Abstract
Both apoptosis and autophagy are highly conserved processes that besides their role in the maintenance of the organismal and cellular homeostasis serve as a main target of tumor therapeutics. Although their important roles in the modulation of tumor therapeutic strategies have been widely reported, the molecular actions of both apoptosis and autophagy are counteracted by cancer protective mechanisms. While apoptosis is a tightly regulated process that is implicated in the removal of damaged or unwanted cells, autophagy is a cellular catabolic pathway that is involved in lysosomal degradation and recycling of proteins and organelles, and thereby is considered an important survival/protective mechanism for cancer cells in response to metabolic stress or chemotherapy. Although the relationship between autophagy and cell death is very complicated and has not been characterized in detail, the molecular mechanisms that control this relationship are considered to be a relevant target for the development of a therapeutic strategy for tumor treatment. In this review, we focus on the molecular mechanisms of apoptosis, autophagy, and those of the crosstalk between apoptosis and autophagy in order to provide insight into the molecular mechanisms that may be essential for the balance between cell survival and death as well as their role as targets for the development of novel therapeutic approaches.
Keywords: apoptosis, autophagy, crosstalk, cancer, therapy
Introduction
Both apoptosis and autophagy are highly conserved processes that in addition to their role in maintenance of organismal and cellular homeostasis are considered relevant therapeutic targets for tumor treatment.1–9 Although apoptosis and autophagy play an important role in the modulation of tumor therapeutics, their molecular action can be counteracted by cancer protective mechanisms.10–12
Apoptosis is a tightly regulated set of cellular events which are associated with biochemical and morphological changes. However, these characteristic changes of apoptosis present a set of potential targets for cell death and thereby are considered a prognostic marker for early assessment of the effi-ciency of anticancer agents. Thus, in vivo, apoptosis has been reported to be a relevant detector for tumor resistance and response during the course of the treatment with chemotherapeutics.13 Autophagy, which is known to be a catabolic pathway, can be activated by cellular stress in response to environmental changes, chemical agents, starvation, or infection.14 However, the promotion of autophagic process, in response to extra- and intracellular stress, leads to the regulation of different mechanisms that in turn mediate cellular events such as cellular adaptation, cell survival, and cell death.15,16 Therefore, understanding the molecular mechanisms implicated in the regulation of either cell survival or cell death will help to identify a relevant target for tumor prevention and treatment. As a tumor suppressive mechanism, autophagy has been discussed in several studies, in which the distribution of autophagic machinery was found to trigger both cellular transformation and tumor progression.17,18 Additionally, pro- and anti-tumor functions of autophagy have been reported. In addition to its tumor suppressive actions, autophagy can enhance tumor progression of tumor cells exhibiting radio- and chemotherapeutic resistance.19 The resistance of cancer cells to available therapeutics is thought to be mediated mainly via the activation of autophagy-dependent pathways or via the suppression of apoptosis-dependent pathways.2 Thus, functional analysis of the mechanisms of tumor resistance to apoptosis in the context of developing novel cancer therapeutics is thought to be an enormous challenge for researchers and clinicians. Additionally, understanding the molecular mechanisms of the crosstalk between apoptosis and autophagy will help to develop new drugs based on their ability to function as BH3 mimetics, a strategy to trigger autophagy-associated cell death in cells conferring resistance to apoptosis.20–23 While the suppression of apoptosis is linked to the induction of autophagy,2,24 the inhibition of autophagy can cause apoptosis.2,25,26 Thus, identifying the mechanisms thought to be involved in the regulation of the crosstalk between apoptosis- and autophagy-associated cell death is considered an important step for the development of optimal chemotherapeutic approaches for tumor treatment.
Apoptosis
Apoptosis is a form of cell death, also referred to as programmed cell death, in which a ‘suicide’ machinery is activated within the cell, leading to the fragmentation of DNA, shrinkage of the cytoplasm, membrane changes, and finally to cell death without any lysis or damage to neighboring cells.27 Thus, apoptosis is a normal phenomenon occurring frequently in a multicellular organism, thereby playing an essential role in organism survival. Apoptosis is considered a compelling aspect of various cellular processes. The regulation of apoptosis is mediated by an intracellular proteolytic cascade.1,2,28,29 The mechanisms of this cascade are similar in all animal cells30,31 and depend on a family of proteases that possess a cysteine at their active site with the ability to cleave their target proteins at specific aspartic acids.32–36 Accordingly, their target proteins are referred as caspases, which are synthesized in the cell in an inactive form (pro-caspases); their activation is mediated by the cleavage of aspartic acids by other caspases. This downstream activation pathway amplifies the intracellular proteolytic cascade. Some of these activated caspases can cleave other key proteins in the cell, such as the nuclear lamins that mediate irreversible breakdown of the nuclear lamina.28,29 Other caspases have been shown to cleave proteins that functionally govern the DNA-degrading enzyme, DNAase, in an inactive form, a cytoprotective mechanism to avoid DNA damage from freeing DNAase and subsequently prevent the initiation of apoptosis.37–39
Mechanisms of apoptosis
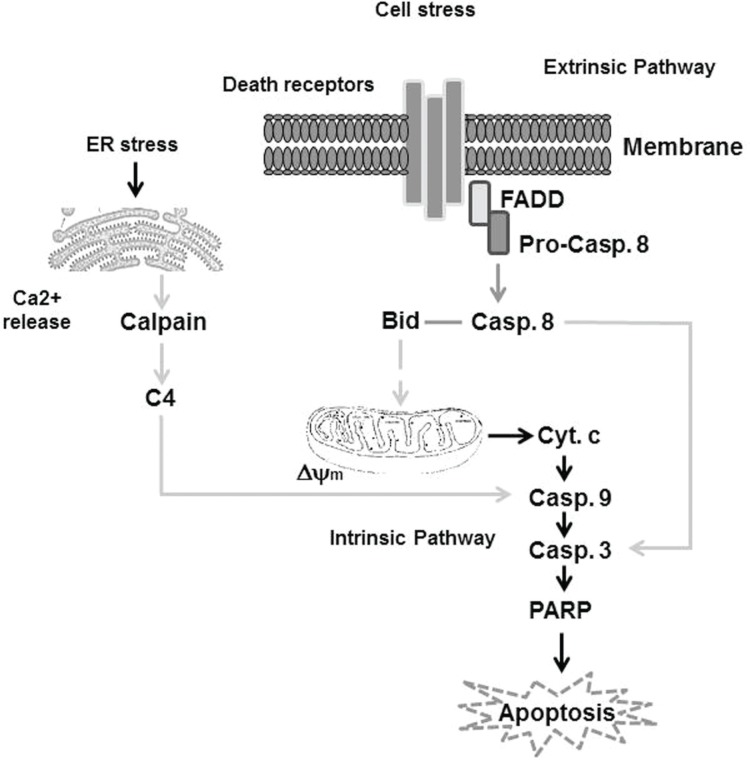
The pathways, which are involved in the regulation of apoptosis, are extremely complicated and appear to be both agent-dependent and tissues-specific.40–42 Thus, based on the inducer and the effector, the apoptotic pathways are either extrinsic or intrinsic pathways.43–45 Besides the major apoptotic pathways, namely, a third apoptotic pathway has been reported. This pathway is specific to cytotoxic T lymphocytes and natural killer (NK) cells and is referred as the perforin/granzyme-A or B pathway.46,47 This perforin pathway is caspase-independent and is mediated through single-stranded DNA damage.48 Although the variation of their induction manner is great, both extrinsic and intrinsic pathways converge by apoptosis through the activation of caspase-3/7.49,50 An outline demonstrates the molecular mechanisms, which are responsible for the regulation of both extrinsic and intrinsic pathways of apoptosis, are shown (Fig. 1).
Figure 1.
Simplified outline showing key proteins within the extrinsic and intrinsic apoptotic pathways in response to extra- and intracellular stress. Generally, death domain-containing receptors including CD95 (APO-1/Fas) can be activated in response to external signals (such as Fas ligand), which triggers the activity of the extrinsic apoptosis pathway through the Fas-associated death domain (FADD). This pathway is mediated by recruitment and activation of caspase-8 (Pro-casp 8), an initiator caspase, in the death-inducing signaling complex followed by direct cleavage of downstream effector caspases such as caspase-3 (casp 3). Thus, casp 8 is an important pro-apoptotic protein for the extrinsic apoptotic pathway. Initiation of the intrinsic apoptosis pathway results from intracellular stress, leading to mitochondrial damage that is characterized by the loss of mitochondrial membrane potential (Δψm) and cytochrome c (cyt c) release. This triggers activation of initiator caspase-9 (casp 9) and ultimately results in activation of effector caspases, such as caspase 3 (casp 3), that mediates the cleavage of poly(ADP-ribose) polymerase (PARP) and finally leads to apoptosis. Additionally, endoplasmic reticulum stress that can be mediated by oxidative stress elevates intracellular calcium (Ca2+) release that subsequently mediates calpain cleavage, leading to the activation of caspase-4 (casp 4), which mediates the cleavage of casp 9, casp 3, and PARP, and finally leads to apoptosis.
Intrinsic pathway
The intrinsic pathway is a non-receptor mediated pathway and functions only via mitochondria associated mechanisms.51 This pathway mediates the intracellular signals that act directly on targets, within the cell, or on those associated with the mitochondrial dysregulation.52 The activation of the intrinsic pathway can act in two opposing patterns.53,54 One of these patterns is mediated by the suppression of the inhibitors of death programs such as growth factors, hormones, and cytokines. However, the other pattern is mediated by direct exposure to several environmental changes such as irradiation, toxins, hypoxia, and viral infections.54,55 In contrast, apoptosis mediated by the intrinsic pathway is characterized by alterations of the inner mitochondrial membrane, that in turn leads to the loss of mitochondrial membrane potential (Δψm).1–4,54 The loss of Δψm leads to the release of the pro-apoptotic proteins, which are characteristic of mitochondrial damage; these molecules include cytochrome c (cyt c), Smac/DIABLO, and the serine protease HtrA2/Omi56,57 apoptosis inducing factors (AIF) endonuclease G and caspase-activated deoxyribonuclease (CAD).54 However, the release of these proteins leads to the activation of caspases such as caspase-9 and caspase-3, whose activation is associated with the loss of Δψm;1,2 the mechanism by which the released cyt c mediates apoptosis is regulated by its binding to both Apaf-1 and pro-caspase-9 to form a protein complex known as the apoptosome.58,59 Through apoptosome formation, caspase-9 becomes active to subsequently mediate the activation of caspase-3, leading to PARP cleavage and finally to apoptosis. The mechanism by which Smac/DIABLO and HtrA2/Omi triggers apoptosis is mediated by the inhibition of inhibitor of apoptosis (IAP) activity.60–62
However, the appearance of AIF, endonuclease G, and CAD proteins is associated with late apoptosis after the cell has decided to die.54 In this case, translocation of AIF to the nucleus leads to DNA fragmentation and subsequently to peripheral chromatin condensation,54,63 a process referred to as “stage I” condensation.64 Additionally, translocation of endonuclease G to the nucleus results in nuclear chromatin cleavage to produce oligonucleosomal DNA fragments.65 In addition, after release from the mitochondria and subsequent cleavage by caspase-3, CAD can translocate into the nucleus where it mediates the fragmentation of the oligonucleosomal DNA,66 a process known as “stage II” condensation.54,66 The control and regulation of mitochondrial dysregulation occur through Bcl-2 family of proteins, which are known to be responsible for mitochondrial integrity and subsequent caspase activation, which in turn leads to apoptotic cell death.67 In this context, the tumor suppressor protein p53 has been widely reported to play a critical role in the regulation of Bcl-2 proteins.68 Bcl-2 proteins include both pro-apoptotic and anti-apoptotic mediators.69 These anti-apoptotic mediators include Bcl-2, Bcl-x, Bcl-XL, Bcl-XS, Bcl-w, BAG, Mcl-1, whereas the pro-apoptotic mediators include Bcl-10, Bax, Bak, Bid, Bad, Bim, Bik, and Blk. Thus, the cellular decision to live or to die is controlled by the balance between both pro- and anti-apoptotic mediators, which can regulate the mitochondrial outer membrane permeabilization (MOMP) and subsequently the regulation of cyt c release from the mitochondria into the cytoplasm.49
The role of Bax and Bak in the regulation of mitochondrial damage has been documented in several studies.1,3 Although the molecular mechanism of both BAK and BAX activation is not well-characterized, the role of Bax has been reported.69–73 However, phosphorylation of both BAK and BAX serves to facilitate their homo-oligomerization and subsequently their localization into mitochondria, leading cyt c release.74,75 However, the involvement of caspase-8 in the activation of Bid-induced activation of Bax as observed in Fas-induced apoptosis76,77 provides an evidence for the “crosstalk” between the death-receptor (extrinsic) pathway and the mitochondrial (intrinsic) pathway.4,50,76 Additionally, the role of Bad in the promotion of mitochondrial dysregulation has been reported. Thus, phosphorylation of Bad on the serine residue enhances the ability of the 14-3-3 protein, a member of a family of multifunctional phosphoserine binding molecules, to capture Bad in the cytosol where it is sequestered.78 However, dephosphorylation of Bad or even its native form can translocate into the mitochondria to cause cyt c release.78 Heterodimerization of Bad with Bcl-Xl or Bcl-2 can lead to enhanced apoptosis by a mechanism that includes neutralizing of the protective effect of both Bcl-Xl or Bcl-2.79 The main function of both Bcl-2 and Bcl-Xl is to inhibit the release of cyt c from the mitochondria and protect the mitochondria from the localization of pro-apoptotic mediators is not well-characterized. However, the mechanism by which both Bcl-2 and Bcl-XL mediate the inhibition of apoptosis is regulated by the activation of caspase proteases.80,81
Both Puma and Noxa are two members of the Bcl-2 family, which are also involved in the promotion of apoptosis in different cell types including melanoma cells.2,82 Puma plays an important role in p53- mediated apoptosis, and its overexpression, in vitro, is correlated with increased expression, conformational change, and translocation of Bax to the mitochondria, that in turn, lead to the loss of Δψm and subsequently the release of cyt c.2,4,82,83 Noxa has also been reported to be a mediator of p53-induced apoptosis and has been shown to localize to both mitochondria and the endoplasmic reticulum (ER);82 its interaction with anti-apoptotic Bcl-2 family members mediates caspase-9 activation.84 Additionally, the regulation of both Puma and Noxa expression by p53 suggests a potential role for both proteins in modulating apoptosis associated with DNA damage.49 Moreover, the potential role of the proto-oncoprotein Myc in modulating apoptosis via p53-dependent and independent mechanisms has been reported.85 Therefore, further elucidation of these pathways should provide new insight into the course of tumorigenesis and tumor therapy.
