Abstract
Objective
To reliably extract two entity types, symptoms and conditions (SCs), and drugs and treatments (DTs), from patient-authored text (PAT) by learning lexico-syntactic patterns from data annotated with seed dictionaries.
Background and significance
Despite the increasing quantity of PAT (eg, online discussion threads), tools for identifying medical entities in PAT are limited. When applied to PAT, existing tools either fail to identify specific entity types or perform poorly. Identification of SC and DT terms in PAT would enable exploration of efficacy and side effects for not only pharmaceutical drugs, but also for home remedies and components of daily care.
Materials and methods
We use SC and DT term dictionaries compiled from online sources to label several discussion forums from MedHelp (http://www.medhelp.org). We then iteratively induce lexico-syntactic patterns corresponding strongly to each entity type to extract new SC and DT terms.
Results
Our system is able to extract symptom descriptions and treatments absent from our original dictionaries, such as ‘LADA’, ‘stabbing pain’, and ‘cinnamon pills’. Our system extracts DT terms with 58–70% F1 score and SC terms with 66–76% F1 score on two forums from MedHelp. We show improvements over MetaMap, OBA, a conditional random field-based classifier, and a previous pattern learning approach.
Conclusions
Our entity extractor based on lexico-syntactic patterns is a successful and preferable technique for identifying specific entity types in PAT. To the best of our knowledge, this is the first paper to extract SC and DT entities from PAT. We exhibit learning of informal terms often used in PAT but missing from typical dictionaries.
Keywords: text mining, online health forums, medical entites extraction, natural language processing
Introduction
In 2013, 59% of adults in the USA sought health information on the internet.1 While these users typically have no formal medical education, they generate large volumes of patient-authored text (PAT) in the form of medical blogs and discussions on online health forums. Their contributions range from rare disease diagnosis to drug and treatment efficacy.
Our eventual goal is to enable open-ended mining and analysis of PAT to improve health outcomes. In particular, PAT can be a great resource for extracting the efficacy and side effects of both pharmaceutical and alternative treatments. Prior work has demonstrated the knowledge value of PAT in mining adverse drug events,2 predicting flu trends3 (although caution is needed4), exploring drug interactions,5 and replicating results of a double-blind medical trial,6 and already websites such as http://www.medify.com and http://www.treato.com aggregate information from PAT on the efficacy of and side effects from drugs. Extraction of sentiment and side effects for drugs and treatments in PAT is only possible on a large scale when we have tools to discover and robustly identify entities such as symptoms, conditions, drugs, and treatments in the text. Most research on extracting such information has focused on clinicians’ notes, and thus most annotation systems are tailored towards them. Unlike expert-authored text, which is composed of terms routinely used by the medical community, PAT contains a great deal of slang and verbose, informal descriptions of symptoms and treatments (eg, ‘feels like a brick on my heart’ or ‘Watson 357’ for Vicodin). Previous research has shown that most terms used by consumers are not in ontologies.7
In this work, we propose inducing lexico-syntactic patterns using seed dictionaries to identify specific medical entity types in PAT. The patterns generalize terms from seed dictionaries to learn new entities. We test our method over two entity types: symptoms and conditions (SCs), and drugs and treatments (DTs) on two of MedHelp's forums: Asthma and Ear, Nose & Throat (ENT). We also report the results of applying our system to three other forums on MedHelp: Adult Type II Diabetes, Acne, and Breast Cancer. Our system is able to extract SC and DT phrases that are not in the seed dictionaries, such as ‘cinnamon pills’ and ‘Opuntia’ as DTs from the Diabetes forum, and ‘achiness’ and ‘lumpy’ as SCs from the Breast Cancer forum.
Objective
Our objective is to learn new SC and DT phrases from PAT without using hand-written rules or any hand-labeled sentences. We define SC as any symptom or condition mentioned in text. The DT label refers to any treatment or intervention performed to improve a symptom or condition. It includes pharmaceutical treatments and drugs, surgeries, interventions (such as ‘getting rid of cat and carpet’ for asthma patients), and alternative treatments (such as acupuncture or garlic). Note that our system ignores negations (eg, in the sentence ‘I don't have asthma’, ‘asthma’ is labeled SC) since it is preferable to extract all SC and DT mentions and handle the negations separately, if required. The labels include all relevant generic terms (eg, ‘meds’, ‘disease’). Devices used to improve a symptom or condition (such as inhalers) are included in DT, but devices that are used for monitoring or diagnosis are not. Some examples of sentences from the Asthma and ENT forums labeled with SC (in italics) and DT (in bold) labels are shown below (more in online supplemental section):
I don't agree with my doctor's diagnostic after research and I think I may have a case of Sinus Mycetoma.
I started using an herbal mixture especially meant for Candida with limited success.
however, with the consistent green and occasional blood in nasal discharge (but with minimal “stuffy” feeling), I wonder if perhaps a problem with chronic sinusitis and or eustachian tubes.
She gave me albuteral and symbicort (plus some hayfever meds and asked me to use the peak flow meter.
My sinus infections were treated electrically, with high voltage million volt electricity, which solved the problem, but the treatment is not FDA approved and generally unavailable, except under experimental treatment protocols.
Background
Medical term annotation is a longstanding research challenge. However, almost no prior work has focused on automatically annotating PAT. Tools such as TerMINE8 and ADEPT9 do not identify specific entity types. Other existing tools such as MetaMap,10 the open biomedical annotator (OBA),11 and Apache cTakes12 perform poorly mainly because they are designed for fine-grained entity extraction on expert-authored text. They essentially perform dictionary matching on text based on source ontologies.10 11 13 Despite being the go-to tools for medical text annotation, previous studies14 comparing OBA and MetaMap with human annotator performance underscore two sources of performance error, which we also notice in our results. The first is ontology incompleteness, which results in low recall, and the second is inclusion of contextually irrelevant terms.9 For example, when restricted to the RxNORM ontology and semantic-type Antibiotic (T195), OBA will extract both ‘Today’ and ‘Penicillin’ from the sentence ‘Today I filled my Penicillin rx’. Other approaches focusing on expert-authored text show improvement in identifying food and drug allergies15 and disease normalization16 with the use of statistical methods. While these statistically-based approaches tend to perform well, they require hand-labeled data, which are both labor intensive to collect and do not generalize across PAT sources.
