Abstract
Recently, a genome-wide association study (GWAS) that identified eight single-nucleotide polymorphisms (SNPs) associated with BMI highlighted a possible neuronal influence on the development of obesity. We hypothesized these SNPs would govern the response of BMI and subcutaneous fat to resistance training in young individuals (age = 24 years). We genotyped the eight GWAS-identified SNPs in the article by Willer et al. in a cohort (n = 796) that undertook a 12-week resistance-training program. Females with a copy of the rare allele (C) for rs17782313 (MC4R) had significantly higher BMIs (CC/CT: n = 174; 24.70 ± 0.33 kg/m2, TT: n = 278; 23.41 ± 0.26 kg/m2, P = 0.002), and the SNP explained 1.9% of overall variation in BMI. Males with a copy of the rare allele (T) for rs6548238 (TMEM18) had lower amounts of subcutaneous fat pretraining (CT/TT: n = 65; 156,534 ± 7,415 mm3, CC: n = 136; 177,825 ± 5,139 mm3, P = 0.019) and males with a copy of the rare allele (A) for rs9939609 (FTO) lost a significant amount of subcutaneous fat with exercise (AT/AA: n = 83; −798.35 ± 2,624.30 mm3, TT: n = 47; 9,435.23 ± 3,494.44 mm3, P = 0.021). Females with a copy of the G allele for a missense variant in the SH2B1 (rs7498665) was associated with less change of subcutaneous fat volume with exercise (AG/GG: n = 191; 9,813 ± 2,250 mm3 vs. AA: n = 126; 770 ± 2,772 mm3; P = 0.011). These data support the original finding that there is an association between measures of obesity and a variant near the MC4R gene and extends these results to a younger population and implicates FTO, TMEM18, and SH2B1 polymorphisms in subcutaneous fat regulation.
The World Health Organization estimates that there are one billion adults who are overweight (BMI >25 kg/m2), with 300 million of these individuals clinically obese (BMI >30 kg/m2). These statistics suggest that Westernized societies are yielding to the global epidemic of obesity (1). Science has progressed to the point where we have begun to understand the biology behind obesity and illustrated the enormous role that genetic–environment interactions (G × E) play in the obese state.
There have been numerous genome-wide association studies (GWAS) in the past 2 years that have discovered loci for BMI (2–6). Previously, we examined the effect of one of these loci (INSIG2; rs7566605) on the interaction between resistance training and subcutaneous fat levels in healthy college-aged individuals (age = 24 years) (7). We found that the INSIG2 polymorphism underlies variation in subcutaneous adiposity in young adult females and suppresses the positive effects of resistance training in young men.
In this study, eight newly revealed GWAS loci (NEGR1 (rs2815752), MTCH2 (rs10838738), TMEM18 (rs6548238), GNPDA2 (rs10938397), SH2B1 (rs7498665), MC4R (rs17782313), FTO (rs9939609), and KCTD15 (rs11084753)) that have been associated with increased BMI in individuals aged 43–65 years were examined for associations with BMI in a younger population (4). We hypothesized that these loci would show a stronger effect in young people due to removal of interactions with age. In addition, we wanted to examine the BMI-associated loci for a potential role in influencing changes in BMI and subcutaneous fat in response to resistance training.
Methods
Subjects
The 796 individuals included in this report were white college-aged adults living within the United States and Ireland who participated in a supervised resistance-training program as part of the Functional SNPs Associated with Muscle Size and Strength (FAMUSS) study (8) (clinical characteristics are reported in Table 1). The study protocol was approved by an ethics committee at Children’s National Medical Center (institutional review board protocol #2449) and at all testing sites.
Table 1.