Extrinsic pathway
The extrinsic pathway is one of the two major apoptotic pathways whose activation is initiated by transmembrane receptor(s) through the ligation to the corresponding ligand(s) or agonist(s) of interest. These receptors include FasL/FasR, TNF-α/TNFR1, Apo3L/DR3, Apo2L/DR4, and Apo2L/DR5.86–91 However, one of the most well-characterized receptors includes the member of the tumor necrosis factor (TNF) receptor gene superfamily.87 These family members share similar cysteine-rich extracellular domains in addition to a cytoplasmic death domain.87 The main function of the death domain is to transmit the external death signal from the cell’s surface to the intracellular signaling pathways. Following binding of the ligand to the corresponding receptor, cytoplasmic adapter proteins can exhibit the corresponding death domains. Ligation of Fas ligand to the Fas receptor mediates its binding to FAS-associated death domain (FADD), whereas ligation of the TNF ligand to the corresponding TNF receptor mediates its binding to TNF-receptor associated death domain (TRADD), which in turn leads to recruitment of FADD and receptor-interacting protein (RIP).92,93 The interaction of FADD with pro-caspase-8 through its death effector domain leads to formation of a death-inducing signaling complex (DISC) that mediates the activation of caspase-8.94 Following caspase-8 activation, death receptor-mediated apoptosis can be initiated.
Execution phase
Both extrinsic and intrinsic pathways lead to the final pathway of apoptosis.49 The execution phase includes activation of execution caspases, which are characteristic of the final phase of apoptosis that is associated with the activation of cytoplasmic endonucleases and proteases. This leads to the degradation of nuclear materials and cytoskeletal proteins.
Both morphological and biochemical changes characteristic of apoptotic cells are mediated by effector “executioner” caspases such as caspase-3, caspase-6, and caspase-7. Activation of these caspases results in cleavage of various substrates such as cytokeratins, PARP, plasma membrane cytoskeletal protein alpha fodrin, and nuclear mitotic apparatus protein (NuMA).95
Caspase-3 is known to be the most important caspase among the executioner caspases and its activation is mediated by initiator caspases such as caspase-8, caspase- 9, or caspase-10.77 In apoptotic cells, activation of caspase-3 mediates activation of various substrates such as endonuclease caspase-activated DNase (CAD), whose activation promotes protein degradation that subsequently causes chromatin condensation in apoptotic cells.96 Additionally, activation of keratin 18 by caspase-3 mediates formation of apoptotic bodies during the course of apoptosis.97 These reports, however, describe the mechanisms responsible for caspase-3-mediated disruption of the cytoskeleton during apoptosis. Moreover, caspase-cleaved gelsolin functions as actin filaments in vitro in a Ca2+-independent manner.98 Expression of the gelsolin cleavage product in multiple cell types triggers the cells to become round, detach from the plate, and undergo nuclear fragmentation,99 suggesting an important role for activated caspase-3-induced cleavage of gelsolin in apoptosis regulation.
After the execution phase is complete, unwanted cellular components should be removed. Therefore, the last step of apoptotic process involves phagocytic uptake of apoptotic cells.49 Phospholipid asymmetry and externalization of phosphatidylserine on the surface of apoptotic cells and membrane fragmentation are characteristic of the execution phase.49 Although the mechanisms responsible for regulating phosphatidylserine translocation to the outer leaflet of the cell has not been described in detail, translocation of phosphatidylserine to the outer side is thought to be mediated by the loss of aminophospholipid translocase activity and nonspecific flip-flop of phospholipids of various classes.100 Accordingly, the roles of Fas, caspase-8, and caspase-3 in regulating phosphatidylserine externalization has been demonstrated.101,102 Although caspase-independent phosphatidylserine exposure was reported to occur during apoptosis of primary T-lymphocytes,101 phosphatidylserine externalization in response to oxidative stress has been reported in erythrocytes.102 Moreover, externalization of phosphatidylserine in the execution phase appears to be an essential cellular mechanism for removing apoptotic cells since the appearance of phosphotidylserine on the outer leaflet of apoptotic cells may facilitate non-inflammatory phagocytic recognition, thereby allowing their early uptake and finally their removal.103
Autophagy
Autophagy is a highly conserved degradation pathway that discards damaged cellular components and is morphologically characterized by the formation of double membrane autophagosomes.2,51 Sequestration of impaired organelles or unwanted cellular components by autophagy facilitates their delivery to lysosomes for degradation and recycling.104–106 In addition to eliminating damaged cellular components, autophagy has been implicated in several physiological and pathological processes, including cell survival and cell death.107 Additionally, the involvement of autophagy in cellular homeostasis and cell and tissue renovation has been reported.108–111 More importantly, the participation of autophagy in the etiopathogenesis of many important human diseases, including metabolic disorders and cancer, has been reported.112 Autophagy has been demonstrated to function as an important cell survival mechanism, particularly when cells are under stress conditions such as oxidative stress and starvation.113–115 Additionally, the involvement of autophagy in regulating cell death processes, in apoptosis,2,116,117 necrotic cell death,118,119 or even autophagic cell death, has been reported.120–122 Thus, the function of autophagy as cell death machinery in different tissue types appears to be established. However, the factors responsible for regulating this mechanism are not well-understood. Although the natural and cellular stress-induced autophagy is crucial processes, the factors responsible in determining whether autophagy mediates cell survival or cell death remain unknown.
Mechanisms of autophagy
Despite its role in maintaining intracellular integrity,123 autophagy is a tightly regulated process that is regulated by distinct cellular mechanisms. One of these mechanisms is the target of rapamycin (TOR) kinase that serves as a control point downstream of signaling responsible for different cellular events such cell growth, metabolism such as growth factor receptor signaling, and insulin signaling.124,125 It was previously shown that activation of TOR kinase is mediated by Akt kinase, PI3-kinase, and growth factor receptor signaling in response to the availability of nutrients to promote growth via induction of ribosomal protein at both transcriptional and translational levels.126 Thus, under cell growth conditions, the TOR pathway can mediate the inhibition of autophagy by suppressing Atg1 kinase activity.127 Additionally, under low levels of adenosine-5′-monophosphate (AMP) or under hypoxia conditions, TOR kinase can be repressed, which blocks the activity of the tumor suppressor proteinsTsc1/Tsc2 as mediated via Rheb, a small GTase required for mTOR activity.126,128 Reduction of Akt activity through inhibition of growth factor receptor or by rapamycin are thought to be possible mechanisms, which are responsible for the repression of TOR kinase.129 Therefore, inhibiting the TOR kinase that is mainly involved in regulating autophagy in response to altered physiological conditions is mediated by the inhibition of growth factor receptors or by inhibitors, leading to increased catabolism, which is considered to be a therapeutic strategy for tumor treatment.
Accordingly, rapamycin and its derivatives, which are known to inhibit mTOR function through a kinase-independent mechanism, have been tested in clinical trials in several tumor types.130,133 However, the action of this inhibitor is mediated by a kinase-independent mechanism that inhibits tumor growth via suppression of protein translation machinery and induction of autophagy.130 In addition to its role in regulating tumor progression and autophagy, TOR has been reported to function as the catalytic component of two distinct complexes, including TORC1 and TORC2.131 Hypoxia can also activate autophagy by hypoxia inducing factor (HIF)-dependent132 or independent mechanisms. However, these mechanisms are regulated via the inhibition of TOR mediated by adenosine monophosphate kinase (AMPK), REDD1, and Tsc1/Tsc2.129,133 Moreover, the most specific targets of autophagy include BNIP3 and BNIP3L; the BNIP3 subfamily of BH3-only proteins can form stable homodimerization complexes that localize to the outer membrane of the mitochondria after cellular stress.134,135 Although these proteins are linked to cell death, their main function is implicated in the regulation of autophagy.134,135 Thus, the main role of BNIP3L/NIX in autophagy is to mediate mitochondrial clearance during the maturation of reticulocytes.136–139 Although the functional role of BNIP3 and BNIP3L in regulating both autophagy and cell death is not well-characterized in detail, various models have been proposed to explain the functional role of BNIP3/BNIP3L in autophagy.140 One of these models describes the role of BNIP3 in deregulation of Beclin-1 by disrupting its interaction with Bcl-2,141 whereas other models of the interaction between BNIP3 and Rheb have been discussed in the context of hypoxia-induced autophagy.142
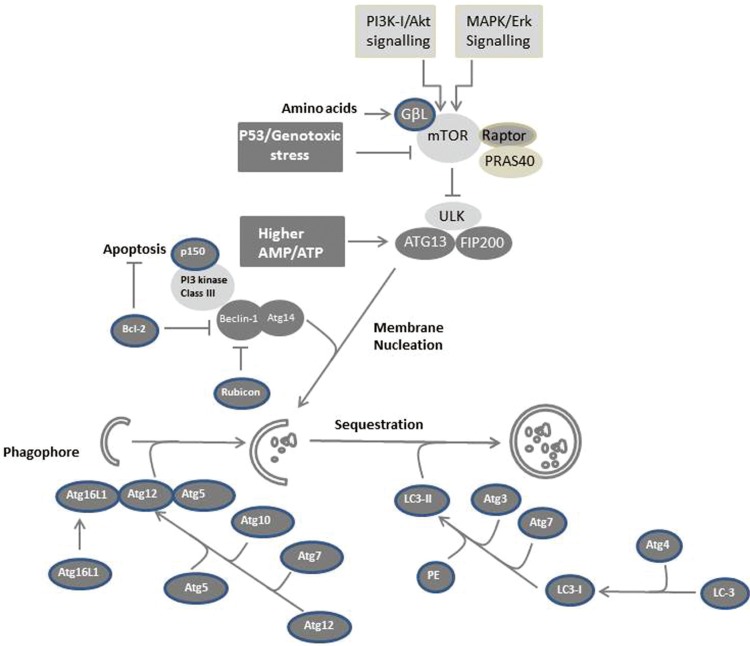
Besides its role in death and cell survival, autophagy is also involved in regulating cell cycle arrest. However, the mechanism responsible for modulating autophagy-induced cell cycle arrest is thought to be regulated by inhibition of the TOR pathway in response to nutrient deprivation by inhibiting the translation machinery of key cell cycle genes, such as cyclin D1.143 The possible mechanisms, which are responsible for the regulation of autophagy in mammalian cells, are outlined in Figure 2.
