Abstract
Objective
Named entity recognition (NER) is one of the fundamental tasks in natural language processing. In the medical domain, there have been a number of studies on NER in English clinical notes; however, very limited NER research has been carried out on clinical notes written in Chinese. The goal of this study was to systematically investigate features and machine learning algorithms for NER in Chinese clinical text.
Materials and methods
We randomly selected 400 admission notes and 400 discharge summaries from Peking Union Medical College Hospital in China. For each note, four types of entity—clinical problems, procedures, laboratory test, and medications—were annotated according to a predefined guideline. Two-thirds of the 400 notes were used to train the NER systems and one-third for testing. We investigated the effects of different types of feature including bag-of-characters, word segmentation, part-of-speech, and section information, and different machine learning algorithms including conditional random fields (CRF), support vector machines (SVM), maximum entropy (ME), and structural SVM (SSVM) on the Chinese clinical NER task. All classifiers were trained on the training dataset and evaluated on the test set, and micro-averaged precision, recall, and F-measure were reported.
Results
Our evaluation on the independent test set showed that most types of feature were beneficial to Chinese NER systems, although the improvements were limited. The system achieved the highest performance by combining word segmentation and section information, indicating that these two types of feature complement each other. When the same types of optimized feature were used, CRF and SSVM outperformed SVM and ME. More specifically, SSVM achieved the highest performance of the four algorithms, with F-measures of 93.51% and 90.01% for admission notes and discharge summaries, respectively.
Keywords: Medical concept recognition, Chinese clinic notes, Natural language processing, Machine learning algorithm
Introduction
Clinical documents are an important type of electronic health record (EHR) data and often contain valuable and detailed patient information for many clinical applications. Natural language processing (NLP) in the medical domain has become an active research area in biomedical informatics, and many studies have successfully demonstrated its uses in clinical practice and research.1 Named entity recognition (NER) in clinical text, which is used to identify the boundary of clinically relevant entities such as diseases and drugs, is one of the fundamental tasks in clinical NLP research and has been extensively studied, including rule-based approaches in early years and machine learning (ML)-based approaches in recent years.2
However, most previous studies on clinical NER have primarily focused on clinical text written in English. With the rapid growth of EHRs in China, information extraction from Chinese clinical text has also become an important task for biomedical informatics researchers in China. In this study, our goal was to assess the performance of ML-based NER approaches that have been developed for English clinical text on Chinese clinical documents. Using manually annotated datasets of admission notes and discharge summaries in Chinese, we evaluated the contributions of different types of feature and ML algorithms for NER in Chinese clinical text. To the best of our knowledge, this is one of the earliest comprehensive studies on features and ML algorithms for Chinese clinical NER.
Background
NER is a fundamental task in NLP research and has been extensively studied in both English and Chinese text.3–5 In the medical domain, early clinical NLP systems often recognize clinical entities using rule-based approaches that rely on dictionary resources.6 7 More recently, ML-based NER approaches have been studied for clinical text, largely because of the availability of annotated clinical corpora. For example, i2b2 (the Center of Informatics for Integrating Biology and the Bedside) at Partners Health Care System has organized a few clinical NER challenges and created annotated corpora for recognizing various clinical entities including medications and signature (the 2009 challenge8) and medical problems, treatments, and laboratory tests (the 2010 i2b2 challenge2). Many top-ranked systems in the 2009 and 2010 i2b2 NLP challenges were primarily based on ML approaches.2 8–10 In ML-based NER approaches, annotated data are typically represented in the BIO format, in which each word is assigned to one of three classes: B, beginning of an entity; I, inside an entity; O, outside of an entity. Therefore, the NER problem now becomes a classification problem to assign one of the three class labels to each word.
Features and ML algorithms appear to be two of the most important factors that affect the performance of ML-based NER systems. In previous clinical NER studies,2 different types of feature, including syntactic (eg, part-of-speech tags) and semantic (eg, semantic classes in UMLS (Unified Medical Language System)) information of context words, as well as word representation information generated from unsupervised analysis,11 12 have been investigated, and all of them conferred beneficial improvement on NER performance. Different ML algorithms have also been used for biomedical NER tasks. Among them, conditional random fields (CRF)13 and support vector machines (SVM)14 are two widely used algorithms. In theory, CRF is a representative sequence-labeling algorithm, which is suitable for the NER problem. SVM is a robust classification algorithm that is based on large margin theory. To include information about neighbor tokens in sequences, researchers have developed methods to incorporate neighbor information into features for SVM-based NER systems.15 16 Another emerging algorithm for NER is the structural SVM (SSVM),17 18 which is an SVM-based discriminative algorithm for structural prediction. Therefore, SSVM combines the advantages of both CRF and SVM and is also suitable for sequence-labeling problems. In one of our recent studies,11 12 we demonstrated that SSVM achieved a slightly better performance on recognizing clinical entities in discharge summaries from US hospitals.
