Abstract
Motivation: The recent advance in high-throughput sequencing technologies is generating a huge amount of data that are becoming an important resource for deciphering the genotype underlying a given phenotype. Genome sequencing has been extensively applied to the study of the cancer genomes. Although a few methods have been already proposed for the detection of cancer-related genes, their automatic identification is still a challenging task. Using the genomic data made available by The Cancer Genome Atlas Consortium (TCGA), we propose a new prioritization approach based on the analysis of the distribution of putative deleterious variants in a large cohort of cancer samples.
Results: In this paper, we present ContastRank, a new method for the prioritization of putative impaired genes in cancer. The method is based on the comparison of the putative defective rate of each gene in tumor versus normal and 1000 genome samples. We show that the method is able to provide a ranked list of putative impaired genes for colon, lung and prostate adenocarcinomas. The list significantly overlaps with the list of known cancer driver genes previously published. More importantly, by using our scoring approach, we can successfully discriminate between TCGA normal and tumor samples. A binary classifier based on ContrastRank score reaches an overall accuracy >90% and the area under the curve (AUC) of receiver operating characteristics (ROC) >0.95 for all the three types of adenocarcinoma analyzed in this paper. In addition, using ContrastRank score, we are able to discriminate the three tumor types with a minimum overall accuracy of 77% and AUC of 0.83.
Conclusions: We describe ContrastRank, a method for prioritizing putative impaired genes in cancer. The method is based on the comparison of exome sequencing data from different cohorts and can detect putative cancer driver genes.
ContrastRank can also be used to estimate a global score for an individual genome about the risk of adenocarcinoma based on the genetic variants information from a whole-exome VCF (Variant Calling Format) file. We believe that the application of ContrastRank can be an important step in genomic medicine to enable genome-based diagnosis.
Availability and implementation: The lists of ContrastRank scores of all genes in each tumor type are available as supplementary materials. A webserver for evaluating the risk of the three studied adenocarcinomas starting from whole-exome VCF file is under development.
Contact: emidio@uab.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
In the past few years, next-generation sequencing (NGS)-based screening has become an important tool for the detection of genetic variants associated with many genetic disorders (Bamshad et al., 2011). Its application on the study of cancer genomes allowed the discovery of several cancer-related genes (Imielinski et al., 2012; Kandoth et al., 2013; Tamborero et al., 2013). The Cancer Genome Atlas (TCGA) Consortium (Cancer Genome Atlas Research Network, 2008) is producing a huge amount of cancer genome sequencing data for >30 cancer types. This enables the detection of an increasing number of variants potentially involved in cancer (Futreal et al., 2004; Imielinski et al., 2012; Kandoth et al., 2013; Lawrence et al., 2014; Stratton et al., 2009; Tamborero et al., 2013). However, the interpretation of genetic variants is a challenging problem (Capriotti et al., 2012; Fernald et al., 2011), and it is difficult to differentiate variation in specific genes responsible for the development and progression of cancer (drivers) from the background (passengers). In general, the prediction of cancer driver mutations is based on the conservation analysis of mutated sites (Capriotti and Altman, 2011; Carter et al., 2009; Kaminker et al., 2007).
The accurate detection of driver mutations is important to define cancer driver genes that play a causative role in oncogenesis through exerting a selective advantage to the cancer cells. So far, several methods for identifying cancer driver genes have been reported (Cerami et al., 2012; Cheng et al., 2014; Dees et al., 2012; Gonzalez-Perez and Lopez-Bigas, 2012; Khurana et al., 2013; Lawrence et al., 2013; Youn and Simon, 2011). The prevalent strategy to identify cancer driver genes works by detecting significantly over-mutated genes in tumors, which are more likely the drivers (Alexandrov et al., 2013; Dees et al., 2012; Khurana et al., 2013). Most of the methods compare the frequency of mutations in an individual gene with the mutation frequency of other genes in the same or related tumors after correction for sequence context and gene size (Meyerson et al., 2010; Vogelstein et al., 2013). Using this approach, a considerable number of driver genes have been discovered in a variety of cancer types. However, many seemingly unrelated genes have also been identified in recent cancer genome sequencing studies (Garraway and Lander, 2013; Watson et al., 2013). The heterogeneity of mutational processes within individuals and cancer types could explain this anomaly (Lawrence et al., 2013). Therefore, there is a pressing need for robust methods to identify cancer driver genes (Meyerson et al., 2010).
We describe here a new probabilistic approach (ContrastRank) to identify putative cancer driver genes based on the estimation of the variation rates of genes in 1000 Genome Project (1000 Genomes Project Consortium, 2010) and TCGA normal samples. ContrastRank can assign a score to each genome, which can discriminate normal from the tumor samples and amongst different tumor types. We tested the method on three whole-exome sequencing data of adenocarcinomas from TCGA. Our method performs highly in discriminating normal from tumor samples, as well as, different types of adenocarcinomas.