The most relevant work to ours is in building the consumer health vocabularies (CHVs). CHVs are ontologies designed to bridge the gap between patient language and the Unified Medical Language System (UMLS) Metathesaurus. We are aware of two CHVs: the open access collaborative (OAC) CHV17 and the MedlinePlus CHV.18 To date, most work in this area has focused on identifying candidate terms of general medical relevance, and not specific entity types, for the OAC CHV.19 We use the OAC CHV to construct our seed dictionaries.
In this paper, we extract SC and DT terms by inducing lexico-syntactic word patterns. The general approach has been shown to be useful in learning different semantic lexicons.20–22 The technique involves first identifying a handful of examples of interest (eg, {countries, Cuba} for finding hyponyms), and then extracting the lexico-syntactic patterns of words typically surrounding these terms in a large corpus of text (eg, ‘X such as Y’). These patterns are then used to identify new examples, and the cycle repeats until no new examples or patterns are discovered.
Materials and methods
Dataset
We used discussion forum text from MedHelp,23 one of the biggest online health community websites. A MedHelp forum consists of thousands of threads; each thread is a sequence of posts by users. The dataset includes some medical research material posted by users but has no clinical text. We excluded from our dataset sentences from one user who had posted very similar posts several thousand times. We tested the performance of our system in extracting DT and SC phrases on sentences from two forums: the Asthma forum and the ENT forum. The Asthma and ENT forums consisted of 39 137 and 215 123 sentences, respectively, in our dataset. In addition, we present qualitative results of our system run on three other forums: the Adult Type II Diabetes forum (63 355 sentences), the Acne forum (65 595 sentences), and the Breast Cancer forum (296 861 sentences). We used the Stanford CoreNLP toolkit24 to tokenize text, split it into sentences, and label the tokens with their part-of-speech (POS) tags and lemma (ie, canonical form). We converted all text into lowercase because PAT usually contains inconsistent capitalization.
Initial labeling using dictionaries
As the first step, we ‘partially’ labeled data using matching phrases from our DT and SC dictionaries. Our DT dictionary, comprising 38 684 phrases, was sourced from: Wikipedia's list of drugs, surgeries, and delivery devices; RxList25; MedlinePlus26; Medicinenet27; phrases with semantic-type ‘procedures’ from MedDRA28; and phrases with relevant semantic types from the NCI Thesaurus.29
Our SC dictionary comprises 100 879 phrases, and was constructed using phrases from MedlinePlus,26 Medicinenet,27 and MedDRA28 (with semantic-type ‘disorders’). We expanded both dictionaries using the OAC CHV17 by adding all synonyms of the phrases previously added. Because the dictionaries are automatically constructed with no manual editing, they might have some incorrect phrases. However, the results show that they perform effectively.
We labeled a phrase with the dictionary label when the sequence of non-stop words (or their lemmas) matched an entry in the dictionary. To match spelling mistakes and morphological variations (such as ‘tickly’), which are common in PAT, we performed fuzzy matching. A token matches a word in the dictionary if the token is longer than six characters and the token and the word are one edit distance away. We ignored the words ‘disease’, ‘disorder’, ‘chronic’, and ‘pre-existing’ in the dictionaries when matching phrases. We removed phrases that are very common on the internet by compiling a list of the 2000 most common words from Google N-grams30 (called GoogleCommonList henceforth). This helps exclude words such as ‘today’ and ‘AS’, which are also names of medicines. Tokens labeled as SC by the SC dictionary were not labeled DT, to avoid labeling ‘asthma’ as DT in the phrase ‘asthma meds’, in case ‘asthma meds’ was in the DT dictionary.
Inducing lexico-syntactic patterns
Our system to learn new SC and DT terms using lexico-syntactic patterns (similar to Thelen and Riloff21) can be summarized as:
Label data using dictionaries
Create patterns using the labeled data and choose the top K patterns
Extract phrases using the learned patterns and choose the top N words
Add new phrases to the dictionaries
Repeat steps 1–4 T times or until converged
We experimented with different phrase and pattern weighting schemes (eg, applying log sublinear scaling in the weighting formulations below) and parameters for our system. We selected the ones that performed best on the Asthma forum test sentences.
Below, we explain the algorithm using DT as an example label for ease of explanation.
Creating patterns
We create potential patterns by looking at two to four words before and after the labeled tokens. We discard contexts that consist of only two or fewer stop words. Words that are labeled with one of the dictionaries (seed as well as learned) are generalized with the class of the dictionary. For example, consider the labeled sentence, ‘I take Advair::DT and Albuterol::DT for asthma::SC’, where ‘::’ indicates the label of the word. The patterns created around the DT word ‘Albuterol’ will be ‘DT and X’, ‘DT and X for SC’, ‘X for SC’, and so on. We create flexible patterns by ignoring the words {‘a’, ‘an’, ‘the’, ‘,’, ‘.’} while matching the patterns and by allowing at most two stop words between the context and the term to be extracted. We create two sets of the above patterns—with and without the POS restriction of the target phrase (eg, that it only contains nouns). Since many symptoms and drugs tend to be more than just one word, we allow matching of one to two tokens. In our experiments, matching three or more consecutive terms extracted noisy phrases, mostly by patterns without the POS restriction. Figure 1 shows an example of two patterns and how they match to two sentences.
Figure 1.

Examples of how patterns match to sentences. X means one token that will be matched, tag means the part of speech tag restriction, {1,2} means one to two words are allowed, FW* means two or fewer words from {a, an, the}, SW* means two or fewer stop words, .* means zero or more characters can match, and lemma means the lemma of the token. Target phrase match is shown in bold, and context match is shown in italics.
