Abstract
Objective
We describe experiments designed to determine the feasibility of distinguishing known from novel associations based on a clinical dataset comprised of International Classification of Disease, V.9 (ICD-9) codes from 1.6 million patients by comparing them to associations of ICD-9 codes derived from 20.5 million Medline citations processed using MetaMap. Associations appearing only in the clinical dataset, but not in Medline citations, are potentially novel.
Methods
Pairwise associations of ICD-9 codes were independently identified in both the clinical and Medline datasets, which were then compared to quantify their degree of overlap. We also performed a manual review of a subset of the associations to validate how well MetaMap performed in identifying diagnoses mentioned in Medline citations that formed the basis of the Medline associations.
Results
The overlap of associations based on ICD-9 codes in the clinical and Medline datasets was low: only 6.6% of the 3.1 million associations found in the clinical dataset were also present in the Medline dataset. Further, a manual review of a subset of the associations that appeared in both datasets revealed that co-occurring diagnoses from Medline citations do not always represent clinically meaningful associations.
Discussion
Identifying novel associations derived from large clinical datasets remains challenging. Medline as a sole data source for existing knowledge may not be adequate to filter out widely known associations.
Conclusions
In this study, novel associations were not readily identified. Further improvements in accuracy and relevance for tools such as MetaMap are needed to realize their expected utility.
Keywords: Data Mining, Electronic Health Records, International Classification of Diseases, Medline, Unified Medical Language System, Natural Language Processing
Introduction
The age of ‘Big Data’ has arrived.1–3 Studies using data collected as part of routine clinical care from hundreds of thousands, or even millions, of patients are becoming increasingly common.4–8 Other large datasets (eg, adverse event reports) are also being linked to these clinical data to accelerate discovery,9 10 leading to new findings of intriguing and potentially clinically relevant associations that could aid in the understanding of disease processes.10 11 For example, Tatonetti et al12 recently discovered an association between elevated blood glucose levels and the co-administration of paroxetine and pravastatin, neither of which raised blood glucose when given alone.
Association analysis methods have been widely used to aid in knowledge discovery, where associations are usually determined by finding pairwise relationships among entities that co-occur at a statistically significant rate compared to the overall population. We previously reported on two separate association analyses, one utilizing 1.5 million free text problem list entries from over 300 000 patients5 and the other using 41.2 million International Classification of Disease, V.9 (ICD-9) codes from over 1.6 million patients.4 The latter dataset represented virtually all possible diagnosis-based clinical associations known to our health system, a large tertiary academic medical center with over 1.8 million outpatient and emergency visits and 44 000 hospital stays annually, derived from over a decade of patient encounters.
In both studies,4 5 we noted that a very large number of statistically significant associations resulted from the analyses, making it impossible to identify all novel ones via manual review alone. We also noted that many of the associations, especially the most statistically significant ones, are already widely known. For example, in the study using free text diagnoses, the well-known associations we found included one between obesity and hypertension and one between Turner syndrome and ovarian failure. Lesser known associations that we confirmed with a manual literature review included hypothyroidism and fibromyalgia as well as gout and cardiomyopathy.5 An example of a well-known association from the second study using ICD-9 codes included end stage renal disease and kidney transplant, whereas an unusual association with no supporting evidence in the literature was between depression and animal bites (primarily cats).4 This latter association was confirmed with a manual chart review.11 However, manually validating the potential novelty of all associations against the literature, from such large-scale analyses, is not feasible. Developing automated approaches that can effectively distinguish known from unknown clinical associations is thus imperative. Such automated methods could be beneficial for a variety of applications, including surveillance of electronic health record data to detect previously unknown or newly arising patterns.
A potential approach to automating the identification of novel associations is through comparing those found in clinical datasets against comprehensive repositories of known associations. The National Library of Medicine's (NLM) Medline/PubMed database is the world's largest indexed repository of biomedical literature, with over 19 million citations from over 5500 journals.13 It might therefore be possible to use Medline as a basis from which such a knowledge repository of associations could be assembled and then compared to associations derived from large clinical datasets. Such literature-based discovery techniques, also known as literature mining, are not new.14–18 However, prior studies have often focused on specific clinical areas, such as psychiatry19 20 or diabetes.21 22 It is unclear how well such an approach might work with an ‘all versus all’ comparison—that is, all patient data from a large health system versus all abstracts in Medline.
Recently, the NLM computationally processed the entire Medline database using their natural language processing (NLP) based named entity recognition software tool, MetaMap.23 MetaMap identifies clinical concepts (eg, ‘type 2 diabetes’) from unstructured biomedical text and then maps them to concept unique identifiers (CUIs) in the Unified Medical Language System (UMLS) Metathesaurus; for example, C0865162 is a CUI for ‘diabetes’. These CUIs can, in turn, be mapped to various taxonomies, vocabularies, and ontologies including ICD-9; for example, the CUI above, C0865162, maps to the ICD-9 code ‘250.0’. MetaMap has been used for a variety of tasks including extracting information from drug labels,24 coding death certificates,25 conducting biosurveillance,26 parsing documents from electronic health records,27 28 supporting Medical Subject Heading (MeSH) assignments through use of the NLM Medical Text Indexer (MTI),29–32 improving information retrieval from Medline,33 and even assigning ICD-9 codes from clinical text.34–36
We hypothesized that clinical associations derived from administrative ICD-9 codes assigned during patient encounters can be compared against associations extracted from the literature to distill novel associations from known ones. That is, if an association resulting from a clinical dataset is not found in Medline, then it may be potentially novel and warrant further investigation. We also hypothesized that among a collection of associations derived from the same large dataset, clinical associations ranked higher (ie, with smaller p values or larger χ2 statistics) would be more likely to be identified in the literature than lower ranked ones, based on the assumption that common problems are more likely to have been studied and published. In this study, we developed and empirically validated a set of computational analyses to assess the feasibility of verifying novel associations identified through mining clinical data against the results from mining the literature. Our primary goal in this method development paper is to explore the feasibility of this approach rather than making new clinical discoveries.
Methods
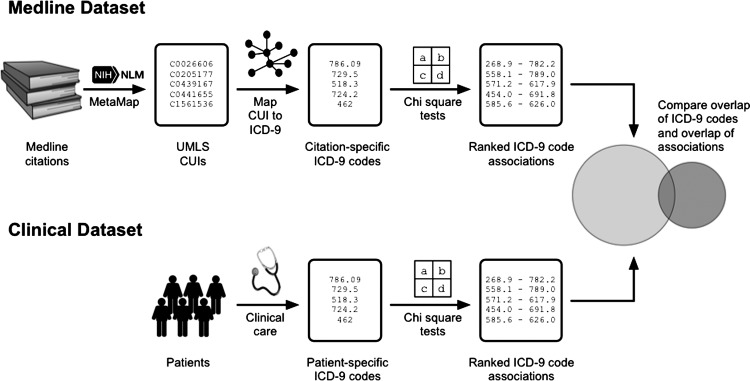
A high-level overview of the analytic approach used in this paper is illustrated in figure 1. Below, we describe the data sources and each of the analytic steps in-depth.
Figure 1.
A high-level overview of the analytic approach, including the data sources and processes.
Description of datasets
Two datasets were used in the analysis. The clinical associations (referred to as ‘Clinical’) were obtained from the dataset processed for a prior study.4 These associations encompass ICD-9 administrative data from both inpatient and outpatient encounters for 1.6 million patients at our health system, and spanned more than a decade. A description of how the clinical associations were computed can be found in the ‘Association analyses’ subsection, below.
The second dataset (referred to as ‘Medline’) was provided by the NLM and included all citations in Medline/PubMed as of November 18, 2011. The NLM has named this the ‘2012 MetaMapped Medline Baseline Results’.37 The dataset contains a total of 20.5 million citations processed by the NLM using MetaMap. It is comprised of 3.3 billion lines of text in the MetaMap Machine Output (MMO) Format, taking over 1.5 terabytes disk space.38 39
Extraction of CUIs
During the named entity extraction process, MetaMap generates a normalized score estimating the degree of match between a candidate term and the Metathesaurus based on four components: centrality, variation, coverage, and cohesiveness.23 40 From the Medline dataset, we parsed all CUIs with a ‘perfect’ MetaMap score of 1000, noting the specific PubMed Identifier (PMID) of the paper(s) from which each of the CUIs was from and whether a CUI was identified in the title or in the abstract of the paper(s). This is referred to as the ‘Medline 1000’ dataset.
Using CUIs with only a perfect score will exclude alternative ways to express a given concept because it essentially requires an exact match and does not allow for synonomy or slight word variations. Therefore, because it was unclear how much information might be lost by including only perfect mapping scores, we also created another dataset consisting of extracted CUIs where candidate concepts had mapping scores of 600 or higher. This allowed us to broaden the scope of our search to ensure a larger capture rate for concepts, since a CUI mapped to an ICD-9 code might otherwise be ‘hidden’ by a higher scoring CUI from a different vocabulary. We refer to the second dataset as ‘Medline 600’. This threshold was chosen because it was lower, and thus more inclusive, than thresholds commonly used in prior studies (see Discussion).