2 METHODS
2.1 Definitions and assumptions
We assume that rare variants are more likely to have functional effect than common variants and among the rare variants the non-synonymous single nucleotide variants (nsSNVs) have the strongest impact. This assumption is supported by the analysis of annotated variants in dbSNP (Sherry et al., 2001), which shows that the fraction of annotated pathogenic variants is significantly higher for nsSNVs (Supplementary Fig. S1A). Moreover, among the nsSNVs, the rare ones harbor significantly higher fraction of deleterious variants (Supplementary Fig. S1B). Thus, we define a putative deleterious variant (PDV) as the nsSNV with allele frequency <0.5%. This frequency threshold for filtering nsSNVs has been recently used to estimate genomic regions under purifying selection (Khurana et al., 2013). We also define putative impaired genes (PIGs) as those genes that carry at least one PDV. For each gene in a set of samples, we can calculate its putative defective rate (PDR) as the fraction of samples in which a given gene carries at least one PDV.
2.2 Datasets
We used three datasets of whole-exome sequence made available by TCGA consortium. We selected three types of adenocarcinomas for which there are >200 pairs of normal/tumor samples that we consider to be the minimum number of samples to perform a 2-fold cross-validation test. For each tumor type, we selected the largest datasets of samples for colon adenocarcinoma (COAD) produced by the Baylor College of Medicine and lung and prostate adenocarcinomas (LUAD and PRAD, respectively) produced by the Broad Institute of MIT and Harvard. The three selected datasets are composed by 220, 625 and 309 matching pairs of normal and tumor samples from patients respectively affected by COAD, LUAD and PRAD.
We also analyzed the genomes of 1092 individuals made available by the 1000 Genomes Consortium. (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20110521). In our analysis, the variation data from eight genes in the chromosome Y were not considered because of the lower number of individuals for which the data are available and missing genotype data for some of the alleles in the samples. We used ANNOVAR (Wang et al., 2010) to annotate the effect of the genetic variants in each VCF file from TCGA and 1000 genomes using the human genome build 19 (hg19).
For tumor datasets specific filtering procedures have been adopted to extract the genetic variants from the Variant Calling Format (VCF) files (Supplementary Methods 1). The filtering procedure applied to COAD, LUAD and PRAD samples allowed us to select an average number of nsSNVs per sample that is comparable with the recently estimated value (∼10 000) (Bamshad et al., 2011). Average values of nsSNVs for the normal and tumor samples in COAD, LUAD, PRAD and 1000 Genomes samples are reported in Supplementary Table S1.
In our analysis, we only focus on PDVs with minor allele frequency (MAF) <0.5%. The MAF is derived from the genomes of 1092 individuals in 1000 Genomes Consortium. All the nsSNVs found in TCGA samples, but not in 1000 Genomes, were considered to have even lower frequencies, and therefore, assumed to be PDVs.
After filtering, the number of PDVs in normal and tumor samples is between 10–16% of the whole set of nsSNVs. The average number of PDVs per individual in 1000 Genome is 318. This is in agreement with the previous published result (1000 Genomes Project Consortium et al., 2012). We mapped all the PDVs to their corresponding genes and calculated the average number of PIGs for each sample. We found that on average the PDVs are affecting ∼700 and 900 PIGs in normal and tumor samples, respectively (Supplementary Table S1). The distributions of the nsSNVs and PDVs across different samples of the three tumor types are shown in Supplementary Figure S2. In addition, a flow chart summarizing the procedure used for preprocessing our datasets is provided in Supplementary Figure S3.
2.3 Gene prioritization score
To discriminate between cancer and normal samples, we adopted a gene prioritization approach based on the analysis of PIGs in normal and tumor subsets. The basic idea behind our statistical approach is that the lower the probability of observing a gene mutated in multiple normal samples the higher the probability of it being a cancer driver gene, when frequently mutated in tumor samples. We estimated the probability of a gene g of being classified k times as a PIG in a set of N tumor samples using a binomial distribution,
| (1) |
where, πg is the probability of having at least one PDV on the gene g. Therefore, the probability Pg of observing x mutated samples where gene g is potentially impaired and with x ≥ k is as follows:
| (2) |
where k > 0. Using this modified version of the cumulative distribution, we can estimate the probability that a gene g is k or more times classified as a PIG on our dataset.
The missing variable for the estimation of Pg is the probability of having at least one PDV in gene g (πg). In our approach, we derived this parameter from the analysis of the occurrence of PIGs in TCGA normal and 1000 Genomes samples. Assuming that rare PDVs have strong functional impact with respect to other types of variants, we classify a gene as a PIG only if it contains at least one PDV with MAF ≤ 0.5% in 1000 Genomes (Khurana et al., 2013). Therefore, given a set of samples I = {I1,I2, … .IN}, where N is the total number of samples, the probability πg can be estimated by calculating the PDR of the gene g in the dataset I composed by N samples. In our analysis, we defined πg as the maximum PDR value for the gene g in TCGA normal and 1000 Genomes samples. We consider this value as the background PDR of each gene.
