Abstract
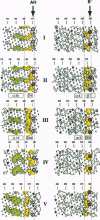
The regions surrounding the catalytic amino acids previously identified in a few "retaining" O-glycosyl hydrolases (EC 3.2.1) have been analyzed by hydrophobic cluster analysis and have been used to define sequence motifs. These motifs have been found in more than 150 glycosyl hydrolase sequences representing at least eight established protein families that act on a large variety of substrates. This allows the localization and the precise role of the catalytic residues (nucleophile and acid catalyst) to be predicted for each of these enzymes, including several lysosomal glycosidases. An identical arrangement of the catalytic nucleophile was also found for S-glycosyl hydrolases (myrosinases; EC 3.2.3.1) for which the acid catalyst is lacking. A (beta/alpha)8 barrel structure has been reported for two of the eight families of proteins that have been grouped. It is suggested that the six other families also share this fold at their catalytic domain. These enzymes illustrate how evolutionary events led to a wide diversification of substrate specificity with a similar disposition of identical catalytic residues onto the same ancestral (beta/alpha)8 barrel structure.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baird S. D., Hefford M. A., Johnson D. A., Sung W. L., Yaguchi M., Seligy V. L. The Glu residue in the conserved Asn-Glu-Pro sequence of two highly divergent endo-beta-1,4-glucanases is essential for enzymatic activity. Biochem Biophys Res Commun. 1990 Jun 29;169(3):1035–1039. doi: 10.1016/0006-291x(90)91998-8. [DOI] [PubMed] [Google Scholar]
- Chen L., Fincher G. B., Høj P. B. Evolution of polysaccharide hydrolase substrate specificity. Catalytic amino acids are conserved in barley 1,3-1,4- and 1,3-beta-glucanases. J Biol Chem. 1993 Jun 25;268(18):13318–13326. [PubMed] [Google Scholar]
- Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992 Jun 18;357(6379):543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- Cupples C. G., Miller J. H., Huber R. E. Determination of the roles of Glu-461 in beta-galactosidase (Escherichia coli) using site-specific mutagenesis. J Biol Chem. 1990 Apr 5;265(10):5512–5518. [PubMed] [Google Scholar]
- Davies G. J., Dodson G. G., Hubbard R. E., Tolley S. P., Dauter Z., Wilson K. S., Hjort C., Mikkelsen J. M., Rasmussen G., Schülein M. Structure and function of endoglucanase V. Nature. 1993 Sep 23;365(6444):362–364. doi: 10.1038/365362a0. [DOI] [PubMed] [Google Scholar]
- Derewenda U., Swenson L., Green R., Wei Y., Morosoli R., Shareck F., Kluepfel D., Derewenda Z. S. Crystal structure, at 2.6-A resolution, of the Streptomyces lividans xylanase A, a member of the F family of beta-1,4-D-glycanases. J Biol Chem. 1994 Aug 19;269(33):20811–20814. [PubMed] [Google Scholar]
- Divne C., Ståhlberg J., Reinikainen T., Ruohonen L., Pettersson G., Knowles J. K., Teeri T. T., Jones T. A. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science. 1994 Jul 22;265(5171):524–528. doi: 10.1126/science.8036495. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C., Bissery V., Benchetrit T., Mornon J. P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987 Nov 16;224(1):149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräbnitz F., Seiss M., Rücknagel K. P., Staudenbauer W. L. Structure of the beta-glucosidase gene bglA of Clostridium thermocellum. Sequence analysis reveals a superfamily of cellulases and beta-glycosidases including human lactase/phlorizin hydrolase. Eur J Biochem. 1991 Sep 1;200(2):301–309. doi: 10.1111/j.1432-1033.1991.tb16186.x. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991 Dec 1;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B., Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993 Aug 1;293(Pt 3):781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. Structural similarity of plant chitinase and lysozymes from animals and phage. An evolutionary connection. FEBS Lett. 1994 Feb 28;340(1-2):129–132. doi: 10.1016/0014-5793(94)80187-8. [DOI] [PubMed] [Google Scholar]