CUI to ICD-9 mapping
From the UMLS Metathesaurus (V.2013AA), we extracted two vocabularies, namely ‘ICD9CM’ and ‘MTHICD9’, in order to map Metathesaurus CUIs to ICD-9 entries. ICD9CM is the clinical modification of the International Classification of Diseases developed by the US Department of Health and Human Services. MTHICD9 provides additional synonyms for ICD9CM terms and was developed for the Metathesaurus by the NLM. Both vocabularies were extracted from the MRCONSO.RRF file that contains concepts, concept names, and their sources (MR: metathesaurus relational; CON: concept; SO: source; RRF: rich release format). For CUIs mapped to more than one ICD-9 code, we included only those that mapped to 50 or fewer distinct ICD-9 codes. This was done because some CUIs mapped to so many different codes that they were considered too non-specific for our analysis. The resulting mapping set contains 36 988 distinct mappings representing 33 999 and 22 198 unique CUIs and ICD-9 codes, respectively. We then processed the CUIs from the MetaMap-prepared Medline citations, retaining only those CUIs that mapped to at least one ICD-9 code.
Association analyses
Leveraging all ICD-9 codes mapped from clinical concepts in Medline citations, we then conducted a large-scale association analysis based on the methodology we had previously developed for discovering clinical associations from patient care data.4 In short, we determined the probability and statistical significance of two codes co-occurring in the same patient profile or Medline citation using the χ2 test. This was done by constructing 2×2 tables for every pair of concepts. For example, in the Medline dataset we determined the χ2 statistic based on the following conditions: (1) citations that mentioned both diagnosis A and diagnosis B; (2) citations that mentioned diagnosis A but not diagnosis B; (3) citations that mentioned diagnosis B but not diagnosis A; and (4) citations that mentioned neither diagnosis A nor B. While there are a number of ways to quantify the relationship between two binary variables, the χ2 test is most popularly used. We used the χ2 statistic to rank order the associations within each dataset to provide a basis for inferring the ‘relative strength’ among the associations in the dataset. Note that temporal aspects were not considered in the experiments reported in this paper.
For the Clinical dataset, in accordance with our prior analysis, only codes that appeared at least 30 times and where at least 10 patients shared the same pair of two codes were considered to be potentially associated. For codes under these thresholds a χ2 test was not performed. By contrast, the association analysis for the Medline dataset considered any pair of codes that appeared together in at least one citation. This was because some citations could represent meta-studies (eg, systematic reviews) as well as studies based on data collected from tens or thousands of patients.41 These datasets (both ‘Medline’ and ‘Clinical’) that included all ICD-9 codes in their original format are referred to as ‘Original’.
Additionally, because coding variations may occur among both clinicians and professional coders,42–45 and because these variations could affect the nature of associations discovered, we also conducted an additional association analysis by collapsing the hierarchical structure of the ICD-9 taxonomy so that all ‘subcategory codes’ were merged into their parent ‘category code’. For example, codes such as ‘250.0’, ‘250.23’, and ‘250.80’ were converted to simply ‘250’, a high-level code used to designate the entire family of diabetes. These collapsed datasets (both ‘Medline’ and ‘Clinical’) are referred to in this paper as ‘Simplified’.
High-level dataset comparisons
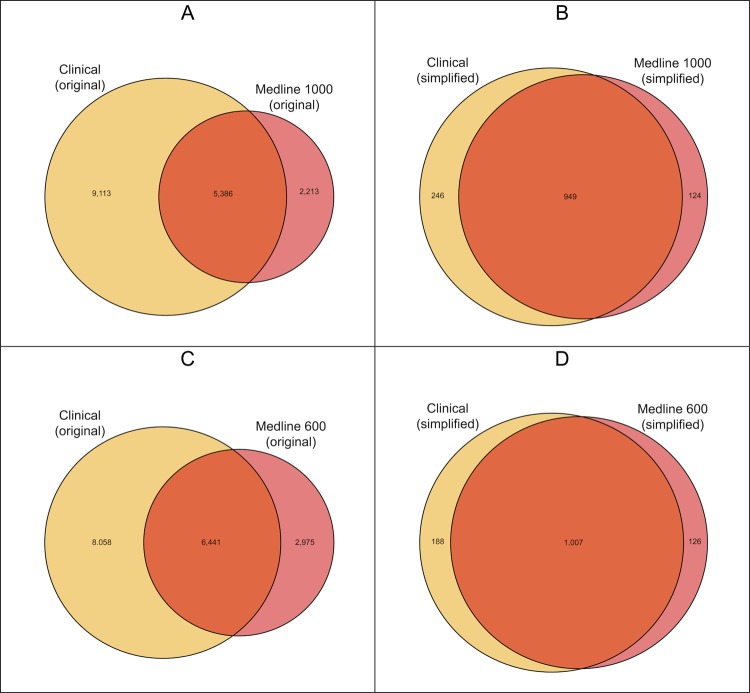
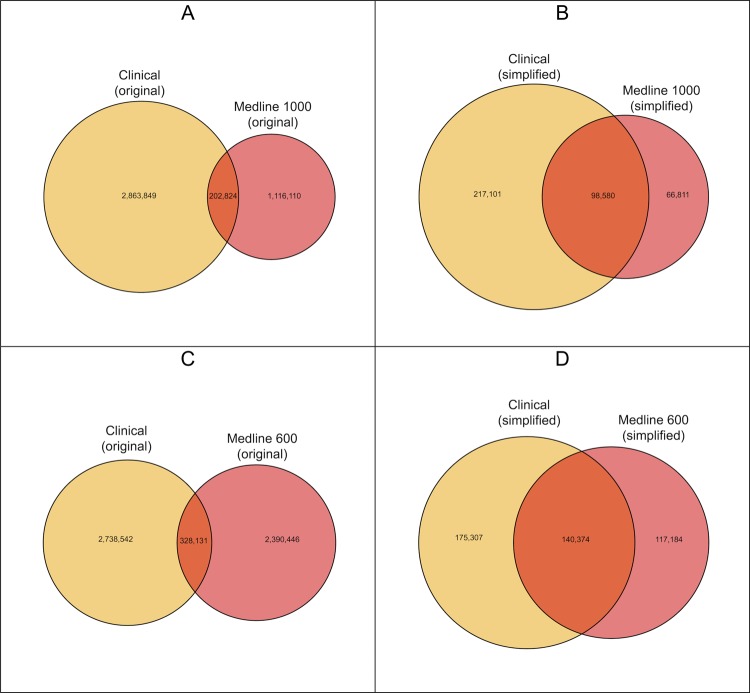
The primary objective of this study was to determine whether associations identified in the literature could be used to filter out known associations identified from patient care data, thus revealing potentially novel clinical associations. Therefore, we compared the Medline and Clinical datasets to determine how much overlap existed in terms of the ICD-9 codes used and the clinical associations discovered in each. This overlap was quantified and visualized using Venn diagrams (figures 2 and 3).46
Figure 2.
Venn diagrams showing overlap of International Classification of Disease, V.9 (ICD-9) codes found in the Clinical and Medline datasets. Panels A and C represent the original codes, whereas Panels B and D represent the simplified codes. Codes that do not overlap between the two datasets have no chance of becoming a pairwise association found in both sets.
Figure 3.
Venn diagrams showing overlap of pairwise associations found in the Clinical and Medline datasets. Panels A and C display the Original datasets, where as Panels B and D represent the Simplified datasets. The left most area in each panel is the one most likely to contain novel associations not previous described in the literature as they are found in the Clinical dataset but not the Medline dataset.