The πg values described above allow us to calculate the probability Pg that each gene g is classified as a PIG in k or more genomes in a given set of tumor samples. We derive a final score for each gene as follows:
| (3) |
In the case, where a gene does not harbor any PDV neither in normal TCGA nor in 1000 Genomes samples, an arbitrary PDR of 5 × 10−4 is assigned to this gene. This smoothing of probability is about half of the probability that could be observed by Laplace correction (add one when a value is missing) in 1000 Genomes (1/1092).
2.4 Exome scoring method
We used the gene scores described above to discriminate between normal and tumor samples. For each genome, we extracted the list of M putative impaired genes (PIGs) G = {g1,g2, … .. ,gM} with at least one PDV with allele frequency <0.5% in 1000 Genomes and calculated the average score S as follows:
| (4) |
where Pg and sg are defined in Equations (2) and (3).
2.5 Benchmarking
To verify the quality of the prioritization score implemented in ContastRank, we compared our results with that of MutSigCV (Lawrence et al., 2013). For this comparison, we assumed three manually curated lists, namely Bushman (Bushman, 2013), COSMIC Census (Forbes et al., 2011) and Vogelstein (Vogelstein et al., 2013) as true positive cancer-related genes (Supplementary Methods 2.1).
We tested the performance of our method (ContrastRank) for its ability to discriminate between normal and tumor samples. We used a simple binary classifier based on the score threshold to separate normal from tumor samples. To test ContastRank, we used a 2-fold cross-validation procedure that maximizes the level of variability among different training sets. The performance of our method has been compared with those of the two alternative approaches namely ContastLow and ContastDiff (see Supplementary Methods 2.3). Finally, we also tested the ability of ContastRank approach to discriminate between the three different tumor types considered in this paper (COAD, LUAD and PRAD).
A detailed description of the methods, the benchmark procedures and the definitions the standard performance measures used in this work are reported in Sections 2 and 3 of the Supplementary Methods.
3 RESULTS
In this paper, we describe ContrastRank, a method that relies on the analysis of PIGs in the TCGA normal and cancer samples for prioritizing potential cancer driver genes.
For each gene in each cancer type (COAD, LUAD and PRAD), we calculated probabilistic scores that allow us to discriminate TCGA normal from tumor samples. In the following sections, we described the results of our analysis of three TCGA adenocarcinoma datasets and the performance of ContrastRank algorithm on each tumor type.
3.1 Analysis of TCGA samples and gene prioritization
3.1.1 Analysis of the cancer prioritization lists
The COAD dataset is composed of 220 normal–tumor pairs. After extracting the PDVs from all the samples, we identified on average 996 and 1276 PDVs in normal and tumor samples, respectively. These variants correspond to an average value of 643 and 880 PIGs for normal and tumor genomes. Analyzing the occurrence of PDVs in the normal samples, we found 14 449 genes with at least one PDV, 50% of which have ≥3 PDVs. This corresponds to an average PDR of 1.4%. In the cancer samples, we observed 17 006 genes with at least one PDV and a median value of 5 PDVs for each gene and an average PDR of 2.3%. By calculating the distributions of all the PDRs in our datasets, the percentage of genes with PDR ≤ 0.05 are about 95, 92 and 82%, respectively, for 1000 Genomes, TCGA normal and tumor samples (Supplementary Fig. S4A).
The LUAD dataset is composed of 625 normal–tumor pairs. The average PDVs are 1202 and 1599 PDVs in normal and tumor samples, respectively which affect on average 751 and 1041 PIGs. In the whole LUAD dataset, we found 16 891 PIGs of which half with ≥3 PDVs and an average PDR of 1.1%. In the cancer samples, there are 18 213 PIGs with a median value of 14 PDVs and an average PDR of 2.2%. Finally, the percentages of PIGs with PDR ≤ 0.05 are about 96, 91 and 81% respectively for 1000 Genomes, TCGA normal and tumor samples (Supplementary Fig. S4B).
Similar results are observed for the PRAD dataset, which is composed of 309 normal–tumor sample pairs. On average we found 1321 and 1540 PDVs respectively in normal and tumor genomes, which correspond to 819 and 953 PIGs. In the whole set of normal samples, there are 15 457 PIGs 50% of which with ≥4 PDVs, and average PDR of 1.3%. In the cancer samples, in total we observed 16 765 PIGs with a median value of 6 PDVs for each PIG and an average PDR of 1.9%. In the case of PRAD, the percentage of PIGs with PDR ≤ 0.05 are about 95, 88 and 84% respectively for 1000 Genomes, TCGA normal and tumor samples (Supplementary Fig. S4C).