D emographic characteristics of the Functional SNPs Associated with Muscle Size and Strength cohort
| BMI cohort | Volumetric fat cohort | |||||||
|---|---|---|---|---|---|---|---|---|
| Females | Males | Females | Males | |||||
| Characteristic | N | Mean ± s.d. | N | Mean ± s.d. | N | Mean ± s.d. | N | Mean ± s.d. |
| Age (years) | 475 | 23.12 ± 5.48 | 321 | 23.72 ± 5.47 | 335 | 22.81 ± 5.17 | 211 | 23.91 ± 5.66 |
| Height (cm) | 478 | 165.02 ± 6.80 | 323 | 177.97 ± 6.88 | 338 | 165.02 ± 7.00 | 212 | 178.19 ± 6.64 |
| Pre-exercise weight (kg) | 478 | 65.23 ± 12.81 | 323 | 79.77 ± 15.98 | 338 | 64.53 ± 12.17 | 212 | 78.19 ± 14.44 |
| Postexercise weight (kg) | 479 | 65.75 ± 12.79 | 323 | 80.22 ± 15.89 | 338 | 65.05 ± 12.29 | 213 | 78.73 ± 14.62 |
| Pre-exercise BMI (kg/m2) | 478 | 23.95 ± 4.53 | 323 | 25.15 ± 4.70 | 338 | 23.68 ± 4.18 | 212 | 24.57 ± 4.02 |
| Postexercise BMI (kg/m2) | 479 | 24.14 ± 4.51 | 323 | 25.30 ± 4.65 | 338 | 23.86 ± 4.25 | 213 | 24.76 ± 4.00 |
| Difference in BMI (kg/m2) | 478 | 0.19 ± 0.87 | 322 | 0.13 ± 0.80 | 338 | 0.19 ± 0.82 | 212 | 0.18 ± 0.82 |
| Pre-exercise subcutaneous fat volume (mm3) | 338 | 256,784 ± 114,086 | 213 | 170,126 ± 88,886 | ||||
| Postexercise subcutaneous fat volume (mm3) | 337 | 263,082 ± 116,803 | 210 | 174,163 ± 93,611 | ||||
| Difference in subcutaneous fat volume (mm3) | 337 | 6,181 ± 30,619 | 210 | 3,996 ± 23,857 | ||||
Measures of obesity
Anthropometric measures were collected using a protocol standardized across all study centers. Height and weight were measured using a calibrated wall-mounted stadiometer and scale, respectively. BMI was calculated as weight in kilogram divided by height in meters squared (kg/m2). In addition to BMI, magnetic resonance imaging measurements of subcutaneous fat were used as another measure of adiposity (7). We were not able to obtain magnetic resonance images for all FAMUSS study participants (see Table 1); therefore, data from only 546 of the 796 total subjects were used for analysis of subcutaneous fat volume.
Exercise training program
A 12-week resistance-training program was performed with the nondominant arm (trained arm). The protocol has been described elsewhere (7–9).
Genotyping
Genomic DNA was isolated from peripheral blood lymphocytes with the Gentra Puregene DNA extraction kit (Qiagen, Valencia, CA) and genotyping was completed using Taqman assays from ABI (Foster City, CA) according to the manufacturer’s instructions (see Table 2 for assay IDs). The end point fluorescent readings were performed on an ABI 7900HT Sequence Detection System (SDS V 2.3 software; Applied Biosystems, Foster City, CA).
Table 2.
Genotyping assays for GWAS-discovered SNPs
| dBSNP ID | Assay IDa |
|---|---|
| rs2815752 | C__26668839_10 |
| rs10838738 | C____432493_10 |
| rs6548238 | C__29311887_10 |
| rs10938397 | C___1594245_10 |
| rs7498665 | C__25999166_10 |
| rs17782313 | C__32667060_10 |
| rs9939609 | C__30090620_10 |
| rs11084753 | C__31497814_10 |
GWAS, genome-wide association study; SNP, single-nucleotide polymorphism.
From Applied Biosystems website.
