84% were reported to be Caucasians and 49% were female; their mean age was 2.0 years (SD 1.03). We also retrieved ICD-9 codes (along with dates of code assignment) for each of the patients. Despite its small size, we used this patient population because of its practical relevance and connections to previous epidemiological research. For training our system, we used a different set of 125 Mayo Clinic pediatric patients, as done previously.12 This cohort was obtained by convenience sample from the Rochester, Minnesota population and excluded two patients whose records were at institutions other than Mayo Clinic. The documents themselves contained HL7-compliant section headings and come from multiple clinical note types, such as admission notes and discharge summaries, from many of Mayo Clinic's service areas.
Table 2 shows some of the statistics of the training and test data sets. While this data set is limited in size, it typifies the setting of patient-level temporal aggregation, which is especially relevant in a chronic disease like asthma. The use of this data set also facilitates comparison with existing work.
Table 2.
Training and test set statistics for the pediatric asthma data used in this study
| Training set | Test set | |
|---|---|---|
| Number of patients | 125 (after excluding 2) | 112 (after excluding 3) |
| Patients positive for asthma | 28.00% (35/125) | 23.21% (26/112) |
| Total clinical documents | 7098 | 2938 |
| Documents positive for asthma | 23.84% (1692/5406) | 20.66% (607/2331) |
Single versus multiple statuses per patient
We arranged the training and testing sets for two different informatics problem settings: a retrospective epidemiological setting and a clinical decision support setting.
The retrospective epidemiological setting looks at a fixed set of patients from a specific time point (the abstraction date), and determines the asthma status. Since there is only a single status per patient (SSP), we will term this SSP training or classification. A model trained on the training set would have 125 instances (patients) to train on; testing would be done on 112 instances (patients).
Additionally, we train and evaluate our machine learning models for a clinical decision support setting. For each patient, we consider every time point at which we have observed evidence of clinical care (ie, a document was generated), thus generating as multiple statuses per patient. We will refer to this as multiple statuses per patient (MSP) training or classification. Note that this is different from document-level classification, since each decision is made based on the patient's full document history up to the observation date, rather than on a single document at that date.
To make an MSP perspective possible, we consider all documents before a gold standard index date to have negative asthma status, and all documents after the index date to have positive asthma status. Then, because we have an aggregation strategy for each observation at time point t, every observation can be classified for asthma status, and compared with this gold standard. Thus, the training set of 125 patients would correspond to 7098 instances (document histories) to train on; testing on 112 patients would amount to 2938 instances (document histories). Testing on MSP also allows us to record the earliest positive asthma status as an estimated index date.
Results
Retrospective epidemiological setting
In table 3, we compare the six aggregation strategies used in machine learning patient classification against two previously reported baselines12: ICD-9 codes (commonly used in practice) and the logic-based patient classification scheme (direct implementation of the criteria of figure 2). The standard statistical tests in epidemiological research are sensitivity (recall), specificity, positive predictive value (precision) (PPV), and negative predictive value (NPV) at the SSP level; as with standard NLP evaluations, we will primarily be concerned with the F1 score. We define these in terms of true positives (TP), false negatives (FN), false positives (FP), and true negatives (TN):
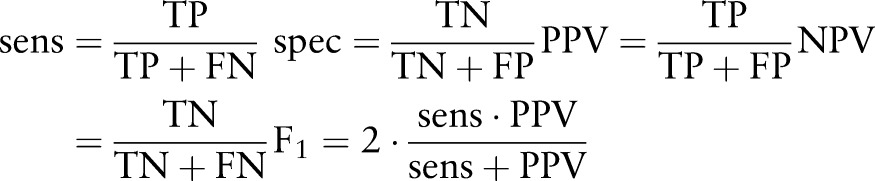 |
Table 3.
