Abstract
Summary: Skyline is a Windows client application for targeted proteomics method creation and quantitative data analysis. The Skyline document model contains extensive mass spectrometry data from targeted proteomics experiments performed using selected reaction monitoring, parallel reaction monitoring and data-independent and data-dependent acquisition methods. Researchers have developed software tools that perform statistical analysis of the experimental data contained within Skyline documents. The new external tools framework allows researchers to integrate their tools into Skyline without modifying the Skyline codebase. Installed tools provide point-and-click access to downstream statistical analysis of data processed in Skyline. The framework also specifies a uniform interface to format tools for installation into Skyline. Tool developers can now easily share their tools with proteomics researchers using Skyline.
Availability and implementation: Skyline is available as a single-click self-updating web installation at http://skyline.maccosslab.org. This Web site also provides access to installable external tools and documentation.
Contact: brendanx@u.washington.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The Skyline software for quantitative targeted proteomics has become increasingly popular since its original publication (MacLean et al., 2010), with >3000 registered users and >6000 instances started most weeks. The rich Skyline document schema contains extensive data generated from targeted proteomics experiments. This combination has motivated researchers and instrument vendors developing software tools with applications in targeted quantitative proteomics to seek seamless integration with Skyline. The raw material for this integration has long been available through custom reports, designed inside Skyline and exported to comma-separated value files for further analysis. The impact of third-party tools created to analyze these reports, however, has been limited by the difficulty with which they could be shared among researchers. Existing tools have been developed outside of Skyline in R, Python and Java or exist as Web sites (Röst et al., 2012). Without detailed and specific knowledge of these tools and their running environments outside Skyline, researchers using Skyline have found them difficult to install and use. The Skyline external tools framework was designed to address these issues by providing a mechanism for researchers to create extensible, shareable and user-friendly tools for use with Skyline.
2 DESIGN AND IMPLEMENTATION
The prior existence of tools for analysis of Skyline results required the creation of an extensible interface that could integrate a wide variety of tool implementation approaches into a uniform medium. The other significant goal of the framework was to enable Skyline users to easily use these tools. These two requirements motivated decisions to support the following:
Point-and-click integration within the Skyline graphical user interface (GUI)
Different tool implementation platforms, including but not limited to R and Python scripts, executable files and Web sites
Guided installation of tools from a single ZIP file
Tool versioning and installation of upgrades
Inclusion in the web-based Tool Store http://skytools.maccosslab.org
Integration of the Tool Store with Skyline
Striving to achieve these goals motivated advances in Skyline that include per replicate annotations, forms support for collecting arguments to command-line programs, posting report data to a web page, automated installation of R, Python and required packages and report caching for repeated tool use.
To demonstrate the usefulness and broad applicability of this software framework, we integrated the following statistical tools implemented in R and Python: MSstats (Choi et al., 2014), QuaSAR and MS1 Probe (Supplementarty Note S1). Others have added more tools (Supplementarty Note S2), available from the Skyline External Tool Store.
2.1 Integration
Integrating external tools within the existing Skyline application provided an opportunity to greatly improve the user experience when running tools. The initial points of integration are accomplished by adding new menu items to a Tools menu inside Skyline (Fig. 1, Supplementary Fig. S1), a Tool Store from which they can be installed and upgraded (Supplementary Figs S2–S4), and a form for configuring them (Fig. 2, Supplementary Fig. S5), modeled after similar User Interface (UI) in Microsoft Visual Studio (Supplementary Fig. S6). These menu items allow users to run each tool through a simple point-and-click interface. To gain more than simply launching separate software, however, the framework must provide these tools with data and context from the open Skyline document. This transfer of information is achieved by allowing each menu item to be associated with a specific Skyline custom report format, giving tool creators a wide range of data values to select from as input for their tools. In the process of integrating our three initial tools, it became clear that each required extra information about the mass spectrometric acquisitions being processed, such as concentration, condition, time point and subject ID. Support for this level of annotation had been implemented in the tools by parsing file names or, alternatively, by requiring a separate sample annotation file. To formalize entry of these values and alleviate the need for an extra file, we implemented per replicate annotations—specified as text, numbers, true/false values and value lists—that can be exported with any custom report (Supplementary Figs S7–S8). Additionally, context-sensitive macros—using the format $(DocumentDir), $(SelPeptide)—may be specified on the tool command line allowing tools to target highly specific data elements within a Skyline document, such as the document directory or the amino acid sequence of the selected peptide.
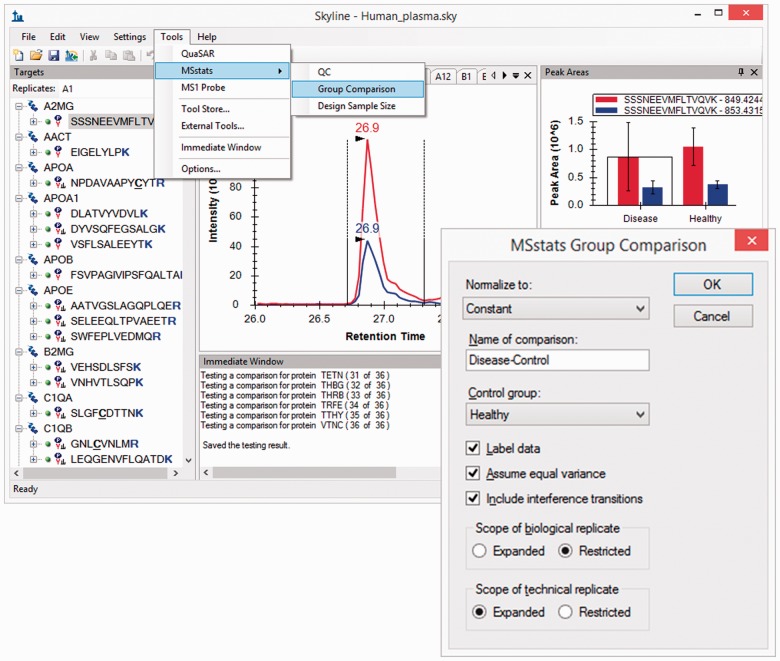
Fig. 1.
