Abstract
Objectives
To evaluate factors affecting performance of influenza detection, including accuracy of natural language processing (NLP), discriminative ability of Bayesian network (BN) classifiers, and feature selection.
Methods
We derived a testing dataset of 124 influenza patients and 87 non-influenza (shigellosis) patients. To assess NLP finding-extraction performance, we measured the overall accuracy, recall, and precision of Topaz and MedLEE parsers for 31 influenza-related findings against a reference standard established by three physician reviewers. To elucidate the relative contribution of NLP and BN classifier to classification performance, we compared the discriminative ability of nine combinations of finding-extraction methods (expert, Topaz, and MedLEE) and classifiers (one human-parameterized BN and two machine-parameterized BNs). To assess the effects of feature selection, we conducted secondary analyses of discriminative ability using the most influential findings defined by their likelihood ratios.
Results
The overall accuracy of Topaz was significantly better than MedLEE (with post-processing) (0.78 vs 0.71, p<0.0001). Classifiers using human-annotated findings were superior to classifiers using Topaz/MedLEE-extracted findings (average area under the receiver operating characteristic (AUROC): 0.75 vs 0.68, p=0.0113), and machine-parameterized classifiers were superior to the human-parameterized classifier (average AUROC: 0.73 vs 0.66, p=0.0059). The classifiers using the 17 ‘most influential’ findings were more accurate than classifiers using all 31 subject-matter expert-identified findings (average AUROC: 0.76>0.70, p<0.05).
Conclusions
Using a three-component evaluation method we demonstrated how one could elucidate the relative contributions of components under an integrated framework. To improve classification performance, this study encourages researchers to improve NLP accuracy, use a machine-parameterized classifier, and apply feature selection methods.
Objective
This study evaluated factors affecting performance of influenza detection, including accuracy of natural language processing (NLP), discriminative ability of Bayesian network (BN) classifiers, and feature selection. Utilizing free-text emergency department (ED) medical reports as input, our influenza detection system comprises a finding-extraction component—the Topaz1 and MedLEE2 3 NLP parsers—and a BN classifier. Our evaluation measured the classification performance over different finding-extraction methods, over different parameterizations of the BNs, and over different sets of findings.
Background and significance
There is a growing interest in leveraging routinely collected electronic health records (EHRs) for patient cohort identification to facilitate biomedical research.4–9 However, many cohorts—or ‘phenotypes’—of interest are defined in part by information that is often (though not always) recorded in clinical notes. This constraint is especially true in early detection of epidemics and in the elucidation of yet-to-be-named diseases. The semi-structured free text of clinical notes must be transformed into structured representations prior to phenotype detection.
The earliest attempt to leverage free-text EHR data to detect phenotype dates to the work of Hripcsak et al,10 who used MedLEE and a rule-based classifier to detect tuberculosis cases from chest radiograph reports, obtaining positive predictive values (PPVs) in the range 0.03–0.96 and sensitivity in the range 0.36–0.93. Aronsky and Haug made the first use of a probabilistic classifier in conjunction with NLP to detect community-acquired pneumonia from data in an EHR. In that study, which showed discriminative ability as measured by area under the receiver operating characteristic (AUROC) curve of 0.98,11 six findings were parsed from clinical documents.
The combination of NLP and classification algorithms have subsequently been applied to the automatic detection of additional phenotypes from EHR data, including inhalational anthrax (AUROC: 0.677),12 cataracts (PPV: 0.95),7 peripheral arterial disease (precision: 0.67–1; recall: 0.84–1),13 and rheumatoid arthritis (PPV: 0.94).14
Automatic influenza detection from EHR data is of particular importance because of the threat of pandemic influenza. Elkin measured the discriminative ability of an NLP parser (in the Multithreaded Clinical Vocabulary Server system at Mayo Clinic) and a regression classifier on Mayo Clinic records, obtaining an AUROC=0.76415 to discriminate between PCR or culture-positive influenza cases and PCR or culture-negative non-influenza controls.
As part of a larger system16 17 that detects and characterizes outbreaks in the Real-time Outbreak and Disease Surveillance Laboratory (RODS) at University of Pittsburgh, we developed a Bayesian Case Detector (BCD) that uses an NLP parser to extract the influenza-related findings from ED reports and a BN classifier to compute the probability that a patient has influenza given the set of NLP extracted findings.18 19 Tsui demonstrated high discrimination between influenza cases and non-influenza controls drawn from a low-influenza summer period (AUROC: 0.973; 95% CI 0.955 to 0.992).18
In this study, we evaluated the individual components along a processing pipeline that starts with free-text ED reports and ends with a probability estimation of the presence of influenza. These components are an NLP parser used for extracting influenza-related findings from free-text, a BN classifier utilized for performing probability estimation, and the findings selected for inferring the presence of influenza.
Materials and methods
In this section, we describe the NLP parsers, the BN classifiers, the testing dataset, and the experiments.
Natural language processing parsers
Topaz
Topaz was developed by Chapman, Chu, and colleagues in our laboratory for use in influenza and shigellosis related finding extraction from ED reports. For this reason, the output of Topaz can be used to directly set the values of nodes in the BNs that we studied. Topaz uses a pipeline of processing components to (1) find and annotate targeted clinical findings, (2) determine whether a finding is mentioned as being present or absent; historical, recent, or hypothetical; and experienced by the patient or someone else,20 and (3) assign a single value of ‘present’, ‘absent’, or ‘missing’ (not mentioned) to each finding taking into account synonyms and multiple possibly contradictory mentions of the finding in the report. Topaz's heuristic resolution of contradictory mentions of a clinical finding within a report includes the following rules: labeling a finding as ‘present’ in summary when Topaz identified at least one positive mention in a report, labeling a finding as ‘absent’ for a report when all mentions identified by Topaz were negative, and labeling ‘missing’ otherwise (when it found no mentions positive or negative).