3.1.2 Cancer gene prioritization scores
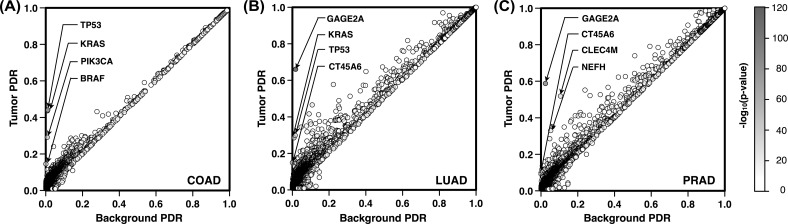
In the next step, we used the PDRs, to prioritize PIGs for each tumor type. We report in Figure 1 a scatter plot representing for each PIG the background PDR on the x-axis (maximum PDR between TCGA normal and 1000 Genomes samples) and the PDR in tumor samples on the y-axis. The points closer to the diagonal have low score because the background PDR and the tumor PDR are similar. Genes with higher impact correspond to the darker points, which are far from the diagonal. In COAD samples, KRAS and TP53 on average have at least one PDV in 48 tumor samples out of 110. Accordingly, these highly scored genes, are almost overlapping, showing PDR values of ∼0.44 in tumor samples and ∼0.01 as background (Fig. 1A). For LUAD, the top genes GAGE2A and KRAS have PDR values of 0.66 and 0.29, respectively (Fig. 1B).
Fig. 1.
(A–C) Scatter plot of tumor versus background PDRs for all PIGs. On the x-axis is reported the background PDR of each gene which corresponds to the maximum PDR in normal and 1000 Genomes samples. On the y-axis are reported the PDRs calculated on the tumor samples. The gray scale on the side assigns darker colors to highly scored PIGs
These values are significantly higher than the background PDR (∼0.01). GAGE2A is also the highest scoring gene in PRAD (Fig. 1C), with at least one PDV in 65 tumor samples out of 110. This corresponds to a PDR of 0.59 in tumor samples, which is significantly higher than the background PDR (0.02). On average, CLEC4M has been observed to be a PIG respectively in 12 and 57 normal and tumor samples over 110. This difference makes CLEC4M the second highest ranked gene for PRAD.
To take into account the possible variability in terms of PDVs across different samples, we analyzed the 10 bootstap samples composed of 110 pairs of cancer and normal samples. This number of pairs corresponds to half the size of the smallest cohort (COAD) among our TCGA datasets. For each one of the subsets, we calculated the PDR of each PIG in tumor samples and compared it against the background PDR value (maximum PDR value observed in TCGA normal and 1000 Genomes samples). Using the equations in Section 2.3, we were able to assign to each gene a logarithmic score that represents its probability of being a PIG in k samples over a set of N tumor samples. The final score calculated for each PIG is the average value of the results obtained on the 10 sampling experiments. Performing this analysis we found 139, 318 and 96 genes with scores larger than 3 for COAD, LUAD and PRAD, respectively. The first four high-ranking genes for COAD are KRAS, TP53, PIK3CA and BRAF. For LUAD, the highest scoring genes are GAGE2A, KRAS, CT45A6 and TP53. For PRAD, these genes are GAGE2A, CLEC4M, CT45A6 and NEFH. The complete list of all the genes and their average scores are included in the Supplementary Files.
3.1.3 Comparing ContrastRank prioritization score
We compared the performance of ContastRank against MutSigCV (Lawrence et al., 2013). We used three manually curated lists of cancer-related genes (Supplementary Methods 2.1) as benchmark datasets and calculated the area under the receiver operating characteristic (ROC) curve (AUC). The AUC (Supplementary Methods 2.3) is obtained by evaluating the false- and true-positive rates at different P-value threshold from the prioritization lists returned by ContastRank and MutSigCV. Although the lack of standard benchmark set for estimating the quality of predictions can potentially affect our results, nevertheless, the ROC curves and the AUCs in Supplementary Figure S5 and Supplementary Table S2 show that ContrastRank consistently performs better than MutSigCV. Considering the Bushman list as benchmark, both methods results in lower AUC values, but ContrastRank shows 3–7% better accuracy than MutSigCV. For the Vogelstein list, both methods perform better and ContrastRank shows even better AUCs (4–15%) than MutSigCV. An intermediate level of improvement is observed in the case of COSMIC cancer gene list (Supplementary Table S2).
3.2 Discriminating TCGA normal and tumor samples
We used ContrastRank to classify normal and cancer samples by scoring the whole genome using the scores derived from individual PIGs. The score assigned to the whole genome is the average values of the scores of all PIGs in a sample (Supplementary Methods 2.3). We used a simple binary classifier that could discriminate between normal and tumor. This method has been applied to each cancer type in our dataset. To evaluate the accuracy of our gene prioritization score, we estimated the performance of ContrastRank using an increasing subset of high scoring genes.