- Imai K., Nakamura M., Yamada M., Asano A., Yokoyama S., Tsuji S., Ginns E. I. A novel transcript from a pseudogene for human glucocerebrosidase in non-Gaucher disease cells. Gene. 1993 Dec 22;136(1-2):365–368. doi: 10.1016/0378-1119(93)90497-q. [DOI] [PubMed] [Google Scholar]
- Jacobson R. H., Zhang X. J., DuBose R. F., Matthews B. W. Three-dimensional structure of beta-galactosidase from E. coli. Nature. 1994 Jun 30;369(6483):761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- Jenkins J., Lo Leggio L., Harris G., Pickersgill R. Beta-glucosidase, beta-galactosidase, family A cellulases, family F xylanases and two barley glycanases form a superfamily of enzymes with 8-fold beta/alpha architecture and with two conserved glutamates near the carboxy-terminal ends of beta-strands four and seven. FEBS Lett. 1995 Apr 10;362(3):281–285. doi: 10.1016/0014-5793(95)00252-5. [DOI] [PubMed] [Google Scholar]
- Laine R. A. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 x 10(12) structures for a reducing hexasaccharide: the Isomer Barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology. 1994 Dec;4(6):759–767. doi: 10.1093/glycob/4.6.759. [DOI] [PubMed] [Google Scholar]
- Lemesle-Varloot L., Henrissat B., Gaboriaud C., Bissery V., Morgat A., Mornon J. P. Hydrophobic cluster analysis: procedures to derive structural and functional information from 2-D-representation of protein sequences. Biochimie. 1990 Aug;72(8):555–574. doi: 10.1016/0300-9084(90)90120-6. [DOI] [PubMed] [Google Scholar]
- MacLeod A. M., Lindhorst T., Withers S. G., Warren R. A. The acid/base catalyst in the exoglucanase/xylanase from Cellulomonas fimi is glutamic acid 127: evidence from detailed kinetic studies of mutants. Biochemistry. 1994 May 24;33(20):6371–6376. doi: 10.1021/bi00186a042. [DOI] [PubMed] [Google Scholar]
- McCarter J. D., Withers S. G. Mechanisms of enzymatic glycoside hydrolysis. Curr Opin Struct Biol. 1994 Dec;4(6):885–892. doi: 10.1016/0959-440x(94)90271-2. [DOI] [PubMed] [Google Scholar]
- Neufeld E. F. Lysosomal storage diseases. Annu Rev Biochem. 1991;60:257–280. doi: 10.1146/annurev.bi.60.070191.001353. [DOI] [PubMed] [Google Scholar]
- Orengo C. A., Jones D. T., Thornton J. M. Protein superfamilies and domain superfolds. Nature. 1994 Dec 15;372(6507):631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- Py B., Bortoli-German I., Haiech J., Chippaux M., Barras F. Cellulase EGZ of Erwinia chrysanthemi: structural organization and importance of His98 and Glu133 residues for catalysis. Protein Eng. 1991 Feb;4(3):325–333. doi: 10.1093/protein/4.3.325. [DOI] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Varghese J. N., Garrett T. P., Colman P. M., Chen L., Høj P. B., Fincher G. B. Three-dimensional structures of two plant beta-glucan endohydrolases with distinct substrate specificities. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2785–2789. doi: 10.1073/pnas.91.7.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Tull D., Meinke A., Gilkes N. R., Warren R. A., Aebersold R., Withers S. G. Glu280 is the nucleophile in the active site of Clostridium thermocellum CelC, a family A endo-beta-1,4-glucanase. J Biol Chem. 1993 Jul 5;268(19):14096–14102. [PubMed] [Google Scholar]
- White A., Withers S. G., Gilkes N. R., Rose D. R. Crystal structure of the catalytic domain of the beta-1,4-glycanase cex from Cellulomonas fimi. Biochemistry. 1994 Oct 25;33(42):12546–12552. doi: 10.1021/bi00208a003. [DOI] [PubMed] [Google Scholar]
- Woodcock S., Mornon J. P., Henrissat B. Detection of secondary structure elements in proteins by hydrophobic cluster analysis. Protein Eng. 1992 Oct;5(7):629–635. doi: 10.1093/protein/5.7.629. [DOI] [PubMed] [Google Scholar]