Learning patterns
We learn new patterns by weighting them using normalization measures and selecting the top patterns. In essence, we want to trade off precision and recall of the patterns to extract the correct phrases. The weighting scheme for a pattern i is
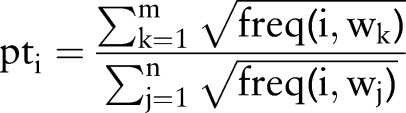 |
where m is the number of words with the label DT that match the pattern, n is the number of all words that match the pattern, and freq (i, wk) is the number of times pattern i matched the phrase wk. Sublinear scaling of the frequency prevents high-frequency words from overshadowing the contribution of low-frequency words. We discard patterns that have weight less than a threshold (=0.5 in our experiments). We also discard patterns when m is equal to n since adding them would be of no benefit for learning new phrases. We remove patterns that occur in the top 500 patterns for the other label. After calculating weights for all the remaining patterns, we choose the top K (=50 in our experiments) patterns.
Learning phrases
We apply the patterns selected by the above process to all the sentences and extract the matched phrases. The phrase-weighting scheme is a combination of term frequency-inverse document frequency (TF-IDF) scoring, weight of the patterns, and relative frequency of the phrases in different dictionaries. The latter weighting term assigns higher weight to words that are sub-phrases of phrases in the entity's dictionary. The weighting function for a phrase p for the label DT is
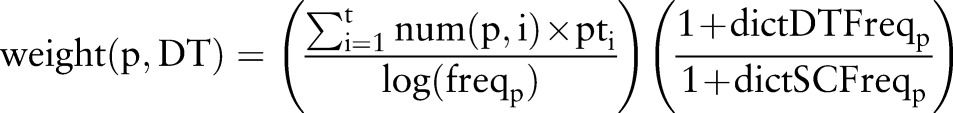 |
where t is the number of patterns that extract the phrase p, num (p,i) is the number of times phrase p is extracted using pattern i, pti is the weight of the pattern i from the previous equation, freqp is frequency of phrase p in the corpus, and dictDTFreqp and dictSCFreqp are the frequency of phrase p in the n-grams of the phrases from the DT dictionary and the SC dictionary, respectively. We discard phrases with weight less than a threshold (=0.2 in our experiments). We also discard phrases that are matched by less than two patterns to improve precision of the system—phrases extracted by multiple patterns tend to be more accurate.
We remove the following kinds of phrases from the set of potential phrases: (1) list of specialists and physicians downloaded from WebMD31; (2) words in the GoogleCommonList; (3) 5000 most frequent tokens from around 1 million tweets from Twitter to avoid learning slang words such as ‘asap’; (4) phrases that are already in any of the dictionaries. We then extract up to the top N (=10 in our experiments) words and label those phrases in the sentences. We also remove body part phrases (198 phrases that were curated from Wikipedia and manually expanded by us) from the set of potential DT phrases.
We repeat the cycle of learning patterns and learning phrases T times (=20 in our experiments) or until no more patterns and words can be extracted.
Evaluation
Test data
We tested our system and the baselines on two forums—Asthma and ENT. For each forum, we randomly sampled 500 sentences, and two authors annotated 250 sentences each. The test sentences were removed from the data used in the learning system. The labeling guidelines for the annotators for the test sentences were to include the minimum number of words to convey the medical information. To calculate the inter-annotator agreement, the annotators labeled 50 sentences from the 250 sentences assigned to the other annotator; the agreement is thus calculated on 100 sentences out of the 500 sentences. The token-level agreement was 96% with Cohen's κ=0.781 for the Asthma test sentences and 96.2% with Cohen's κ=0.801 for the ENT test sentences. We used the Asthma forum as a development forum to select parameters, such as the maximum number of patterns and phrases added in an iteration, total number of iterations, and the thresholds for learning patterns and phrases. We discuss the effect of varying these parameters on our system's performance in the online supplemental section. We used ENT as a test forum; no parameters were tuned on the ENT forum test set.
Metrics
We used token-level precision, recall, and F1 metrics to evaluate our system and the baselines. We chose token-level measures over entity-level measures because labeling entities partially is still useful in this domain. We discuss the difference between the two types of metrics and present results using the entity-level measures in the online supplemental section. The entity-level evaluation results show similar trends to the token-level results. Note that accuracy is not a good measure of the task because most of the tokens are labeled ‘none’, and thus labeling everything as ‘none’ achieves very high accuracy and zero recall. Precision (or positive predictive value) is the percentage of correct tokens among all the tokens that are extracted. Recall (or sensitivity) is the percentage of correct tokens in the test set that are extracted by a system. F1 score is the harmonic mean of precision and recall. We ignore about 200 very common words (such as ‘i’, ‘am’), 26 very common medical terms and their derivatives (such as ‘disease’, ‘doctor’), and words that do not start with a letter (see online supplemental section for the full list) when evaluating the systems.
Baselines
We compared our system with the OBA annotator11 and the MetaMap annotator.10 We evaluated both the baselines with the default settings. We also compared our algorithm with the pattern learning system proposed by Xu et al.22 We describe the details of these systems below.
MetaMap
We used the Java API of MetaMap 2013v2. We used the semantic types {Antibiotic, Clinical Drug, Drug Delivery Device, Steroid, Therapeutic or Preventive Procedure, Vitamin, Pharmacologic Substance} for DT and {Disease or Syndrome, Sign or Symptom, Congenital Abnormality, Experimental Model of Disease, Injury or Poisoning, Mental or Behavioral Dysfunction, Finding} for SC.
OBA
We used the web service provided by OBA to label the sentences. We used the semantic types {Pharmacologic Substance, Steroid, Vitamin, Antibiotic, Therapeutic or Preventive Procedure, Medical Device, Substance, Clinical Drug, Drug Delivery Device, Biomedical or Dental Material} for DT and the semantic types {Sign or Symptom, Injury or Poisoning, Disease or Syndrome, Mental or Behavioral Dysfunction, Rickettsia or Chlamydia} for SC.
Xu et al22 learned surface patterns for extracting diseases from Medline paper abstracts. They ranked patterns based on overlap of words extracted by potential patterns with a seed pattern. Potential words were ranked by the scores of the patterns that extracted them. We compared our system with their best performing ranking measures: BalancedRank for patterns and Best-pattern-based rank for words. Since they focus only on extracting diseases from research paper abstracts, their seed pattern ‘patients with X’ will not perform well on our dataset. Thus, for each label, we created patterns according to their algorithm and chose the pattern weighted highest by our system as their seed pattern. The seed patterns were the same as the top patterns shown in table 3.