3.2.1 Analysis of ContrastRank results
The performance of ContrastRank in discriminating COAD samples from the normal was calculated dividing the whole COAD dataset in 2 subsets of 110 pairs of tumor/normal samples. Depending on the random sampling, the highest scored gene was either KRAS or TP53. Using only the score of the highest-ranking gene, the method showed an accuracy of 71% (Matthews correlation 0.51 and AUC 0.71). The same method based on all the genes but the first (ContrastLow) attained an accuracy of 70% (correlation 0.38 and AUC 0.61). Expectedly, using only the first ranking gene, the performances of ContrastRank and ContrastDiff were identical. Including more genes in the analysis made ContastRank to perform better than ContrastDiff (Supplementary Fig. S5 panels A and C). The main accuracy measures (Q2, C and AUC) began to diverge after the use of the first four genes.
In this case, ContastRank showed 92% accuracy (Matthews correlation coefficient 0.84 and AUC 0.92). In comparison with ContrastDiff, this corresponds to 6% higher accuracy, 0.1 higher correlation, and 0.03 higher AUC. The quality of our cancer gene prioritization approach is also confirmed by the decreasing performance of ContrastLow when removing a significant number of highly scored genes (Supplementary Fig. S5B). If the score threshold for selecting high scoring gene was arbitrarily set to 3, we found on average 239 genes passed this threshold. Thus, using genes scoring >3, ContastRank showed higher performance than ContrastLow and ContrastDiff (Table 2).
Table 2.
Performance of the method in discriminating tumor types
| Tumor | Q2 | PPV | TPR | NPV | TNR | C | AUC | NG |
|---|---|---|---|---|---|---|---|---|
| COAD | 0.98 | 0.97 | 0.99 | 0.99 | 0.97 | 0.96 | 0.99 | 107/128 |
| LUAD | 0.77 | 0.74 | 0.83 | 0.82 | 0.71 | 0.55 | 0.83 | 274/28 |
| PRAD | 0.84 | 0.78 | 0.95 | 0.94 | 0.72 | 0.69 | 0.89 | 59/199 |
Note: Q2, overall accuracy; PPV and NPV, positive and negative predicted values; TPR and TNR, true positive and negative rates; MCC, Matthew’s correlation; AUC, area under the (ROC) curve. NG is the number of top positive/ lowest negative genes with score higher than 3/ lower than −3.
For LUAD samples, we preformed a 2-fold cross-validation on 625 pairs of normal/tumor samples. The highest scoring PIG resulting from the analysis of randomly selected subsets of samples was GAGE2A. Using only the GAGE2A score to discriminate between normal and tumor samples, ContrastRank method resulted in 82% overall accuracy (Matthews correlation 0.67 and AUC 0.81). On the same subset, ContrastLow ranking method that used all the gene scores but GAGE2A resulted in 74% overall accuracy (Matthews correlation 0.55 and AUC 0.62). As the PDR of GAGE2A in tumor sample was significantly higher than that of normal samples, the accuracy of ContrastDiff method based on the first gene was the same as ContrastRank. The large number of genes with high PDRs in LUAD made the performance of ContrastRank to significantly diverge from ContrastDiff after considering the first 50 highly scored PIGs (Supplementary Fig. S6 panels A and C). Indeed, when the whole genome was scored using the 50 highest ranked genes ContrastRank reached an overall accuracy of 94% (correlation 0.88 and AUC 0.98). When compared with ContrastDiff, these were 5% higher overall accuracy, 0.1 higher correlation and 0.03 higher AUC. The performance of ContrastLow is reported in Supplementary Figure S6B. When a score threshold of 3 was used for selecting highly ranked genes, we found 494 high scoring PIGs. Using PIGs with score >3, ContrastRank performed at 97% overall accuracy with correlation coefficient 0.93 and AUC 0.99 (Table 2).
Finally, in the case of PRAD, we performed 2-fold cross-validation procedure randomly selecting normal/tumor samples from a cohort of 309 matching pairs. Similar to LUAD, also for PRAD the highest scored PIG was GAGE2A. Using only the first ranking gene ContrastRank reached 78% overall accuracy with 0.61 correlation and 0.78 AUC. In comparison with ContrastDiff, the performance of ContrastRank began to be significantly better after selecting the first 30 highest-ranking PIGs (Supplementary Fig. S7 panels A and C). In that case, ContrastRank reached 90% overall accuracy with 0.81 correlation and 0.96 AUC. Compared with ContrastDiff, these values were of 6% higher overall accuracy, 0.13 higher correlation and 0.04 better AUC. When removing the first 1000 highly scored genes (∼6%), the performance of ContrastLow dropped down to 0.54 overall accuracy, 0.09 correlation and 0.52 AUC (Supplementary Fig. S7B). Using PIGs with score higher than 3, ContrastRank method resulted in 91% overall accuracy with 0.83 correlation coefficient and 0.97 AUC (Table 1).