Statistical analyses
Hardy–Weinberg equilibrium was tested for each single-nucleotide polymorphism (SNP) using a one-degree of freedom χ2-test. Baseline BMI and subcutaneous fat volume were analyzed as continuous quantitative traits. Due to large gender differences in baseline values, all analyses were performed separately for males and females. Bivariate correlation analyses of BMI and fat volume showed statistically significant correlations with age and baseline weight; therefore, genotype–phenotype associations were assessed using analysis of covariance, with BMI adjusted for age and fat volume adjusted for age and body weight. All significant associations from the main analysis of covariance model were subjected to pair-wise statistical tests among the three genotype groups. Linear tests were performed between each of the genotype groups to determine which groups were significantly different from one another. Linear regression analysis, including likelihood ratio tests between full (containing genotype and covariates) and constrained (containing covariates only) models were performed to estimate the proportion of variance in volumetric measurements that was attributable to genotype. Analyses used a nominal P value of 0.05 as significant. The resulting P values from these tests were adjusted for multiple comparisons using the Sidak post hoc test. To address to some extent the problem of multiple testing, we compared our unadjusted P values to a significance level of 0.006 (0.05/8 SNPs) for BMI, subcutaneous fat, and changes in BMI and subcutaneous fat with unilateral-resistance training on the nondominant arm.
Results
We studied participants in the FAMUSS study of young adults (average age 24 years), enrolled into a unilateral upper arm 12-week supervised resistance-training program. The demographics of our population of European-descent (white) individuals are provided in Table 1. We have divided the study population into two groups (BMI cohort and volumetric fat cohort) because we were not able to measure subcutaneous fat volumes from the magnetic resonance images of some subjects.
The weight gains in the university-based population studied are well documented (10). After the 12-week study period, females gained 0.87 ± 3.21% kg, (pre/postexercise: 65.23 ± 12.81/65.75 ± 12.80 kg, P < 0.0001 and males gained 0.55 ± 2.69% kg of weight (pre/postexercise: 79.81 ± 15.94/80.22 ± 15.89 kg, P = 0.003). Therefore females added 0.9% to their BMI and males added 0.7%. Both the trained and untrained arms in females and males as a whole gained subcutaneous fat volume after 12 weeks of resistance training, with the gains significant in females (3%, P = 0.005; 2.8%, P = 0.13 females and males, respectively).
We tested eight GWAS loci previously associated with BMI in older adults (NEGR1 (rs2815752), MTCH2 (rs10838738), TMEM18 (rs6548238), GNPDA2 (rs10938397), SH2B1 (rs7498665), MC4R (rs17782313), FTO (rs9939609), and KCTD15 (rs11084753)) (4). All eight variants were in Hardy–Weinberg equilibrium in our population of European-descent individuals. The allele frequencies for the SNPs are provided in Table 3. The common allele frequencies for our population (whites) were very similar to the allele frequencies for the HapMap CEPH population (Utah residents with ancestry from northern and western Europe).
Table 3.
Genotype frequencies of SNPs for participants in FAMUSS
| dBSNP ID | Alleles (+/−) | Effect Allelea | Frequency in FAMUSS (+) |
Published frequency for CEU (+)b |
Allele (+) (%) | Heteros (%) | Allele (−) (%) |
|---|---|---|---|---|---|---|---|
| rs2815752 | A/G | A | 0.639 | 0.637 | 40.1 | 47.5 | 12.4 |
| rs10838738 | G/A | G | 0.663 | 0.637 | 43.7 | 45.3 | 11.0 |
| rs6548238 | C/T | C | 0.818 | 0.850 | 67.1 | 29.4 | 3.5 |
| rs10938397 | A/G | G | 0.571 | 0.554 | 31.1 | 51.9 | 17.0 |
| rs7498665 | A/G | G | 0.612 | 0.619 | 37.7 | 47.0 | 15.3 |
| rs17782313 | T/C | T | 0.781 | 0.735 | 61.1 | 33.9 | 5.0 |
| rs9939609 | T/A | A | 0.602 | 0.540 | 35.1 | 50.1 | 14.8 |
| rs11084753 | G/A | G | 0.667 | 0.690 | 44.6 | 44.1 | 11.4 |
CEU, European Caucasian; FAMUSS, Functional SNPs Associated with Muscle Size and Strength; SNP, single-nucleotide polymorphism.
According to Willer et al. (4).
Utah residents with northern and western European ancestry from the CEPH collection used in HAPMAP.