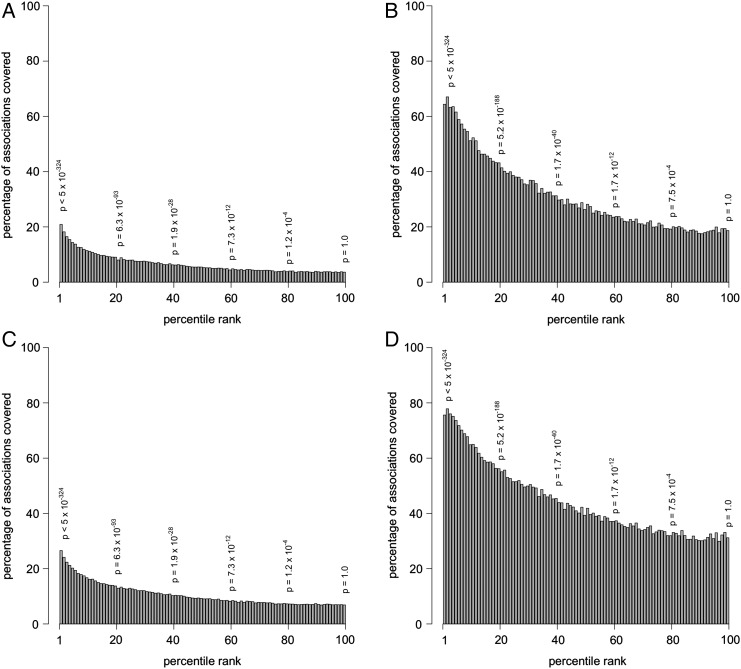
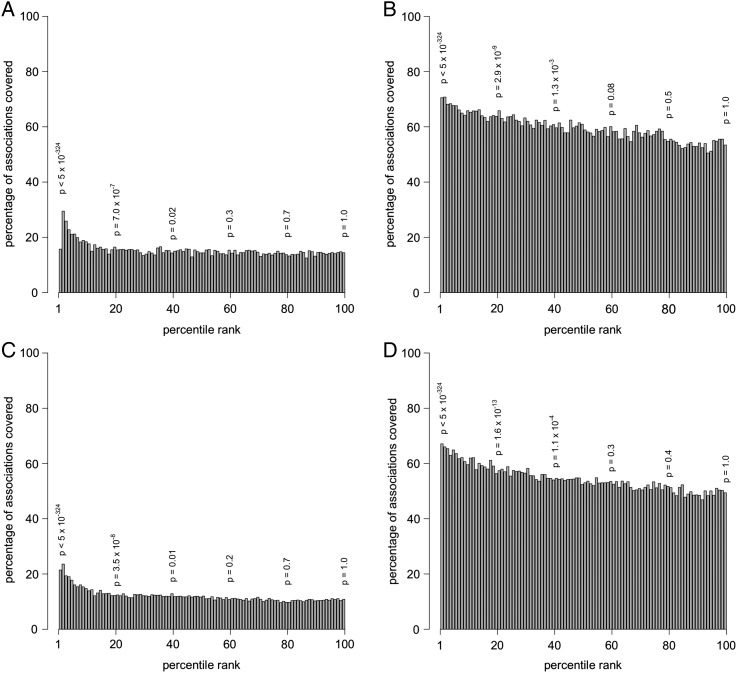
We then modeled the probability of an association being in the literature as a function of its ranking in the clinical dataset. Associations in the clinical dataset were ranked according to their χ2 statistic, and each association was assigned a percentile rank ranging from 1st (highest) to 100th (lowest). For all of the clinical associations assigned to the same percentile, we determined which ones had corresponding associations in the Medline dataset. These results were visualized using bar plots (figure 4). We also modeled the converse—that is, the probability of an association being in the Clinical dataset as a function of its χ2 ranking in the Medline dataset.
Figure 4.
Bar plots showing the relationship between the rank of an association in the Clinical dataset and the probability of the association appearing in the Medline dataset. (A) Clinical (original) appearing in Medline 1000 (original); (B) Clinical (simplified) appearing in Medline 1000 (simplified); (C) Clinical (original) appearing in Medline 600 (original); (D) Clinical (simplified) appearing in Medline 600 (simplified). The p-values shown represent the significance of the Clinical associations at that percentile.
Association-specific comparisons
We also looked at individual associations derived from our clinical dataset to better estimate the utility and reliability of using the literature as a screening tool to validate novelty. We examined this through two steps. First, to determine the reliability of the associations found in both the Clinical and Medline datasets, we randomly selected Clinical associations at several specific levels of significance (eg, 50th percentile, 75th percentile) for which there were corresponding Medline associations. ICD-9 ‘V’ codes were not considered in this part of the analysis because they are often less directly related to specific diagnoses. Two practicing, informatics-trained physicians (DAH and MS), with 14 and 6 years of postgraduate experience, respectively, independently reviewed the titles and abstracts from which the literature-based associations were derived. Each citation was judged to either support the association identified from the clinical dataset or not to support it. Additionally, associations were judged to be clinically ‘surprising’ if there was no clear explanation for why the two diagnoses might be related. Agreements in the two reviewers’ judgments were quantified using the κ statistic.
We then randomly sampled clinical associations for which there were no corresponding associations found in the Medline dataset to determine if they might represent novel associations, and to compare the performance of MetaMap against manual strategies inspecting for novelty. The same two physicians reviewed these associations to determine if they were clinically surprising. They also conducted a manual literature search in Medline in an attempt to find out whether, for each of the associations, at least one citation could be found that provides evidence confirming its validity.
Additional analyses
We performed several additional analyses and report the results in online appendices A, B, and C. Online supplementary appendix A provides several groups of scatter plots exhibiting the relationship between the frequency of an ICD-9 code appearing in each of the datasets analyzed, and the corresponding number of associations to which it belonged. In online supplementary appendix B, we explored the distribution of association rankings as a function of an ICD-9 code's frequency in the original clinical dataset. We selected 15 ICD-9 codes that appeared with varying frequencies in the clinical dataset for this experiment. Finally, in online supplementary appendix C, we explored the potential for using the UMLS relationships as defined in the UMLS relational table MRREL.RRF as another source of associations that could be used to help filter known from unknown associations. Additional methodological details can be found in each of the online supplementary appendices. Note that in this study, we did not explore the use of Semantic MEDLINE47 48 because the relationships contained in Semantic MEDLINE are directly derived from MetaMap-processed Medline citations, where are therefore not expected to generate substantially different results from our primary analysis.
All statistical analyses reported in this paper were conducted using R V.2.15.3. Venn diagrams were created using the VennDiagram Package for R.46 While we used the χ2 statistic to rank the associations, the corresponding p values are also reported to aid in interpretation. For computation of the datasets, we used a 2010 Apple Mac Mini equipped with a 2.66 GHz Core 2 Duo processor and 8 GB RAM, and for storage we used an external 4 TB hard drive connected via USB 2.0.
Results
Characteristics of the datasets
Processing the large dataset of all Medline citations yielded 333.3 million non-distinct CUIs with a perfect score of 1000. About 28.5 million of these CUIs (8.5%) were identified in the titles, with the remaining 304.9 million (91.5%) identified in the abstracts. There were approximately 16.3 million unique citations represented in this dataset. However, only a subset of the CUIs, approximately 23.1 million CUIs representing 5.1 million unique citations, mapped to at least one of 7599 distinct ICD-9 codes. Additional characteristics of the dataset are shown in table 1.
Table 1.
Characteristics of the datasets used in the analysis
| Original datasets | Simplified datasets | |||||
|---|---|---|---|---|---|---|
| Clinical | Medline 1000 | Medline 600 | Clinical | Medline 1000 | Medline 600 | |
| Rows of data | 41 192 825 | 23 051 034 | 147 994 791 | 41 192 825 | 23 051 034 | 147 994 791 |
| Patients (clinical data)/citations (Medline data) in full dataset | 1 620 280 | 5 069 886 | 9 975 979 | 1 620 280 | 5 069 886 | 9 975 979 |
| Patients/citations included in final pairwise comparisons dataset | 1 619 785 | 5 069 347 | 9 975 705 | 1 620 271 | 5 069 851 | 9 975 966 |
| Distinct ICD-9 codes | 14 499 | 7599 | 9416 | 1195 | 1073 | 1133 |
| Possible pairwise comparisons* | 105 103 251 | 28 868 601 | 44 325 820 | 713 415 | 575 128 | 641 278 |
| Actual pairwise comparisons† | 3 066 673 | 1 318 933 | 2 718 577 | 315 681 | 165 391 | 257 558 |
| Unique ICD-9 codes meeting criteria to be used in a pairwise comparison‡ | 8601 | 7309 | 9233 | 1099 | 1057 | 1126 |
*Possible pairwise comparisons is determined by (n2−n)/2, where n is the number of distinct ICD-9 codes in the dataset.
†A pairwise comparison was only calculated under the following conditions: (1) for the clinical data if (a) each code was assigned to at least 30 patients and (b) the pair of codes were present together in at least 10 patients; (2) for the Medline data if (a) each code was assigned to at least 1 citation and (b) the pair of codes was present together in at least 1 citation.
‡The actual number of distinct ICD-9 codes that were used in the pairwise comparisons in each dataset. Not all possible codes contributed to a pairwise comparison.
ICD-9, International Classification of Disease, V.9
The ‘Medline 600’ dataset used less stringent criteria for selecting CUIs for inclusion, and thus 3.5 billion non-distinct CUIs were identified, with 348.2 million (9.8%) in the titles and 3.2 billion (90.2%) in the abstracts. This dataset contained 20.4 million unique citations, representing all but 0.4% of the citations included in the original dataset. Only CUIs that mapped to an ICD-9 code were retained, resulting in 148.0 million CUIs, 10.0 million citations, and 9416 unique ICD-9 codes. Table 1 also summarizes the characteristics of this dataset.
Only one ICD-9 code (401.9, ‘unspecified essential hypertension’) appeared in the top 30 most frequently appearing codes in the Original Clinical dataset as well as the top 30 of the Original Medline datasets. By reducing the coding variation in the Simplified datasets, six of the top 30 codes in the Clinical dataset can also be found in the top 30 of at least one of the corresponding Medline datasets (table 2). For the Clinical datasets, this list represents the most common diseases (diagnoses) treated by our health system. For the Medline datasets, it shows the most common diagnoses discussed in the literature, based on MetaMap.
Table 2.