Performance of various asthma ascertainment methods in the retrospective research setting, that is, testing on single status per patient (SSP)
| SSP training | MSP training F1 | |||||
|---|---|---|---|---|---|---|
| Sens. (%) | Spec. (%) | PPV (%) | NPV (%) | F1 (%) | ||
| ICD-9 codes | 30.8 | 93.2 | 57.1 | 82.2 | 40.0 | |
| NLP-Logic | 80.8 | 95.3 | 84.0 | 94.3 | 82.4 | |
| NLP-ML-Sum | 84.62 | 95.35 | 84.62 | 95.35 | 84.62 | 83.33 |
| NLP-ML-Or | 57.69 | 95.35 | 78.95 | 88.17 | 66.67 | 74.42 |
| NLP-ML-PDF-IN | 92.31 | 93.02 | 80.00 | 97.56 | 85.71 | 81.63 |
| NLP-ML-CDF | 84.62 | 94.19 | 81.48 | 95.29 | 83.02 | 83.33 |
| NLP-ML-PDF-ST | 84.62 | 95.35 | 84.62 | 95.35 | 84.62 | 80.00 |
| NLP-ML-PDF-NC | 92.31 | 93.02 | 80.00 | 97.56 | 85.71 | 66.67 |
CDF, cumulative distribution function; ML,machine learning; MSP,multiple statuses per patient; NLP, natural language processing;NPV, negative predictive value; PDF, probability density function;PPV, positive predictive value; Sens., sensitivity; Spec., specificity.
Most of the aggregation methods enable machine learning classifiers to outperform the baseline ICD-9 codes (significant, with p<0.05, by two-tailed paired t test). This is most likely because ICD-9 codes and any defined primary and secondary features are noisy, with discrepancies more easily overcome in a machine learning algorithm. However, it is not clear whether machine learning classifiers outperform the baseline rule-based NLP-Logic system (increases not significant at the p < 0.05 level).
In this test, PDF-IN aggregation was the most effective in detecting patients who have asthma, according to F1 score. (Note that the non-causal PDF-IN aggregation method is equivalent to causal PDF-IN aggregation for the SSP setting, since we did not consider documents after the abstraction date.) The PDF-IN strategy has excellent recall (92.31%), but the Sum and PDF-ST aggregation methods have higher precision (84.62%). The Or aggregation method is clearly less accurate (significant at p < 0.05 compared to each other machine learning method); it likely requires too many symptoms before it will classify a patient as positive for asthma.
In this retrospective cohort identification setting, the test is on the SSP setting; thus the standard training data to use would be SSP data. Because these data are limited (here, only 125 cases), we may be interested to know whether we can utilize the MSP perspective to expand the training set to 7098 ‘patients’ (ie, treat each MSP as a separate patient). The final column of table 3 shows us that this is not necessarily beneficial except in the worst classifiers (ie, Or aggregation), and perhaps overfits the data set (eg, for 66.67% F1 for PDF-NC).
Clinical decision support setting
In table 4, we consider the clinical decision support setting, in which we will evaluate the same patient multiple times based on their history at each point in time (MSP testing). In this MSP testing, we consider each time point for each patient as a separate instance in the evaluation (ie, 2938 test instances); however, we adjust for the fact that some patients have more documents than others. To do so, we scale TP, FN, FP, and TN counts by the number of documents per patient 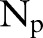 so that each patient contributes a proportional amount to the overall metric:
so that each patient contributes a proportional amount to the overall metric:
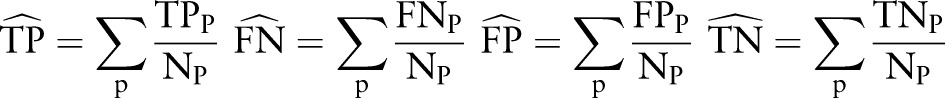 |
The resulting sensitivity, specificity, PPV, NPV, and F1 scores consider each patient equally. (Note that these adjusted metrics are equivalent to the standard metrics if the setting is SSP rather than MSP, since the numerators are 0 or 1, and Np is 1.)
Table 4.