However, most previous work on clinical NER tasks has focused on text written in English. With the rapid growth of EHRs in China, there is a huge need to extract information from clinical notes written in Chinese. However, there is very limited work on NER in Chinese clinical text. Wang et al19 applied CRF, SVM, and maximum entropy (ME) to recognize symptoms and pathogenesis in ancient Chinese medical records and showed that CRF achieved a better performance. Wang et al20 conducted a preliminary study on symptom name recognition in clinical notes of traditional Chinese medicine. A more recent and related study by Xu et al21 proposed a joint model that integrates segmentation and NER simultaneously to improve the performance of both tasks in Chinese discharge summaries.
Despite the important contributions of previous studies on Chinese clinical NER, none has systematically evaluated the effects of different features and different ML algorithms on NER in Chinese clinical text. It is important to compare the differences between Chinese and English clinical text and to investigate whether the NER methods that we have developed for English clinical text are also effective with Chinese clinical text. For example, one major difference between English and Chinese is that segmentation of Chinese text is more difficult because you cannot rely on white spaces to separate words. In this study, we created large annotated datasets of Chinese admission notes and discharge summaries, then systematically evaluated different types of feature (eg, syntactic, semantic, and segmentation information) and four ML algorithms, CRF, SVM, SSVM, and ME. To the best of our knowledge, this is one of the earliest comprehensive studies in Chinese clinical NER research and we believe it will provide valuable insights into NLP research in Chinese clinical text.
Methods
Datasets and annotation
One month (March 2011) of admission notes and discharge summaries were collected from the EHR database of Peking Union Medical College Hospital in China. After excluding very short notes (incomplete notes), we randomly selected 400 admission notes and 400 discharge summaries for this study. All patient identifiers were manually removed before annotation. Two Chinese medical doctors (XL and KG) were recruited to annotate four types of clinical entity—problems, tests, procedures, and medications—by following annotation guidelines developed in this study. The annotation guidelines were similar to those used in the 2010 i2b2 NLP challenge (https://www.i2b2.org/NLP/Relations/Documentation.php), but were translated into Chinese. One main difference is that we broke down the ‘treatment’ category in the i2b2 challenge into two categories: ‘procedures’ and ‘medications’. Thus, we had four types of entity in this study instead of three as in the i2b2 challenge. In addition, we also specified some rules for determining entity boundaries in Chinese text. To calculate the inter-rater agreement for annotation, 40 notes were annotated by both annotators. The remaining 360 notes were annotated by a single annotator only.
ML-based NER
To convert the NER task into an ML-based classification problem, we used the ‘BIO’ tags to represent the boundaries of entities. As we have four types of entity in this study, we generated nine different tags in total: B-problem, B-procedure, B-test, B-medication, I-problem, I-procedure, I-test, I-medication, and O. Figure 1 shows examples of annotated entities labeled with BIO tags.
Figure 1.
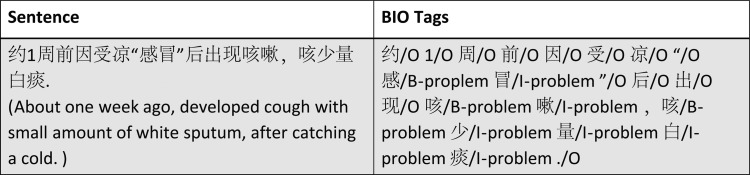
Examples of Chinese medical named entity recognition (NER) representation.
Features
As shown in table 1, we used four types of feature: (1) bag-of-characters; (2) bag-of-words (based on two segmentation approaches); (3) part-of-speech (POS) tags (only available for one segmentation approach); and (4) section information. One major difference between Chinese and English text is that words in Chinese are formed by continuous Chinese characters without any space. For example, the word ‘cough’  is formed by two Chinese characters: “
is formed by two Chinese characters: “ ” and “
” and “ ”. Figure 2 shows the output of a sentence after segmentation. The bag-of-characters approach simply used individual Chinese characters as features. If word segmentation is applied to Chinese text, we can use identified word segments as features (called ‘bag-of-words’ here). We implemented two word segmentation methods in this study: (1) the Stanford Word Segmenter (http://nlp.stanford.edu/software/segmenter.shtml), which supports general Chinese text, but not clinical Chinese text; and (2) a simple dictionary lookup approach, which uses the forward maximum match algorithm to search the New dictionary of medicine and drugs, a clinical dictionary containing 704 896 medical concepts in Chinese. When the Stanford Word Segmenter was used, POS tags were generated by the system as well, which were also used as features in this study. In addition, we manually reviewed some notes and defined 35 different section headers (eg, ‘history of illness’) as additional features.