Table 3.
Top 10 patterns and top 15 phrases extracted for the Asthma and the ENT forums
| DT | SC |
|---|---|
| Asthma | |
| i be put on (X|pos:noun) i have be on (X|pos:noun) use DT and (X|pos:noun) put he on (X|pos:noun) prescribe DT and (X|pos:noun) mg of X he put I on X to give he (X|pos:noun) i have be use X and put I on X |
(X|pos:noun) SC etc. reduce SC (X|pos:noun) first SC (X|pos:noun) have history of (X|pos:noun) develop SC (X|pos:noun) really bad SC (X|pos:noun) not cause SC (X|pos:noun) symptom be (X|pos:noun) (X|pos:noun) SC feel and be diagnose with X |
| inhaler, inhalers, steroid inhaler, albuterol inhaler, b5, preventive inhaler, ventolin inhaler, advar, seritide, steroid inhalers, symbicort trubohaler, agumentin, pantoloc, inahler, puffs | flare, flare-up, rad, congestion, mucus, tightness, sinuses, exces mucus, cataracts-along, athsma, vcd, sensation, mites, nasal, ashtma |
| ENT | |
| have endoscopic (X|pos:noun) include DT (X|pos:noun) and put I on (X|pos:noun) (X|pos:noun) 500 mg 2 round of (X|pos:noun) and be put on (X|pos:noun) have put I on (X|pos:noun) (X|pos:adj) DT and use ent put I on X (X|pos:noun) and nasal rinse |
persistent SC (X|pos:noun) have have problem with (X|pos:noun) diagnose I with SC (X|pos:noun) morning with SC (X|pos:noun) (X|pos:noun) SC cause SC have be treat for (X|pos:noun) year SC (X|pos:noun) (X|pos:noun) SC even though (X|pos:noun) SC like SC daughter have SC X |
| otic, z-pack, z-pac, predneson, tylenol sinus, amoxillin, saline nasal, eardrops, regimen, inhaler, peroxide, rinse, amoxcilyn, rinses, anti-nausea, saline, mucodyn, flixonase, vertin, amocicillan | dysfunction, sinus, sinuses, lymph, gland, tonsilitus, sinues, sensation, congestion, pharynx, tightness, mucus, tonsil, onset, ethmoid sinus |
To improve readability we have shown only the sequences of lemmas from the patterns. X indicates the target phrase and ‘pos:’ indicates the part-of-speech restriction.
DT, drugs and treatments; ENT, Ear, Nose & Throat; SC, symptoms and conditions.
Conditional random field (CRF) classifier
A CRF is a Markov random field-based classifier that uses word features and context features such as the words and labels of nearby words. Even though the data are only partially labeled using dictionaries, CRFs can learn correct labels using the context features. We experimented with many different features and settings and report the best results. We removed sentences in which none of the words were labeled and fixed the label of words that are labeled by dictionaries. We used distributional similarity features, which were computed using the Brown clustering method,32 33 on all sentences of the MedHelp forums. We built the classifier using the Stanford NER toolkit.34 We also present results of CRFs with self-training (‘CRF-2’ and ‘CRF-20’ for 2 and 20 iterations, respectively), in which a CRF is trained on the sentences labeled by dictionaries and predictions using the trained CRF from the previous iteration.
Results
Table 1 shows F1 scores for our system across different dictionary labeling schemes. ‘Dictionary’ refers to the seed dictionary without fuzzy matching or removing common words. Fuzzy matching (indicated by ‘-F’) and removing common words (indicated by ‘-C’) increase the F1 scores by 3–5%. Table 2 shows precision, recall, and F1 scores of our system and the baselines. The suffix ‘-C’ indicates post-processing the output of MetaMap and OBA by removing common words.
Table 1.
F1 scores for labeling with dictionaries using different types of labeling schemes
| F1 Asthma-DT | F1 Asthma-SC | F1 ENT-DT | F1 ENT-SC | |
|---|---|---|---|---|
| Dictionary | 58.21 | 71.39 | 49.66 | 60.32 |
| Dictionary-C | 60.29 | 73.32 | 53.93 | 61.88 |
| Dictionary-F-C | 62.50 | 74.59 | 54.74 | 63.13 |
‘-F’ means using fuzzy matching, and ‘-C’ means pruning words that are in GoogleCommonList.
DT, drugs and treatments; ENT, Ear, Nose & Throat; SC, symptoms and conditions.
Table 2.
Precision, recall, and F1 scores for the two forums and the two labels
| System | DT | SC | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | |
| Asthma | ||||||
| OBA | 52.25 | 56.50 | 54.25* | 78.87 | 60.08 | 68.20* |
| OBA-C | 62.06 | 53.15 | 57.25* | 83.62 | 58.24 | 68.66* |
| MetaMap | 68.42 | 57.56 | 62.52* | 58.63 | 80.24 | 67.75* |
| MetaMap-C | 77.60 | 54.98 | 64.36* | 70.28 | 75.15 | 72.63* |
| Dictionary-F-C | 89.65 | 47.97 | 62.50* | 78.73 | 70.87 | 74.59* |
| Xu et al.-25 | 89.57 | 53.87 | 67.28* | 77.29 | 72.09 | 74.60* |
| Xu et al.-50 | 85.96 | 54.24 | 66.51* | 76.28 | 72.70 | 74.45* |
| CRF | 87.09 | 49.81 | 63.38* | 77.68 | 73.72 | 75.65* |
| CRF-2 | 87.74 | 50.18 | 63.84* | 77.63 | 73.52 | 75.52* |
| CRF-20 | 86.53 | 49.81 | 63.23* | 76.64 | 73.52 | 75.05* |
| Our system | 86.88 | 58.67 | 70.04 | 78.10 | 75.56 | 76.81 |
| ENT | ||||||
| OBA | 43.22 | 55.73 | 48.68* | 67.51 | 50.52 | 57.79* |
| OBA-C | 49.73 | 51.36 | 50.53* | 70.55 | 46.18 | 55.82* |
| MetaMap | 56.39 | 53.00 | 54.64 | 57.01 | 64.23 | 60.40* |
| MetaMap-C | 64.08 | 49.72 | 55.99 | 67.40 | 58.50 | 62.63* |
| Dictionary-F-C | 82.41 | 40.98 | 54.74* | 74.35 | 54.86 | 63.13* |
| Xu et al.-25 | 76.50 | 40.98 | 53.37* | 73.48 | 54.86 | 62.82* |
| Xu et al.-50 | 62.80 | 41.53 | 49.99* | 73.88 | 57.46 | 64.64* |
| CRF | 79.38 | 42.07 | 55.71 | 72.06 | 56.42 | 63.29* |
| CRF-2 | 79.20 | 43.71 | 56.33 | 71.39 | 55.90 | 62.70* |
| CRF-20 | 67.79 | 43.71 | 53.15* | 70.61 | 55.90 | 62.40* |
| Our system | 82.82 | 44.80 | 58.15 | 71.65 | 61.45 | 66.16 |
The horizontal line separates systems that do not learn new phrases from the systems that do. The suffix ‘-C’ indicates post-processing the output of MetaMap and OBA by removing common words from GoogleCommonList. An asterisk denotes that our system is statistically significantly better (for two-tailed p value <0.05) than the system using approximated randomization (see the Supplemental section for more details). Scores in bold indicate the highest score for the metric. Xu et al. −25 and Xu et al. −50 are results of the Xu et al system for 25 and 50 iterations, respectively.