MedLEE
MedLEE was developed by Friedman and colleagues at Columbia University. It has a pipeline of programming components, each of which is guided by a corresponding knowledge component such as lexicon and grammar. MedLEE's pre-processor component and phrase regularization component execute tasks similar to Topaz's first step. MedLEE's parser assigns element modifiers (eg, certainty, severity) that are similar to Topaz's second step. However, the version (64-bit, 2012 release) of MedLEE that we used does not resolve contradictory mentions of findings that it extracts from a report. Since our BN classifiers require features to be ‘present’, ‘absent’, or ‘missing’ in summary, we applied Topaz's heuristic resolution rules to MedLEE output. In addition, we mapped some MedLEE output, which was represented by Unified Medical Language System Concept Unique Identifiers (UMLS CUIs) to a few influenza-related findings without corresponding UMLS CUIs (CUIs). For examples, we mapped C0021400 (influenza) to a ‘suspected flu’ finding.
BN classifiers
A BN classifier represents probabilistic knowledge related to a classification task in the form of an acyclic graph whose structure represents dependent and independent relationships between random variables and a set of conditional probability tables (CPTs). A BN structure is the set of nodes and arcs between nodes denoting the probabilistic dependencies among the represented variables. The set of CPTs comprises a table for each random variable in the BN structure conditioned on its parents. The structure and CPTs of a BN can be manually elicited from an expert21 or automatically estimated from training data by machine learning algorithms.22 23 We will refer to the process of specifying CPTs for a BN as parameterization in the following sections.
Three BN classifiers
To study the effects of different parameterization methods for BN classifiers (expert, machine-parameterization) on influenza detection performance, we created three BN classifiers that differed only in how their CPTs were determined.
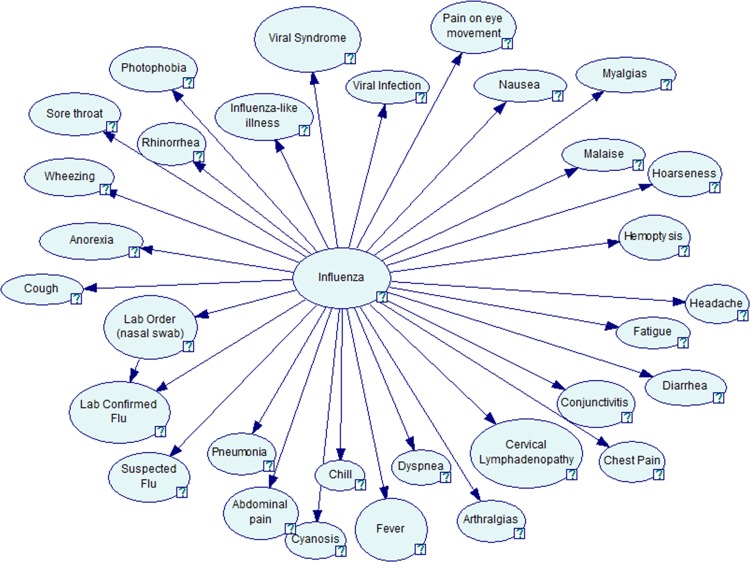
Author FT and two physicians defined a simple BN structure for all three classifiers (figure 1). They first identified a set of 31 clinical findings used by clinicians in diagnosing influenza. One physician is a board-certified infectious disease specialist who has over 40 years’ clinical experience and more than 5 years’ research experience in biomedical informatics.
Figure 1.
Bayesian network for influenza detection (GeNIe24 visualization).
They defined a near naïve BN, assuming that all of the influenza-related findings were conditionally independent given influenza status, with the exception of the ‘lab confirmed flu’ finding that depended on both influenza status and whether the report mentioned a nasal swab order. Naïve BN has been shown to have reasonable discriminative ability25 26 and its CPTs were easier for physicians to estimate than CPTs of a more complicated model.
Expert-defined BN classifier
Author FT elicited conditional probabilities for each finding given its parent node(s) in the network from the infectious disease physician mentioned above. He elicited 64 conditional probabilities, including two conditional probabilities for each of 30 nodes that are assumed to be conditionally independent and four conditional probabilities for the ‘lab confirmed flu’ node. For example, two questions during elicitation for the ‘cough’ node were ‘what is the probability that an influenza patient has cough?’ and ‘what is the probability that a non-influenza patient has cough?’ We refer to the resulting classifier as the expert-defined-BN classifier.
Machine-learned classifiers
We created a set of training data with which to parameterize another two classifiers.
Training data for machine-learned classifiers
Influenza cases: We obtained 468 ED reports of PCR-positive influenza patients from the period January 1, 2008 to August 31, 2010. The reports were de-identified by the De-ID tool.27
Non influenza controls: We obtained 29 004 de-identified ED reports of patients whose visits were not associated with a positive influenza PCR test from the period July 1, 2010 to August 31, 2010.
Using these reports, we created two training sets to parameterize the BN classifiers. One training set contained findings that were extracted using Topaz and the other training set contained findings that were extracted using MedLEE. Both training sets comprised 29 472 instances where a single instance had 31 influenza-related findings.