”. Figure 2 shows the output of a sentence after segmentation. The bag-of-characters approach simply used individual Chinese characters as features. If word segmentation is applied to Chinese text, we can use identified word segments as features (called ‘bag-of-words’ here). We implemented two word segmentation methods in this study: (1) the Stanford Word Segmenter (http://nlp.stanford.edu/software/segmenter.shtml), which supports general Chinese text, but not clinical Chinese text; and (2) a simple dictionary lookup approach, which uses the forward maximum match algorithm to search the New dictionary of medicine and drugs, a clinical dictionary containing 704 896 medical concepts in Chinese. When the Stanford Word Segmenter was used, POS tags were generated by the system as well, which were also used as features in this study. In addition, we manually reviewed some notes and defined 35 different section headers (eg, ‘history of illness’) as additional features.
Table 1.
Features used for Chinese medical entity recognition
| Feature type | Explanation |
|---|---|
| Bag-of-characters | Individual Chinese characters in a window |
| Bag-of-words | Individual Chinese words in a window. Two methods were used for word segmentation: the Stanford Word Segmenter and a dictionary lookup program. |
| Part-of-speech (POS) | POS tags, only available from the Stanford Word Segmenter |
| Section information | Section headers from a predefined list |
Figure 2.

An example of word segmentation in Chinese.
ML algorithms
NER problems can be considered as either a pure classification problem or a sequence-labeling problem. In this study, we compared four state-of-the-art ML algorithms: two for classification problems (SVM14 and ME22) and two for sequence-labeling problems (CRF13 and SSVM17 18). SVM and SSVM are discriminative statistical algorithms based on large margin theory, while ME and CRF are discriminative statistical algorithms based on probability theory. All of them have been widely used in NLP.
Assume there is a sequence-labeling problem of independent and identically distributed training samples 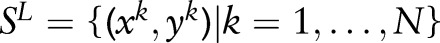 . We use
. We use 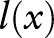 to denote the sequence length of input
to denote the sequence length of input 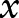 ,
, 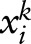 denotes the i-th subinput of
denotes the i-th subinput of 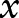 ,
, 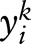 denotes the i-th sublabel of
denotes the i-th sublabel of 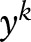 , and
, and 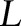 denotes the sublabel set. This problem can be treated as a classification problem of training samples
denotes the sublabel set. This problem can be treated as a classification problem of training samples 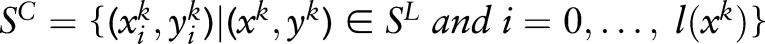 if we assume all sublabels are independent of each other.
if we assume all sublabels are independent of each other.
SVM uses a linear discriminative function to model the score of an observation 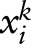 and a random variable yik:
and a random variable yik: 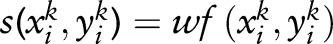 , where
, where 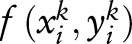 are features. The total loss function on the training samples
are features. The total loss function on the training samples 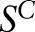 can be written as:
can be written as:
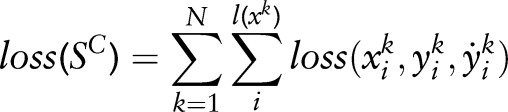 |
1 |
where 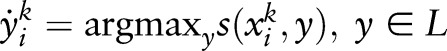 and loss(xik,yik,y˙ik) =
and loss(xik,yik,y˙ik) = 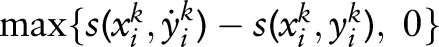 . This problem can be transformed into a quadratic programming problem as follows:
. This problem can be transformed into a quadratic programming problem as follows:
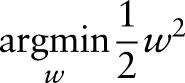 |
2 |
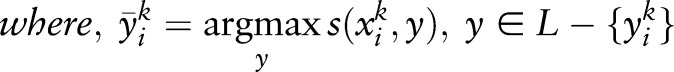 |
Many algorithms have been proposed to optimize equation (2), such as sequential minimal optimization (SMO)23 and cutting plane (CP).23–25 In our experiments, we used liblinear (http://www.csie.ntu.edu.tw/~cjlin/liblinear/) as an implementation of SVM, which optimizes equation (2) by CP.