Figure 2.
Possible mechanisms of autophagy in mammalian cells. The formation of autophagosomes is mediated by a conserved pathway in all mammalian cells in response to variable cellular stresses. Cellular stress that results from the availability of nutrients (starvation) or from the exposure to growth factors such as endothelial growth factor (EGF) lead to activation of PI3K-I/Akt and MAPK/ERK signaling, respectively. ULK-Atg13-FIP200 complexes mediate mTOR signaling downstream of the autophagy machinery. Starvation or growth factor treatment suppresses mTOR-mediated phosphorylation of ULK and Atg13 resulting in ULK-mediated phosphorylations of Atg13, FIP200, and ULK itself, and subsequently to membrane nucleation. Downstream of the TOR-suppressive signaling pathway, Atg proteins function to form the autophagosome. The PI3 kinase complex (class III) including Beclin-1/Atg6 is required to initiate autophagosome formation. Elongation of the isolation membrane requires Atg12 and Atg8 (LC3) ubiquitin-like modification systems. A ubiquitin-like protein, Atg12, is conjugated with Atg5 and forms a large complex with Atg16L1. LC3, a ubiquitin-like protein, associates with both the isolation membrane and the completed autophagosome as a conjugate with phosphatidylethanolamine.
Crosstalk Between Apoptosis and Autophagy
Based on their central role in cell survival and cell death, mitochondria have been shown to play an essential role in the regulation of apoptosis in response to extensive cellular stress leading to the loss of Δψm, and to the subsequent release of pro-apoptotic molecules such as cyt c,2–4 and AIF.1 The accumulation of both cyt c and AIF in the cytoplasm results in the activation of caspases and, finally, apoptotic cell death.1,2 However, if the loss of Δψm is limited to only a subset of mitochondria, selective autophagosomal machinery will eliminate the limited subset of depolarized mitochondria, a cellular function that serves as cytoprotective mechanism to prevent mitochondrial damage.144
Apart from their molecular authenticity and induction manner, the involvement of several proteins that regulate both apoptosis and autophagy has been reported.145 Accordingly, the anti-apoptotic protein Bcl-2 has been shown to play an essential role in regulating both autophagy and apoptosis as mediated by the binding of the pro-autophagic protein Beclin1 with pro-apoptotic proteins such as Bax. Thus, extensive cellular stress can lead to the loss of Δψm that mainly results in the release of mitochondrial proteins such as cyt c and AIF and subsequently initiates apoptosis.1,146,147 Additionally, the depletion of nutrients leads first to the release of Beclin1, that in turn induces activation of PI3K and subsequently the induction of autophagy.148,149 The extension of nutrient deprivation can lead to release of the pro-apoptotic protein Bax from Bcl-2, which mediates the loss of Δψm and subsequently initiates the apoptotic machinary.116,150 Thus, the molecular coupling of both autophagy and apoptosis is determined by the type and longevity of cellular stress.151,152 Additionally, the pattern and fashion of the subcellular localizations of Bcl-2 at the ER and/or to mitochondria is thought to be the main factor that determines the switch between autophagy and apoptosis.153,154 However, the role of Beclin-1 in regulating autophagy is mediated by the by localization of Bcl-2 at the ER.155,156 The regulation of apoptosis can be mediated through the localization of Bcl-2 to mitochondria.157 Accordingly, the cellular outcome between autophagy or apoptosis appears to depend on the entity of the mitochondria rather than the molecular mechanism induced.1,2 Therefore, mitochondrial damage can overpower the pro-survival autophagic pathway, leading to apoptosis via a mechanism mediated by the release of pro-apoptotic proteins such as Bax from anti-apoptotic proteins such as Bcl-2. Moreover, the involvement of a number of proteins in regulating the crosstalk between autophagy and apoptosis has been reported. These include the autophagy-regulating proteins Beclin1, the class III PI3K, and ATG4D.158–160 Thus, the cleavage of these proteins by caspases facilitates their localization to mitochondria where they serve new functions such as promoting mitochondria-mediated apoptosis. Therefore, the destruction of autophagic function of Beclin-1 and PI3K through caspase-dependent cleavage is considered to be an alternative strategy for tumor treatment. The autophagy-regulating protein 5 (ATG5) is a member of autophagy-regulating protein family. This protein is characterized by its ability to link apoptosis and autophagy.161 Upon autophagy induction, ATG5 becomes active and subsequently initiates autophagosomic formation.161 Additionally, cleavage of ATG5 by calpain in response to cellular stress promotes localization of cleaved ATG5 products to the mitochondria, where they bind to Bcl-XL, leading to the loss of Δψm, and finally to apoptosis.162–164 Thus, unlike cleaved Beclin1, class III PI3K, or ATG4D, cleaved ATG5 appears to functionally trigger apoptosis without any additional apoptotic mediators.
Although the functional role of Beclin-1 as Bcl-2-interacting protein, in addition to be an initiator factor for the development of autophagic formation,165 its precise role in regulating the crosstalk between autophagy and apoptosis has not completely characterized. The anti-apoptotic protein Bcl-2 is the most thoroughly described prototypic member of the Bcl-2 family of apoptosis-regulating proteins,165 in addition to being characteristic for their Bcl-2 homology (BH) domains.165 Thus, based on the number of functions of these BH domains, three groups are recognized. One of these groups includes the multi-domain anti-apoptotic Bcl-2 family proteins such as Bcl-2, Bcl-xL, Bcl-w, A1, and Mcl-1, the second one includes the multi-domain pro-apoptotic Bcl-2 family proteins such as Bax, Bak, and Bok, and the third group includes pro-apoptotic Bcl-2 family members that possess only the BH3 domain and are therefore referred to as BH3-only proteins such as Bim, Bid, Bad, Bmf, Puma, Noxa, Bik, Hrk, and Mule. The main function of these multidomain pro-apoptotic Bcl-2 family members (Bax and Bak) is to induce the loss of Δψm that subsequently leads to the release of cyt c and, in turn, the activation of downstream caspases such as caspase-3 and 9.1–3,76,82 Anti-apoptotic Bcl-2 family members serve mainly to inhibit pro-apoptotic proteins, such as Bax and Bak.166 Despite the similarity of their pattern and their function, the manner and function of the BH3-only proteins in modulating their anti-apoptotic function is different. Some BH3-only proteins exert their function by their binding to the anti-apoptotic Bcl-2 family members, and thereby block their function as repressor for the multi-domain of the pro-apoptotic Bcl-2 proteins.167,168 Otherwise, these BH3-only proteins exert their function by the direct interaction with the multi-domain of pro-apoptotic proteins and thereby block their pro-apoptotic function.169,170
In addition its function as interacting partner of Bcl-2, Beclin-1 has been reported to play an important role in antiviral host defense in some studies,165 whereas in another study it was shown to function as a tumor suppressor protein.171 In other studies, Beclin-1 has been reported to function as an ortholog of the yeast autophagy protein Atg6 and thereby is implicated in the regulation of autophagic formation in mammalian cells.172 Accordingly, the expression of Beclin-1 was found to be essential for promoting autophagic formation in Atg6-deficient yeast.172 Additionally, reduction of the tumorigenicity of Beclin-1-expressing cells confirmed further the antitumor function of Beclin-1.172 Functional analysis of Beclin-1 suggested a molecular mechanism whereby Beclin-1 initiates autophagy.173,174 These mechanisms appear to be mediated through the interaction of Atg6/Beclin-1 with the phosphoinositide 3-kinase (PI3K).173,174
However, the identification of Beclin-1 as a Bcl-2 binding partner revealed that the interaction between Bcl-2 and Beclin-1 is involved in the formation of a complex that is thought to play a central role in modulating the crosstalk between signaling pathways of apoptosis and autophagy.175,176 As with other BH3-only proteins, Beclin-1 was shown to interact with Bcl-2, Bcl-xL, and Mcl-1 through the BH3 domain; evidence for this is the inhibition of the observed interaction in response to mutations in BH3 domain or even in the receptor domain in anti-apoptotic Bcl-2 family members.175–176
More importantly, the interaction between Beclin-1 and the BH3 domain of anti-apoptotic Bcl-2 family members is thought to be the primary mechanism responsible for the inhibition of Beclin-1-mediated induction of autophagy under nutrient-adequate conditions.177,178 Although the Bcl-2, Bcl-xL, and Mcl-1 proteins are mainly localized to the mitochondria as a protective mechanism for mitochondria, the localization of Bcl-2 family members to the ER has been shown to inhibit starvation-induced autophagy.177,178 Although extensive studies on the role of Bcl-2 family members have examined inhibition of the autophagic function of Beclin-1, the precise role of Beclin-1 in the processes of autophagic formation is not fully understood. However, the identification of the novel Bcl-2 interacting partner, nutrient-deprivation autophagy factor-1 (NAF-1),179 provided insight into the mechanism of the interaction of Bcl-2 with Beclin-1. This provided insight into the role of Beclin-1 in the process of autophagic formation. Accordingly, the function of NAF-1 appears to be implicated in the stabilization of the Bcl-2-Beclin-1 complex at the ER;179 therefore, the loss of NAF-1 is responsible for disrupting the Bcl-2-Beclin-1 complex and subsequently inducing autophagy, which is evidence for the inhibitory role of Bcl-2 family members in Beclin-1-mediated autophagy.179
The mechanism by which Beclin-1 mediates the induction of autophagy under starvation or stress conditions is thought to be regulated by the release of Bcl-2 and Bcl-XL from the formed complex with Beclin-1.178,180 Therefore, the dissociation of Bcl-2/Bcl-XL-Beclin-1 has been reported to be regulated by c-Jun-N-terminal kinase-mediated phosphorylation of Bcl-2181–184 death-associated protein kinase (DAPK)-mediated phosphorylation of Beclin-1,188–190 translocation of the nuclear protein high-mobility group box 1 (HMGB1) to the cytosol,188,189 or competition with other BH3-only proteins for Bcl-2 binding.180,185,186,188–190
Induction of mitogen activated protein kinase (MAPK), c-Jun-N-terminal kinase (JNK), in response to cellular stress results in the phosphorylation of the three residues in the regulatory loop of Bcl-2, leading to disruption of its binding to Beclin-1,180 an essential step in the induction of autophagy in nutrient-shortage conditions. Thus, cells with either mutations in the phosphorylation sites of the regulatory loop of Bcl-2 or even deficiency in JNK activation appear to be unable to undergo starvation-induced autophagy.180 Therefore, the constitutive activation of JNK can induce autophagy under nutrient-sufficient conditions in Bcl-2-expressing cells.180 The involvement of JNK in modulation of autophagy in response to ER stress, oxidative stress, cancer drugs, and stimulation also occurs through the death receptor Fas.181–184
In addition, the disruption of Bcl-2-Beclin-1 complex is thought to be a target for pro-apoptotic BH3-only proteins such as Bad and Bax, which can mediate the inhibition of Beclin-1-mediated induction of autophagy.187,190
Although the essential role of Beclin-1 in the link between autophagy and apoptosis has been reported in several studies, complex formation of Beclin-1/Vps34 is not necessary for the modulation of autophagic formation in all cell types.191–194
Besides the ability of Bcl-2 family proteins to regulate autophagy through the interaction with Beclin-1, caspases have been shown to inhibit autophagy via a mechanism mediated by the cleavage of autophagy-related proteins.2,195 Accordingly, the role of apoptosis-associated proteases in the regulation of the balance between apoptosis and autophagy has been reported.164 Additionally, the detection of N-terminal cleavage products of Atg5 in multiple cell types and its translocation to the mitochondria leads to mitochondrial dysregulation and subsequently induction of cyt c release164 suggests an essential role for caspases in the inhibition of autophagy. More importantly, the promotion of apoptosis by overexpression of N-terminal Atg5 cleavage products is considered evidence for the ability of the cleaved Atg5 to trigger apoptosis.164 Although N-terminal cleavage products of Atg5 are unable to promote autophagy,164 the pro-apoptotic function of Atg5 seems to be limited and can perhaps sensitize only tumor cells to anti-cancer agents-induced apoptosis.164 Moreover, overexpression of LC3 did not appear to exert any effect on apoptosis since the enhanced sensitivity to apoptosis observed in cells expressing cleaved Atg5 is linked to the pro-apoptotic function of the N-terminal Atg5 fragment164 rather than to enhanced autophagy.