Table 1.
Performance of the methods on different tumor types
| Tumor | Method | Q2 | PPV | TPR | NPV | TNR | C | AUC |
|---|---|---|---|---|---|---|---|---|
| COAD | CRank | 0.92 | 0.97 | 0.86 | 0.87 | 0.97 | 0.84 | 0.94 |
| CLow | 0.72 | 0.78 | 0.66 | 0.72 | 0.78 | 0.47 | 0.79 | |
| CDiff | 0.76 | 0.78 | 0.77 | 0.80 | 0.74 | 0.55 | 0.83 | |
| LUAD | CRank | 0.97 | 0.97 | 0.96 | 0.96 | 0.97 | 0.93 | 0.99 |
| CLow | 0.84 | 0.88 | 0.79 | 0.81 | 0.89 | 0.69 | 0.91 | |
| CDiff | 0.89 | 0.89 | 0.90 | 0.90 | 0.88 | 0.79 | 0.96 | |
| PRAD | CRank | 0.91 | 0.92 | 0.91 | 0.91 | 0.92 | 0.83 | 0.97 |
| CLow | 0.70 | 0.66 | 0.75 | 0.72 | 0.74 | 0.43 | 0.77 | |
| CDiff | 0.79 | 0.83 | 0.74 | 0.77 | 0.83 | 0.58 | 0.87 |
Note: Performance of ContrastRank, ContrastLow and ContastDiff (respectively, CRank, CLow and CDiff) calculated using an average number 239, 494 and 127 genes with score >3 respectively for COAD, LUAD and PRAD. Q2, overall accuracy; PPV and NPV, positive and negative predicted values; TPR and TNR, true positive and negative rates; MCC, Matthew’s correlation; AUC = area under the (ROC) curve.
3.2.2 Performances with unrelated normal samples
To estimate the lower bound performance of ContrastRank in the discrimination between normal and tumor samples, we tested our method using a 2-fold cross-validation approach (CV Unseen) where the normal samples are swapped between the two subsets (Supplementary Methods 2.2). With this procedure, none of the normal samples in one subset were from the same patient as the tumor samples. The results in Supplementary Table S3 show an average decrease of the overall accuracy from 2–15% and 1–16% in AUC with respect to the standard cross-validation procedure (CV Identifier). The decrease of performance is inversely proportional to the number of samples of each tumor type.
3.3 Discriminating adenocarcinomas samples
We evaluated the ability of ContrastRank to discriminate tumor samples from different types of adenocarcinomas. For this purpose, we built three dataset containing 50% of the samples from the tumor type under study (positive cases) and the other 50% equally divided between the two remaining tumor types (negative cases). Next, we calculated the score associated with each PIG in both the halves of the dataset. We used the difference between the scores obtained for the subset of positive cases (adenocarcinoma under study) and negative cases (mixture of the other two tumor types). According to this definition, we had highly positive scores that correspond to genes with high PDR, significantly higher in tumor type under study, and negative scores, which are associated to genes with high PDR in remaining tumor types.
We used both set of genes in our classification scheme and calculated the performance for each tumor types. We first analyzed the scheme to discriminate COAD from the other two tumor types (LUAD and PRAD). The gene with observed highest score for COAD were TP53 and KRAS, and the lowest negative scores were for CT45A6 and GAGE2A. This is in agreement with the previous results where KRAS and TP53 were highly ranked gene for discrimination of COAD tumors from normal samples. On the other hand, GAGE2A and CT45A6 were highly scored genes for LUAD and PRAD.
When discriminating LUAD samples from the others, the highest positively discriminating genes were GAGE2A and CT45A6 and highest negatively discriminating genes were PIK3CA and SPOP. Interestingly, KRAS, a high scoring PIG in LUAD, was not a high scoring discriminator because it is highly scored in a negative tumor type, namely COAD. In contrast, PIK3CA, a gene with low PDR in LUAD, became a strong negative discriminator because of its high PDR in a negative case, again COAD. Finally, for PRAD, the highest positive discriminators were GAGE2A and CLEC4M and the highest negative discriminators were KRAS and TP53. These data were in agreement with the previous results on PRAD dataset that assigned high PDR scores to GAGE2A and CLEC4M. The lowest negative discriminating scores assigned to KRAS and TP53 were justified because of their being among the top four PIGs for the negative cases, i.e. COAD and LUAD. The discrimination scores used in these tests are reported in the Supplementary Files.