The 30 most common codes for the Simplified versions of the three datasets
| Simplified Clinical dataset | Simplified Medline 1000 dataset | Simplified Medline 600 dataset | ||||||
|---|---|---|---|---|---|---|---|---|
| Code | Description | Freq. | Code | Description | Freq. | Code | Description | Freq. |
| 786 | Symptoms involving respiratory syststem/chest | 323,562 | 780 | General symptom | 279,960 | 89 | Interview, evaluation, consultation, and examination | 684,766 |
| 780 | General symptom | 286,381 | 89 | Interview, evaluation, consultation, and examination | 168,522 | 780 | General symptom | 669,453 |
| 789 | Symptoms involving abdomen, pelvis | 228,190 | 239 | Neoplasms of unspecified nature | 164,581 | 239 | Neoplasms of unspecified nature | 491,658 |
| 719 | Joint disorders | 226,585 | 88 | Other diagnostic radiology and related techniques | 164,304 | 99 | Other nonoperative procedures | 452,538 |
| 427 | Cardiac dysrhythmias | 206,499 | 99 | Other nonoperative procedures | 151,387 | 88 | Other diagnostic radiology and related techniques | 425,485 |
| V72 | Special investigations and exams | 195,803 | 199 | Malignant neoplasm | 134,119 | 199 | Malignant neoplasm | 370,794 |
| 518 | Diseases, lung, other | 183,058 | 250 | Diabetes mellitus | 116,196 | 279 | Disorders of the immune mechanism | 316,417 |
| 465 | Acute infections of upper respiratory tract | 171,964 | 401 | Essential hypertension | 101,952 | E904 | Accident due to hunger/thirst/exposure | 311,337 |
| V70 | General medical examination | 171,947 | 997 | Complication affecting body | 95,460 | 759 | Congenital anomalies | 254,442 |
| 724 | Back disorders | 152,960 | 402 | Hypertensive heart disease | 93,712 | 39 | Operations on vessels | 215,326 |
| 729 | Disorders, soft tissues | 151,388 | 405 | Secondary hypertension | 93,431 | 87 | Diagnostic radiology | 203,444 |
| V04 | Need for prophylactic vaccination | 149,835 | 403 | Hypertensive chronic kidney disease | 93,270 | 042 | HIV disease | 193,453 |
| V07 | Need for prophylactic measures | 148,123 | 404 | Hypertensive heart and chronic kidney disease | 93,148 | 250 | Diabetes mellitus | 187,375 |
| 787 | Symptoms involving digestive system | 141,223 | 338 | Pain | 80,803 | 695 | Erythematous conditions | 176,305 |
| 959 | Injury, not otherwise specified | 139,420 | 042 | HIV disease | 78,847 | 782 | Symptoms involving skin, other tissue | 172,965 |
| V67 | Follow-up examination | 139,155 | 278 | Overweight, obesity and other hyperalimentation | 70,312 | 338 | Pain | 172,190 |
| 782 | Symptoms involving skin, other tissue | 138,882 | 429 | Ill-defined heart disease | 69,354 | 997 | Complication affecting body | 168,875 |
| V06 | Need for combination vaccination | 135,206 | 427 | Cardiac dysrhythmias | 68,396 | 401 | Essential hypertension | 168,857 |
| 401 | Essential hypertension | 133,645 | 410 | Acute myocardial infarction | 67,988 | 799 | Morbidity/mortality, ill-defined | 162,701 |
| 784 | Symptoms involving head and neck | 129,325 | E904 | Accident due to hunger/thirst/exposure | 67,891 | 92 | Nuclear medicine | 159,785 |
| V76 | Screening for malignant neoplasms | 118,977 | 311 | Depressive disorder | 66,878 | 429 | Ill-defined heart disease | 158,653 |
| V20 | Health supervision of infant/child | 114,588 | 787 | Symptoms involving digestive system | 66,615 | 402 | Hypertensive heart disease | 157,874 |
| 785 | Symptoms involving cardiovascular system | 113,977 | 414 | Chronic ischemic heart disease | 65,600 | 405 | Secondary hypertension | 157,495 |
| 733 | Bone and cartilage disorders | 112,034 | 782 | Symptoms involving skin, other tissue | 62,834 | 403 | Hypertensive chronic kidney disease | 157,374 |
| 599 | Urethra/urinary tract disorders | 102,919 | 39 | Actinomycotic infections | 61,703 | 404 | Hypertensive heart and chronic kidney disease | 156,992 |
| 367 | Refraction/accommodation disorder | 102,395 | 995 | Adverse effects | 59,973 | 368 | Visual disturbances | 155,964 |
| 709 | Disorders of skin & sbcutn tissue | 101,979 | 786 | Symptoms involving respiratory system/chest | 58,738 | 995 | Adverse effects | 155,379 |
| V05 | Need for prophylactic vaccination | 98,882 | 300 | Anxiety, dissociative and somatoform disorders | 58,397 | 427 | Cardiac dysrhythmias | 149,674 |
| 530 | Esophagus diseases | 98,170 | 759 | Congenital anomalies | 56,048 | 268 | Vitamin D deficiency | 142,584 |
| 382 | Suppurative otitis media | 95,640 | 277 | Unspecified metabolism disorder | 55,589 | 269 | Other nutritional deficiencies | 142,075 |
Codes that appear in the top 30 of the Clinical dataset and in the top 30 of least one of the Medline datasets are highlighted to show the concordance.
Figure 2 shows the overlap of the ICD-9 codes that were included in the datasets being compared. Slightly more than a third (37.1%) of the ICD-9 codes from the Original Clinical dataset were found at least once in the Original Medline 1000 dataset, and only 44.4% were found in the more inclusive Original Medline 600 dataset. When the datasets were simplified by merging subcategory codes, more overlap was evident: four-fifths (79.4%) of the codes in the Simplified Clinical dataset were in the Simplified Medline 1000 dataset, and 84.3% were in the Simplified Medline 600 dataset.
Clinical associations also found in Medline
We hypothesized that many of the associations discovered in the Clinical dataset would have been reported in Medline, achieving a more manageable number of potentially novel associations to be explored further for clinical significance. However, this was not the case according to the results of our analyses. As shown in the Venn diagrams illustrating the overlap between the Medline and Clinical datasets (figure 3), in the original datasets, only 6.6% of the Clinical associations had a correlate in the Medline 1000 dataset, with slightly more (10.7%) found in the Medline 600 dataset. The simplified datasets, where the ICD codes of the same family were merged into parent categories, displayed higher coverage, with 31.2% of the Clinical associations also present in the Medline 1000 data, and almost half (44.5%) of the Clinical associations also found in Medline 600.
We anticipated that higher ranked Clinical associations (based on larger χ2 statistics or, conversely, smaller p values) would be more likely to be found in Medline, and this was evident in the bar plots shown in figure 4, where A/C and B/D show the trends for the Clinical datasets, Original versus Simplified, respectively. In figure 4, it can clearly be observed that higher ranked associations in the Clinical set in general were more likely to be found in the Medline dataset. However, our results did not suggest as strong a correlation for the converse. That is, the relationship between the rank of an association found in Medline and the likelihood of finding that association in the Clinical dataset was not as strong (figure 5).
Figure 5.
Bar plots showing the relationship between the rank of an association in the Medline dataset and the probability of the association appearing in the Clinic dataset. (A) Medline 1000 (original) appearing in Clinical (original); (B) Medline 1000 (simplified) appearing in Clinical (simplified); (C) Medline 600 (original) appearing in Clinical (original); (D) Medline 600 (simplified) appearing in Clinical (simplified). The p-values shown represent the significance of the Medline associations at that percentile.
Specific associations
Table 3 shows associations identified from both the Clinical and Medline 1000 datasets at varying levels of significance. As shown in the table, the lower-ranked associations tended to be more clinically ‘surprising’. However, our manual review of the citations showed that many of the titles/abstracts mentioning two diagnoses together did not truly suggest an actual association between the diagnoses. That is, the mere mention of two diagnoses in the same citation is not a reliable indicator of a clinically meaningful association. Table 4 displays associations in the Clinical dataset that were not present in Medline 1000. Again, clinically surprising associations tended to be less significant (lower-ranked), and they were also less commonly found in Medline even with a manual search.
Table 3.