In addition to the ability of apoptosis-associated proteases in regulating the balance between apoptosis and autophagy, a link between the upstream signals that induce apoptosis through the extrinsic pathway has been reported.196 Extrinsic apoptosis is initiated by the ligation of death receptors with their cognate ligands, which results in formation of a death-inducing signaling complex (DISC) from Fas-associated protein with the death domain (FADD) and the initiator caspase, pro-caspase-8. Formation of this complex results in the cleavage of pro-caspase 8 and subsequently the release of active caspase 8, which cleaves and activates downstream targets, including effector caspases.197,198 Thus, in addition to its roles in regulating apoptosis, a key component of DISC has been also reported in several studies to be involved in modulating autophagy.199–201 Additionally, the deficiency or even the inhibition of caspase-8 was found to result in excessive autophagic formation in different cell types.199 Moreover, mutations in FADD were found to prevent its interaction with pro-caspase-8, resulting in the inhibition of apoptosis and a switch to autophagy.200,201
Although Atg5 has been shown to interact with FADD through its death domain (DD), the deficiency of FADD does not appear to influence autophagic formation.202 In contrast, the loss of FADD was found to recover the increased caspase-dependent cell death induced by overexpression of Atg5, evidence for the important role of the Atg5-FADD interaction in regulating apoptosis rather than autophagy.202
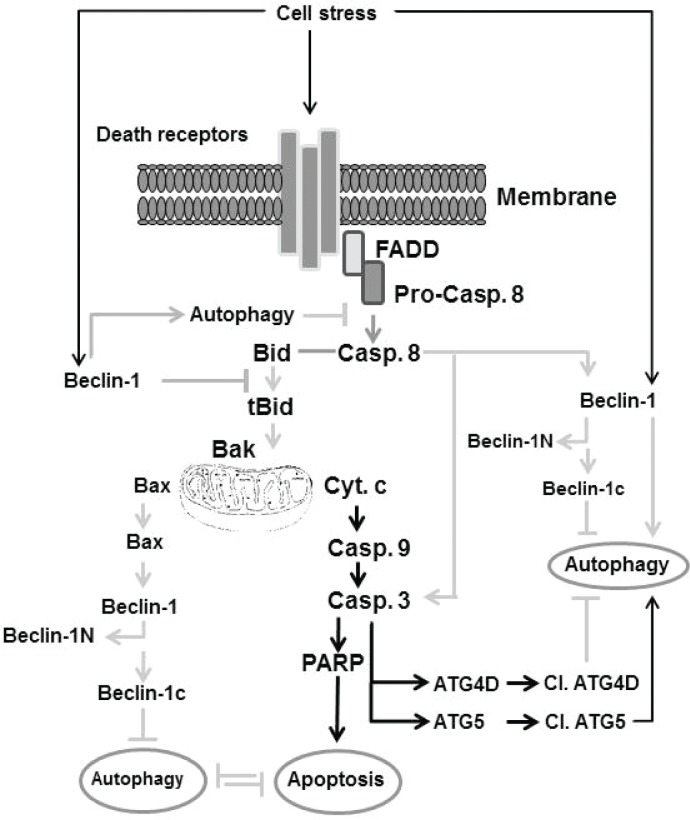
Interestingly, the induction of autophagy in cells by the inhibition of extrinsic apoptotic pathways is thought to be associated mostly with an increased induction of cell death.202,203,205 Although the role of Atg5 is essential for both autophagosomic formation and cell death by exposure to IFN-γ, the ability of N-terminal cleavage product of Atg5 to induce apoptosis indicates that Atg5-mediated cell death is not associated with the loss of the autophagic function of Atg5.164,202 The possible pathways, which are thought to be involved in the modulation of the cross talk between apoptosis and autophagy, are outlined in Figure 3.
Figure 3.
Outline of the molecular mechanisms thought to be involved in the regulation of the cross-talk between apoptosis and autophagy. Some extracellular stress can trigger both apoptosis and autophagy via receptor-dependent and independent mechanisms. One of these mechanism includes the induction of Beclin-1 that triggers autophagy, that in turn inhibits apoptosis by a mechanism mediated by the inhibition of caspase-8 cleavage, and subsequently blocks the cleavage of Bid. Another mechanism includes the activation of death receptor, leading to cleavage of caspase-8 via the Fas-associated death domain. The cleavage of caspase-8 leads to cleavage of Bid, which triggers the loss of mitochondrial membrane potential characterized by cyt c release, caspase-9, caspase-3, and PARP cleavage, the hallmark of apoptosis. Cleavage of caspase-3 triggers cleavage of ATG40, leading to the inhibition of autophagy, whereas the cleavage of ATG5 by caspase-3 results in induction of autophagy. Additionally, accumulation of phosphorylated Bax triggers cleavage of Beclin-1 into Beclin-1N and Beclin-1c. Accumulation of Beclin-1N leads to inhibition of induced autophagy.
Therapeutic Strategies
Based on intensive studies focusing mainly on the modulation of autophagy through apoptotic signaling pathways, it is becoming clear that autophagy is involved in the regulation of apoptosis. Thus, the inhibition of apoptosis by autophagy has been shown to be regulated by the degradation of pro-apoptotic proteins including caspases.203 More important, autophagy is involved in regulating cancer development and progression in addition to being a key factor for determining tumor cell sensitivity to anticancer therapy.204,205 Accordingly, several conventional and experimental antitumor strategies have been reported as relevant strategies for tumor therapy. These strategies include the combination of conventional therapies with autophagy regulators.204,205 These autophagic regulators include rapamycin,206,207 arsenic trioxide,208,209 temozolomide,210,211 kringle domains of plasminogen,212 phenethyl isothiocyanate,213 OSU03012 (derivative of celecoxib),214 NVPBEZ235,215,216 and cell wall skeleton of Mycobacterium bovis Bacillus Calmette Guerin.217 Additionally, other autophagic regulators such as phenethyl isothiocyanate, the celecoxib derivative OSU03012 and kringle domains of plasminogen (endostatin kringle 5, K5) have emerged as promising cancer chemopreventive agents based on their ability to induce autophagic cell death.213 Moreover, several cancer therapies, including DNA-damaging chemotherapeutic temozolomide218 and radiation219 have been shown to induce autophagy both in vitro and in vivo.220 However, this induced autophagy has been shown to serve as a protective mechanism against the toxicity of the applied anticancer agents.221 Additionally, radiation therapy has been reported to promote autophagy by a mechanism mediated through the upregulation of autophagy mediators including Beclin-1, ATG3, ATG4, ATG5, and ATG12.219,220 Furthermore, some chemotherapeutic agents such as histone deacetylase inhibitors221 and cisplatin222 have been shown to induce autophagy through the accumulation of reactive oxygen species in mitochondria. Preclincal studies have also addressed a powerful therapeutic potential for curcumin in tumor treatment; however, the therapeutic potential of curcumin due the ability of curcumin to trigger both apoptosis223 and autophagy.224
The reliability of proteasome inhibitors alone or in combination with other drugs has been reported to induce apoptosis as well as autophagy in different tumor types including melanoma.2,225–227 However, the induction of apoptosis by proteasome inhibitors is linked to the suppression of transcription of anti-apoptotic factors228–230 and is associated with the upregulation of pro-apoptotic proteins.2,225–227 While induction of autophagy by proteasome inhibitor regulated by the stabilization of anti-apoptotic proteins including the protein Mcl-1, as shown in melanoma cells.2
Based on previous and current findings, autophagy and apoptosis appear to play opposing roles in cancer cells since the inhibition of autophagy by chloroquine was found to sensitize tumor resistance to anti-cancer agents-induced apoptosis.231–234 Additionally, the role of autophagy in the resistance of tumor cells to the pro-apoptotic effects of TRAIL has been reported in several studies.234–236 Unlike most superfamily members, TRAIL as a cancer therapeutic possesses pro-apoptotic effects that is specific to tumor cells.237,238 However, the advantage of TRAIL as a tumor therapeutic agent becomes unattractive based on the development of TRAIL resistance in many tumors.239 Therefore, intensive studies on the tumor resistance mechanisms of TRAIL revealed that the failure of TRAIL to trigger apoptosis of tumor cells is associated with the cyto-protective effects of autophagy. Correspondingly, c-FLIP-expression is resistant to TRAIL-induced apoptosis and is characterized by the increase of Beclin-1 expression and a marked autophagosome formation in response to the activation of TRIAL or Fas receptors.240 Accordingly, the inhibition of the autophagic machinery in cells expressing c-FLIP or in Bax-deficient cells enhances their sensitivity to TRAIL-mediated cell death.234,235 Thus, based on the findings of widely reported studies, apoptosis and autophagy appear to work together. While autophagy mediates the degradation of caspases, caspases can also target the cleavage of autophagy-related proteins.
However, the use of autophagy by tumor cells as a cytoprotective mechanism to block chemotherapeutic agents-induced apoptosis seems to be a general phenomenon. Cytoprotective effects associated with autophagy were reported in different tumor types following treatment with anticancer agents including gefitinib, erlotinib, and celecoxib.240–242 Despite these ongoing clinical efforts, the use of autophagy inhibitors as a therapeutic strategy in cancer, thus far, has not been established. Therefore, further preclinical evaluation to optimize the efficacy of the suggested therapies is urgently needed.
Taken together, in addition to the elucidation of molecular mechanisms, whereby autophagy counteracts or synergizes apoptosis, or vice versa, understanding of the details of these mechanisms provides insight into the potentials and limitations of the combination of cancer therapies targeting both apoptosis and autophagy.
Conclusion
Although the relationship between apoptosis and autophagy has been widely reported, the identification of the molecular mechanisms which govern this relationship have not been described in detail. Thus, understanding the molecular mechanism of the crosstalk between apoptosis and autophagy may help to develop a relevant therapeutic strategy for tumor treatment. In this review, we provided insight into the molecular mechanisms of the crosstalk between apoptosis and autophagy. In addition, molecular targets, which are thought to be essential for the balance between autophagy and apoptosis, have been discussed. Additionally, the relevant pathways of both apoptosis and autophagy, which are thought to be targets for novel therapeutic approaches, were discussed.
Footnotes
Author Contributions
Conceived and designed the experiments: MH, AE. Analyzed the data: MH. Wrote the first draft of the manuscript: MH, AE, YH, and DS. Contributed to the writing of the manuscript: MH, YH. Agree with manuscript results and conclusions: MH. Jointly developed the structure and arguments for the paper: MH, AE. Made critical revisions and approved final version: MH, YH. All authors reviewed and approved of the final manuscript.
Funding
This work was supported by grants from German Research Foundation (HA 5081/3–1), from L’Alsace Contre le Cancer, France, and from German Cancer Research Foundation (10-2202-Ha 1) to M. Hassan.
Competing Interests
Authors disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests. Provenance: the authors were invited to submit this paper.