Skyline UI with populated tools menu and immediate window output. A GUI form to collect user input
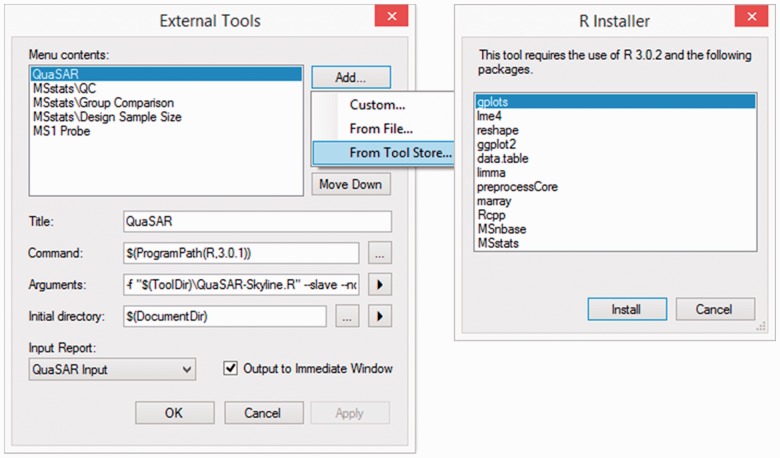
Fig. 2.
The External Tools form for adding a zipped tool. Skyline support for automated R and package installation
The framework allows installed tools to complete a full analysis and provide feedback without ever leaving the Skyline application. When running a tool, Skyline will automatically generate a required custom report and provide it directly to the tool itself, through standard input, a temporary file on disk or a post to a web page. Exported reports are cached to support multiple runs of a given tool quickly.
2.2 Ease of use
When a user clicks a tool menu item, Skyline invokes the external tool itself, whether that tool is an executable file, a Web site or a script for another program like R or Python. Tools can be specified so that the output from the tool run is redirected into the Skyline’s immediate window, a text pane docked in the Skyline application main display (Fig. 1). As a result, the tool can be run entirely within Skyline, without requiring the user to open or switch to another program.
Our three initial tools were implemented in R and Python, with many options but also with interfaces that limited naïve user interaction. Before full integration with Skyline, MSstats and MS1 Probe both used command window input prompts and QuaSAR used a web form on a page that required user sign-in, preventing direct integration with Skyline. To improve ease of use for Skyline users, we introduced GUI forms that can be used to specify input values to each tool and change settings and options associated with each tool. A tool creator with basic knowledge of any Microsoft .NET language can create a small .NET project implementing a simple form to collect user input. The project can be compiled to a dynamically linked library file and included in the tool installation ZIP file, described below. Skyline will check the existence of such a form when a tool is run and display it to the user (Fig. 1).
All three initial tools benefited greatly from adding .NET forms to provide tool users with a more intuitive interface. The forms provide default values and settings that allow the user to run a basic analysis by simply clicking an OK button, or changing just a few values for a custom analysis. The forms also verify input, display information about what certain settings do and save settings changes from run to run.
2.3 Sharing installable tools
The external tools framework was designed to be compatible with existing tools and also flexible enough to support many future tools and tool version upgrades. External tools are packaged as compressed ZIP files that contain all the necessary components required to install and use the tools (Supplementary Fig. S9). Once users have obtained the ZIP file of an external tool, they can simply navigate to the External Tools form (Fig. 2) within Skyline and begin the installation process by selecting the file location on the local directory. Skyline will then perform a number of steps to install the tool. For example, Skyline will verify whether the ZIP file is structured properly and handle conflicts if the tool requires the use of the existing custom reports or custom annotations (Supplementary Fig. S10).
The installable tools framework has extra support for tools using R or Python scripts (Supplementary Fig. S11). If necessary, Skyline will download these programs and guide the user through their installation and the installation of any associated packages required to use a tool (Fig. 2). Ultimately, once a tool is fully installed into Skyline, the user will not have to perform any other steps to use the tool.
3 SUMMARY AND FUTURE DIRECTION
Through the Skyline external tools framework, Skyline users will gain broader access to tools developed within the scientific community that can provide meaningful analyses of the data generated in their experiments, and tool creators will reach a large audience of proteomics researchers. We have established an External Tool Store web page (http://skytools.maccosslab.org) to connect the Skyline user base with tool creators, so that users may easily search for and install new tools and tool developers may push changes in existing tools to researchers using them.
Funding: This work was supported in part by a grant from National Institutes of Health (R01 GM103551). Additional support was provided by Agilent Technologies through an industry gift to the Skyline project.
Conflicts of Interest: none declared.
Supplementary Material
REFERENCES
- MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röst H, et al. A computational tool to detect and avoid redundancy in selected reaction monitoring. Mol. Cell. Proteomics. 2012;11:540–549. doi: 10.1074/mcp.M111.013045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.