Expectation–Maximization maximum a posteriori (EM-MAP) algorithm: Besides the expert-defined-BN classifier, we created two additional BN classifiers—both initially parameterized by expert and further trained using the Topaz training set (BN-EM-Topaz) or the MedLEE training set (BN-EM-MedLEE). We used the EM-MAP algorithm28 as the machine learning method with a stopping criterion of |Δ(P(model|data))|<0.01. We selected the EM-MAP algorithm because it can handle missing features in the training instances (eg, findings that an NLP parser labels as ‘missing’) and it can use CPTs elicited by experts as prior knowledge. These BN classifiers are parameterized using both expert's knowledge and training data, and are especially useful when the occurrence of certain findings is rare.
Testing dataset
The testing dataset comprised reports for both ED patients with influenza and ED patients without influenza (shigellosis). This testing dataset was used in all experiments described in the remaining sections of Methods.
Influenza cases: We obtained 124 de-identified ED reports of all PCR-positive influenza patients seen in four EDs in Allegheny County, Pennsylvania between December 1, 2010 and June 30, 2011.
Non-influenza (shigellosis) controls: We used a convenience sample of 87 shigellosis cases from the same EDs for the period January 1, 2010 to June 30, 2010. This set represents all ED patients that have positive culture results for shigellosis. In using this non-representative sample, we recognized that shigellosis and influenza have symptomatic overlap (eg, both diseases can cause fever and diarrhea) and our BN classifier did not represent special shigellosis-related findings (eg, rectal bleeding and stool order) that might help to discriminate between the diagnoses.
Annotation method: Three board-certified physicians annotated the 211 ED reports in the testing dataset. To ensure that all physicians could reach a similar annotation baseline standard, we first asked them to review 10 sample reports together, and then we measured Cohen's κ value, a measure of inter-annotator agreement. When Cohen's κ value reached 0.8 or greater, we considered each physician to have reached the annotation standard. Then, each physician was assigned overlapping subsets of the reports. To ensure annotation quality, 12% of reports were reviewed by at least two annotators and their agreement was measured during the annotation process. If there was discrepancy between two physicians’ annotations, the third physician would review the discrepancy and make the final decision after discussion with the other two physicians.
Using this testing dataset, we measured the NLP parsers’ finding-extraction performance and classifiers’ influenza-detection performance. Our primary analyses used 31 findings mentioned in the section introducing BN classifiers (figure 1), while the secondary analyses used the 17 most influential findings as we will discuss later.
Metrics of NLP-finding-extraction performance
We measured the performance of Topaz and MedLEE using accuracy, recall, and precision. We calculated CIs using bootstrap percentiles with SAS V.9.3.29
Measurement of the classification performance
We used the AUROC as a measure of classification performance. Since we had three finding-extraction methods (expert, Topaz, or MedLEE) and three BN classifiers (expert-defined-BN, BN-EM-Topaz, or BN-EM-MedLEE), we performed nine experiments. The ‘true’ disease status (gold standard) was defined by laboratory test results. To compare AUROCs, we calculated p values using DeLong's two-sided comparisons30 as implemented in the pROC31 package of the R statistical software. To elucidate the relative contribution of the finding-extraction method (human-annotated findings vs Topaz/MedLEE-extracted finding) and parameterization method (human-parameterized classifier vs machine-parameterized classifiers) on classification performance, we compared AUROCs in groups using Friedman’s two-way non-parametric analysis of variance model (ANOVA)32 with SAS V.9.3.
Secondary analyses: measurement of the effect of feature selection on performance
To determine the effect of feature selection on influenza detection, we studied a subset of influenza-related findings. In a naïve BN, the posterior odds (eg, P(disease=True|findings)/P(disease=False|findings)) equals the product of prior odds (ie, P(disease=True)/P(disease=False)) and the likelihood ratios (LR) of each finding. Therefore, we defined a subset of influential findings as those findings (in the BN-EM-Topaz classifier) that had LR positive (LR+) greater than three or LR negative (LR−) less than 0.33 (one third).
We then measured the accuracy, precision, and recall of Topaz and MedLEE for each of these findings. We further assessed AUROCs of classifiers only using these influential findings and compared them with classifiers leveraging the complete finding set using a one-sided paired Wilcoxon signed-rank test.
Results
Inter-annotator agreement
The κ values measuring inter-annotator agreement for each pairwise comparison of annotations of three physicians were 0.8346, 0.8592, and 0.8933, indicating reliable agreement.