CRF, conditional random field; DT, drugs and treatments; ENT, Ear, Nose & Throat; SC, symptoms and conditions.
Table 3 shows the top 10 patterns and top 15 phrases extracted from the Asthma and ENT forums by our system. Table 4 shows the phrases extracted by our system from the following three forums: Acne, Breast Cancer, and Adult Type II Diabetes. We can broadly group the extracted phrases into four categories, which are described below.
Table 4.
Top 50 SC and DT phrases extracted by our system for three different forums
| Acne | Diabetes | Breast Cancer | |
|---|---|---|---|
| DT | |||
| New terms | diane35, retinoid, dianette, retinoids, topical retinoids, femodene, ginette, cilest, dalacin-t, dalacin, piriton, freederm, byebyeblemish, non-hormonal anti-androgen, sudocrem, byebye blemish, dermatologists, dian-35, canneston, microdermabrasions, isotrexin, noxema, proactiv, derm, cleansers, concealer, proactive, creme, microdermabrasion, moisturizer, minocylin | ambulance, basil, bedtime, c-peptide, cinnamon, diaformin, glycomet, glycomet-gp-1, hydrochlorothiazide-quinapril, hydrochlorothiazide-reserpine, lipoic, minidiab, neurotonin, opuntia, rebuilder, sometime, tritace | hormonal fluctuations, rads, ayurveda, ameridex, tram flap, bilateral mastectomy, flaps, incision, thinly, taxanes, bisphosphonates, bisphosphonate, mammosite, rad, imagery, stimulation, relicore, bezielle, wle (wide local excision), lymph spread-wle, moisturising, lymphnode, lympe, her2 neu, hormone-suppressing |
| Sub-phrases | topical, depo, contraceptives, contraceptive, aloe vera, topicals, salicylic, d3, peroxide, androgens-male, cleanser | asprin, bolus, carb, carbohydrate, carbohydrates, ovulation, regimen | hormonal, topical, antagonists, excision, vit, sentinel, cmf, primrose, augmentation, depo, flap |
| Abbreviations | a1c, a1c cutoff, a1cs, endo (endocrinology), ob gyn, ogtt (oral glucose tolerance test), xr (extended release) | recon (reconstruction), neu | |
| Misspellings | oxytetrcaycline, contracpetive, anitbiotics, oxytetracylcine, oxytracycline, lymecylcine, sprionolactone, benzol peroxide, depot-shot, tetracylcines, shampo, dorxy, steriod, moisturising, perscription | actoplusmet, awhile, basil-known, birth-control, blood-cholesterol, condrotin, darvetcet, diabix, excercise, fairley, htis, inslin, klonopin-i, metforim, metform, metformun100 mg, metmorfin, omigut40, pils, sutant | homonal, steriod, horonal, releif, ibubofrin, tamoxofin, tomoxphen, reloxifen, tamoxafin, tomoxifin, steriods, tamixofin |
| SC | |||
| New terms | squeeze blackheads, squeeze, breakouts, teenager, itchiness, coldsores, blemishes, blemish, breakout, chin, break-outs, re-appearing, outbreaks, poke, puss, flares, bum, outbreak, coldsore, acneic, armpit, teenagers | borderline diabetic, c-peptide, calories, checkup, educator, harden, rarer, sugary, thorough, type2 | armpit, grandmother, aunt, cancer-grandmother, cancer-having, survivor, aunts, morphologies, diagnosing |
| Sub-phrases | lesions, bumps, irritation, glands, bump, forehead, lumps, scalp, cheeks, follicles, dryness, gland, flare-up, pilaris rubra, puberty, cystic, follicular, inflamed, follicle, pcos, soreness, groin, occurrence, discoloration, relapse, oily | abdomen, blockage, bowel, calfs, circulatory, cirrhosis, disruptions, dryness, fibro, flour, fluctuations, foggy, lesion, lumps, masturbation, menopause, onset, pcos, precursor, sensations, spike, spikes, thighs, urine | lesions, lump, soreness, lumps, phyllodes, situ, ducts, lesion, sensations, needle, menopause, manifestations, variant, mutation, manifestation, onset, duct, lymph, gland, benign, irritation, abnormality, glands, mutations, asymmetry, occurrence, leaking, parenchymal, bump, unilateral, thighs, menstrual, subtypes, ductal, colon, bumps |
| Abbreviations | a-fib (atrial fibrillation), carbs (carbohydrates), cardio, ha1c, hep (hepatitis), hgba1c, hypo, oj (orange juice), t2 (type 2) | hx (history), ibc (inflammatory breast cancer) | |
| Spelling mistakes | becuase, forhead | allegeries, energyless, jsut, neurothapy, tyoe, vomiting-more, weezyness | caner, posibility, tratment |
Erroneous phrases (as determined by us) are shown in gray. Full forms of some abbreviations are in italics. Note that abbreviations are also new terms but are categorized separately because of their frequency in PAT.