SSVM uses a similar method to model the sequence-labeling problems. The discriminative function for a sequence-labeling sample 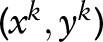 can be represented by a first-order Markov chain in the following form:
can be represented by a first-order Markov chain in the following form:
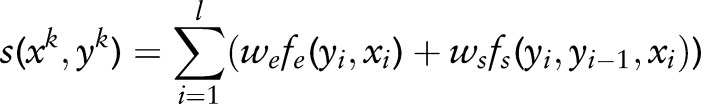 |
3 |
where 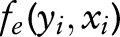 are emission features, and
are emission features, and 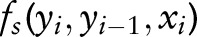 are transmission features. The sequence-labeling problem can be formatted as a quadratic programming problem as follows:
are transmission features. The sequence-labeling problem can be formatted as a quadratic programming problem as follows:
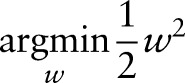 |
4 |
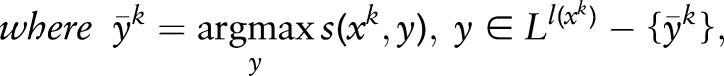 |
There are several types of loss function, and the Hamming window distance is usually used for sequence-labeling problems. It can be written as 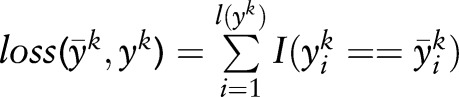 , where
, where 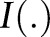 means whether the condition in the parentheses is satisfied. Equation (4) can also be solved by SMO and CP. In our experiments, we used SVMhmm (http://www.cs.cornell.edu/people/tj/svm_light/svm_hmm.html) as an implement of SSVM, which solved equation (4) by CP.
means whether the condition in the parentheses is satisfied. Equation (4) can also be solved by SMO and CP. In our experiments, we used SVMhmm (http://www.cs.cornell.edu/people/tj/svm_light/svm_hmm.html) as an implement of SSVM, which solved equation (4) by CP.
ME uses an exponential distribution to model the conditional distribution of a random variable 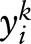 on an observation
on an observation 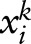 :
: 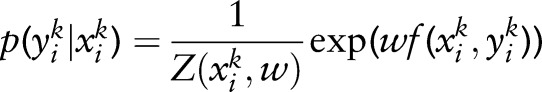 , where Z(xik,w)=Σy
, where Z(xik,w)=Σy
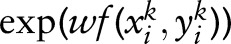 . The maximum log-likelihood estimation function on the training samples
. The maximum log-likelihood estimation function on the training samples  can be written as:
can be written as:
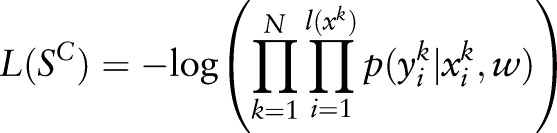 |
5 |
which can be solved by Generalized Iterative Scaling (GIS),26 Broyden–Fletcher–Goldfarb–Shanno (BFGS),27 limited-memory BFGS (L-BFGS),28 stochastic gradient (SG),29 and so on. In our experiments, we used maxent (http://homepages.inf.ed.ac.uk/lzhang10/maxent_toolkit.html) as an implement of ME, and set L-BFGS as its training algorithm.
CRF uses an undirected graph to model the conditional distribution of random variables Y conditioned on observations X: p(Y|X). For example, given a sample of length l, (x,y), the conditional probability p(y|x) can be represented by a first-order Markov chain in the following form:
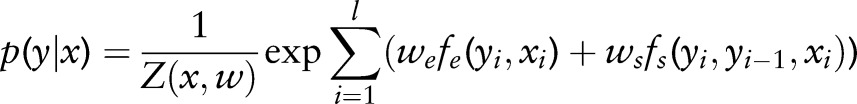 |
6 |
where 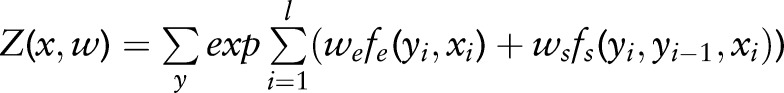 is a normalization factor. The maximum log-likelihood estimation function on the training samples
is a normalization factor. The maximum log-likelihood estimation function on the training samples 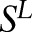 can be written as:
can be written as:
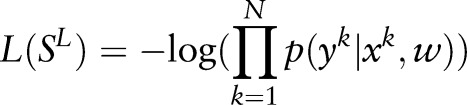 |
7 |
Equation (7) can be solved by the same algorithms as used for ME, such as GIS, BFGS, L-BFGS, SG, and so on. In our experiments, we used CRF++ (http://crfpp.googlecode.com/svn/trunk/doc/index.html) as an implementation of CRF, which optimizes equation (7) by L-BFGS.
Experiments and evaluation
For either discharge summaries or admission notes, we divided 400 notes into two subsets: two-thirds (266 notes) for training and one-third (134 notes) for testing. The parameters of classifiers were optimized using the training set via a 10-fold cross-validation method. Then we evaluated and reported the performance using the independent test set. As CRF is the most widely used algorithm for NER, we first investigated the effects of different types of feature based on the CRF classifier. We started with the baseline system that used bag-of-character features only, and then progressively added bag-of-word features based on different segmentation methods, POS tags, and section information. Once the optimized feature combination was identified on the basis of the CRF classifier, we evaluated the performance of other ML algorithms (SVM, ME and SSVM) using the same sets of features.