Influenza-related findings in the testing dataset
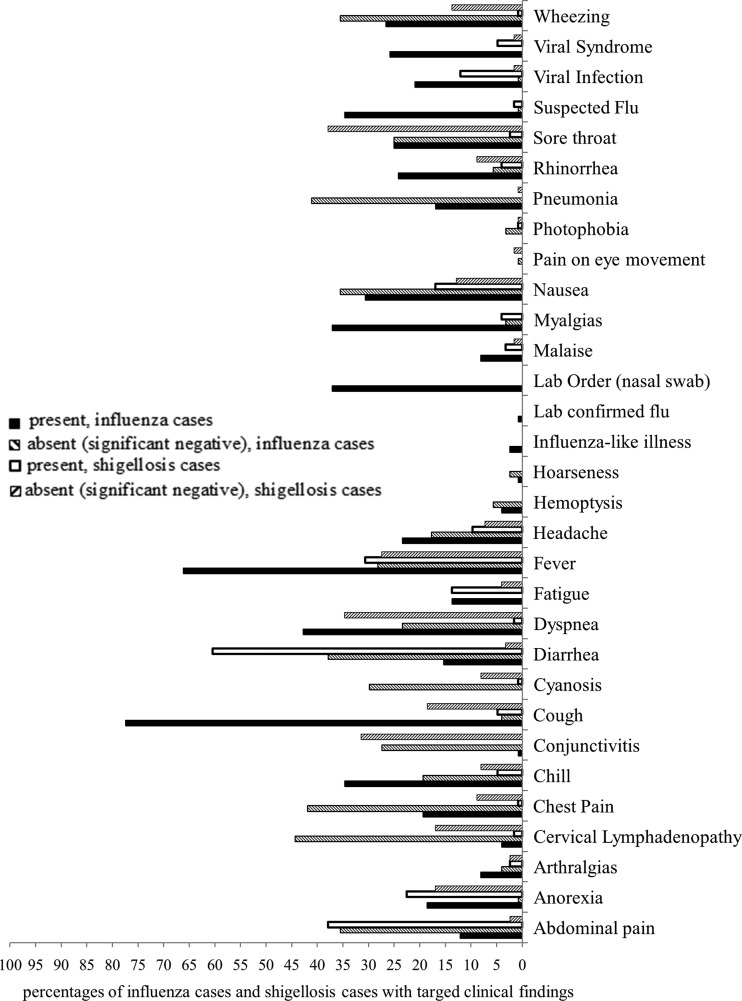
Figure 2 shows the frequency at which the treating clinicians documented 31 influenza-related findings in the influenza and non-influenza (shigellosis) patients in the testing dataset. The most frequently documented positive finding in the influenza cases was cough (77.42%), while the most frequently documented negative finding was cervical lymphadenopathy (44.35%). The most frequently documented positive finding in the shigellosis cases was diarrhea (60.48%), while the most frequently documented negative finding was sore throat (37.90%). On average, the clinicians documented about 11 influenza-related findings (six positive findings and five negative findings) in the influenza cases and about seven influenza-related findings (three positive findings and four negative findings) in the shigellosis controls.
Figure 2.
Percentages of influenza cases and shigellosis cases with targeted influenza-related findings.
NLP accuracy for influenza-related findings
Table 1 shows the overall accuracy, recall, and precision of Topaz and MedLEE for the entire set of influenza-related findings (left-hand side). The right-hand side of table 1 presents the results for a subset of influential findings and will be described in the secondary analyses section.
Table 1.
Summary of performance measures for Topaz and MedLEE
| Measures | Performance for primary analyses (use all 31 findings) | Performance for secondary analyses (use 17 influential findings) | ||||
|---|---|---|---|---|---|---|
| Topaz | MedLEE | p Value (Topaz vs MedLEE) | Topaz | MedLEE | p Value (Topaz vs MedLEE) | |
| Accuracy | 0.91 (0.90 to 0.91) | 0.90 (0.89 to 0.91) | 0.1650 | 0.92 (0.91 to 0.92) | 0.90 (0.89 to 0.91) | 0.0537 |
| Accuracy (absent and present) | 0.78 (0.76 to 0.80) | 0.71 (0.70 to 0.73) | <0.0001* | 0.75 (0.73 to 0.78) | 0.70 (0.67 to 0.73) | 0.0047* |
| Recall for present | 0.80 (0.77 to 0.82) | 0.79 (0.77 to 0.82) | 0.7499 | 0.72 (0.69 to 0.76) | 0.77 (0.74 to 0.80) | 0.0453* |
| Recall for absent | 0.76 (0.73 to 0.78) | 0.62 (0.59 to 0.65) | <0.0001* | 0.81 (0.77 to 0.84) | 0.58 (0.53 to 0.63) | <0.0001* |
| Precision for present | 0.85 (0.83 to 0.87) | 0.90 (0.88 to 0.92) | 0.0002* | 0.92 (0.90 to 0.94) | 0.92 (0.89 to 0.94) | 0.8100 |
| Precision for absent | 0.87 (0.85 to 0.90) | 0.90 (0.88 to 0.93) | 0.0852 | 0.89 (0.86 to 0.92) | 0.87 (0.83 to 0.91) | 0.5144 |
95% CIs in parentheses.
For each report, physicians and NLP parsers labeled values (present, absent, or missing) for each of the 31 influenza-related findings. With physician annotations as gold standard, we calculated accuracy, recall, and precision as measurements of NLP accuracy as follows:
Accuracy: A+E+I/(A+B+C+D+E+F+G+H+I).
Accuracy (absent and present): A+E/(A+B+D+E+G+H).
Recall for present (sensitivity): A/(A+D+G).
Recall for absent (specificity): E/(B+E+H).
Precision for present (positive predictive value): A/(A+B+C).
Precision for absent (negative predictive value): E/(D+E+F).
where A stands for number of findings with both expert and NLP labeled present; B stands for number of findings with expert labeled absent but NLP labeled present; C stands for number of findings with expert labeled missing but NLP labeled present; D stands for number of findings with expert labeled present but NLP labeled absent; E stands for number of findings with both expert and NLP labeled absent; F stands for number of findings with expert labeled missing but NLP labeled absent; G stands for number of findings with expert labeled present but NLP labeled missing; H stands for number of findings with expert labeled absent but NLP labeled missing; and I stands for number of findings with both expert and NLP labeled missing.
95% CI of the empirical distribution is obtained by bootstrapping with replacement (2000 times, sample size is 31 features per report×211 report=6541 each time).