To prove the ability of our procedure based on the difference of scores obtained with the ContrastRank method, we evaluated the accuracy of a binary classifier able to discriminate between the tumor under study and the remaining types. The performances were calculated considering an increasing number of highly discriminative PIGs. Because in the new scoring scale negative scores are present, we included in the list of selected PIGs an equal numbers of highest positive and lowest negative scoring genes. In Supplementary Figure S8 we report the average values of the main accuracy measures (Q2, C and AUC) for each tumor type. At first sight, it is possible to notice that LUAD is the most difficult tumor to distinguish from the other types. This observation is confirmed by the results in Table 2 where we report the accuracy of our ContrastRank method using PIG genes with scores higher than 3 and lower than −3. With this cutoff score, COAD tumor samples could be discriminated with an overall accuracy of 98% (correlation 0.96 and AUC 0.99) using an average number of 107 top positive and 128 lowest negative genes. With the same threshold the performance on LUAD samples was the poorest, reaching 77% overall accuracy (correlation 0.55 and AUC 0.83) using an average number of 274 positively and 28 negatively scored genes. In the case of PRAD, the method showed an intermediate level of accuracy, reaching 84% (correlation 0.69 and AUC 0.89) using on average number of 59 top and 199 lowest ranking genes.
4 DISCUSSION
Accurate variant calling and appropriate filtering procedures are important prerequisites for the analysis of whole-exome sequencing data. Although alternative variant calling procedures can result in different number of variants, the consistency of our datasets composition (Supplementary Table S1), in terms of total SNVs, PDVs and PIGs, makes us confident about the average quality of nsSNVs used in our analysis. This is also confirmed by similar distributions of PDVs and nsSNVs in each sample (Supplementary Fig. S2). The general idea of using rare nsSNVs is supported by the analysis of variants annotated in dbSNP (Supplementary Fig. S1) Furthermore, the selection of 0.5% as a threshold for defining PDVs is consistent with previously large-scale analysis of human genetic variants (Khurana et al., 2013). The analysis of selected PDVs and the distribution of PDRs for PIGs in each dataset (1000 Genomes, TCGA normal and tumor samples) shows an increasing percentage of PIGs with PDR > 0.05 from 1000 Genomes to TCGA tumor samples (Supplementary Fig. S4). On average, for the three tumor types (COAD, LUAD and PRAD), the number of PIGs with PDRs > 0.05 are 4.5, 9.5 and 17.7% in 1000 Genomes, TCGA normal and tumor samples, respectively. This observation is in agreement with the idea that samples for normal tissues in patients affected by cancer can contain a high rate of putative functionally deleterious variants than samples from healthy individuals.
ConstrastRank allows us to prioritize possible cancer driver genes. We first compared the performance of ContrastRank against MutSigCV using three different manually curated lists of cancer-related genes, namely Bushman, COSMIC Census and Vogelstein (see Section 2.5). Although a real evaluation of the performance is still a difficult task in absence of a true benchmark set, ContrastRank performs better than MutSigCV for all three tumor types under study (Supplementary Fig. S5 and Supplementary Table S2).
Looking at ContrastRank results, for COAD, we find 139 genes with ContrastRank with average score >3. Comparing these genes with the Bushman, COSMIC Census and Vogelstein gene lists we found between 11 and 27% possible cancer-causing genes in common (Supplementary Table S4). Using the Fisher’s exact test to compare the number of oncogenes in highly ranked PIGs and the number of oncogenes over the remaining genes, we found P-values lower than 10−7 for all the benchmark sets. In addition, we found that eight of our highly ranked genes are included in the list of 11 high significantly mutated colorectal cancer genes provided by Broad Institute (http://cancergenome.broadinstitute.org/). The same analysis on LUAD dataset detected 319 PIGs, between 5 to 15% of which are in the three cancer gene lists. Comparing the fraction of oncogene in the highly ranked genes against the number of oncogenes on the remaining genes, we found a significant P-value lower than 0.01. Comparing our list of LUAD genes with score >3 with the list provided by Broad Institute, we found 11 common genes over 14. Similar results are obtained for prostate cancer for which we found a total number of 96 highly scored genes, 5 to 19% of which are also in the benchmark lists of cancer-related genes. Also in this case, the comparison of the distributions of cancer genes in low and high scored subset resulted in Fisher’s exact test P-values lower than 0.01. In the case of PRAD, the Broad Institute only reports one gene that is also included in the list of our highly scored genes. All the numbers about the comparison between the gene lists obtained using ContrastRank and those in Bushman, COSMIC Census and Vogelstein gene lists are summarized in Supplementary Table S4.