The performance of NER systems was measured by standard micro-averaged precision, recall, and F-measure for all entities.2 We developed an evaluation tool to calculate their values based on the official evaluation program developed in the 2010 i2b2 NLP challenge. The evaluation program provides two sets of measures—exact match and inexact match—where exact match means that an entity is correctly predicted if, and only if, the starting and ending offsets are exactly the same as those in the gold standard; the inexact match means that an entity is correctly predicted if it overlaps with any entity in the gold standard.
Results
Table 2 shows the statistics of the corpora of Chinese discharge summaries and admission notes used in this study. There were 30 793 sentences and 38 973 entities in 400 admission notes, and 22 838 sentences and 39 334 entities in 400 discharge summaries. The proportion of each type of concept in 800 notes (both admission and discharge summaries) was 56.95% for problems, 29.73% for tests, 8.54% for procedures and 4.78% for medications. The problems and tests were almost equally distributed in admission and discharge summaries, while procedures and medications were mainly in discharge summaries. Based on the annotations on 40 notes, the inter-annotation agreements using kappa statistics30 on admission and discharge summaries were 0.957 and 0.924, respectively, which indicates that the annotation was reliable.
Table 2.
Summary statistics of annotated datasets of Chinese discharge summaries and admission notes
| Dataset | Notes | Type | Sentences | Characters | NER tasks | ||||
|---|---|---|---|---|---|---|---|---|---|
| Problems | Procedures | Tests | Medications | Total | |||||
| Training | 266 | Admission | 20 506 | 277 701 | 16 253 | 1500 | 7414 | 840 | 26 007 |
| Discharge | 15 140 | 243 069 | 13 308 | 2995 | 8093 | 1757 | 26 153 | ||
| All | 35 646 | 520 770 | 29 561 | 4495 | 15 507 | 2597 | 52 160 | ||
| Test | 134 | Admission | 10 287 | 139 885 | 8180 | 671 | 3754 | 361 | 12 966 |
| Discharge | 7698 | 125 335 | 6851 | 1522 | 4021 | 787 | 13 181 | ||
| All | 17 985 | 265 220 | 15 031 | 2193 | 7775 | 1148 | 26 147 | ||
| Total | 400 | Admission | 30 793 | 417 586 | 24 433 | 2171 | 11 168 | 1201 | 38 973 |
| Discharge | 22 838 | 368 404 | 20 159 | 4517 | 12 114 | 2544 | 39 334 | ||
| All | 53 631 | 785 990 | 44 592 | 6688 | 23 282 | 3745 | 78 307 | ||
NER, named entity recognition.
Table 3 shows the performance of the CRF-based systems on test sets when different features were used for admission and discharge summaries, respectively. The numbers in columns 2–5 are F-measures followed by corresponding recall and precision values in parentheses for all entities using the exact-matching or inexact-matching criterion. Both word segmentation approaches slightly improved the NER performance, and the dictionary lookup method seemed to give a better performance. For example, on discharge summaries, the Stanford Word Segmenter improved the F-measure from 88.89% to 89.01%, while the dictionary lookup approach improved the F-measure from 88.89% to 89.19%. The POS tag information following Stanford segmentation did not further improve the NER performance. The section information also helped the NER system slightly (F-measure 88.95% vs 88.89% at baseline on discharge summaries). However, this improvement is minimal, as the 95% CIs for the F-measure (%) of ‘BOC+SECTION’ were (88.46 to 89.42) (88.94±0.48) using the two-tailed t test based on bootstrapping sampling, which randomly selected 5000 sentences with replacement 200 times.
Table 3.
Performance of the CRF-based NER systems on Chinese admission and discharge notes when different features were used
| Feature | Admission notes | Discharge summaries | ||
|---|---|---|---|---|
| Exact-match | Inexact-match | Exact-match | Inexact-match F-measure (R/P) | |
| BOC | 93.18 (93.70/92.66) | 94.32 (94.85/93.80) | 88.89 (89.80/87.99) | 90.75 (91.68/89.83) |
| BOC+BOW-STAN | 93.19 (93.59/92.79) | 94.40 (94.81/94.00) | 89.01 (89.87/88.16) | 90.95 (91.83/90.08) |
| BOC+BOW-STAN+POS | 93.14 (93.46/92.81) | 94.37 (94.70/94.04) | 88.89 (89.59/88.21) | 90.86 (91.57/90.16) |
| BOC+BOW-DICT | 93.30 (93.66/92.94) | 94.50 (94.87/94.13) | 89.19 (90.16/88.24) | 90.97 (91.96/90.00) |
| BOC+SECTION | 93.28 (93.63/92.93) | 94.40 (94.76/94.05) | 88.95 (89.96/87.96) | 90.71 (91.74/89.70) |
| BOC+BOW-STAN+SECTION | 93.22 (93.61/92.83) | 94.45 (94.85/94.06) | 89.02 (89.95/88.12) | 90.89 (91.83/89.96) |
| BOC+BOW-DICT+SECTION | 93.52 (93.77/93.26) | 94.69 (94.95/94.43) | 89.23 (90.29/88.20) | 91.00 (92.08/89.94) |
Values are F-measure (recall/precision) (%).