The 17 influential findings indicated in BN-EM-Topaz were arthralgia, cervical lymphadenopathy, chill, cough, fever, hoarseness, influenza-like illness, lab confirmed influenza, lab order (nasal swab), malaise, myalgia, rhinorrhea, sore throat, suspected flu, viral infection, viral syndrome, and wheezing. 95% CI of the empirical distribution is obtained by bootstrapping with replacement (2000 times, sample size is 17 influential findings per report×211 report=3587 each time).
Each p value was calculated with a two-sided z test for comparison of two proportions. *p<0.05.
BN, Bayesian network; NLP, natural language processing.
Because we post-processed the MedLEE output, the following evaluation results only reflect the accuracy of MedLEE for influenza findings when coupled with the post-processing that we employed.
We found that Topaz was more accurate than MedLEE at identifying influenza-related findings documented by treating clinicians in their reports (accuracy: 0.78 vs 0.71, p<0.0001). Topaz and MedLEE had similar recall (ie, sensitivity) for positive findings (0.80 vs 0.79, p=0.7499), but Topaz had better recall for negative findings (specificity) (0.76 vs 0.62, p<0.0001). MedLEE's precision for positive findings (positive predictive values) was significantly better than Topaz (0.90 vs 0.85, p=0.0002), while its precision for negative findings (negative predictive value) was similar to Topaz (0.90 vs 0.87, p=0.0852).
Classification performance for nine combinations
Table 2 shows the classification performance for all nine combinations of the finding-extraction method (expert, Topaz, or MedLEE) and classifier (expert-defined BN, BN-EM-Topaz, or BN-EM-MedLEE), with the upper half of the table showing for classifier using 31 findings and the lower half listing for classifier using 17 influential findings. Table 3 shows the p values for each two-sided comparison of AUROCs of two combinations of finding-extraction method and classifier.
Table 2.
AUROCs (95% CIs) of nine possible combinations of finding-extraction method and BN classifier
| BN classifiers | Finding-extraction methods | ||
|---|---|---|---|
| Expert | Topaz | MedLEE | |
| Primary analyses: BN classifiers using 31 influenza-related findings | |||
| Expert-defined BN | 0.70 (0.63 to 0.77) | 0.64 (0.57 to 0.71) | 0.64 (0.57 to 0.72) |
| BN-EM-Topaz | 0.79 (0.73 to 0.85) | 0.73 (0.66 to 0.79) | 0.71 (0.64 to 0.78) |
| BN-EM-MedLEE | 0.77 (0.70 to 0.83) | 0.70 (0.63 to 0.77) | 0.66 (0.59 to 0.74) |
| Secondary analyses: BN classifiers using 17 influential findings* | |||
| Expert-defined BN | 0.80 (0.74 to 0.86) | 0.76 (0.69 to 0.82) | 0.73 (0.66 to 0.80) |
| BN-EM-Topaz | 0.82 (0.76 to 0.88) | 0.75 (0.69 to 0.82) | 0.74 (0.68 to 0.81) |
| BN-EM-MedLEE | 0.79 (0.73 to 0.85) | 0.73 (0.66 to 0.80) | 0.70 (0.63 to 0.77) |
*The 17 influential findings indicated in BN-EM-Topaz were arthralgia, cervical lymphadenopathy, chill, cough, fever, hoarseness, influenza-like illness, lab confirmed influenza lab order (nasal swab), malaise, myalgia, rhinorrhea, sore throat, suspected flu, viral infection, viral syndrome, and wheezing.
AUROC, area under the receiver operating characteristic; BN, Bayesian network.
Table 3.
Paired two-sided DeLong tests for comparison among AUROCs of Bayesian case detectors with different combinations of finding-extraction method and Bayesian network (BN) classifier
| e+E | e+T | e+M | t+E | t+T | t+M | m+E | m+T | m+M | |
|---|---|---|---|---|---|---|---|---|---|
| Primary analyses: BN classifiers using 31 influenza-related findings | |||||||||
| e+E | 0.0001* | 0.0042* | 0.0044* | 0.4231 | 0.9399 | 0.0083* | 0.9321 | 0.1437 | |
| e+T | 0.0195* | <0.0001* | 0.0007* | <0.0001* | <0.0001* | <0.0001* | <0.0001* | ||
| e+M | <0.0001* | 0.0229* | 0.0006* | <0.0001* | 0.0029* | <0.0001* | |||
| t+E | 0.0011* | 0.0182* | 0.8959 | 0.0265* | 0.4805 | ||||
| t+T | 0.0012* | 0.0052* | 0.2914 | 0.0004* | |||||
| t+M | 0.0526 | 0.8289 | 0.0337* | ||||||
| m+E | 0.0045* | 0.4466 | |||||||
| m+T | <0.0001* | ||||||||
| m+M | |||||||||
| Secondary analyses: BN classifiers using 17 influenza-related findings | |||||||||
| e+E | 0.1151 | 0.4847 | 0.0345* | 0.0362* | 0.0038* | 0.0005* | 0.0157* | 0.0001* | |
| e+T | 0.0005* | 0.0037* | 0.0005* | <0.0001* | <0.0001* | 0.0001* | <0.0001* | ||
| e+M | 0.1490 | 0.0438* | 0.0019* | 0.0065* | 0.0251* | <0.0001* | |||
| t+E | 0.6497 | 0.0496* | 0.1426 | 0.4892 | 0.0166* | ||||
| t+T | 0.0012* | 0.2962 | 0.6471 | 0.0107* | |||||
| t+M | 0.9391 | 0.5289 | 0.1460 | ||||||
| m+E | 0.3206 | 0.1110 | |||||||
| m+T | <0.0001* | ||||||||
| m+M | |||||||||
Each combination of finding-extraction method and BN classifier is represented as a lower case letter plus a upper case letter. The lower case letters are abbreviations of finding-extraction methods: e, expert findings; t, Topaz findings; m, MedLEE findings. The upper case letters are abbreviations of BN classifiers: E, expert-defined BN; T, BN-EM-Topaz; M, BN-EM-MedLEE.