Clinical associations also found in the ‘Original’ Medline 1000 dataset
| ICD-9 code | Description | ICD-9 code | Description | Association p value derived from Clinical dataset |
Clinically surprising? | Number of Medline citations with both concepts mentioned | Supporting citations (PMID)† |
Citation could be interpreted as describing an association? |
|---|---|---|---|---|---|---|---|---|
| 1st percentile (highest ranked associations) | ||||||||
| 290.0 | Senile dementia | 331.0 | Alzheimer's disease | <5×10−324 | No | 1139 | 11138345 2614500 7056516 |
No Yes No |
| 344.61 | Cauda equina syndrome with neurogenic bladder | 596.54 | Neurogenic bladder | <5×10−324 | No | 11 | 11880062 16813905 3400548 |
* No No |
| 184.4 | Malignant neoplasm of vulva | 233.3 | Carcinoma in situ, unspecified female genital organs | <5×10−324 | No | 1 | 2082867 | * |
| 396.2 | Mitral valve insufficiency and aortic valve stenosis | 424.1 | Aortic valve disorders | <5×10−324 | No | 2 | 11163732 9665226 |
Yes Yes |
| 362.21 | Retrolental fibroplasia | 769 | Respiratory distress syndrome in newborn | <5×10−324 | No | 92 | 12709796 19568962 15716610 |
* * * |
| 25th percentile | ||||||||
| 268.9 | Unspecified vitamin D deficiency (268.9) | 782.2 | Localized superficial swelling, mass, or lump | 3.8×10−68 | Yes | 3 | 11233710 21806909 9339283 |
No No No |
| 558.1 | Gastroenteritis and colitis due to radiation | 789.0 | Abdominal pain | 7.1×10−68 | No | 1 | 8140765 | Yes |
| 279.5 | Graft-versus-host disease | E888.9 | Accidental fall | 2.4×10−67 | Yes‡ | 1 | 9250172 | No |
| 131.9 | Trichomoniasis | 590.80 | Pyelonephritis | 3.0×10−67 | * | 1 | 9286064 | * |
| 078.19 | Viral warts | 622.11 | Mild dysplasia of cervix | 8.4×10−67 | No | 1 | 7559948 | Yes |
| 50th percentile | ||||||||
| 276.52 | Hypovolemia | 403.9 | Hypertensive renal disease | 9.8×10−23 | No | 73 | 11602456 17113396 365408 |
Yes Yes No |
| 255.41 | Glucocorticoid deficiency | 788.3 | Functional urinary incontinence | 4.8×10−22 | Yes | 1 | 9836036 | Yes |
| 413.9 | Angina pectoris | 596.51 | Hypertonicity of bladder | 7.0×10−22 | Yes | 1 | 17689623 | No |
| 286.9 | Coagulation defect | 344.00 | Quadriplegia | 8.0×10−22 | * | 4 | 11403538 16958632 3508705 |
No Yes No |
| 229.9 | Benign neoplasm of unspecified site | 354.0 | Carpal tunnel syndrome | 4.9×10−21 | Yes | 1 | 1672719 | * |
| 75th percentile | ||||||||
| 416.9 | Chronic pulmonary heart disease | 783.41 | Failure to thrive | 4.6×10−6 | No | 24 | 12597677 8165079 2657582 |
Yes Yes Yes |
| 351.0 | Bell's palsy | 742.59 | Congenital anomalies of spinal cord | 4.7×10−6 | No | 1 | 18756840 | No |
| 302.72 | Psychosexual dysfunction with inhibited sexual excitement | 576.2 | Obstruction of bile duct | 4.6×10−6 | Yes | 1 | 16402030 | No |
| 259.9 | Endocrine disorder | 368.00 | Amblyopia | 4.6×10−6 | Yes | 1 | 15105955 | Yes |
| 611.72 | Lump or mass in breast | 759.6 | Hamartoses | 4.7×10−6 | No | 1 | 19737912 | * |
| 100th percentile (lowest ranked associations) | ||||||||
| 571.2 | Alcoholic cirrhosis of liver | 617.9 | Endometriosis | 0.99 | Yes | 1 | 8834254 | * |
| 153.9 | Malignant neoplasm of colon | 487.1 | Influenza with other respiratory manifestations | 0.99 | Yes | 3 | 12889684 18544745 20813181 |
No No No |
| 454.0 | Varicose veins of lower extremities with ulcer | 691.8 | Other atopic dermatitis and related conditions | 0.98 | Yes | 1 | 3442079 | No |
| 585.6 | End stage renal disease | 626.0 | Absence of menstruation | 0.99 | No | 23 | 3130865 16619340 9593608 |
Yes * Yes |
| 110.5 | Dermatophytosis of the body | 135 | Sarcoidosis | 0.99 | Yes | 1 | 11476274 | No |
These were selected from the area of overlap in figure 3A. The lower-ranked associations tend to be more clinically surprising. Many of the citations found in the literature did not actually suggest a true association after manual review. Whether an association was clinically surprising was independently determined by two physicians (κ statistic 0.84; 95% CI 0.63 to 1.05). The κ statistic for agreement on whether the citations found by our approach supported the association was 0.55 (95% CI 0.30 to 0.79).
*Opinions for which the physicians differed.
†For associations with more than three citations, three were randomly selected for this analysis.
‡The abstract stated, ‘donor stem cells become tolerant to host antigens and fall to cause GVHD’. The word ‘fall’ was coded into the ICD-9 code for a fall. Not only was this the wrong context for a fall, but the word ‘fall’ in this abstract is actually a typographic error and should have been ‘fail’.
ICD-9, International Classification of Disease, V.9; GVHD, graft-versus-host disease; PMID, PubMed Identifier.
Table 4.
Clinical associations not found in the Original Medline 1000 dataset
| ICD-9 code | Description | ICD-9 code | Description | Association p value derived from clinical dataset |
Clinically surprising? | Citations found (PMID)† |
|---|---|---|---|---|---|---|
| 1st percentile (highest ranked associations) | ||||||
| 719.46 | Joint pain, lower leg | 848.9 | Sprain and Strain, unspecified site | <5×10−324 | No | 21549978 9343643 |
| 172.1 | Malignant melanoma of skin and eyelid | 190.3 | Malignant neoplasm of conjunctiva | <5×10−324 | No | 21478094 10811089 |
| 250.71 | Diabetes with peripheral circulatory disorders | 337.1 | Peripheral autonomic neuropathy | <5×10−324 | No | 20724598 2779736 |
| 153.0 | Malignant neoplasm of colon | 230.4 | Carcinoma in situ of rectum | <5×10−324 | No | 21125511 6894080 |
| 309.28 | Adjustment disorder with mixed anxiety and depressed mood | 724.2 | Lumbago | <5×10−324 | No | 21665125 18673099 |
| 25th percentile | ||||||
| 535.50 | Gastritis and gastroduodenitis | 787.6 | Fecal incontinence | 4.8 x10 −68 | * | 16712555 |
| 518.0 | Pulmonary collapse | 876.1 | Open wound of back | 9.8×10−68 | No | 17554992 |
| 474.0 | Chronic tonsillitis and adenoiditis | 558.9 | Non-infectious gastroenteritis and colitis | 7.6×10−67 | Yes | 12080166 |
| 377.39 | Optic neuritis | 432.1 | Subdural hemorrhage | 7.8×10−67 | Yes | |
| 307.59 | Eating disorder | 564.01 | Slow transit constipation | 9.9×10−68 | No | 19139750 10925980 |
| 50th percentile | ||||||
| 600 | Hyperplasia of prostate | 747.61 | Gastrointestinal vessel anomaly | 1.89×10−23 | Yes | |
| 296.20 | Major depressive affective disorder | 734 | Pes planus | 3.5×10−23 | Yes | |
| 164.8 | Malignant neoplasm of mediastinum | 427.89 | Cardiac dysrhythmia | 3.7×10−22 | No | 15284266 21387697 |
| 227.0 | Benign neoplasm of adrenal gland | 788.41 | Urinary frequency | 5.3×10−22 | Yes | |
| 250.51 | Type 1 diabetes with ophthalmic manifestations | 959.5 | Finger injury | 1.9×10−21 | * | 18820219 |
| 75th percentile | ||||||
| 217 | Benign neoplasm of breast | 569.85 | Angiodysplasia of intestine with hemorrhage | 4.7×10−06 | Yes | |
| 362.31 | Central retinal artery occlusion | 836.0 | Tear of medial cartilage or meniscus of knee | 4.7×10−06 | Yes | |
| 373.32 | Contact and allergic dermatitis of eyelid | 656.90 | Unspecified fetal and placental problem, affecting management of mother, unspecified as to episode of care or not applicable | 4.7×10−06 | Yes | |
| 110.5 | Dermatophytosis of the body | 246.2 | Thyroid cyst | 4.7×10−06 | Yes | 1607406 |
| 011.90 | Pulmonary tuberculosis | 701.1 | Keratoderma, acquired | 4.8×10−06 | Yes | 9828554 |
| 100th percentile (lowest ranked associations) | ||||||
| 459.9 | Circulatory system disorder | 684 | Impetigo | 0.99 | * | 22642914 |
| 171.9 | Malignant neoplasm of connective and other soft tissue | 333.1 | Essential tremor | 0.99 | Yes | |
| 189.0 | Malignant neoplasm of kidney | 795.5 | Nonspecific reaction to test for tuberculosis | 0.99 | Yes | 20623161 |
| 225.3 | Benign neoplasm of spinal cord | 558.9 | Non-infectious gastroenteritis and colitis | 0.99 | Yes | |
| 378.31 | Hypertropia | 722.10 | Displacement of lumbar intervertebral disc without myelopathy | 0.99 | Yes | |
These were selected from the left-most area in figure 3A. Associations become more clinically surprising as their ranking decreases. Whether an association was clinically surprising was independently determined by two physicians (κ statistic 0.75; 95% CI 0.48 to 1.01). A manual search for citations by each physicians revealed potential associations that were not detected with the automated approach using MetaMap.
*Opinions for which the physicians differed.
†If only one citation is listed, it means that only one reviewer found a citation that supported the association. If two are listed, each reviewer found at least one citation to support the association. If none are listed, neither reviewer was able to find a citation to support the association.
ICD-9, International Classification of Disease, V.9; PMID, PubMed Identifier.