DT, drugs and treatments; PAT, patient-authored text; SC, symptoms and conditions.
New terms
One goal of extracting medical information from PAT is to learn new treatments that patients are using or symptoms they are experiencing. Our system extracted phrases such as ‘stabbing pain’, ‘flakiness’, ‘plaque buildup’, which are not in the seed dictionaries. It also extracted alternative and preventative treatments such as ‘HEPA’ for high-efficiency particulate absorption air filter, ‘cinnamon pills’, ‘vinegar pills’, ‘basil’, and ‘opuntia’. Effects of alternative and new treatments are usually studied in small-scale clinical trials (eg, the effects of the Opuntia plant and cinnamon on diabetes patients in clinical trials have been studied by Frati-Munari et al35 and Khan et al 36). In contrast, our system enables the discovery and extraction of new DT and SC phrases in PAT and the study of their effects reported by patients on a larger scale in online forums.
Abbreviations
Patterns leverage context to extract abbreviations from PAT, despite the fact that, unlike in well-formed text, abbreviations in PAT tend to lack identifying structure such as capitalization and periods. Some examples of abbreviations that our system extracted are: ‘neb’ for nebulizer, ‘labas’ for long-acting β agonists, and ‘lada’ for latent autoimmune diabetes of adults.
Sub-phrases
Patients frequently do not use full names of diseases and drugs in PAT. For example, it would not be unusual for patients to refer to ‘vitamin b12’ simply as ‘b12’. These partial phrases did not get labeled by dictionaries because dictionaries contain long precise phrases and we label a phrase only when it fully matches a dictionary phrase. When we ran trial experiments that labeled phrases even when they partially matched a dictionary phrase, it resulted in low precision. Our pattern learning system learns relevant sub-phrases of the dictionary phrases without sacrificing much precision. For example, the system is able to learn that ‘large’ is not a relevant word by itself even though it occurs frequently in the SC dictionary, but ‘deficiency’ is. More examples include ‘inhaler’, ‘b5’, and ‘puffer’.
Spelling mistakes
Spelling mistakes are very common in PAT, especially for DT mentions. Context-sensitive patterns allow us to extract a wider range of spelling mistakes than would be possible with typical edit distance metrics (as in Dictionary-F-C). For example, the system extracts ‘neurothapy’ for neuropathy, ‘ibubofrin’ for ibuprofen, and ‘metforim’ for metformin.
To compare the efficacy of our system for extracting relevant phrases apart from spelling mistakes with MetaMap, we clustered all the strings from both the systems that were one edit distance away and normalized them to their most frequent spelling variation. We compare the top most frequent phrases extracted from the Diabetes forum in figure 2. We can see that our system extracts more relevant phrases.
Figure 2.
Top drugs and treatments (DT) and symptoms and conditions (SC) phrases extracted by our system, MetaMap, and MetaMap-C for the Diabetes forum. Numbers in parentheses indicate the number of times the phrase was extracted by the system. Erroneous phrases (as determined by us) are shown in gray. The reason we do not extract insulin is because it exists (incorrectly) in the automatically curated SC dictionary and we do not label DT phrases that are in the SC dictionary. For our system, we concatenated all consecutive words with the same label as one phrase, in contrast with MetaMap, which many times extracted consecutive words as different phrases (leading to the difference in the frequency of some phrases). For example, our system extracted ‘diabetes drug dependency’, but MetaMap extracted it as ‘diabetes’ and ‘drug dependency’. Similarly, our system extracted ‘latent autoimmune diabetes in adults’, whereas MetaMap extracted ‘latent’ and ‘autoimmune diabetes’.
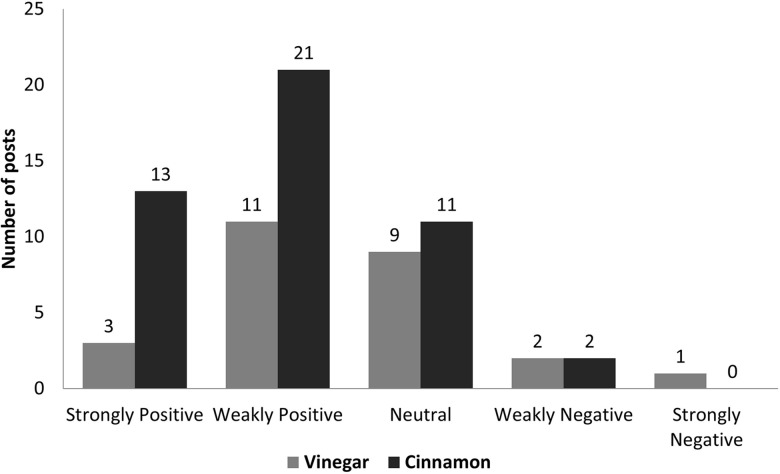
Our system can be used to explore different (possibly previously unknown) treatments that people are using for a condition. In turn, this can lead to novel insights, which can be further explored by the medical community. For example, from the Diabetes forum, our system extracted ‘cinnamon’ and ‘vinegar’ as DTs. An informal analysis of the posts reveals that ‘cinnamon’ was generally considered helpful by the community, and ‘vinegar’ had mixed reviews (figure 3).
Figure 3.
To study the anecdotal efficacy of ‘cinnamon’ and ‘vinegar’ for managing diabetes, we manually labeled the posts that mentioned the terms as treatment for diabetes (47 out of 49 posts for ‘cinnamon’ and 26 out of 30 posts for ‘vinegar’) with the sentiment towards that treatment. Both terms were extracted as drugs and treatment (DT) by our system for the Diabetes forum. ‘Strongly positive’ means the treatment helped the person. ‘Weakly positive’ means the person is using the treatment or has heard positive effects of it. ‘Neutral’ means the user is not using the treatment and did not express an opinion in the post. ‘Weakly negative’ means the person has heard that the treatment does not work. ‘Strongly negative’ means the treatment did not work for the person. More details are in the online supplemental section.