Each p value was calculated with a DeLong two-sided comparison of AUROC. The *p<0.05.
The 17 influential findings indicated in BN-EM-Topaz were arthralgia, cervical lymphadenopathy, chill, cough, fever, hoarseness, influenza-like illness, lab confirmed influenza lab order (nasal swab), malaise, myalgia, rhinorrhea, sore throat, suspected flu, viral infection, viral syndrome, and wheezing.
AUROC, area under the receiver operating characteristic.
The combination of the finding-extraction method and classifier with the highest performance was the combination of expert findings with BN-EM-Topaz (AUROC: 0.79; 95% CI 0.73 to 0.85), suggesting that NLP misclassification contributed to less accurate influenza case identification. The pairing with the lowest performance was the pairing of Topaz findings with expert-defined BN (AUROC: 0.64; 95% CI 0.57 to 0.71), suggesting that parameters in expert-defined BN may not well represent correlations between NLP extracted clinical findings and the disease.
Effect of the finding-extraction method on classification performance
In the primary analyses, all three classifiers achieved better discriminative ability when associated with expert findings than with NLP (Topaz/MedLEE) findings (average AUROC: 0.75 vs 0.68, p=0.0113). Specifically, expert-defined BN classifier using expert findings was better than the same classifier using Topaz findings (AUROC: 0.70 vs 0.64, p=0.0044) or MedLEE findings (AUROC: 0.70 vs 0.64, p=0.0083). This pattern also held for the secondary (influential findings) analyses.
Effect of classifier on classification performance
In the primary analyses, all three finding-extraction methods worked best when the classifier was BN-EM-Topaz, followed by BN-EM-MedLEE, then expert-defined BN. The machine-learned classifiers had greater discriminative ability than the expert-defined BN classifier (average AUROC: 0.73 vs 0.66, p=0.0059). For example, associated with expert findings, expert-defined BN classifier (AUROC: 0.70) did not perform as well as either the BN-EM-Topaz classifier (AUROCs: 0.79, p<0.0001) or the BN-EM-MedLEE classifier (AUROCs: 0.77, p=0.0042). However, these differences largely disappeared in the secondary analyses using the most influential findings.
When comparing the two machine-learned classifiers in the primary analyses, we found that the BN-EM-Topaz classifier yielded greater accuracy than the BN-EM-MedLEE classifier: AUROCexpert-findings+BN-EM-Topaz=0.79 vs AUROCexpert-findings+BN-EM-MedLEE=0.77, p=0.0195; AUROCTopaz-findings+BN-EM-Topaz=0.73 vs AUROCTopaz-findings+BN-EM-MedLEE=0.70, p=0.0012; AUROCMedLEE-findings+BN-EM-Topaz=0.71 vs AUROCMedLEE-findings+BN-EM-MedLEE=0.66, p<0.0001. The superiority of BN-EM-Topaz over BN-EM-MedLEE remained in the secondary analyses.
Secondary analyses of NLP and classifier performance for 17 influential findings
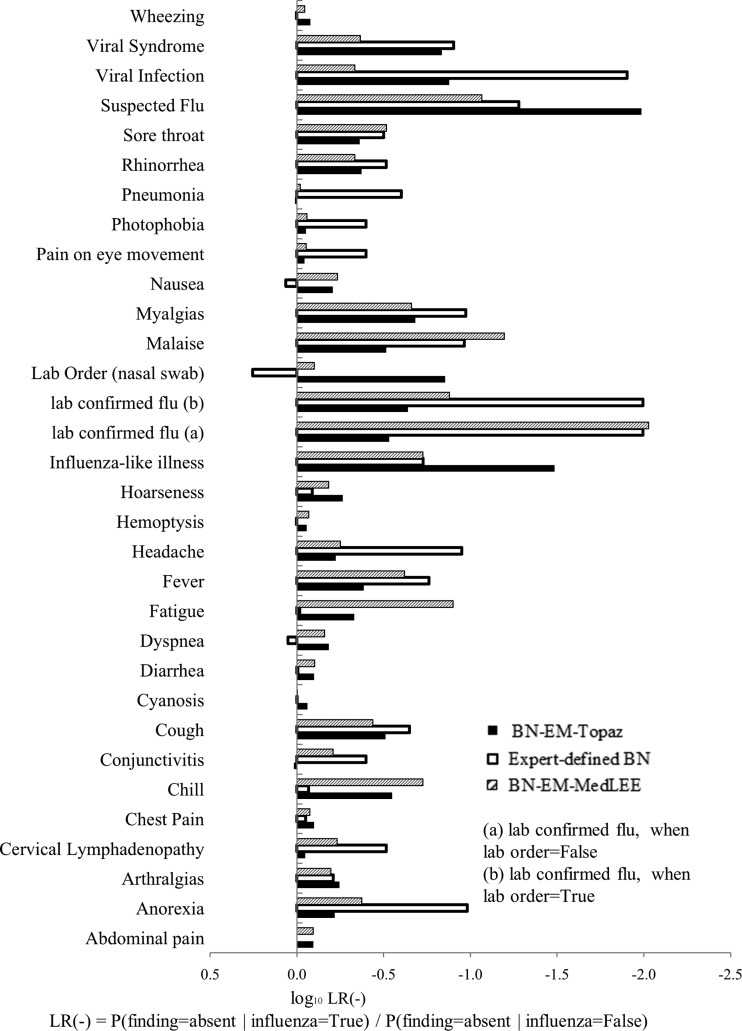
The 17 influential findings indicated in BN-EM-Topaz were arthralgia, cervical lymphadenopathy, chill, cough, fever, hoarseness, influenza-like illness, lab confirmed influenza, lab order (nasal swab), malaise, myalgia, rhinorrhea, sore throat, suspected flu, viral infection, viral syndrome, and wheezing (figures 3 and 4).