Additional results
The experiments reported in online supplementary appendices A–C revealed several additional insights. First, ICD-9 codes that occur more frequently in the Clinical dataset tend to belong to more associations than those codes that occur rarely (see online supplementary appendix A). Second, for many of the ICD-9 codes, the distribution of association rankings to which each code belongs demonstrated greater spread in the Clinical dataset compared to the Medline dataset (see online supplementary appendix B). Finally, relationships defined within the UMLS relational table can be an additional resource for known associations that are not found within Medline (see online supplementary appendix C). However, the Clinical dataset still contained many associations not included in the UMLS relational table.
Discussion
The results of the experiments reported in this paper show that while combining literature mining and clinical data mining could aid in the discovery of novel associations, the overlap between the Clinical and Medline datasets was surprisingly low. Considering that many of the concurrent mentions of diagnoses in the same abstract were likely due to chance rather than as a result of true associations, we would have expected these false positives to have increased the amount of overlap. We did find a general trend that higher ranked clinical associations were more likely to also be found in Medline citations (figure 4), but this was not as pronounced as we had expected it to be. Even among the highest ranked clinical associations, and using the more inclusive Medline 600 data, only 27% of the clinical associations were also found in the Medline dataset (figure 4C). Some of this difference may be attributable to the fact that our dataset included historical, deprecated codes that are no longer identified or mapped by current systems such as MetaMap. An example from our previous work was code V72.3 (gynecological examination) which was no longer used after 2005, and was replaced by the more granular codes V72.31 and V72.32.4 Our clinical dataset had 87 420 patients with the older code V72.3, and V72.3 was present in 6005 associations. By contrast, there were 55 212 patients with the newer code V72.31 that was present in 5105 associations.
It is possible that many of the associations found in the patient care data are not clinically meaningful, or are related to one another due to confounding factors such as age or gender. And, many widely and historically known associations might not have been discussed in the literature indexed in Medline (the earliest citation in Medline is from 1809). There may also be an inherent bias in Medline because not every clinical association is necessarily an interesting research topic worth publishing, and the research literature does not necessarily discuss clinical conditions proportionally to their prevalence in the population. Other online resources such as Wikipedia might also provide relevant, supplemental clinical coverage not otherwise included in Medline. Further, our study only utilized titles and abstracts of Medline citations, leaving out the majority of details present in the main body of each publication.
We did find a trend that the lower-ranked associations tended to be more clinically surprising, with reasonable inter-rater agreement between the two human reviewers (tables 3 and 4). However, the low inter-rater agreement on whether or not citations were in support of the associations (table 3) demonstrates that identifying supporting scientific evidence is a challenge even among trained clinicians. Further, the fact that the reviewers were able to locate citations in support of 15 out of the 25 associations that were not identified in our Medline data (table 4) suggests that many relationships may be described in the literature in a manner that is not readily interpretable by computational tools such as MetaMap. While consolidating ICD-9 codes of the same disease category resulted in improved coverage, it might come with a price of potentially losing clinical meaning, especially when concepts that were combined should truly remain distinct. In the future, it may be beneficial to group codes according to their clinical relatedness, rather than hierarchically, as has been done for disorders such as stroke49 50 or depression.11
Of course, there may be clinically valid associations that simply have not yet been reported in the literature, as we had discussed in our prior work.11 As an example, table 4 exhibits a clinical association between vitamin D deficiency and non-specific swellings or lumps. The citations we manually reviewed did not seem to directly support this association, but two of the citations did describe a potential relationship between lumps/nodules and malnutrition/malnourishment,51 52 the latter of which could result in a vitamin D deficiency.53
It is also worth comparing our current study to other similar work. Holmes et al, for example, used MedLEE, another medical NLP tool,54 to extract concepts and map them to ICD-9 codes from Medline abstracts, Wikipedia articles, and discharge summaries for several rare diseases.55 The study also used administrative data consisting of ICD-9 billing codes. This approach was able to identify associations found in the clinical data not found in the published literature, and vice versa. The authors also noted that ICD-9 is limited in its coverage of concepts, which likely limited their ability to detect additional associations, just as its use was likely a limiting factor of our study.
The literature-based discovery approach has also been used to identify other clinical relationships. For example, Vos et al20 used the lack of citations in Medline as a source of information to identify novel associations between psychiatric and somatic disorders. Experts reviewed candidates to determine which had clear explanations for their relatedness, or could not be readily explained (thus suggesting truly novel relationships). Another similar study extracted concepts from free text patient records and mapped them to ICD-10 codes in order to discover disease associations.19 The findings were then linked to the Online Mendelian Inheritance in Man (OMIM) resource which describes genetic disorders and their phenotypes.56 An experienced clinician manually reviewed the top candidates to identify ‘interesting’ associations that were not previously known. In our study we only used ICD-9 codes from administrative data, but future work could include codes derived from clinical documents using NLP tools, as some other studies have done.
MeSH concepts from Medline have also been used to aid in the discovery of associations between clinical concepts.57 For example, Avillach et al58 used MeSH concepts to confirm associations with adverse drug events, defined as ‘drug safety signals’. In the study, a threshold of an association being mentioned in three or more citations was used, whereas in our study we included even a single citation mentioning both diagnoses.
Other approaches could also be used to improve the performance of similar data mining methods, some of which could be applied when the original data are collected. For example, if authors were given the option to manually annotate Medline citations with additional coded data (eg, diagnoses, procedures, drugs), the need for complex post-hoc NLP could be diminished. Additionally, development of a curated knowledge base of known associations could be useful in a variety of contexts. Ontologies could also be leveraged to find associations that may not be explicitly defined but may be discoverable through the ontological relationships. There are a multitude of association measures,59 and selecting the right measure to capture a subjective notion of ‘interestingness’ is a research topic of future investigation. Association measures differ in what they are aiming to capture but many of them use the same quantities, for example, marginals, conditionals, and sizes of intersections.
It is possible that the validity of our study findings is contingent on the performance (ie, accuracy and relevance) of MetaMap. Processing free text is complex and MetaMap, similar to many other NLP-based named entity extraction systems, can introduce systematic errors in its classification of clinical concepts.28 60–63 One recent study, for example, reported an F-measure of 61% when MetaMap was applied to a biomedical corpus including Medline abstracts.64 Nevertheless, with the continued support from the NLM, the performance of MetaMap is expected to improve over time,23 and in certain use scenarios it has been demonstrated that MetaMap outperforms human annotators.65 Further, there have been studies showing that MetaMap is able to identify a broader range of concepts than other NLP systems.66
It is also worth pointing out that in our experiments we were not attempting to compare the coding accuracy of MetaMap to other named entity recognition systems, nor were we trying to compare the accuracy of MetaMap to the accuracy of ICD-9 coding in clinical encounters. The goal of converting the concepts in the Medline citations into ICD-9 codes was to have a common coding framework with which to compare both the clinical and Medline datasets. Named entity recognition and ICD-9 coding are very different tasks, and often follow different ‘rules’ for converting text into their respective coded counterparts. The clinically assigned ICD-9 codes themselves represent an abstraction of the diagnoses in clinical text and it may not always be the case that the levels of granularity of codes from a clinical dataset would match those identified from the literature. We attempted to address the potential granularity issue by merging the codes in our ‘Simplified’ datasets. Still, it is important to note that the two datasets were not created with the intention of being compared in this manner, and the base entities (abstracts vs patients) from which the data were extracted are also not directly comparable.
The identification of falls related to graft-versus-host disease (GVHD) due to a typographic error (‘fail’→‘fall’) in an abstract we reviewed (table 3) demonstrates that more work is warranted to accurately identify concepts in the correct context. Yet, this spurious literature support does not rule out the existence of this relationship, which was found in our Clinical dataset. Indeed, patients with GVHD can experience balance impairments67 which could, in turn, result in falls, even if this association is not explicitly mentioned in the citations we analyzed. This general assertion is supported by the results shown in table 4 which demonstrate that in multiple cases experienced clinicians could identify citations supporting the clinical associations while the automated approach was unable to.
In the experiments reported in this paper, we used two distinct score thresholds (1000 and 600) to process the results generated by MetaMap. The use of the MetaMap score of 1000 may have been too restrictive, not allowing the system to detect variations of a concept, and could have excluded many potential concepts that were present in the Medline citations. This is why we also used the threshold of 600, which was lower than scores commonly used in prior studies, to make sure the results achieved were more inclusive. One risk of reducing the threshold to 600 is a higher likelihood of incorrect text classifications yielding more clinically irrelevant associations. Yet, even with that potential loss of accuracy, there were still many clinical associations not identified in the Medline dataset.
In prior literature, studies have either not reported the specific MetaMap scores used24 28 68 69 or have reported seemingly arbitrary thresholds for including concepts in their work. Thresholds of 700,70 800,71–73 850,74 900,75 and 95076 77 have all been used in the past, as well as perfect scores of 1000.78 79 Further, some studies have used the highest ranking score among all candidate concepts for inclusion, with no mention of a minimum threshold.80–82 While it has been stated that the threshold can ‘usually be determined simply by examining MetaMap output for typical text in a given application’,83 there appears to be no consensus about the optimal MetaMap score above which results should be retained. This lack of a consensus may affect the generalizability of research using such tools, including our current study.