References
- 1.Selimovic D, Badura HE, El-Khattouti A, et al. Vinblastine-induced apoptosis of melanoma cells is mediated by Ras homologous A protein (Rho A) via mitochondrial and non-mitochondrial-dependent pathways. Apoptosis. 2013;18(8):980–997. doi: 10.1007/s10495-013-0844-4. [DOI] [PubMed] [Google Scholar]
- 2.Selimovic D, Porzig BB, El-Khattouti A, et al. Bortezomib/proteasome inhibitor triggers both apoptosis and autophagy-dependent pathways in melanoma cells. Cell Signal. 2013;25(1):308–318. doi: 10.1016/j.cellsig.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Selimovic D, Sprenger A, Hannig M, Haïkel Y, Hassan M. Apoptosis related protein-1 triggers melanoma cell death via interaction with the juxta membrane region of p75 neurotrophin receptor. J Cell Mol Med. 2012;16(2):349–361. doi: 10.1111/j.1582-4934.2011.01304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cetindere T, Nambiar S, Santourlidis S, Essmann F, Hassan M. Induction of indoleamine 2,3-dioxygenase by death receptor activation contributes to apoptosis of melanoma cells via mitochondrial damage-dependent ROS accumulation. Cell Signal. 2010;22(2):197–211. doi: 10.1016/j.cellsig.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Hassan M, Alaoui A, Feyen O, et al. The BH3-only member Noxa causes apoptosis in melanoma cells by multiple pathways. Oncogene. 2008;27(33):4557–4568. doi: 10.1038/onc.2008.90. [DOI] [PubMed] [Google Scholar]
- 6.Meng S, Xu J, Wu Y, Ding C. Targeting autophagy to enhance oncolytic virus-based cancer therapy. Expert Opin Biol Ther. 2013;13(6):863–873. doi: 10.1517/14712598.2013.774365. [DOI] [PubMed] [Google Scholar]
- 7.Bristol ML, Emery SM, Maycotte P, Thorburn A, Chakradeo S, Gewirtz DA. Autophagy inhibition for chemosensitization and radiosensitization in cancer: do the preclinical data support this therapeutic strategy? J Pharmacol Exp Ther. 2013;344(3):544–552. doi: 10.1124/jpet.112.199802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 2013;73(1):3–7. doi: 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 9.Hu YL, Jahangiri A, Delay M, Aghi MK. Tumor cell autophagy as an adaptive response mediating resistance to treatments such as antiangiogenic therapy. Cancer Res. 2012;72(17):4294–4299. doi: 10.1158/0008-5472.CAN-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J, Gong Z, Shen HM. The role of autophagy in liver cancer: Molecular mechanisms and potential therapeutic targets. Biochim Biophys Acta. 2013;1836(1):15–26. doi: 10.1016/j.bbcan.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Zhang N, Qi Y, Wadham C, et al. FTY720 induces necrotic cell death and autophagy in ovarian cancer cells: a protective role of autophagy. Autophagy. 2010;6(8):1157–1167. doi: 10.4161/auto.6.8.13614. [DOI] [PubMed] [Google Scholar]
- 12.Singhapol C, Pal D, Czapiewski R, Porika M, Nelson G, Saretzki GC. Mitochondrial telomerase protects cancer cells from nuclear DNA damage and apoptosis. PLoS ONE. 2013;8(1):e52989. doi: 10.1371/journal.pone.0052989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Zhu J, Tang Y, et al. X-linked inhibitor of apoptosis positive nuclear labeling: a new independent prognostic biomarker of breast invasive ductal carcinoma. Diagn Pathol. 2011;6:49. doi: 10.1186/1746-1596-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(19):1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- 15.King JS. Mechanical stress meets autophagy: potential implications for physiology and pathology. Trends Mol Med. 2012;18(10):583–588. doi: 10.1016/j.molmed.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Ding F, Shao ZW, Xiong LM. Cell death in intervertebral disc degeneration. Apoptosis. 2013;18(7):777–785. doi: 10.1007/s10495-013-0839-1. [DOI] [PubMed] [Google Scholar]
- 17.Roesly HB, Khan MR, Chen HD, et al. The decreased expression of Beclin-1 correlates with progression to esophageal adenocarcinoma: the role of deoxycholic acid. Am J Physiol Gastrointest Liver Physiol. 2012;302(8):G864–G872. doi: 10.1152/ajpgi.00340.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Outschoorn UE, Whitaker-Menezes D, Lin Z, et al. Cytokine production and inflammation drive autophagy in the tumor microenvironment: role of stromal caveolin-1 as a key regulator. Cell Cycle. 2011;10(11):1784–1793. doi: 10.4161/cc.10.11.15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tseng HC, Liu WS, Tyan YS, Chiang HC, Kuo WH, Chou FP. Sensitizing effect of 3-methyladenine on radiation-induced cytotoxicity in radio-resistant HepG2 cells in vitro and in tumor xenografts. Chem Biol Interact. 2011;192(3):201–208. doi: 10.1016/j.cbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Ahn JH, Jang GH, Lee M. Defective autophagy in multidrug resistant cells may lead to growth inhibition by BH3-mimetic gossypol. J Cell Physiol. 2012 doi: 10.1002/jcp.24305. [DOI] [PubMed] [Google Scholar]
- 21.Saleem A, Dvorzhinski D, Santanam U, et al. Effect of dual inhibition of apoptosis and autophagy in prostate cancer. Prostate. 2012;72(12):1374–1381. doi: 10.1002/pros.22487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brem EA, Thudium K, Khubchandani S, et al. Distinct cellular and therapeutic effects of obatoclax in rituximab-sensitive and -resistant lymphomas. Br J Haematol. 2011;153(5):599–611. doi: 10.1111/j.1365-2141.2011.08669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M, Hossain MS, Tan W, et al. Inhibition of isoprenylcysteine carboxylmethyltransferase induces autophagic-dependent apoptosis and impairs tumor growth. Oncogene. 2010;29(35):4959–4970. doi: 10.1038/onc.2010.247. [DOI] [PubMed] [Google Scholar]
- 24.Zhao C, Yin S, Dong Y, et al. Autophagy-dependent EIF2AK3 activation compromises ursolic acid-induced apoptosis through upregulation of MCL1 in MCF-7 human breast cancer cells. Autophagy. 2013;9(2):196–207. doi: 10.4161/auto.22805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirzoeva OK, Hann B, Hom YK, et al. Autophagy suppression promotes apoptotic cell death in response to inhibition of the PI3K-mTOR pathway in pancreatic adenocarcinoma. J Mol Med. 2011;89(9):877–889. doi: 10.1007/s00109-011-0774-y. [DOI] [PubMed] [Google Scholar]
- 26.Ding WX, Chen X, Yin XM. Tumor cells can evade dependence on autophagy through adaptation. Biochem Biophys Res Commun. 2012;425(3):684–688. doi: 10.1016/j.bbrc.2012.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edelmann B, Bertsch U, Tchikov V, et al. Caspase-8 and caspase-7 sequentially mediate proteolytic activation of acid sphingomyelinase in TNF-R1 receptosomes. EMBO J. 2011;30(2):379–394. doi: 10.1038/emboj.2010.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Liu R, Li J, et al. Quercetin induces protective autophagy in gastric cancer cells: involvement of Akt-mTOR- and hypoxia-induced factor 1 α-mediated signaling. Autophagy. 2011;7(9):966–978. doi: 10.4161/auto.7.9.15863. [DOI] [PubMed] [Google Scholar]
- 29.Muñoz-Pinedo C. Signaling pathways that regulate life and cell death: evolution of apoptosis in the context of self-defense. Adv Exp Med Biol. 2012;738:124–143. doi: 10.1007/978-1-4614-1680-7_8. [DOI] [PubMed] [Google Scholar]
- 30.Merz S, Hammermeister M, Altmann K, Dürr M, Westermann B. Molecular machinery of mitochondrial dynamics in yeast. Biol Chem. 2007;388(9):917–926. doi: 10.1515/BC.2007.110. [DOI] [PubMed] [Google Scholar]
- 31.Yuan S, Akey CW. Apoptosome structure, assembly, and procaspase activation. Structure. 2013;21(4):501–515. doi: 10.1016/j.str.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chae SS, Yoo CB, Jo C, Yun SM, Jo SA, Koh YH. Caspases-2 and -8 are involved in the presenilin1/gamma-secretase-dependent cleavage of amyloid precursor protein after the induction of apoptosis. J Neurosci Res. 2010;88(9):1926–1933. doi: 10.1002/jnr.22356. [DOI] [PubMed] [Google Scholar]
- 33.Park K, Yi SY, Kim UL, et al. Monitoring of cleavage preference for caspase-3 using recombinant protein substrates. J Microbiol Biotechnol. 2009;19(9):911–917. doi: 10.4014/jmb.0902.088. [DOI] [PubMed] [Google Scholar]
- 34.Shen J, Yin Y, Mai J, et al. Caspase-1 recognizes extended cleavage sites in its natural substrates. Atherosclerosis. 2010;210(2):422–429. doi: 10.1016/j.atherosclerosis.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van de Craen M, Declercq W, Van den brande I, Fiers W, Vandenabeele P. The proteolytic procaspase activation network: an in vitro analysis. Cell Death Differ. 1999;6(11):1117–1124. doi: 10.1038/sj.cdd.4400589. [DOI] [PubMed] [Google Scholar]
- 36.Zhang F, Lau SS, Monks TJ. A dual role for poly (ADP-ribose) polymerase-1 during caspase-dependent apoptosis. Toxicol Sci. 2012;128(1):103–114. doi: 10.1093/toxsci/kfs142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu WH, Luo SJ, Chen CL, et al. Vinca alkaloids cause aberrant ROS-mediated JNK activation, Mcl-1 downregulation, DNA damage, mitochondrial dysfunction, and apoptosis in lung adenocarcinoma cells. Biochem Pharmacol. 2012;83(9):1159–1171. doi: 10.1016/j.bcp.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Herzog N, Hartkamp JD, Verheugd P, et al. Caspase-dependent cleavage of the mono-ADP-ribosyltransferase ARTD10 interferes with its pro-apoptotic function. FEBS J. 2013;280(5):1330–1343. doi: 10.1111/febs.12124. [DOI] [PubMed] [Google Scholar]
- 39.Liu YQ, Hu XY, Lu T, et al. Retigeric acid B exhibits antitumor activity through suppression of nuclear factor-κB signaling in prostate cancer cells in vitro and in vivo. PLoS ONE. 2012;7(5):e38000. doi: 10.1371/journal.pone.0038000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jehle J, Staudacher I, Wiedmann F, et al. Regulation of apoptosis in HL-1 cardiomyocytes by phosphorylation of the receptor tyrosine kinase EphA2 and protection by lithocholic acid. Br J Pharmacol. 2012;167(7):1563–1572. doi: 10.1111/j.1476-5381.2012.02117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunner T, Arnold D, Wasem C, Herren S, Frutschi C. Regulation of cell death and survival in intestinal intraepithelial lymphocytes. Cell Death Differ. 2001;8(7):706–714. doi: 10.1038/sj.cdd.4400854. [DOI] [PubMed] [Google Scholar]
- 42.Ng KB, Bustamam A, Sukari MA, et al. Induction of selective cytotoxicity and apoptosis in human T4-lymphoblastoid cell line (CEMss) by boesenbergin a isolated from boesenbergia rotunda rhizomes involves mitochondrial pathway, activation of caspase 3 and G2/M phase cell cycle arrest. BMC Complement Altern Med. 2013;13:41. doi: 10.1186/1472-6882-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen KC, Willmore WG, Tayabali AF. Cadmium telluride quantum dots cause oxidative stress leading to extrinsic and intrinsic apoptosis in hepatocellular carcinoma HepG2 cells. Toxicology. 2013;306:114–123. doi: 10.1016/j.tox.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Thomas SA, Vasudevan S, Thamkachy R, et al. Upregulation of DR5 receptor by the diaminothiazole DAT1 [4-amino-5-benzoyl-2-(4-methoxy phenyl amino) thiazole] triggers an independent extrinsic pathway of apoptosis in colon cancer cells with compromised pro and antiapoptotic proteins. Apoptosis. 2013;18(6):713–726. doi: 10.1007/s10495-013-0826-6. [DOI] [PubMed] [Google Scholar]
- 45.Scavullo C, Servida F, Lecis D, et al. Single-agent Smac-mimetic compounds induce apoptosis in B chronic lymphocytic leukaemia (B-CLL) Leuk Res. 2013;37(7):809–815. doi: 10.1016/j.leukres.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 46.Pandey MK, Sung B, Ahn KS, Kunnumakkara AB, Chaturvedi MM, Aggarwal BB. Gambogic acid, a novel ligand for transferrin receptor, potentiates TNF-induced apoptosis through modulation of the nuclear factor-kappaB signaling pathway. Blood. 2007;110(10):3517–3525. doi: 10.1182/blood-2007-03-079616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinvalet D, Zhu P, Lieberman J. Granzyme A induces caspase-independent mitochondrial damage, a required first step for apoptosis. Immunity. 2005;22(3):355–370. doi: 10.1016/j.immuni.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer. 2002;2(4):277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 50.Cui Z, Song E, Hu DN, Chen M, Rosen R, McCormick SA. Butein induces apoptosis in human uveal melanoma cells through mitochondrial apoptosis pathway. Curr Eye Res. 2012;37(8):730–739. doi: 10.3109/02713683.2012.671436. [DOI] [PubMed] [Google Scholar]