BOC, bag-of-characters; BOW-DICT, bag-of-words from dictionary lookup; BOW-STAN, bag-of-words from the Stanford Word Segmenter; CRF, conditional random fields; NER, named entity recognition; POS, part-of-speech information from Stanford Word Segmenter; SECTION, section information.
The best performance, F-measures of 89.23% and 93.52% for discharge and admission notes, respectively, was achieved when bag-of-characters and bag-of-words from the dictionary lookup, and section information were combined. In addition, we noticed that the NER systems always achieved a better performance on admission notes than discharge summaries when the same features were used. For example, when only the bag-of-character features were used, the F-measure of the CRF-based NER system was 93.18% on admission notes vs 88.89% on discharge summaries.
The detailed results of the best CRF-based NER system for each entity type are shown in table 4. F-measures ranged from 82.89% to 95.06% for admission notes and 78.51% to 91.82% for discharge summaries among four types of entity. Performance was best for tests and worst for procedures. For each type of entity, precision was always higher than recall.
Table 4.
Detailed results of the best CRF-based NER system on admission and discharge summaries for each entity type
| Entity | Admission notes | Discharge summaries | ||
|---|---|---|---|---|
| Exact-match | Inexact-match | Exact-match | Inexact-match | |
| Overall | 93.52 (93.77/93.26) | 94.69 (94.95/94.43) | 89.23 (90.29/88.20) | 91.00 (92.08/89.94) |
| Problems | 93.96 (93.99/93.92) | 95.35 (95.39/95.32) | 90.19 (90.61/89.77) | 92.20 (92.63/91.77) |
| Procedures | 82.89 (85.44/80.48) | 85.34 (87.97/82.86) | 78.51 (82.80/74.64) | 81.48 (85.93/77.46) |
| Tests | 95.06 (95.22/94.91) | 95.41 (95.56/95.26) | 91.82 (92.22/91.42) | 92.89 (93.30/92.49) |
| Medications | 86.44 (88.18/84.76) | 88.98 (90.78/87.26) | 87.41 (90.82/84.24) | 88.33 (91.78/85.13) |
Values are F-measure (recall/precision) (%).
CRF, conditional random fields; NER, named entity recognition.
Using the optimized feature sets (bag-of-characters, bag-of-words from the dictionary lookup, and section information), we compared the four ML algorithms on admission and discharge notes. Results are reported in table 5. The sequence-labeling algorithms (CRF and SSVM) were superior to the classification algorithms (ME and SVM). For example, SSVM outperformed SVM by 2.99% and 4.45% in F-measure for admission notes and discharge summaries, respectively. The best performance was achieved by SSVM, which was similar to CRF on admission notes (93.53% vs 93.52%), but was better than CRF on discharge summaries (90.01% vs 89.23%).
Table 5.
Comparison of four state-of-the-art machine learning algorithms on Chinese admission and discharge summaries when optimized features were used
| Algorithm | Admission notes | Discharge summaries | ||
|---|---|---|---|---|
| Exact-match | Inexact-match | Exact-match | Inexact-match | |
| SVM | 90.54 (90.81/90.27) | 93.70 (93.99/93.42) | 85.56 (85.89/85.21) | 89.87 (90.23/89.52) |
| ME | 90.43 (91.07/89.80) | 93.49 (94.15/92.84) | 85.15 (86.01/84.30) | 89.70 (90.61/88.80) |
| CRF | 93.52 (93.77/93.26) | 94.69 (94.95/94.43) | 89.23 (90.29/88.20) | 91.00 (92.08/89.94) |
| SSVM | 93.53 (92.93/94.15) | 95.35 (94.72/95.97) | 90.01 (89.19/90.84) | 92.65 (91.91/93.51) |
Values are F-measure (recall/precision) (%).
CRF, conditional random fields; ME, maximum entropy; SVM, support vector machines; SSVM, structural support vector machines.
Discussion
In this study, we investigated ML-based approaches for NER in Chinese clinical text. We manually created annotated datasets of 400 admission notes and 400 discharge summaries in Chinese, and systematically evaluated the contributions of different types of features and ML algorithms for NER in Chinese clinical text. Our results showed that word segmentation information based on a Chinese medical dictionary and section information was beneficial to NER tasks in Chinese clinical text. When the same features were used, we also demonstrated that SSVM achieved the best performance of the four different ML algorithms. This was consistent with a previous study on NER in English clinical text.11 12 These findings will all be useful for future Chinese clinical NLP research.