Performance of various aggregation methods in a clinical decision support setting, that is, testing on multiple statuses per patient (MSP)
| SSP training | MSP training | |||||
|---|---|---|---|---|---|---|
| Sens. (%) | Spec. (%) | PPV (%) | NPV (%) | F1 (%) | F1 (%) | |
| NLP-ML-Sum | 83.03 | 93.69 | 63.99 | 97.61 | 72.28 | 71.62 |
| NLP-ML-Or | 49.56 | 81.56 | 26.62 | 92.29 | 34.64 | 65.33 |
| NLP-ML-PDF-IN | 88.74 | 91.52 | 58.55 | 98.37 | 70.55 | 70.40 |
| NLP-ML-CDF | 82.54 | 93.18 | 62.03 | 97.53 | 70.83 | 72.85 |
| NLP-ML-PDF-ST | 81.37 | 95.05 | 68.93 | 97.42 | 74.63 | 71.09 |
| NLP-ML-PDF-NC | 90.94 | 82.82 | 41.68 | 98.54 | 57.16 | 56.57 |
CDF, cumulative distribution function; ML, machine learning; NLP, natural language processing; PDF, probability density function; NPV, negative predictive value; PPV, positive predictive value; Sens., sensitivity; Spec., specificity; SSP, single status per patient.
First, we notice that PPV (column 4) and F1 (column 6) are overall lower when testing on MSP rather than on SSP. This demonstrates that the MSP setting is more difficult overall. In addition to the missed diagnoses and FP diagnoses that are penalized in SSP, early or late diagnoses are penalized in MSP evaluation.
In this setting, the most balanced strategy (according to F1) is now PDF-ST aggregation (74.63%; p<0.001, two-tailed paired t test compared to each other machine learning approach). PDF-ST is a technique that considers the timing of asthma status  rather than the timing of the index date, so it is a good fit for the problem of MSP classification. The PDF-IN and PDF-NC strategies were successful in the SSP setting but are somewhat less so here. They still have relatively good recall (88.74% and 90.94%), but suffer in terms of precision. In particular, the non-causal version is highly likely to make early estimates of asthma status (see ‘Timing estimation’ section below) and is penalized for this. Also, the Or aggregation method again performs poorly; the discriminating evidence in the features appears to have been lost due to the coarse granularity of feature representation.
rather than the timing of the index date, so it is a good fit for the problem of MSP classification. The PDF-IN and PDF-NC strategies were successful in the SSP setting but are somewhat less so here. They still have relatively good recall (88.74% and 90.94%), but suffer in terms of precision. In particular, the non-causal version is highly likely to make early estimates of asthma status (see ‘Timing estimation’ section below) and is penalized for this. Also, the Or aggregation method again performs poorly; the discriminating evidence in the features appears to have been lost due to the coarse granularity of feature representation.
Similarly to the retrospective epidemiological setting, we sought to determine whether using MSP training data would help alleviate the problem of a small training data set. While precision of many of the aggregation methods improved (not pictured), this was offset by lower recall, and the F1 scores (column 7) of promising aggregators did not improve significantly. This is somewhat surprising, given that the training and testing data were from the same MSP distribution. However, on closer examination, an MSP training set over-counts the instances early in a patient's timeline, and this likely adds to the model as much noisy information as it does useful information.
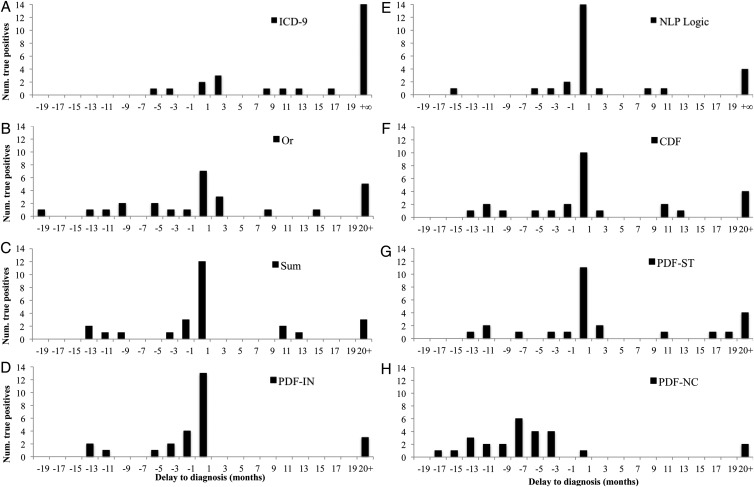
Timing estimation
We characterized the timing estimation accuracy of automatic diagnosis according to the histograms of figure 6. In these plots, only gold standard positive asthma cases in the SSP setting are considered; thus, they depict recall and timing, but do not depict precision. The time 0 on the x-axis represents the index date, and the bars indicate how many estimated index dates were delayed by the same amount of time. In figure 5A,E, +∞ corresponds to FN (ie, true positive asthma status was never detected), whereas in figure 5B–D, F–H, these FN are conflated with any estimated index dates that were 20+ months after the index date.