- 51.Serrano FA, Matsuo AL, Monteforte PT, et al. A cyclopalladated complex interacts with mitochondrial membrane thiol-groups and induces the apoptotic intrinsic pathway in murine and cisplatin-resistant human tumor cells. BMC Cancer. 2011;11:296. doi: 10.1186/1471-2407-11-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chipuk JE, Fisher JC, Dillon CP, Kriwacki RW, Kuwana T, Green DR. Mechanism of apoptosis induction by inhibition of the anti-apoptotic BCL-2 proteins. Proc Natl Acad Sci U S A. 2008;105(51):20327–20332. doi: 10.1073/pnas.0808036105. [Pubmed: 19074266] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elmore S. Apoptosis: A Review of Programmed Cell Death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [Pubmed: 17562483] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saelens X, Festjens N, Vande Walle L, van Gurp M, van Loo G, Vandenabeele P. Toxic proteins released from mitochondria in cell death. Oncogene. 2004;23(16):2861–2874. doi: 10.1038/sj.onc.1207523. [DOI] [PubMed] [Google Scholar]
- 55.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102(1):33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 56.Garrido C, Kroemer G. Life’s smile, death’s grin: vital functions of apoptosis-executing proteins. Curr Opin Cell Biol. 2004;16(6):639–646. doi: 10.1016/j.ceb.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 57.Chinnaiyan AM. The apoptosome: heart and soul of the cell death machine. Neoplasia. 1999;1(1):5–15. doi: 10.1038/sj.neo.7900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hill MM, Adrain C, Duriez PJ, Creagh EM, Martin SJ. Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes. EMBO J. 2004;23(10):2134–2145. doi: 10.1038/sj.emboj.7600210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Loo G, Saelens X, Matthijssens F, et al. Caspases are not localized in mitochondria during life or death. Cell Death Differ. 2002;9(11):1207–1211. doi: 10.1038/sj.cdd.4401101. [DOI] [PubMed] [Google Scholar]
- 60.Schimmer AD. Inhibitor of apoptosis proteins: translating basic knowledge into clinical practice. Cancer Res. 2004;64(20):7183–7190. doi: 10.1158/0008-5472.CAN-04-1918. [DOI] [PubMed] [Google Scholar]
- 61.Ekert PG, Vaux DL. The mitochondrial death squad: hardened killers or innocent bystanders? Curr Opin Cell Biol. 2005;17(6):626–630. doi: 10.1016/j.ceb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 62.Joza N, Susin SA, Daugas E, et al. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410(6828):549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 63.Susin SA, Daugas E, Ravagnan L, et al. Two distinct pathways leading to nuclear apoptosis. J Exp Med. 2000;192(4):571–580. doi: 10.1084/jem.192.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 65.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391(6662):43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 66.Cory S, Adams JM. The Bcl-2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2(9):647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 67.Schuler M, Green DR. Mechanisms of p53-dependent apoptosis. Biochem Soc Trans. 2001;29(Pt 6):684–648. doi: 10.1042/0300-5127:0290684. [DOI] [PubMed] [Google Scholar]
- 68.70-Yip KW, Reed JC. Bcl-2 family proteins and cancer. Oncogene. 2008;27:6398–406. doi: 10.1038/onc.2008.307. [Pubmed: 18955968] [DOI] [PubMed] [Google Scholar]
- 69.Korsmeyer SJ, Wei MC, Saito M, Weiler S, Oh KJ, Schlesinger PH. Pro-apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ. 2000;7(12):1166–1173. doi: 10.1038/sj.cdd.4400783. [DOI] [PubMed] [Google Scholar]
- 70.Kuwana T, Bouchier-Hayes L, Chipuk JE, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17(4):525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 71.Kuwana T, Mackey MR, Perkins G, et al. Bid, Bax lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111(3):331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 72.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2(3):183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 73.Lindsten T, Ross AJ, King A, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6(6):1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei MC, Zong WX, Cheng EH, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292(5517):727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hassan M, Feyen O, Grinstein E. Fas-induced apoptosis of renal cell carcinoma is mediated by apoptosis signal-regulating kinase 1 via mitochondrial damage-dependent caspase-8 activation. Cell Oncol. 2009;31(6):437–456. doi: 10.3233/CLO-2009-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27(48):6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 77.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87(4):619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 78.Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80(2):285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 79.Newmeyer DD, Bossy-Wetzel E, Kluck RM, Wolf BB, Beere HM, Green DR. Bcl-xL does not inhibit the function of Apaf-1. Cell Death Differ. 2000;7(4):402–407. doi: 10.1038/sj.cdd.4400665. [DOI] [PubMed] [Google Scholar]
- 80.Chau BN, Cheng EH, Kerr DA, Hardwick JM. Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Mol Cell. 2000;6(1):31–40. [PubMed] [Google Scholar]
- 81.Hassan M, Alaoui A, Feyen O, et al. The BH3-only member Noxa causes apoptosis in melanoma cells by multiple pathways. Oncogene. 2008;27(33):4557–4568. doi: 10.1038/onc.2008.90. [DOI] [PubMed] [Google Scholar]
- 82.Liu FT, Newland AC, Jia L. Bax conformational change is a crucial step for PUMA-mediated apoptosis in human leukemia. Biochem Biophys Res Commun. 2003;310(3):956–962. doi: 10.1016/j.bbrc.2003.09.109. [DOI] [PubMed] [Google Scholar]
- 83.Oda E, Ohki R, Murasawa H, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288(5468):1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 84.Meyer N, Kim SS, Penn LZ. The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol. 2006;16(4):275–287. doi: 10.1016/j.semcancer.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 85.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104(4):487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 86.Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281(5381):1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 87.Chicheportiche Y, Bourdon PR, Xu H, et al. TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. J Biol Chem. 1997;272(51):32401–32410. doi: 10.1074/jbc.272.51.32401. [DOI] [PubMed] [Google Scholar]
- 88.Peter ME, Krammer PH. Mechanisms of CD95 (APO-1/Fas)-mediated apoptosis. Curr Opin Immunol. 1998;10(5):545–551. doi: 10.1016/s0952-7915(98)80222-7. [DOI] [PubMed] [Google Scholar]
- 89.Suliman A, Lam A, Datta R, Srivastava RK. Intracellular mechanisms of TRAIL: apoptosis through mitochondrial-dependent and -independent pathways. Oncogene. 2001;20(17):2122–2133. doi: 10.1038/sj.onc.1204282. [DOI] [PubMed] [Google Scholar]
- 90.Rubio-Moscardo F, Blesa D, Mestre C, et al. Characterization of 8p21.3 chromosomal deletions in B-cell lymphoma: TRAIL-R1 and TRAIL-R2 as candidate dosage-dependent tumor suppressor genes. Blood. 2005;106(9):3214–3222. doi: 10.1182/blood-2005-05-2013. [DOI] [PubMed] [Google Scholar]
- 91.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappaB activation. Cell. 1995;81(4):495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 92.Wajant H, Pfizenmaier K, Scheurich P. TNF-related apoptosis inducing ligand (TRAIL) and its receptors in tumor surveillance and cancer therapy. Apoptosis. 2002;7(5):449–459. doi: 10.1023/a:1020039225764. [DOI] [PubMed] [Google Scholar]
- 93.Kischkel FC, Hellbardt S, Behrmann I, et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14(22):5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Slee EA, Adrain C, Martin SJ. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276(10):7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- 95.Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 1998;391(6662):96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 96.Martin DN, Baehrecke EH. Caspases function in autophagic programmed cell death in Drosophila. Development. 2004;131(2):275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- 97.Kothakota S, Azuma T, Reinhard C, et al. Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science. 1997;278(5336):294–298. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 98.Bratton DL, Fadok VA, Richter DA, Kailey JM, Guthrie LA, Henson PM. Appearance of phosphatidylserine on apoptotic cells requires calcium-mediated nonspecific flip-flop and is enhanced by loss of the aminophospholipid translocase. J Biol Chem. 1997;272(42):26159–26165. doi: 10.1074/jbc.272.42.26159. [DOI] [PubMed] [Google Scholar]
- 99.Ferraro-Peyret C, Quemeneur L, Flacher M, Revillard JP, Genestier L. Caspase-independent phosphatidylserine exposure during apoptosis of primary T lymphocytes. J Immunol. 2002;169(9):4805–4810. doi: 10.4049/jimmunol.169.9.4805. [DOI] [PubMed] [Google Scholar]
- 100.Mandal D, Mazumder A, Das P, Kundu M, Basu J. Fas-, caspase 8-, and caspase 3-dependent signaling regulates the activity of the aminophospholipid translocase and phosphatidylserine externalization in human erythrocytes. J Biol Chem. 2005;280(47):39460–39467. doi: 10.1074/jbc.M506928200. [DOI] [PubMed] [Google Scholar]
- 101.Fadok VA, Chimini G. The phagocytosis of apoptotic cells. Semin Immunol. 2001;13(6):35–72. doi: 10.1006/smim.2001.0333. [DOI] [PubMed] [Google Scholar]
- 102.Mackeh R, Perdiz D, Lorin S, Codogno P, Poüs C. Autophagy and microtubules—new story, old players. J Cell Sci. 2013;126(Pt 5):1071–1080. doi: 10.1242/jcs.115626. [DOI] [PubMed] [Google Scholar]
- 103.Legakis JE, Yen WL, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy. 2007;3(5):422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- 104.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139(7):1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khan IA, Lu JP, Liu XH, Rehman A, Lin FC. Multifunction of autophagy-related genes in filamentous fungi. Microbiol Res. 2012;167(6):339–345. doi: 10.1016/j.micres.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 106.Christian P, Sacco J, Adeli K. Autophagy: Emerging roles in lipid homeostasis and metabolic control. Biochim Biophys Acta. 2013;1831(4):819–824. doi: 10.1016/j.bbalip.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 107.Markaki M, Tavernarakis N. Metabolic Control by Target of Rapamycin and Autophagy during Ageing—A Mini-Review. Gerontology. 2013;59(4):340–348. doi: 10.1159/000348599. [DOI] [PubMed] [Google Scholar]
- 108.Quan W, Jung HS, Lee MS. Role of autophagy in the progression from obesity to diabetes and in the control of energy balance. Arch Pharm Res. 2013;36(2):223–229. doi: 10.1007/s12272-013-0024-7. [DOI] [PubMed] [Google Scholar]
- 109.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 110.Gómez-Suaga P, Fdez E, Blanca Ramírez M, Hilfiker S. A Link between Autophagy and the Pathophysiology of LRRK2 in Parkinson’s Disease. Parkinsons Dis. 2012;2012:324521. doi: 10.1155/2012/324521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dutta D, Calvani R, Bernabei R, Leeuwenburgh C, Marzetti E. Contribution of impaired mitochondrial autophagy to cardiac aging: mechanisms and therapeutic opportunities. Circ Res. 2012;110(8):1125–1138. doi: 10.1161/CIRCRESAHA.111.246108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu H, Mi S, Li Z, Hua F, Hu ZW. Interleukin 17A inhibits autophagy through activation of PIK3CA to interrupt the GSK3B-mediated degradation of BCL-2 in lung epithelial cells. Autophagy. 2013;9(5):730–742. doi: 10.4161/auto.24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Luo YH, Wu SB, Wei YH, et al. Cadmium-based quantum dot induced autophagy formation for cell survival via oxidative stress. Chem Res Toxicol. 2013;26(5):662–673. doi: 10.1021/tx300455k. [DOI] [PubMed] [Google Scholar]
- 114.Chang Z, Shi G, Jin J, et al. Dual PI3K/mTOR inhibitor NVP-BEZ235–induced apoptosis of hepatocellular carcinoma cell lines is enhanced by inhibitors of autophagy. Int J Mol Med. 2013;31(6):1449–1456. doi: 10.3892/ijmm.2013.1351. [DOI] [PubMed] [Google Scholar]
- 115.Zhou ZR, Zhu XD, Zhao W, et al. Poly (ADP-ribose) polymerase-1 regulates the mechanism of irradiation-induced CNE-2 human nasopharyngeal carcinoma cell autophagy and inhibition of autophagy contributes to the radiation sensitization of CNE-2 cells. Oncol Rep. 2013;29(6):2498–2506. doi: 10.3892/or.2013.2382. [DOI] [PubMed] [Google Scholar]
- 116.Chan SH, Kikkawa U, Matsuzaki H, Chen JH, Chang WC. Insulin receptor substrate-1 prevents autophagy-dependent cell death caused by oxidative stress in mouse NIH/3T3 cells. J Biomed Sci. 2012;19:64. doi: 10.1186/1423-0127-19-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hammerová J, Uldrijan S, Táborská E, Vaculová AH, Slaninová I. Necroptosis modulated by autophagy is a predominant form of melanoma cell death induced by sanguilutine. Biol Chem. 2012;393(7):647–658. doi: 10.1515/hsz-2011-0279. [DOI] [PubMed] [Google Scholar]