Discussion and future work
In this work, we induce lexico-syntactic patterns on data labeled using dictionaries, but no hand-labeled data, to identify DT and SC phrases in PAT. The performance of the system increases by removing common words from dictionaries and matching words fuzzily.
In most cases, our system significantly outperforms current standard tools in medical informatics. MetaMap and OBA have lower computational time since they do not match words fuzzily or learn new dictionary phrases, but have lower performance. All systems extracted SC terms with higher recall than DT terms because many simple SC terms (such as ‘asthma’) occurred frequently and were present in the dictionary. The improvement in performance of our system over the baselines is higher for DT than for SC, mainly because SC terms are usually verbose and descriptive and hence harder to extract using patterns. In addition, the performance is higher on the Asthma than the ENT forum for two reasons. First, the system was tuned on the Asthma forum. Second, the Asthma test set had many easy to label DT and SC phrases, such as ‘asthma’ and ‘inhaler’. On the other hand, many ENT phrases were longer and not present in seed dictionaries, such as ‘milk free diets’ and ‘smelly nasal discharge’.
One of the reasons that the CRF does not perform so well, despite being very popular for extracting entities from human-labeled text data, is that the data is partially labeled using dictionaries. Thus, the data is noisy and lack the full supervision provided in human-labeled data, making the word-level features not very predictive. CRF missed extracting some common terms such as ‘inhaler’ and ‘inhalers’ as DT (‘inhaler’ occurred only as a sub-phrase in the seed dictionary), and extracted some noisy terms, such as ‘afraid’ and ‘icecream’. In addition, CRF uses context for labeling data; we show in the online supplemental section that using context in the form of patterns performs worse than dictionary matching for labeling data. Our system, on the other hand, learned new dictionary phrases by exploiting context, but labeled data by dictionary matching. Self-training the CRF initially increased the F1 score for DT but performed worse in subsequent iterations. The system of Xu et al22 performed worse because of its overdependence on the seed patterns: it gave low scores to patterns that extracted phrases that had low overlap with the phrases extracted by the seed patterns, which resulted in lower recall.
Our results show that bootstrapping using patterns gives effective in-domain dictionary expansion for SC and DT phrases. As we can see from the top extracted phrases for the three MedHelp forums, our system uncovers novel terms for SCs and DTs, some of which refer to lesser-known home remedies (such as ‘basil’ and ‘cinnamon’ for diabetes) and components of daily care and management. The system extracts some incorrect phrases, which can be discarded by manual supervision. Such discoveries are valuable on two fronts: first, they may comprise a useful candidate set for future research into alternative treatments; second, they can be used to suggest candidate terms for various dictionaries and ontologies. There are two reasons for the overall lower recall and precision on this dataset than for extracting some other types of medical entities on clinical text. First, DT and SC definitions are broad, encompassing any symptom, condition, treatment, or intervention. Second, PAT contains slang and verbose descriptions that are usually not present in dictionaries. One limitation of our system is that it does not identify long descriptive phrases, such as ‘olive leaf nasal extract nasal spray’ and ‘trouble looking straight ahead’. More research is needed to robustly identify these to increase recall of the system. In addition, incorrect phrases in the dictionaries, which were curated automatically, reduced the precision of our system. Further research into automatically removing incorrect entries in the dictionaries will help to improve the precision.
Future improvements to performance would allow us to reap enhanced benefits from automatic medical term extraction. Improving precision, for example, would reduce the manual effort required to verify extracted terms to perform an analysis similar to the one shown in figure 3. Improving recall would increase the range of terms that we extract. For example, at present, our system still misses relevant terms, such as ‘oatmeal’ as a DT for diabetes.
Our results open several avenues for future work on mining and analyzing PAT. Extraction of DT and SC entities allows us to investigate connections and relationships between drug pairs and between drugs and symptoms. Prior work has successfully identified adverse drug events in electronic medical records37; using self-report patient data (such as found on MedHelp), we might uncover novel information on how particular drug combinations affect users. One such case study to identify side effects of drugs was presented by Leaman et al.2 Our system can also help to analyze sentiment towards various treatments, including home remedies and alternative treatments, for a particular disease; manually enumerating all treatments, along with their morphological variations, is difficult. Finally, we note that our system does not require any labeled data of sentences and thus can be applied to many different types of PAT (such as patient emails) and entity types (such as diagnostic tests).
Conclusion
We demonstrate a method for identifying medical entity types in PAT. We induce lexico-syntactic patterns using a seed dictionary of desirable terms. Annotating specific types of medical terms in PAT is difficult because of lexical and semantic mismatches between experts’ and consumers’ description of medical terms. Previous ontology-based tools such as OBA and MetaMap are good at fine-grained concept mapping on expert-authored text, but they have low accuracy on PAT.
We demonstrate that our method improves performance for the task of extracting two entity types—DTs and SCs—from MedHelp's Asthma and ENT forums by effectively expanding dictionaries in context. Our system extracts: new entities that are missing from the seed dictionaries; abbreviations; relevant sub-phrases of seed dictionary phrases; and spelling mistakes. In evaluation, our system significantly outperformed in most cases MetaMap, OBA, an existing system that uses word patterns for extracting diseases, and a CRF classifier. We believe that the ability to effectively extract specific entities is the key first step towards deriving novel findings from PAT.
Supplementary Material
Acknowledgments
The authors thank Bethany Percha for her comments on the manuscript. We thank MedHelp for sharing their anonymized data with us.
Footnotes
Contributors: Conception and design: all authors; data acquisition: DLM and SG; algorithm design: SG and CDM; experiment design and execution: SG and CDM; inter-annotator data: SG and DLM; drafting of manuscript: SG and DLM; critical revision of the paper for important intellectual content: all authors; final approval of the paper: all authors.
Funding: This work was supported by the Defense Advanced Research Projects Agency (DARPA) Deep Exploration and Filtering of Text (DEFT) Program under Air Force Research Laboratory (AFRL) contract No FA8750-13-2-0040 and by NSF grant CCF-0964173. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the view of the DARPA, AFRL, or the US government.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Fox S, Duggan M.2013. Health Online. Pew Internet and American Life Project. http://www.pewinternet.org/Reports/2013/Health-online.aspx.