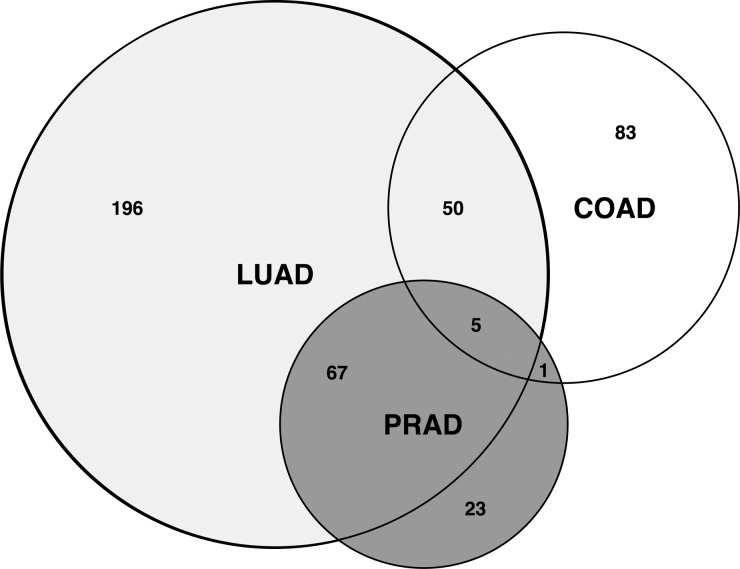
We showed that ContrastRank scores could discriminate TCGA normal from tumor samples, as well as different types of adenocarcinomas from each other. Using genes with score higher than 3, we showed that our approach could discriminate tumor from normal samples with an overall accuracy >90% and AUC >0.95 for each tumor type (see Table 1). Good levels of performances are also obtained in a more stringent test (CV Unseen) in which the normal and tumor samples with same identifier were kept disjoint (Supplementary Table S3). Furthermore, the difference of ContrastRank scores could discriminate different tumor types. Using a cutoff score of 3, the results show that in the worst case (LUAD), our method reaches an average overall accuracy of 77% and AUC 0.83. Better performances are obtained for PRAD and COAD (Table 2). These results are in agreement with the analysis of highly scored genes for each tumor type. Using common size datasets composed by 220 samples equally distributed between positive and negative cases, we found 139, 318 and 96 high scored genes (average score > 3) for COAD, LUAD and PRAD, respectively. Comparing these lists of genes we found that 75 and 40% of highly scored PIGs in PRAD and COAD respectively are overlapping with highly scored genes in LUAD (Fig. 2). In this scenario, LUAD seems to involve a larger spectrum of PIGs with respect to COAD and PRAD. The observed heterogeneity could be explained by larger sample size for the LUAD dataset, this reasoning is not true for COAD that in spite of having around 2/3 of the samples in the PRAD dataset (Supplementary Table S1) results in higher number of high scored PIGs. Our analysis also revealed that TP53, BRAF, AR, NBEA, RNF145 are the five common PIGs across the three adenocarcinomas.
Fig. 2.
Venn diagram representing the number of PIGs with average scores >3 for colon, lung and prostate adenocarcinomas (respectively COAD, LUAD and PRAD). Among those genes TP53, BRAF, NBEA, AR, RNF145 are in common in the three adenocarcinomas
In conclusion, in this paper we presented ContastRank, a new method for prioritizing cancer-related genes. According to our analysis, our method is able to detect already well-known genes and identify new genes potentially involved in the insurgence and progression of tumor. Although we showed that ContastRank reaches good performance even when compared against MutSigCV, a further calibration of the method is needed before it could be applied to larger sets of genomic data. The main hurdles are standardization of the variant calling procedure, optimization of the method for selecting PDVs, inclusion of other types of variants other than nonsynonymous and selection of representative set of normal samples that capture genetic heterogeneity of each tumor type.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge The Cancer Genome Atlas Consortium for allowing access to the restricted whole-exome sequencing data for colon, lung and prostate adenocarcinomas. We thank the anonymous reviewers for their helpful comments that allowed us to improve the quality of this paper.
Funding: E.C. and M.K.B. were supported by start-up funds from the Department of Pathology at the University of Alabama, Birmingham.
Conflicts of Interest: none declared.
REFERENCES
- 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Bushman F. Cancer gene list. 2013 http://www.bushmanlab.org/links/genelists. [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Altman RB. A new disease-specific machine learning approach for the prediction of cancer-causing missense variants. Genomics. 2011;98:310–317. doi: 10.1016/j.ygeno.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, et al. Bioinformatics for personal genome interpretation. Brief. Bioinform. 2012;13:495–512. doi: 10.1093/bib/bbr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WC, et al. DriverDB: an exome sequencing database for cancer driver gene identification. Nucleic Acids Res. 2014;42:D1048–D1054. doi: 10.1093/nar/gkt1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees ND, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernald GH, et al. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011;27:1741–1748. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, et al. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic Acids Res. 2012;40:e169. doi: 10.1093/nar/gks743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, et al. Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res. 2007;67:465–473. doi: 10.1158/0008-5472.CAN-06-1736. [DOI] [PubMed] [Google Scholar]
- Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana E, et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, et al. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, et al. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborero D, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci. Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, et al. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson IR, et al. Emerging patterns of somatic mutations in cancer. Nat. Rev. Genet. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn A, Simon R. Identifying cancer driver genes in tumor genome sequencing studies. Bioinformatics. 2011;27:175–181. doi: 10.1093/bioinformatics/btq630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.