Conclusion
This work demonstrates the potential utility but persisting challenges of using large biomedical knowledge repositories for identifying novel relationships derived from clinical datasets. At a broad scale, additional filtering approaches will likely be needed to reduce the size of the set to a reasonable number for expert review, and the addition of other resources beyond Medline could help to distinguish novel associations from well-known ones. Improved NLP and concept extraction capabilities would also be expected to play a significant role in improving the performance of such an approach. Informatics researchers should consider the implications of these promising, but potentially limited, data mining approaches when exploring ‘big data’, and how the findings should be presented and interpreted.
Supplementary Material
Footnotes
Contributors: All authors provided substantial contributions to this work and agree to be held accountable for all parts of the work. Specific contributions include: DAH: conception, design, data acquisition, interpretation, drafting and revising the work, and final approval of the version to be published. MS: data acquisition, interpretation, drafting and revising the work, and final approval of the version to be published. KZ: interpretation, drafting and revising the work, and final approval of the version to be published. QM: statistical consultation, interpretation, drafting and revising the work, and final approval of the version to be published. KS: statistical consultation, interpretation, drafting and revising the work, and final approval of the version to be published. ARA: interpretation, drafting and revising the work, and final approval of the version to be published. NR: statistical consultation, interpretation, drafting and revising the work, and final approval of the version to be published.
Funding: The research reported in this publication was supported in part by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR000433. Additional support was provided by the University of Michigan Cancer Center's (UMCC) Biomedical Informatics Core with partial support from the National Institutes of Health through the UMCC Support Grant (CA46592), and from National Science Foundation grants IIS-0905313, CCF-0937133, IIS-1054199, and CCF-1048168. This work was supported in part by the Intramural Research Program of the NIH, National Library of Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the supporting agencies.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Geetha Ramani R, GraciaJacob S. Data mining in clinical data sets: a review. Int J Appl Info Syst 2012;4:15–26 [Google Scholar]
- 2.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012;13:395–405 [DOI] [PubMed] [Google Scholar]
- 3.Ohno-Machado L. Big science, big data, and a big role for biomedical informatics. J Am Med Inform Assoc 2012;19(e1):e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanauer DA, Ramakrishnan N. Modeling temporal relationships in large scale clinical associations. J Am Med Inform Assoc 2013;20:332–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanauer DA, Rhodes DR, Chinnaiyan AM. Exploring clinical associations using ‘-omics’ based enrichment analyses. PLoS ONE 2009;4:e5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leeper NJ, Bauer-Mehren A, Iyer SV, et al. Practice-based evidence: profiling the safety of cilostazol by text-mining of clinical notes. PLoS ONE 2013;8:e63499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullins IM, Siadaty MS, Lyman J, et al. Data mining and clinical data repositories: insights from a 667,000 patient data set. Comput Biol Med 2006;36:1351–77 [DOI] [PubMed] [Google Scholar]
- 8.Wright A, Chen ES, Maloney FL. An automated technique for identifying associations between medications, laboratory results and problems. J Biomed Inform 2010;43:891–901 [DOI] [PubMed] [Google Scholar]
- 9.Pathak J, Kiefer RC, Chute CG. Using linked data for mining drug-drug interactions in electronic health records. Stud Health Technol Inform 2013;192:682–6 [PMC free article] [PubMed] [Google Scholar]
- 10.Tatonetti NP, Fernald GH, Altman RB. A novel signal detection algorithm for identifying hidden drug-drug interactions in adverse event reports. J Am Med Inform Assoc 2012;19:79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanauer DA, Ramakrishnan N, Seyfried LS. Describing the relationship between cat bites and human depression using data from an electronic health record. PLoS ONE 2013;8:e70585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 2011;90:133–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fact Sheet Medline. 2013 [November 4, 2013]. http://www.nlm.nih.gov/pubs/factsheets/medline.html
- 14.Gabetta M, Larizza C, Bellazzi R. A Unified Medical Language System (UMLS) based system for literature-based discovery in medicine. Stud Health Technol Inform 2013;192:412–16 [PubMed] [Google Scholar]
- 15.Hristovski D, Stare J, Peterlin B, et al. Supporting discovery in medicine by association rule mining in Medline and UMLS. Stud Health Technol Inform 2001;84(Pt 2):1344–8 [PubMed] [Google Scholar]
- 16.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet 2006;7:119–29 [DOI] [PubMed] [Google Scholar]
- 17.Weeber M, Klein H, de Jong-van den Berg LTW, et al. Using concepts in literature-based discovery: Simulating Swanson's Raynaud-fish oil and migraine-magnesium discoveries. J Am Soc Inf Sci Technol 2001;52:548–57 [Google Scholar]
- 18.Weeber M, Kors JA, Mons B. Online tools to support literature-based discovery in the life sciences. Brief Bioinform 2005;6:277–86 [DOI] [PubMed] [Google Scholar]
- 19.Roque FS, Jensen PB, Schmock H, et al. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput Biol 2011;7:e1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vos R, Aarts S, van Mulligen E, et al. Finding potentially new multimorbidity patterns of psychiatric and somatic diseases: exploring the use of literature-based discovery in primary care research. J Am Med Inform Assoc 2014;21:139–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fechete R, Heinzel A, Perco P, et al. Mapping of molecular pathways, biomarkers and drug targets for diabetic nephropathy. Proteomics Clin Appl 2011;5:354–66 [DOI] [PubMed] [Google Scholar]
- 22.Rebholz-Schuhmann D, Grabmuller C, Kavaliauskas S, et al. A case study: semantic integration of gene-disease associations for type 2 diabetes mellitus from literature and biomedical data resources. Drug Discov Today. [epub ahead of print 4 Nov 2013] [DOI] [PubMed] [Google Scholar]
- 23.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010;17:229–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fung KW, Jao CS, Demner-Fushman D. Extracting drug indication information from structured product labels using natural language processing. J Am Med Inform Assoc 2013;20:482–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis K, Staes C, Duncan J, et al. Identification of pneumonia and influenza deaths using the Death Certificate Pipeline. BMC Med Inform Decis Mak 2012;12:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman WW, Fiszman M, Dowling JN, et al. Identifying respiratory findings in emergency department reports for biosurveillance using MetaMap. Stud Health Technol Inform 2004;107(Pt 1):487–91 [PubMed] [Google Scholar]
- 27.Meystre S, Haug PJ. Natural language processing to extract medical problems from electronic clinical documents: performance evaluation. J Biomed Inform 2006;39:589–99 [DOI] [PubMed] [Google Scholar]
- 28.St-Maurice J, Kuo MH, Gooch P. A proof of concept for assessing emergency room use with primary care data and natural language processing. Methods Inf Med 2013;52:33–42 [DOI] [PubMed] [Google Scholar]
- 29.Sharma V, Sarkar IN. Leveraging concept-based approaches to identify potential phyto-therapies. J Biomed Inform 2013;46:602–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aronson AR, Mork JG, Gay CW, et al. The NLM indexing initiative's medical text indexer. Stud Health Technol Inform 2004;107(Pt 1):268–72 [PubMed] [Google Scholar]
- 31.Jimeno-Yepes A, Wilkowski B, Mork JG, et al. A bottom-up approach to MEDLINE indexing recommendations. AMIA Annu Symp Proc 2011;2011:1583–92 [PMC free article] [PubMed] [Google Scholar]
- 32.Jimeno-Yepes AJ, Plaza L, Mork JG, et al. MeSH indexing based on automatically generated summaries. BMC Bioinformatics 2013;14:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aronson AR, Rindflesch TC. Query expansion using the UMLS Metathesaurus. Proc AMIA Annu Fall Symp 1997:485–9 [PMC free article] [PubMed] [Google Scholar]
- 34.Aronson AR, Bodenreider O, Demner-Fushman D, et al., eds From indexing the biomedical literature to coding clinical text. Workshop BioNL'07; Prague, Czech Republic, 2007 [Google Scholar]
- 35.Kavuluru R, Han S, Harris D. Unsupervised extraction of diagnosis codes from EMRs using knowledge-based and extractive text summarization techniques. In: Proceedings of the 26th Canadian Conference on Artificial Intelligence, Canadian AI 2013:77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suominen H, Ginter F, Pyysalo S, et al. Machine learning to automate the assignment of diagnosis codes to free-text radiology reports: a method description. In: Proceedings of the ICML/UAI/COLT 2008 Workshop on Machine Learning for Health-Care Applications; Helsinki, Finland, 2008 [Google Scholar]
- 37. MetaMapped Results Information. [November 4, 2013]. http://skr.nlm.nih.gov/resource/MetaMappedBaselineInfo.shtml.
- 38.Lang FM. MetaMap 2012 Machine Output Explained. 2012 [November 4, 2013]. http://metamap.nlm.nih.gov/2012_MMO.pdf.
- 39. Machine Output (2008) Explained. 2008 [November 4, 2013]. http://skr.nlm.nih.gov/Help/MMO_08_Info.html.