- 118.Zhang X, Wei H, Liu Z, et al. A novel protoapigenone analog RY10-4 induces breast cancer MCF-7 cell death through autophagy via the Akt/mTOR pathway. Toxicol Appl Pharmacol. 2013;270(2):122–128. doi: 10.1016/j.taap.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 119.García-Zepeda SP, García-Villa E, Díaz-Chávez J, Hernández-Pando R, Gariglio P. Resveratrol induces cell death in cervical cancer cells through apoptosis and autophagy. Eur J Cancer Prev. 2013 doi: 10.1097/CEJ.0b013e328360345f. [DOI] [PubMed] [Google Scholar]
- 120.Lamy L, Ngo VN, Emre NC, et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell. 2013;23(4):435–449. doi: 10.1016/j.ccr.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jin S. Autophagy, mitochondrial quality control, and oncogenesis. Autophagy. 2006;2(2):80–84. doi: 10.4161/auto.2.2.2460. [DOI] [PubMed] [Google Scholar]
- 122.Mazure NM, Pouysségur J. Atypical BH3-domains of BNIP3 and BNIP3L lead to autophagy in hypoxia. Autophagy. 2009;5(6):868–869. doi: 10.4161/auto.9042. [DOI] [PubMed] [Google Scholar]
- 123.Bellot G, Garcia-Medina R, Gounon P, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29(10):2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6(9):729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 125.Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4(7):851–865. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 126.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221(1):3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shaw RJ. LKB1 and AMP-activated kinase control of mTOR signalling and growth. Acta Physiol. 2009;196(1):65–80. doi: 10.1111/j.1748-1716.2009.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 129.Rubio-Moscardo F, Blesa D, Mestre C, et al. Characterization of 8p21.3 chromosomal deletions in B-cell lymphoma: TRAIL-R1 and TRAIL-R2 as candidate dosage-dependent tumor suppressor genes. Blood. 2005;106(9):3214–3222. doi: 10.1182/blood-2005-05-2013. [DOI] [PubMed] [Google Scholar]
- 130.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20(1):51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22(2):239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16(7):939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27(17):6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454(7201):232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schweers RL, Zhang J, Randall MS, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104(49):19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mammucari C, Milan G, Romanello V, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 137.Hamacher-Brady A, Brady NR, Gottlieb RA, Gustafsson AB. Autophagy as a protective response to Bnip3-mediated apoptotic signaling in the heart. Autophagy. 2006;2(4):307–309. doi: 10.4161/auto.2947. [DOI] [PubMed] [Google Scholar]
- 138.Tracy K, Macleod KF. Regulation of mitochondrial integrity, autophagy and cell survival by BNIP3. Autophagy. 2007;3(6):616–619. doi: 10.4161/auto.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang H, Bosch-Marce M, Shimoda LA, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283(16):10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Li Y, Wang Y, Kim E, et al. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J Biol Chem. 2007;282(49):35803–35813. doi: 10.1074/jbc.M705231200. [DOI] [PubMed] [Google Scholar]
- 141.Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21(4):521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Walsh CM, Edinger AL. The complex interplay between autophagy, apoptosis, and necrotic signals promotes T-cell homeostasis. Immunol Rev. 2010;236:95–109. doi: 10.1111/j.1600-065X.2010.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Landau G, Ran A, Bercovich Z, et al. Expression profiling and biochemical analysis suggest stress response as a potential mechanism inhibiting proliferation of polyamine-depleted cells. J Biol Chem. 2012;287(43):35825–35837. doi: 10.1074/jbc.M112.381335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yao F, Wang G, Wei W, Tu Y, Tong H, Sun S. An autophagy inhibitor enhances the inhibition of cell proliferation induced by a proteasome inhibitor in MCF-7 cells. Mol Med Rep. 2012;5(1):84–88. doi: 10.3892/mmr.2011.590. [DOI] [PubMed] [Google Scholar]
- 145.Wang S, Xia P, Shi L, Fan Z. FADD cleavage by NK cell granzyme M enhances its self-association to facilitate procaspase-8 recruitment for auto-processing leading to caspase cascade. Cell Death Differ. 2012;19(4):605–615. doi: 10.1038/cdd.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Díaz GD, Li Q, Dashwood RH. Caspase-8 and apoptosis-inducing factor mediate a cytochrome c-independent pathway of apoptosis in human colon cancer cells induced by the dietary phytochemical chlorophyllin. Cancer Res. 2003;63(6):1254–1261. [PubMed] [Google Scholar]
- 147.Xi H, Barredo JC, Merchan JR, Lampidis TJ. Endoplasmic reticulum stress induced by 2-deoxyglucose but not glucose starvation activates AMPK through CaMKKβ leading to autophagy. Biochem Pharmacol. 2013;85(10):1463–1477. doi: 10.1016/j.bcp.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 148.Dou Z, Pan JA, Dbouk HA, et al. Class IA PI3K p110β subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell. 2013;50(1):29–42. doi: 10.1016/j.molcel.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu W, Otkur W, Li L, et al. Autophagy induced by silibinin protects human epidermoid carcinoma A431 cells from UVB-induced apoptosis. J Photochem Photobiol B, Biol. 2013;123:23–31. doi: 10.1016/j.jphotobiol.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 150.Castino R, Bellio N, Follo C, Murphy D, Isidoro C. Inhibition of PI3k class III-dependent autophagy prevents apoptosis and necrosis by oxidative stress in dopaminergic neuroblastoma cells. Toxicol Sci. 2010;117(1):152–162. doi: 10.1093/toxsci/kfq170. [DOI] [PubMed] [Google Scholar]
- 151.Sasnauskiene A, Kadziauskas J, Vezelyte N, Jonusiene V, Kirveliene V. Damage targeted to the mitochondrial interior induces autophagy, cell cycle arrest and, only at high doses, apoptosis. Autophagy. 2009;5(5):743–744. doi: 10.4161/auto.5.5.8701. [DOI] [PubMed] [Google Scholar]
- 152.Bhutia SK, Dash R, Das SK, et al. Mechanism of autophagy to apoptosis switch triggered in prostate cancer cells by antitumor cytokine melanoma differentiation-associated gene 7/interleukin-24. Cancer Res. 2010;70(9):3667–3676. doi: 10.1158/0008-5472.CAN-09-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Leber B, Andrews DW. Closing in on the link between apoptosis and autophagy. F1000 Biol Rep. 2010;2:88. doi: 10.3410/B2-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen R, Jin R, Wu L, et al. Reticulon 3 attenuates the clearance of cytosolic prion aggregates via inhibiting autophagy. Autophagy. 2011;7(2):205–216. doi: 10.4161/auto.7.2.14197. [DOI] [PubMed] [Google Scholar]
- 155.Thomenius MJ, Wang NS, Reineks EZ, Wang Z, Distelhorst CW. Bcl-2 on the endoplasmic reticulum regulates Bax activity by binding to BH3-only proteins. J Biol Chem. 2003;278(8):6243–6250. doi: 10.1074/jbc.M208878200. [DOI] [PubMed] [Google Scholar]
- 156.Yu J, Li X, Tashiro S, Onodera S, Ikejima T. Bcl-2 family proteins were involved in pseudolaric acid B-induced autophagy in murine fibrosarcoma L929 cells. J Pharmacol Sci. 2008;107(3):295–302. doi: 10.1254/jphs.08091fp. [DOI] [PubMed] [Google Scholar]
- 157.Wang W, Fan H, Zhou Y, Duan P, Zhao G, Wu G. Knockdown of autophagy-related gene BECLIN1 promotes cell growth and inhibits apoptosis in the A549 human lung cancer cell line. Mol Med Rep. 2013;7(5):1501–1505. doi: 10.3892/mmr.2013.1379. [DOI] [PubMed] [Google Scholar]
- 158.Li X, Yan J, Wang L, et al. Beclin1 inhibition promotes autophagy and decreases gemcitabine-induced apoptosis in Miapaca 2 pancreatic cancer cells. Cancer Cell Int. 2013;13(1):26. doi: 10.1186/1475-2867-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Castino R, Bellio N, Follo C, Murphy D, Isidoro C. Inhibition of PI3k class III-dependent autophagy prevents apoptosis and necrosis by oxidative stress in dopaminergic neuroblastoma cells. Toxicol Sci. 2010;117(1):152–162. doi: 10.1093/toxsci/kfq170. [DOI] [PubMed] [Google Scholar]
- 160.Ouyang DY, Xu LH, He XH, et al. Autophagy is differentially induced in prostate cancer LNCaP, DU145 and PC-3 cells via distinct splicing profiles of ATG5. Autophagy. 2013;9(1):20–32. doi: 10.4161/auto.22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.An CH, Kim MS, Yoo NJ, Park SW, Lee SH. Mutational and expressional analyses of ATG5, an autophagy-related gene, in gastrointestinal cancers. Pathol Res Pract. 2011;207(7):433–437. doi: 10.1016/j.prp.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 162.Shi M, Zhang T, Sun L, et al. Calpain, Atg5 and Bak play important roles in the crosstalk between apoptosis and autophagy induced by influx of extracellular calcium. Apoptosis. 2013;18(4):435–451. doi: 10.1007/s10495-012-0786-2. [DOI] [PubMed] [Google Scholar]
- 163.Yousefi S, Perozzo R, Schmid I, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8(10):1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 164.Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72(11):8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Heath-Engel HM, Wang B, Shore GC. Bcl-2 at the endoplasmic reticulum protects against a Bax/Bak-independent paraptosis-like cell death pathway initiated via p20Bap31. Biochim Biophys Acta. 2012;1823(2):335–347. doi: 10.1016/j.bbamcr.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 166.Kazi A, Sun J, Doi K, et al. The BH3 alpha-helical mimic BH3-M6 disrupts Bcl-X(L), Bcl-2, and MCL-1 protein-protein interactions with Bax, Bak, Bad, or Bim and induces apoptosis in a Bax- and Bim-dependent manner. J Biol Chem. 2011;286(11):9382–9392. doi: 10.1074/jbc.M110.203638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Howells CC, Baumann WT, Samuels DC, Finkelstein CV. The Bcl-2-associated death promoter (BAD) lowers the threshold at which the Bcl-2-interacting domain death agonist (BID) triggers mitochondria disintegration. J Theor Biol. 2010;271:114–123. doi: 10.1016/j.jtbi.2010.11.040. [DOI] [PubMed] [Google Scholar]
- 168.Zhang N, Hartig H, Dzhagalov I, Draper D, He YW. The role of apoptosis in the development and function of T lymphocytes. Cell Res. 2005;15(10):749–769. doi: 10.1038/sj.cr.7290345. [DOI] [PubMed] [Google Scholar]
- 169.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9(1):47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 170.Aita VM, Liang XH, Murty VV, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59(1):59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 171.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 172.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2(4):330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152(3):519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL’s target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol. 2007;372(1):223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 175.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282(17):13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 176.Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26(10):2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 178.Chang NC, Nguyen M, Germain M, Shore GC. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 2010;29(3):606–618. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30(6):678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ogata M, Hino S, Saito A, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26(24):9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang Y, Wu Y, Cheng Y, et al. Fas-mediated autophagy requires JNK activation in HeLa cells. Biochem Biophys Res Commun. 2008;377(4):1205–1210. doi: 10.1016/j.bbrc.2008.10.151. [DOI] [PubMed] [Google Scholar]
- 182.Li DD, Wang LL, Deng R, et al. The pivotal role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression during anticancer agents-induced autophagy in cancer cells. Oncogene. 2009;28(6):886–898. doi: 10.1038/onc.2008.441. [DOI] [PubMed] [Google Scholar]
- 183.Wu H, Wang MC, Bohmann D. JNK protects Drosophila from oxidative stress by trancriptionally activating autophagy. Mech Dev. 2009;126:8–9. 624–637. doi: 10.1016/j.mod.2009.06.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zalckvar E, Berissi H, Mizrachy L, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10(3):285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zalckvar E, Berissi H, Eisenstein M, Kimchi A. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy. 2009;5(5):720–722. doi: 10.4161/auto.5.5.8625. [DOI] [PubMed] [Google Scholar]