Figure 3.
Log10 LR+ (likelihood ratios) of features in expert-defined BN, BN-EM-Topaz, and BN-EM-MedLEE.
Figure 4.
Log10 LR− (likelihood ratios) of features in expert-defined BN, BN-EM-Topaz, and BN-EM-MedLEE.
The right-hand side of table 1 compares the finding-extraction accuracy of Topaz and MedLEE for these findings. Topaz still had a significantly higher accuracy than MedLEE for these 17 findings with value of being either present or absent (0.75 vs 0.70, p=0.0047). Similarly, Topaz's recall for negative findings was still significantly higher than MedLEE's (0.81 vs 0.58, p<0.0001). However, Topaz's recall for positive findings became significantly lower than MedLEE's (0.72 vs 0.77, p=0.0453). The accuracy, recall, and precision of Topaz and MedLEE for each finding are listed in online supplemental table S1.
The lower half of table 2 lists AUROCs of nine combinations of finding-extraction method and BN classifier. Comparing them with the upper half, we found that classifiers that only used the 17 influential findings had significantly better performance (average AUROC: 0.76>0.70, p=0.004).
Discussion
Effects of finding-extraction on classification performance
The accuracy of NLP extraction of influenza-related findings from ED reports varies by finding and differs for the determination of positive and significant negative findings. The Topaz and MedLEE parsers accurately determined 71–78% of the findings. Topaz performed significantly better than MedLEE on mentions of absent findings (significant negatives) (0.76 vs 0.62, p<0.0001), and MedLEE had significantly better precision for positive findings (0.90 vs 0.85, p=0.0002). The accuracy of MedLEE could be biased because of the post-processes mentioned in the method section (ie, applying Topaz's heuristic resolution rules and mapping some UMLS CUIs to influenza-related findings).
Both primary and secondary analyses suggested that all three classifiers achieved greater discrimination when combined with expert findings, followed by Topaz findings, then MedLEE findings. This correlation was present even when there was a mismatch between NLP parser and BN classifier. These results suggested the importance of finding-extraction accuracy for influenza detection and encouraged the use of the highest performing NLP system available regardless of the NLP system used to train the classifier.
Effects of classifier on classification performance
Usually, it is not easy for an expert to accurately quantify the correlations between findings and the disease, and using an NLP parser as finding-extraction method could further complicate the situation. In this study, the machine-learned classifiers were shown to have better discriminative abilities than the expert-defined classifier across all finding-extraction methods, indicating the benefit of turning data into knowledge. Starting the machine learning process from BN classifiers that are initially parameterized by experts, the EM-MAP algorithm is especially useful when the occurrences of certain findings are rare.
Effects of feature selection on classification performance
In this study, feature selection based on likelihood ratio values that were calculated with CPTs in a machine-learned BN classifier significantly improved the performance of influenza detection, suggesting that feature selection could be an efficient way to improve classification performance.
Limitations
Although the research has reached its aims, one major limitation is the use of a non-representative sample of non-influenza (shigellosis) controls in the testing dataset. This decision was pragmatic as we did not have the resources to develop additional expert-annotated ED charts. In addition, our use of PCR tests as a gold standard may have biased the testing set with positive cases that are more severe symptomatically and thus easier to distinguish than average influenza cases in EDs. Therefore, the AUROC in the range 0.64–0.82 should not be taken as an indication of the performance of influenza detection that we would expect in an operational ED setting; further evaluation with a randomly selected testing dataset would be more informative.
Significance
Influenza detection is important in both clinical care and public health practice. Automatic influenza detection from EHR data still depends on the ability to extract symptoms and signs from unstructured data. The present paper described a systematic approach for evaluating the relative contributions of the components in a process of extracting symptoms and signs, feature selection, and classification for the disease influenza. Although using a biased control may limit the interpretation of the results of the present study, the three-component evaluation could be applied more generally to a broader problem of detection of any disease phenotype that involves clinical information that is stored in free-text reports.
Conclusion
Using a three-component evaluation method we demonstrated how one could elucidate the relative contributions of components under an integrated framework. To improve classification performance, this study encourages researchers to improve NLP accuracy, use a machine-parameterized classifier incorporating both expert knowledge and data patterns, and apply feature selection methods. This study addresses the concern of using one NLP system to train a classifier and another NLP system in production—using the highest performing NLP system available regardless of the NLP system used to train the classifier is advised.