- 40.Aronson AR. MetaMap evaluation. Bethesda, MD: National Library of Medicine, 2001. [February 15, 2014]. http://skr.nlm.nih.gov/papers/references/mm.evaluation.pdf [Google Scholar]
- 41.Zhang S, Hunter DJ, Hankinson SE, et al. A prospective study of folate intake and the risk of breast cancer. JAMA 1999;281:1632–7 [DOI] [PubMed] [Google Scholar]
- 42.Lorence DP, Ibrahim IA. Disparity in coding concordance: do physicians and coders agree? J Health Care Finance 2003;29:43–53 [PubMed] [Google Scholar]
- 43.Lorence DP, Ibrahim IA. Benchmarking variation in coding accuracy across the United States. J Health Care Finance 2003;29:29–42 [PubMed] [Google Scholar]
- 44.O'Malley KJ, Cook KF, Price MD, et al. Measuring diagnoses: ICD code accuracy. Health Serv Res 2005;40(5 Pt 2):1620–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surjan G. Questions on validity of International Classification of Diseases-coded diagnoses. Int J Med Inform 1999;54:77–95 [DOI] [PubMed] [Google Scholar]
- 46.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 2011;12:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cairelli MJ, Miller CM, Fiszman M, et al. Semantic MEDLINE for discovery browsing: using semantic predications and the literature-based discovery paradigm to elucidate a mechanism for the obesity paradox. AMIA Annu Symp Proc 2013;2013: 164–73 [PMC free article] [PubMed] [Google Scholar]
- 48.Kilicoglu H, Fiszman M, Rodriguez A, et al., eds. Semantic {MEDLINE}: {A} web application to manage the results of {PubMed} searches. Proceedings of the Third International Symposium on Semantic Mining in Biomedicine (SMBM 2008), Turku, Finland: Turku Centre for Computer Science (TUCS), 2008 [Google Scholar]
- 49.Roumie CL, Mitchel E, Gideon PS, et al. Validation of ICD-9 codes with a high positive predictive value for incident strokes resulting in hospitalization using Medicaid health data. Pharmacoepidemiol Drug Saf 2008;17:20–6 [DOI] [PubMed] [Google Scholar]
- 50.Spolaore P, Brocco S, Fedeli U, et al. Measuring accuracy of discharge diagnoses for a region-wide surveillance of hospitalized strokes. Stroke 2005;36:1031–4 [DOI] [PubMed] [Google Scholar]
- 51.Haraoka G, Muraoka M, Yoshioka N, et al. First case of surgical treatment of Farber's disease. Ann Plast Surg 1997;39:405–10 [DOI] [PubMed] [Google Scholar]
- 52.Olczak-Kowalczyk D, Krasuska-Slawinska E, Rokicki D, et al. Case report: Infantile systemic hyalinosis: a dental perspective. Eur Arch Paediatr Dent 2011;12:224–6 [DOI] [PubMed] [Google Scholar]
- 53.Fraser DR. Vitamin D-deficiency in Asia. J Steroid Biochem Mol Biol 2004;89–90:491–5 [DOI] [PubMed] [Google Scholar]
- 54.Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp 2000:270–4 [PMC free article] [PubMed] [Google Scholar]
- 55.Holmes AB, Hawson A, Liu F, et al. Discovering disease associations by integrating electronic clinical data and medical literature. PLoS ONE 2011;6:e21132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamosh A, Scott AF, Amberger JS, et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005;33(Database issue):D514–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srinivasan P, Rindflesch T. Exploring text mining from MEDLINE. Proc AMIA Symp 2002:722–6 [PMC free article] [PubMed] [Google Scholar]
- 58.Avillach P, Dufour JC, Diallo G, et al. Design and validation of an automated method to detect known adverse drug reactions in MEDLINE: a contribution from the EU-ADR project. J Am Med Inform Assoc 2013;20:446–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan P-N, Kumar V, Srivastava J. Selecting the right interestingness measure for association patterns. Proceedings of the eighth ACM SIGKDD international conference on Knowledge discovery and data mining (KDD ‘02); Edmonton, Canada, 2002:32–41 [Google Scholar]
- 60.Ogren PV, Savova GK, Chute CG. Constructing evaluation corpora for automated clinical named entity recognition. Proceedings of the Sixth International Conference on Language Resources and Evaluation (LREC'08); May 28-29-30, 2008; Marrakech, Morocco, 2008:3143–50 [Google Scholar]
- 61.Pratt W, Yetisgen-Yildiz M. A study of biomedical concept identification: MetaMap vs. people. AMIA Annu Symp Proc 2003:529–33 [PMC free article] [PubMed] [Google Scholar]
- 62.Stanfill MH, Williams M, Fenton SH, et al. A systematic literature review of automated clinical coding and classification systems. J Am Med Inform Assoc 2010;17:646–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trieschnigg D, Pezik P, Lee V, et al. MeSH Up: effective MeSH text classification for improved document retrieval. Bioinformatics 2009;25:1412–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang N, Singh B, Afzal Z, et al. Using rule-based natural language processing to improve disease normalization in biomedical text. J Am Med Inform Assoc 2013;20:876–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruau D, Mbagwu M, Dudley JT, et al. Comparison of automated and human assignment of MeSH terms on publicly-available molecular datasets. J Biomed Inform 2011;44(Suppl 1):S39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Milian K, Bucur A, van Harmelen F, et al. Identifying most relevant concepts to describe clinical trial eligibility criteria. 6th International Conference on Health Informatics (HEALTHINF 2013); February 11–14, 2013; Barcelona, Spain [Google Scholar]
- 67.Grauer O, Wolff D, Bertz H, et al. Neurological manifestations of chronic graft-versus-host disease after allogeneic haematopoietic stem cell transplantation: report from the consensus conference on clinical practice in chronic graft-versus-host disease. Brain 2010;133:2852–65 [DOI] [PubMed] [Google Scholar]
- 68.Neves M, Damaschun A, Mah N, et al. Preliminary evaluation of the CellFinder literature curation pipeline for gene expression in kidney cells and anatomical parts. Database (Oxford) 2013;2013:bat020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tran N, Luong T, Krauthammer M. Mapping terms to UMLS concepts of the same semantic type. AMIA Annu Symp Proc 2007:1136. [PubMed] [Google Scholar]
- 70.Mathur S, Dinakarpandian D. Automated ontological gene annotation for computing disease similarity. AMIA Summits Transl Sci Proc 2010;2010:12–16 [PMC free article] [PubMed] [Google Scholar]
- 71.Bedrick S, Edinger T, Cohen A, et al., eds Identifying Patients for Clinical Studies from Electronic Health Records: TREC 2012 Medical Records Track at OHSU. The Twenty-First Text REtrieval Conference (TREC 2012) Proceedings; 2012 November 6–9; Gaithersburg, Maryland, 2012 [Google Scholar]
- 72.Patterson O, Igo S, Hurdle JF. Automatic acquisition of sublanguage semantic schema: towards the word sense disambiguation of clinical narratives. AMIA Annu Symp Proc 2010;2010:612–16 [PMC free article] [PubMed] [Google Scholar]
- 73.Roberts K, Harabagiu SM. A flexible framework for deriving assertions from electronic medical records. J Am Med Inform Assoc 2011;18:568–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.French L, Lane S, Law T, et al. Application and evaluation of automated semantic annotation of gene expression experiments. Bioinformatics 2009;25:1543–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Melton GB, Moon S, McInnes B, et al. Automated identification of synonyms in biomedical acronym sense inventories. Proceedings of the NAACL HLT 2010 Second Louhi Workshop on Text and Data Mining of Health Documents; Los Angeles, California: 1867742: Association for Computational Linguistics, 2010: 46–52 [Google Scholar]
- 76.Gurulingappa H, Müller B, Hofmann-Apitius M, et al., eds Information Retrieval Framework for Technology Survey in Biomedical and Chemistry Literature. The Twentieth Text REtrieval Conference (TREC 2011) Proceedings; 2011 November 15–18; Gaithersburg, Maryland, 2011 [Google Scholar]
- 77.Patel CO, Garg V, Khan SA. What do patients search for when seeking clinical trial information online? AMIA Annu Symp Proc 2010;2010:597–601 [PMC free article] [PubMed] [Google Scholar]
- 78.Hanauer DA, Liu Y, Mei Q, et al. Hedging their mets: the use of uncertainty terms in clinical documents and its potential implications when sharing the documents with patients. AMIA Annu Symp Proc 2012;2012:321–30 [PMC free article] [PubMed] [Google Scholar]
- 79.Yip V, Mete M, Topaloglu U, et al. Concept discovery for pathology reports using an N-gram model. AMIA Summits Transl Sci Proc 2010;2010:43–7 [PMC free article] [PubMed] [Google Scholar]
- 80.Bejan CA, Vanderwende L, Xia F, et al. Assertion modeling and its role in clinical phenotype identification. J Biomed Inform 2013;46:68–74 [DOI] [PubMed] [Google Scholar]
- 81.Friedlin J, Overhage M. An evaluation of the UMLS in representing corpus derived clinical concepts. AMIA Annu Symp Proc 2011;2011:435–44 [PMC free article] [PubMed] [Google Scholar]
- 82.Herskovic JR, Tanaka LY, Hersh W, et al. A day in the life of PubMed: analysis of a typical day's query log. J Am Med Inform Assoc 2007;14:212–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aronson AR. MetaMap: Mapping Text to the UMLS Metathesaurus. 2006. [November 4, 2013]. http://skr.nlm.nih.gov/papers/references/metamap06.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.