- 186.Bialik S, Kimchi A. The death-associated protein kinases: structure, function, and beyond. Annu Rev Biochem. 2006;75:189–210. doi: 10.1146/annurev.biochem.75.103004.142615. [DOI] [PubMed] [Google Scholar]
- 187.Kang R, Livesey KM, Zeh HJ, Loze MT, Tang D. HMGB1: a novel Beclin 1-binding protein active in autophagy. Autophagy. 2010;6(8):1209–1211. doi: 10.4161/auto.6.8.13651. [DOI] [PubMed] [Google Scholar]
- 188.Tang D, Kang R, Livesey KM, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190(5):881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 2010;17(2):268–277. doi: 10.1038/cdd.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Arsov I, Adebayo A, Kucerova-Levisohn M, et al. A role for autophagic protein beclin 1 early in lymphocyte development. J Immunol. 2011;186(4):2201–2209. doi: 10.4049/jimmunol.1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kovacs JR, Li C, Yang Q, et al. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ. 2012;19(1):144–152. doi: 10.1038/cdd.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.McLeod IX, Zhou X, Li QJ, Wang F, He YW. The class III kinase Vps34 promotes T lymphocyte survival through regulating IL-7Rα surface expression. J Immunol. 2011;187(10):5051–5061. doi: 10.4049/jimmunol.1100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204(1):25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Martin P, Pardo J, Schill N, et al. Granzyme B-induced and caspase 3-dependent cleavage of gelsolin by mouse cytotoxic T cells modifies cytoskeleton dynamics. J Biol Chem. 2010;285(24):18918–18927. doi: 10.1074/jbc.M109.056028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Beaudouin J, Liesche C, Aschenbrenner S, Hörner M, Eils R. Caspase-8 cleaves its substrates from the plasma membrane upon CD95-induced apoptosis. Cell Death Differ. 2013;20(4):599–610. doi: 10.1038/cdd.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Thome M, Tschopp J. Regulation of lymphocyte proliferation and death by FLIP. Nat Rev Immunol. 2001;1(1):50–58. doi: 10.1038/35095508. [DOI] [PubMed] [Google Scholar]
- 197.Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol. 2009;10(4):348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- 198.Yu L, Alva A, Su H, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304(5676):1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 199.Bell BD, Leverrier S, Weist BM, et al. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci U S A. 2008;105(43):16677–16682. doi: 10.1073/pnas.0808597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Thorburn J, Moore F, Rao A, et al. Selective inactivation of a Fas-associated death domain protein (FADD)-dependent apoptosis and autophagy pathway in immortal epithelial cells. Mol Biol Cell. 2005;16(3):1189–1199. doi: 10.1091/mbc.E04-10-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pyo JO, Jang MH, Kwon YK, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280(21):20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 202.Singh BN, Kumar D, Shankar S, Srivastava RK. Rottlerin induces autophagy which leads to apoptotic cell death through inhibition of PI3K/Akt/mTOR pathway in human pancreatic cancer stem cells. Biochem Pharmacol. 2012;84(9):1154–1163. doi: 10.1016/j.bcp.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 203.Levine B, Kroemer G. Autophagy in aging, disease and death: the true identity of a cell death impostor. Cell Death Differ. 2009;16(1):1–2. doi: 10.1038/cdd.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Marx J. Autophagy: is it cancer’s friend or foe? Science. 2006;312(5777):1160–1161. doi: 10.1126/science.312.5777.1160. [DOI] [PubMed] [Google Scholar]
- 205.Torguson R, Waksman R. Overview of the 2007 Food and Drug Administration Circulatory System Devices Panel meeting on the Xience V Everolimus-Eluting Coronary Stent. Am J Cardiol. 2008;102(12):1624–1630. doi: 10.1016/j.amjcard.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 206.Wu YT, Tan HL, Huang Q, Ong CN, Shen HM. Activation of the PI3K-Akt-mTOR signaling pathway promotes necrotic cell death via suppression of autophagy. Autophagy. 2009;5(6):824–834. doi: 10.4161/auto.9099. [DOI] [PubMed] [Google Scholar]
- 207.Chiu HW, Ho SY, Guo HR, Wang YJ. Combination treatment with arsenic trioxide and irradiation enhances autophagic effects in U118-MG cells through increased mitotic arrest and regulation of PI3K/Akt and ERK1/2 signaling pathways. Autophagy. 2009;5(4):472–483. doi: 10.4161/auto.5.4.7759. [DOI] [PubMed] [Google Scholar]
- 208.Charoensuk V, Gati WP, Weinfeld M, Le XC. Differential cytotoxic effects of arsenic compounds in human acute promyelocytic leukemia cells. Toxicol Appl Pharmacol. 2009;239(1):64–70. doi: 10.1016/j.taap.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 209.Zustovich F, Lombardi G, Della Puppa A, Rotilio A, Scienza R, Pastorelli D. A phase II study of cisplatin and temozolomide in heavily pre-treated patients with temozolomide-refractory high-grade malignant glioma. Anticancer Res. 2009;29(10):4275–4279. [PubMed] [Google Scholar]
- 210.Addeo R, De Rosa C, Faiola V, et al. Phase 2 trial of temozolomide using protracted low-dose and whole-brain radiotherapy for nonsmall cell lung cancer and breast cancer patients with brain metastases. Cancer. 2008;113(9):2524–2531. doi: 10.1002/cncr.23859. [DOI] [PubMed] [Google Scholar]
- 211.Nguyen TM, Subramanian IV, Kelekar A, Ramakrishnan S. Kringle 5 of human plasminogen, an angiogenesis inhibitor, induces both autophagy and apoptotic death in endothelial cells. Blood. 2007;109(11):4793–4802. doi: 10.1182/blood-2006-11-059352. [DOI] [PubMed] [Google Scholar]
- 212.Bommareddy A, Hahm ER, Xiao D, et al. Atg5 regulates phenethyl isothiocyanate-induced autophagic and apoptotic cell death in human prostate cancer cells. Cancer Res. 2009;69(8):3704–3712. doi: 10.1158/0008-5472.CAN-08-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gao M, Yeh PY, Lu YS, et al. OSU-03012, a novel celecoxib derivative, induces reactive oxygen species-related autophagy in hepatocellular carcinoma. Cancer Res. 2008;68(22):9348–9357. doi: 10.1158/0008-5472.CAN-08-1642. [DOI] [PubMed] [Google Scholar]
- 214.Vázquez-Martín A, Oliveras-Ferraros C, del Barco S, Martín-Castillo B, Menéndez JA. mTOR inhibitors and the anti-diabetic biguanide metformin: new insights into the molecular management of breast cancer resistance to the HER2 tyrosine kinase inhibitor lapatinib (Tykerb) Clin Transl Oncol. 2009;11(7):455–459. doi: 10.1007/s12094-009-0384-0. [DOI] [PubMed] [Google Scholar]
- 215.Liu TJ, Koul D, LaFortune T, et al. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol Cancer Ther. 2009;8(8):2204–2210. doi: 10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yuk JM, Shin DM, Song KS, et al. Bacillus calmette-guerin cell wall cytoskeleton enhances colon cancer radiosensitivity through autophagy. Autophagy. 2010;6(1):46–60. doi: 10.4161/auto.6.1.10325. [DOI] [PubMed] [Google Scholar]
- 217.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68(5):1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 218.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11(4):448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 219.Amaravadi RK, Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res. 2007;13(24):7271–7279. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 220.Carew JS, Nawrocki ST, Kahue CN, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110(1):313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Takahashi A, Kimura T, Takabatake Y, et al. Autophagy guards against cisplatin-induced acute kidney injury. Am J Pathol. 2012;180(2):517–525. doi: 10.1016/j.ajpath.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 222.Tai CJ, Wang CK, Tai CJ, et al. Aqueous Extract of Solanum nigrum Leaves Induces Autophagy and Enhances Cytotoxicity of Cisplatin, Doxorubicin, Docetaxel, and 5-Fluorouracil in Human Colorectal Carcinoma Cells. Evid Based Complement Alternat Med. 2013 doi: 10.1155/2013/514719. [Pubmed: 23843876] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Shinojima N, Yokoyama T, Kondo Y, Kondo S. Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy. Autophagy. 2007;3(6):635–637. doi: 10.4161/auto.4916. [DOI] [PubMed] [Google Scholar]
- 224.Han X, Xu B, Beevers CS, et al. Curcumin inhibits protein phosphatases 2A and 5, leading to activation of mitogen-activated protein kinases and death in tumor cells. Carcinogenesis. 2012;33(4):868–875. doi: 10.1093/carcin/bgs029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Nikrad M, Johnson T, Puthalalath H, Coultas L, Adams J, Kraft AS. The proteasome inhibitor bortezomib sensitizes cells to killing by death receptor ligand TRAIL via BH3-only proteins Bik and Bim. Mol Cancer Ther. 2005;4(3):443–449. doi: 10.1158/1535-7163.MCT-04-0260. [DOI] [PubMed] [Google Scholar]
- 226.Bonner HP, Concannon CG, Bonner C, Woods I, Ward MW, Prehn JH. Differential expression patterns of Puma and Hsp70 following proteasomal stress in the hippocampus are key determinants of neuronal vulnerability. J Neurochem. 2010;114(2):606–616. doi: 10.1111/j.1471-4159.2010.06790.x. [DOI] [PubMed] [Google Scholar]
- 227.Hagenbuchner J, Ausserlechner MJ, Porto V, et al. The anti-apoptotic protein BCL2L1/Bcl-xL is neutralized by pro-apoptotic PMAIP1/Noxa in neuroblastoma, thereby determining bortezomib sensitivity independent of prosurvival MCL1 expression. J Biol Chem. 2010;285(10):6904–6912. doi: 10.1074/jbc.M109.038331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Juvekar A, Manna S, Ramaswami S, et al. Bortezomib induces nuclear translocation of IκBα resulting in gene-specific suppression of NF-κB—dependent transcription and induction of apoptosis in CTCL. Mol Cancer Res. 2011;9(2):183–194. doi: 10.1158/1541-7786.MCR-10-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ri M, Iida S, Ishida T, et al. Bortezomib-induced apoptosis in mature T-cell lymphoma cells partially depends on upregulation of Noxa and functional repression of Mcl-1. Cancer Sci. 2009;100(2):341–348. doi: 10.1111/j.1349-7006.2008.01038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pérez-Galán P, Roué G, López-Guerra M, et al. BCL-2 phosphorylation modulates sensitivity to the BH3 mimetic GX15-070 (Obatoclax) and reduces its synergistic interaction with bortezomib in chronic lymphocytic leukemia cells. Leukemia. 2008;22(9):1712–1720. doi: 10.1038/leu.2008.175. [DOI] [PubMed] [Google Scholar]
- 231.Zang Y, Thomas SM, Chan ET, et al. The next generation proteasome inhibitors carfilzomib and oprozomib activate prosurvival autophagy via induction of the unfolded protein response and ATF4. Autophagy. 2012;8(12):1873–1874. doi: 10.4161/auto.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117(2):326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Han J, Hou W, Goldstein LA, et al. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J Biol Chem. 2008;283(28):19665–19677. doi: 10.1074/jbc.M710169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6(7):891–900. doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Enhancement of tumor-TRAIL susceptibility by modulation of autophagy. Autophagy. 2008;4(7):940–943. doi: 10.4161/auto.6769. [DOI] [PubMed] [Google Scholar]
- 236.French LE, Tschopp J. The TRAIL to selective tumor death. Nat Med. 1999;5(2):146–147. doi: 10.1038/5505. [DOI] [PubMed] [Google Scholar]
- 237.Walczak H, Miller RE, Ariail K, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5(2):157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 238.Bhojani MS, Rossú BD, Rehemtulla A. TRAIL and anti-tumor responses. Cancer Biol Ther. 2003;2(4 Suppl 1):S71–S78. [PubMed] [Google Scholar]
- 239.Leahomschi S, Molinsky J, Klanova M, et al. Multi-level disruption of the extrinsic apoptotic pathway mediates resistance of leukemia cells to TNF-related apoptosis-inducing ligand (TRAIL) Neoplasma. 2013;60(2):223–31. doi: 10.4149/neo_2013_030. [DOI] [PubMed] [Google Scholar]
- 240.Park KJ, Lee SH, Kim TI, et al. A human scFv antibody against TRAIL receptor 2 induces autophagic cell death in both TRAIL-sensitive and TRAIL-resistant cancer cells. Cancer Res. 2007;67(15):7327–34. doi: 10.1158/0008-5472.CAN-06-4766. [DOI] [PubMed] [Google Scholar]
- 241.Han W, Pan H, Chen Y, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS ONE. 2011;6(6):e18691. doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Huang S, Sinicrope FA. Celecoxib-induced apoptosis is enhanced by ABT-737 and by inhibition of autophagy in human colorectal cancer cells. Autophagy. 2010;6(2):256–269. doi: 10.4161/auto.6.2.11124. [DOI] [PMC free article] [PubMed] [Google Scholar]