In this study, the best performance of our NER system for Chinese discharge summaries was an F-measure of 90.01%, which is similar to the best F-measure (90.24%) reported in another recent NER study on Chinese discharge summaries.21 These results are much better than the best result on the 2010 i2b2/VA NLP challenge on clinical entity recognition from English discharge summaries (F-measure 85.23%).2 9 It is difficult to determine exactly why English clinical text is more difficult for NER tasks. We conducted an analysis of entity frequency in both English (the i2b2 corpus) and Chinese discharge summaries. It seems that entities in English clinical text are sparser than those in Chinese clinical text. In Chinese discharge summaries, 53.18% of entities occurred once, whereas in English clinical text 76.02% of entities occurred once. Therefore, the higher percentage of low-frequency entities may be one reason for the performance difference between English and Chinese clinical text. Moreover, the difference between exact-matching and inexact-matching F-measures of our best NER system on Chinese discharge summaries (2.64%) is much smaller than the best result on the i2b2/VA NLP challenge on clinical entity recognition in English discharge summaries (8.39%),2 9 indicating that the boundaries of entities in Chinese clinical text are easier to determine than in English clinical text. This may be another reason for the performance difference between English and Chinese clinical entity recognition.
Word segmentation is one of the major differences between English and Chinese text processing. However, when we used the Stanford Word Segmenter—a state-of-the-art Chinese segmenter in the general domain—the performance of the NER system did not improve much. More improvement was observed when a Chinese medical dictionary was used for word segmentation. This finding suggests that domain knowledge is important for word segmentation in Chinese clinical text. In future, we plan to use domain-specific word segmentation approaches for Chinese clinical text by combining medical knowledge bases with statistical word segmentation methods, to further improve NER performance. It is not surprising that the sequence-labeling algorithms were superior to the classification algorithms for NER in Chinese clinic notes, as sequence-labeling algorithms take the relationships between neighboring labels into consideration. However, it was important to verify that SSVM, a relatively new sequential labeling algorithm, could achieve a slightly better performance than CRF on NER in Chinese clinical text. This finding, together with our previous results,11 12 demonstrated that SSVM could be a competitive alternative to CRF on NER tasks in both English and Chinese clinical texts.
Furthermore, we analysed the errors in our best system. We found that most errors occurred in long entities with combined structures. For example, in a long problem entity “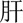 (liver)
(liver) 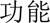 (function)
(function) 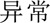 (abnormal)
(abnormal) 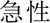 (acute)
(acute)  (exacerbation)” (acute exacerbation of abnormal liver function), only part of it—“
(exacerbation)” (acute exacerbation of abnormal liver function), only part of it—“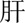 (liver)
(liver) 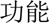 (function)
(function) 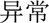 ” (abnormal)—was predicted to be a problem. Information about syntactic structures of Chinese sentences could potentially help in this scenario. However, there is very limited work on syntactic parsing of clinical text in Chinese, which requires extensive resources and effort (eg, building a Treebank), but is probably worth investigating. Another type of error was caused by unseen samples in the training set. For example, a procedure “
” (abnormal)—was predicted to be a problem. Information about syntactic structures of Chinese sentences could potentially help in this scenario. However, there is very limited work on syntactic parsing of clinical text in Chinese, which requires extensive resources and effort (eg, building a Treebank), but is probably worth investigating. Another type of error was caused by unseen samples in the training set. For example, a procedure “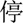 (discontinue)
(discontinue) 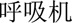 (ventilator)” (discontinue ventilator) was not detected because there were no similar medical concepts in the training dataset. Increasing the sample size may solve this problem.
(ventilator)” (discontinue ventilator) was not detected because there were no similar medical concepts in the training dataset. Increasing the sample size may solve this problem.
Conclusions
In this study, we systematically investigated features and ML algorithms for the NER task in Chinese clinical text using a manually annotated corpus of 400 admission notes and 400 discharge summaries. Our results show that both word segmentation and section information improved NER in Chinese clinical text, and SSVM, a recent sequential labeling algorithm, outperformed CRF and other classification algorithms. Our best system achieved F-measures of 90.01% and 93.52% on Chinese discharge summaries and admission notes, respectively, indicating a promising start for Chinese NLP research.
Footnotes
Contributors: The work was a collaboration of all the authors. JL, BT, MJ, and HX constructed the guidelines for corpus annotation. XL and KG annotated corpora used in the study. JL, BT, and HX designed the methods and experiments. BT carried out the experiments. JL, BT and HX analyzed the data, interpreted the results, and drafted the paper. All the authors have contributed to, seen, and approved the manuscript.
Funding: This study was partly supported by the National Natural Science Foundation of China (NSFC) (Grant 81 171 426) and the China Postdoctoral Science Foundation (CPSF) (Grant 2011M500669).