Figure 6.
Delay in diagnosis for patients with asthma. Higher bars close to 0 indicate that more asthma index dates were estimated correctly. Bars to the left of 0 indicate early automatic diagnosis, bars to the right of 0 indicate delayed diagnosis. The ‘20+’ designation includes each system's false negatives.
In figure 5A,E, it is clear that all NLP-based methods outperform ICD-9 codes, and that the NLP-Logic method produces a solid timing estimate.12 The relative performance of machine learning algorithms (figure 5B–D,F–H) depends on the aggregation method; we consider how many cases were detected within 1 month (the center bar) and the overall distribution. The PDF-IN method is perhaps the most intriguing, in that it is almost never delayed on a diagnosis, and detects 50.0% of positive cases within 1 month. Sum aggregation has a similar characteristic, but estimates the timing later overall than PDF-IN or NLP-Logic. PDF-ST and CDF are also similar to each other, with PDF-ST identifying a few more cases within 1 month. The Or aggregation method is the most distributed of the methods, and leaves more cases undetected than NLP-Logic. While non-causal PDF aggregation has the fewest detected 20+ months late, it tends to pre-empt true index dates. This is expected, given the fact that in a non-causal aggregation, 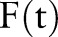 values before the index date get some weighting from events that happen later; this always includes at least the index date and any observations after the index date.
values before the index date get some weighting from events that happen later; this always includes at least the index date and any observations after the index date.
Discussion
We have tested our aggregation methods in the applied setting of asthma cohort discovery. We assert that this is a real-world evaluation scenario, and better aligned with downstream retrospective public health studies than typical evaluations like document classification. This does mean, though, that the presented results are limited in scope by the size and underlying population of the data sets that were used. Thousands of documents seems like a reasonable size for an NLP evaluation; however, because this corresponds to just over a hundred patients, and only tens of positive cases, the test set is not necessarily a generalizable estimate of performance.
Our results should not be interpreted as picking the best aggregation algorithm, but as exploring and evaluating patient-level phenomena, and perhaps eliminating a few options (PDF-NC in the clinical decision support setting, or Or in any setting). There are many factors to consider in choosing how to do temporal aggregation on a given domain or problem. A rule-based method (the baseline NLP-Logic method) may be intuitive, simple, and precise, but it requires an expert to develop rules and refine them on a data set. The Sum and Or aggregation methods are essentially unsupervised, and are thus quite simple to conceive, implement, and describe; however, there may be settings (such as clinical decision support) in which they are more poorly suited. For the PDF-based methods, some of the features suffered from data sparsity, with insufficient data to do reliable kernel estimation of the PDF. However, our results did not clearly show a clear advantage of the unsupervised Sum and Or aggregation methods compared to the kernel-estimated methods, implying that feature sparsity did not occlude the conclusions to be drawn from our results. Rather, it is quite notable that the extra cost in learning a PDF may pay off in the end when such a PDF correctly models the evaluation situation, as we noted with PDF-ST in the clinical decision support setting.
Furthermore, the set of primary features is quite limited compared to a standard document classification feature set (eg, bag-of-words, n-gram, or character-based features) or structured data-based phenotyping algorithm.34 Even the existing primary features could have included negated terms as contraindicating evidence. We analyze our techniques based on the small number of primary features because: (1) the PDF-based aggregation methodologies we have introduced are not optimized for large numbers of features; and (2) mirroring the explicit criteria22 aligns with the realistic future use of any NLP-discovered cohorts in public health. In a production system, other typical machine learning features could accompany the expert-based features, once the dimensionality of the feature space is reduced through feature selection.