Supplementary Material
Acknowledgments
The authors wish to thank Drs John Dowling, Paul Thyvalikakath, and Tatiana Bogdanovich for annotating the reports used in our study. We acknowledge Dr Wendy Chapman, David Chu, Dr Henk Harkema, and Lee Christiansen for developing Topaz software when they worked in the RODS laboratory. We acknowledge Dr Carol Friedman for providing MedLEE software that is partly supported by Grants R01 LM010016 and R01 LM008635 from the National Library of Medicine. We would like to thank Howard Su for medical record retrieval and de-identification, Dr Gregory F Cooper and Dr Shyam Visweswaran for their helpful discussions, and Kevin Bui for his programming skills.
Footnotes
Contributors: YY and FT analyzed data, drafted the article, and took the responsibility for accuracy of data analysis and result formatting. MW and JUE brought insight into study design, data analysis and interpretation, and went through many drafts and revisions. QL did annotator training and gathered the expert findings for shigellosis reports.
Funding: This work is supported in part by Grants P01-HK000086 and U38HK000063 from the Centers for Disease Control and Prevention, Grant SAP #40000012020 from the Pennsylvania Department of Health, and Grant R01LM011370-01A1 from the National Library of Medicine.
Competing interests: None.
Ethics approval: University of Pittsburgh.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Chu D. Clinical feature extraction from emergency department reports for biosurveillance [master's thesis]. Pittsburgh, University of Pittsburgh, 2007 [Google Scholar]
- 2.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004;11:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011:4–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of electronic health record oriented phenotyping algorithms. AMIA Annu Symp Proc 2011:274–83 [PMC free article] [PubMed] [Google Scholar]
- 6.Chute CG, Pathak J, Savova GK, et al. The SHARPn project on secondary use of electronic medical record data: progress, plans, and possibilities. AMIA Annu Symp Proc 2011;2011:248–56 [PMC free article] [PubMed] [Google Scholar]
- 7.Peissig PL, Rasmussen LV, Berg RL, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J Am Med Inform Assoc 2012;19:225–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li DC, Endle CM, Murthy S, et al. Modeling and executing electronic health records driven phenotyping algorithms using the NQF quality data model and JBoss® drools engine. AMIA Annu Symp Proc 2012;2012:532–41 [PMC free article] [PubMed] [Google Scholar]
- 9.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013;20:e147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hripcsak G, Knirsch CA, Jain NL, et al. Automated tuberculosis detection. J Am Med Inform Assoc 1997;4:376–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aronsky D, Haug PJ. Diagnosing community-acquired pneumonia with a Bayesian network. Proc AMIA Symp 1998:632–6 [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman W, Wagner M, Cooper G, et al. Creating a text classifier to detect chest radiograph reports consistent with features of inhalational anthrax. J Am Med Inform Assoc 2003;10:494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kullo IJ, Fan J, Pathak J, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc 2010;17:568–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elkin PL, Froehling DA, Wahner-Roedler DL, et al. Comparison of natural language processing biosurveillance methods for identifying influenza from encounter notes. Ann Intern Med 2012;156:11–18 [DOI] [PubMed] [Google Scholar]
- 16.RODS_Laboratory. Demonstration of Influenza Monitoring System 2013. http://www.youtube.com/watch?v=qOlGbrTsS-A&hd=1
- 17.Wagner MM, Tsui FC, Cooper GF, et al. Probabilistic, decision-theoretic disease surveillance and control. Online J Public Health 2011;3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsui FC, Wagner MM, Cooper GF, et al. Probabilistic case detection for disease surveillance using data in electronic medical records. Online J Public Health 2011;3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsui FC, Espino J, Sriburadej T, et al. Building an automated Bayesian case detection system. 9th Annual Conference of the International Society for Disease Surveillance; Park City, UT: 2010 [Google Scholar]
- 20.Harkema H, Dowling JN, Thornblade T, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009;42:839–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heckerman D. Probabilistic similarity networks. Massachusetts Institute of Technology, 1991 [Google Scholar]
- 22.Cooper GF, Herskovits E. A Bayesian method for the induction of probabilistic networks from data. Mach Learn 1992;9:309–47 [Google Scholar]
- 23.Cooper GF, Hennings-Yeomans P, Visweswaran S, et al. An efficient Bayesian method for predicting clinical outcomes from genome-wide data. AMIA Annu Symp Proc 2010:127–31 [PMC free article] [PubMed] [Google Scholar]
- 24.Druzdzel MJ. SMILE: structural modeling, inference, and learning engine and GeNIe: a development environment for graphical decision-theoretic models (Intelligent Systems Demonstration). In: Proceedings of the Sixteenth National Conference on Artificial Intelligence Menlo Park; CA: AAAI Press/The MIT Press, 1999:902–3 [Google Scholar]
- 25.Domingos P, Pazzani M. On the optimality of the simple Bayesian classifier under zero-one loss. Mach Learn 1997;29:103–30 [Google Scholar]
- 26.Rish I. An empirical study of the naive Bayes classifier. IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence 2001;Vol 3:41–6 [Google Scholar]
- 27.Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol 2004;121:176–86 [DOI] [PubMed] [Google Scholar]
- 28.Jensen FV, Nielsen TD. Bayesian networks and decision graphs. 2nd edn. Springer, 2007 [Google Scholar]
- 29.Efron B, Tibshirani R. An Introduction to the bootstrap. Chapman & Hall/CRC, 1994 [Google Scholar]
- 30.DeLong ER, DeLong DM, Clarke-Pearson D. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837–45 [PubMed] [Google Scholar]
- 31.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman M. The use of ranks to avoid the assumption of normality implicit in the analysis of variance. J Am Statist Assoc 1937;32:675–701 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.