- 2.Leaman R, Wojtulewicz L, Sullivan R, et al. Towards internet-age pharmacovigilance: extracting adverse drug reactions from user posts to health-related social networks. Proceedings of the 2010 Workshop on Biomedical Natural Language Processing Association for Computational Linguistics, 2010:117–25 [Google Scholar]
- 3.Carneiro HA, Mylonakis E. Google trends: a web-based tool for real-time surveillance of disease outbreaks. Clin Infect Dis 2009;49:1557–64 [DOI] [PubMed] [Google Scholar]
- 4.Butler D. When Google got flu wrong. Nature 2013;494:155–6 [DOI] [PubMed] [Google Scholar]
- 5.White RW, Tatonetti NP, Shah NH, et al. Web-scale pharmacovigilance: listening to signals from the crowd. J Am Med Inform Assoc 2013;20:404–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wicks P, Vaughan TE, Massagli MP, et al. Accelerated clinical discovery using self-reported patient data collected online and a patient-matching algorithm. Nat Biotechnol 2011;29:411–14 [DOI] [PubMed] [Google Scholar]
- 7.Smith CA, Wicks PJ. PatientsLikeMe: consumer health vocabulary as a folksonomy. AMIA Annual Symposium Proceedings Vol 2008. American Medical Informatics Association, 2008:682. [PMC free article] [PubMed] [Google Scholar]
- 8.Frantzi K, Ananiadou S, Mima H. Automatic recognition of multi-word terms: the C-value/NC-value method. Int J Digit Libr 2000;3:115–30 [Google Scholar]
- 9.MacLean D, Heer J. Identifying medical terms in patient-authored text: a Crowdsourcing-based approach. J Am Med Inform Assoc 2013;20:1120–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings of the AMIA Symposium American Medical Informatics Association, 2001:17. [PMC free article] [PubMed] [Google Scholar]
- 11.Jonquet C, Shah NH, Musen MA. The open biomedical annotator. Summit Transl Bioinform 2009;2009:56–60 [PMC free article] [PubMed] [Google Scholar]
- 12.Apache cTakes. http://ctakes.apache.org
- 13.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010;17–21:229–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pratt W, Yetisgen-Yildiz M. A study of biomedical concept identification: MetaMap vs. people. AMIA Annual Symposium Proceedings Vol 2003. American Medical Informatics Association, 2003:529–33 [PMC free article] [PubMed] [Google Scholar]
- 15.Epstein RH, St Jacques P, Stockin M, et al. Automated identification of drug and food allergies entered using non-standard terminology. J Am Med Inform Assoc 2013;20:962–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang N, Singh B, Afzal Z, et al. Using rule-based natural language processing to improve disease normalization in biomedical text. J Am Med Inform Assoc 2012;20:876–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Open Access, Collaborative Consumer Health Vocabulary Initiative. http://www.consumerhealthvocab.org (accessed Feb 2013)
- 18.MedlinePlus XML Files. http://www.nlm.nih.gov/medlineplus/xml.html
- 19.Zeng QT, Tse T. Exploring and developing consumer health vocabularies. J Am Med Inform Assoc 2006;13:24–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hearst MA. Automatic acquisition of hyponyms from large text corpora. Proceedings of the 14th International conference on Computational linguistics Association for Computational Linguistics, 1992:539–45 [Google Scholar]
- 21.Thelen M, Riloff E. A Bootstrapping method for learning semantic lexicons using extraction pattern contexts. Proceedings of the Conference on Empirical Methods in Natural Language Processing 2002:214–21 [Google Scholar]
- 22.Xu R, Supekar K, Morgan A, et al. Unsupervised method for automatic construction of a disease dictionary from a large free text collection. AMIA Annual Symposium Proceedings Vol 2008. American Medical Informatics Association, 2008:820–4 [PMC free article] [PubMed] [Google Scholar]
- 23.MedHelp. Data spans from 2007 to May 2011. http://www.medhelp.org
- 24.Stanford CoreNLP Toolkit. http://nlp.stanford.edu/software/corenlp.shtml (accessed Aug 2013)
- 25.RxList. http://www.rxlist.com (accessed Jan 2013)
- 26.MedlinePlus.. http://www.nlm.nih.gov/medlineplus (accessed Jan 2013)
- 27.MedicineNet. http://www.medicinenet.com (accessed Jan 2013)
- 28.MedDRA: Medical Dictionary for Regulatory Activities. http://www.meddra.org (accessed Feb 2013)
- 29. NCI Thesaurus. Semantic types accessed: Antibiotic, Clinical Drug, Laboratory Procedure, Medical Device, Steroid, and Therapeutic or Preventive Procedure. http://ncit.nci.nih.gov (accessed Mar 2013)
- 30.Google N-grams. http://storage.googleapis.com/books/ngrams/books/datasetsv2.html (accessed Jan 2008)
- 31.WebMD. http://www.webmd.com (accessed Oct 2013)
- 32.Liang P. Semi-supervised learning for natural language. MIT EECS, 2005 [Google Scholar]
- 33.Brown PF, deSouza PV, Mercer RL, et al. Class-based n-gram models of natural language. Comput Linguist 1992;18:467–79 [Google Scholar]
- 34.Finkel JR, Grenager T, Manning CD. Incorporating non-local information into information extraction systems by Gibbs sampling. Association of Computational Linguistics, 2005:363–70 [Google Scholar]
- 35.Frati-Munari AC, Gordillo BE, Altamirano P, et al. Hypoglycemic effect of Opuntia streptacantha Lemaire in NIDDM. Diabetes Care 1998;11:63–6 [DOI] [PubMed] [Google Scholar]
- 36.Khan A, Safdar M, Ali Khan M, et al. Cinnamon improves glucose and lipids of people with type 2 diabetes. Diabetes Care 2003;26:3215–18 [DOI] [PubMed] [Google Scholar]
- 37.Tatonetti NP, Fernald GH, Altman RB. A novel signal detection algorithm for identifying hidden drug-drug interactions in adverse event reports. J Am Med Inform Assoc 2012;19:79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.