Conclusion
We have specified the practical problem of patient-level temporal aggregation from clinical text, and defined several probability-based evidence aggregation methods to overcome the challenge of time-distributed evidence. Our evaluations utilized the preliminary test case of EMR text-based identification of a pediatric asthma, for which we made use of an established cohort of pediatric patients. Results in both a retrospective epidemiological setting and in a clinical decision support setting showed that several aggregation algorithms had satisfactory patient classification performance, and also predicted the timing of estimates with promising accuracy.
Acknowledgments
The authors would like to thank Drs Andrew Hashikawa, Robert Voigt, Kavishwar Wagholikar, Ravikumar KE, and Siddhartha Jonnalagadda, as well as Ms Shirley Johnson, for support on previous work.
Footnotes
Contributors: STW led the study design and analysis and drafted the manuscript. YJJ designed and conducted the original study, and assembled the data for this study. SS provided support on machine learning algorithms, and HL provided support with statistics and aggregation methods. All authors edited the manuscript.
Funding: This work was supported in part by National Science Foundation grant ABI:0845523, National Institute of Health grants R01LM009959 and R01GM102282, NIH Roadmap Grant U54 HG004028, SHARP 4 grant 90TR000201 (PI: Chute), the Scholarly Clinician Award from the Mayo Foundation (PI: Juhn), the National Institute of Allergy and Infectious Disease (R21 AI101277, PI: Juhn), and Mayo Clinic NIH Relief Grant (R21 HD075961, PI: Wu).
Competing interests: None.
Ethics approval: The Mayo Clinic institutional review board approved this study.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The data cannot be shared due to patient confidentiality concerns. The NLP algorithm and aggregation methods can be obtained by contacting the authors.
References
- 1.Chapman W, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 2.Uzuner O, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonnalagadda SR, Li D, Sohn S, et al. Coreference analysis in clinical notes: a multi-pass sieve with alternate anaphora resolution modules. J Am Med Inform Assoc 2012;19:867–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rink B, Roberts K, Harabagiu SM. A supervised framework for resolving coreference in clinical records. J Am Med Inform Assoc 2012;19:875–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savova GK, Fan J, Ye Z, et al. Discovering peripheral arterial disease cases from radiology notes using natural language processing. AMIA Annu Symp Proc 2010;2010:722–6 [PMC free article] [PubMed] [Google Scholar]
- 6.Sohn S, Savova GK. Mayo Clinic smoking status classification system: extensions and improvements. AMIA Annu Symp Proc 2009;2009:619–23 [PMC free article] [PubMed] [Google Scholar]
- 7.Harkema H, Dowling JN, Thornblade T, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009;42:839–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao C, Wei WQ, Solbrig HR, et al. CNTRO: a semantic web ontology for temporal relation inferencing in clinical narratives. AMIA Annu Symp Proc 2010;2010:787–91 [PMC free article] [PubMed] [Google Scholar]
- 9.Albright D, Lanfranchi A, Fredriksen A, et al. Towards comprehensive syntactic and semantic annotations of the clinical narrative. J Am Med Inform Assoc 2013;20:922–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hersh WR, Voorhees EM. Overview of the TREC 2012 medical records track. The Twenty-first Text REtrieval Conference Proceedings TREC; Gaithersburg, MD: National Institute of Standards and Technology, 2012 [Google Scholar]
- 11.Voorhees EM, Tong RM. Overview of the TREC 2011 medical records track. The Twentieth Text REtrieval Conference Proceedings TREC; Gaithersburg, MD: National Institute of Standards and Technology, 2011 [Google Scholar]