Competing interests: None.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–44 [PubMed] [Google Scholar]
- 2.Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tjong Kim Sang EF. Introduction to the CoNLL-2002 shared task: language-independent named entity recognition. Proceedings of the 6th conference on Natural language learning Vol 20, Stroudsburg, PA, USA, 2002:1–4 [Google Scholar]
- 4.Tjong Kim Sang EF, De Meulder F. Introduction to the CoNLL-2003 shared task: language-independent named entity recognition. Proceedings of the seventh conference on Natural language learning at HLT-NAACL 2003 Vol 4, Stroudsburg, PA, USA, 2003:142–7 [Google Scholar]
- 5.Nadeau D, Sekine S. A survey of named entity recognition and classification. Lingvisticae Investig 2007;30:3–26 [Google Scholar]
- 6.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010;17:229–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bruijn B, Cherry C, Kiritchenko S, et al. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc 2011;18:557–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang M, Chen Y, Liu M, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc 2011;18:601–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang B, Cao H, Wu Y, et al. Clinical entity recognition using structural support vector machines with rich features. Proceedings of the ACM sixth international workshop on Data and text mining in biomedical informatics, New York, NY, USA, 2012:13–20 [Google Scholar]
- 12.Tang B, Cao H, Wu Y, et al. Recognizing clinical entities in hospital discharge summaries using Structural Support Vector Machines with word representation features. BMC Med Inform Decis Mak 2013;13:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lafferty J, McCallum A, Pereira FCN. Conditional random fields: probabilistic models for segmenting and labeling sequence data. Departmental Papers (CIS), 2001 [Google Scholar]
- 14.Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;20:273–97 [Google Scholar]
- 15.Kudoh T, Matsumoto Y. “Use of support vector learning for chunk identification,” presented at the Proceedings of the 2nd workshop on Learning language in logic and the 4th conference on Computational natural language learning Vol 7, 2000:142–4 [Google Scholar]
- 16.Kudo T, Matsumoto Y. “Chunking with support vector machines,” presented at the NAACL ‘01: Second meeting of the North American Chapter of the Association for Computational Linguistics on Language technologies 2001, 2001:1–8
- 17.Taskar B, Guestrin C, Koller D. Max-margin Markov networks. Proceedings of the Seventeenth Annual Conference on Neural Information Processing Systems, 2003 [Google Scholar]
- 18.Tsochantaridis I, Joachims T, Hofmann T, et al. Large margin methods for structured and interdependent output variables. J Mach Learn Res 2005;6:1453–84 [Google Scholar]
- 19.Wang S, Li S, Chen T. Recognition of Chinese Medicine Named Entity Based on Condition Random Field. J Xiamen University (Natural Science) 2009;48:349–64 [Google Scholar]
- 20.Wang Y, Liu Y, Yu Z, et al. A preliminary work on symptom name recognition from free-text clinical records of traditional chinese medicine using conditional random fields and reasonable features. Proceedings of the 2012 Workshop on Biomedical Natural Language Processing, Stroudsburg, PA, USA, 2012:223–30 [Google Scholar]
- 21.Xu Y, Wang Y, Liu T, et al. Joint segmentation and named entity recognition using dual decomposition in Chinese discharge summaries. J Am Med Inform Assoc 2014;21:e84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller G, Horn D. Maximum entropy approach to probability density estimation. 1998 Second International Conference on Knowledge-Based Intelligent Electronic Systems, 1998. Proceedings KES ‘98, 1998;1:225–30 [Google Scholar]
- 23.Platt J. Sequential minimal optimization: a fast algorithm for training support vector machines. Technique Report 1998:1–21 [Google Scholar]
- 24.Franc V, Sonnenburg S. Optimized cutting plane algorithm for support vector machines. Proceedings of the 25th international conference on Machine learning, New York, NY, USA, 2008:320–7 [Google Scholar]
- 25.Keerthi SS, Sundararajan S, Chang K-W, et al. A sequential dual method for large scale multi-class linear SVMs. the 14th ACM SIGKDD international conference on Knowledge discovery and data mining 2008:408–16. [Google Scholar]
- 26.Darroch J, Ratcliff D. Generalized Iterative Scaling for Log-Linear Models. Ann Math Stat 1972;43:1470–80 [Google Scholar]
- 27.Head JD, Zerner MC. A Broyden—Fletcher—Goldfarb—Shanno optimization procedure for molecular geometries. Chem Phys Lett 1985;122:264–70 [Google Scholar]
- 28.Liu DC, Nocedal J. On the limited memory BFGS method for large scale optimization. Math Programming 1989;45:503–28 [Google Scholar]
- 29.Vishwanathan SVN, Schraudolph NN, Schmidt MW, et al. Accelerated training of conditional random fields with stochastic gradient methods. ICML, 2006:969–76 [Google Scholar]
- 30.Hripcsak G, Rothschild AS. Agreement, the F-Measure, and Reliability in Information Retrieval. J Am Med Inform Assoc 2005;12:296–8 [DOI] [PMC free article] [PubMed] [Google Scholar]