- 12.Wu ST, Sohn S, Ravikumar KE, et al. Automated chart review for asthma cohort identification using natural language processing: an exploratory study. Ann Allergy Asthma Immunol 2013;111:364–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juhn YJ, Kita H, Yawn BP, et al. Increased risk of serious pneumococcal disease in patients with asthma. J Allergy Clin Immunol 2008;122:719–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capili CR, Hettinger A, Rigelman-Hedberg N, et al. Increased risk of pertussis in patients with asthma. J Allergy Clin Immunol 2012;129:957–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch BA, Van Norman CA, Jacobson RM, et al. Impact of delay in asthma diagnosis on health care service use. Allergy Asthma Proc 2010;31:e48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch BA, Fenta Y, Jacobson RM, et al. Impact of delay in asthma diagnosis on chest X-ray and antibiotic utilization by clinicians. J Asthma 2012;49:23–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohn S, Wagholikar KB, Li D, et al. Comprehensive temporal information detection from clinical text: medical events, time, and TLINK identification. J Am Med Inform Assoc 2013;20:836–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun W, Rumshisky A, Uzuner O. Temporal reasoning over clinical text: the state of the art. J Am Med Inform Assoc 2013;20:814–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plaisant C, Mushlin R, Snyder A, et al. LifeLines: using visualization to enhance navigation and analysis of patient records. Proc AMIA Symp 1998:76–80 [PMC free article] [PubMed] [Google Scholar]
- 20.Martins SB, Shahar Y, Galperin M, et al. Evaluation of KNAVE-II: a tool for intelligent query and exploration of patient data. Stud Health Technol Inform 2004;107(Pt 1):648–52 [PubMed] [Google Scholar]
- 21.Bui AA, Aberle DR, Kangarloo H. TimeLine: visualizing integrated patient records. IEEE Trans Inf Technol Biomed 2007;11:462–73 [DOI] [PubMed] [Google Scholar]
- 22.Beard CM, Yunginger JW, Reed CE, et al. Interobserver variability in medical record review: an epidemiological study of asthma. J Clin Epidemiol 1992;45:1013–20 [DOI] [PubMed] [Google Scholar]
- 23.Bauer BA, Reed CE, Yunginger JW, et al. Incidence and outcomes of asthma in the elderly. A population-based study in Rochester, Minnesota. Chest 1997;111:303–10 [DOI] [PubMed] [Google Scholar]
- 24.Hunt LW, Jr, Silverstein MD, Reed CE, et al. Accuracy of the death certificate in a population-based study of asthmatic patients. JAMA 1993;269:1947–52 [PubMed] [Google Scholar]
- 25.Juhn YJ, Qin R, Urm S, et al. The influence of neighborhood environment on the incidence of childhood asthma: a propensity score approach. J Allergy Clin Immunol 2010;125:838–43.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juhn YJ, Sauver JS, Katusic S, et al. The influence of neighborhood environment on the incidence of childhood asthma: a multilevel approach. Soc Sci Med 2005;60:2453–64 [DOI] [PubMed] [Google Scholar]
- 27.Juhn YJ, Weaver A, Katusic S, et al. Mode of delivery at birth and development of asthma: a population-based cohort study. J Allergy Clin Immun 2005;116: 510–16 [DOI] [PubMed] [Google Scholar]
- 28.Silverstein MD, Reed CE, O'Connell EJ, et al. Long-term survival of a cohort of community residents with asthma. N Engl J Med 1994;331:1537–41 [DOI] [PubMed] [Google Scholar]
- 29.Silverstein MD, Yunginger JW, Reed CE, et al. Attained adult height after childhood asthma: effect of glucocorticoid therapy. J Allergy Clin Immunol 1997;99: 466–74 [DOI] [PubMed] [Google Scholar]
- 30.Yawn BP, Yunginger JW, Wollan PC, et al. Allergic rhinitis in Rochester, Minnesota residents with asthma: frequency and impact on health care charges. J Allergy Clin Immunol 1999;103(1 Pt 1):54–9 [DOI] [PubMed] [Google Scholar]
- 31.Yunginger JW, Reed CE, O'Connell EJ, et al. A community-based study of the epidemiology of asthma. Incidence rates, 1964–1983. Am Rev Respir Dis 1992;146:888–94 [DOI] [PubMed] [Google Scholar]
- 32.Juhn Y, Kung A, Voigt R, et al. Characterisation of children's asthma status by ICD-9 code and criteria-based medical record review. Prim Care Respir J 2011;20:79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo KH, Johnson SK, Voigt RG, et al. Characterization of asthma status by parent report and medical record review. J Allergy Clin Immunol 2007;120:1468–9 [DOI] [PubMed] [Google Scholar]
- 34.Pacheco JA, Avila PC, Thompson JA, et al. A highly specific algorithm for identifying asthma cases and controls for genome-wide association studies. AMIA Annu Symp Proc 2009;2009:497–501 [PMC free article] [PubMed] [Google Scholar]