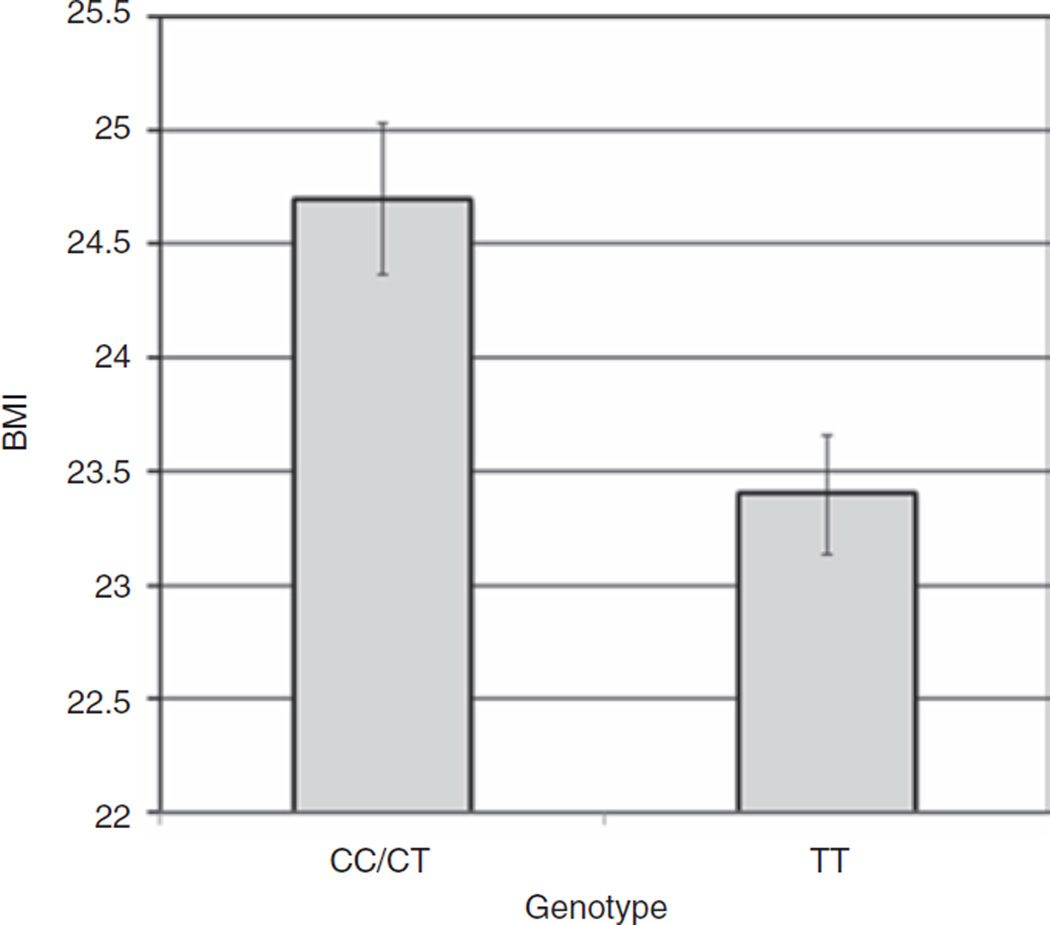
Each locus was then tested for association with BMI and subcutaneous fat volume, and the change in these variables in response to resistance training (see Supplementary Table S1). The allele frequencies for the eight SNPs and the alleles associated with BMI in the Willer et al. GWAS (4) are provided in Table 3. One of the eight SNPs we tested was found to be associated with BMI in our young cohort. Specifically, the MC4R rs17782313 SNP was associated with BMI in females using a dominant model (CC/CT: n = 174; 24.70 ± 0.33 kg/m2, TT: n = 278; 23.41 ± 0.26 kg/m2, P = 0.003). This SNP explained 1.9% (P = 0.002) of the phenotype (Figure 1). There were no significant associations with BMI response to upper arm 12-week resistance training. NEGR1 (rs2815752), MTCH2 (rs10838738), GNPDA2 (rs10938397), and KCTD15 (rs11084753) SNPs did not show any associations with BMI, subcutaneous fat, and their response to resistance training in our young cohort.
Figure 1.
Effect of genotype for the MC4R SNP rs17782313 on BMI levels in females. Females with a copy of the C allele for the MC4R SNP (rs17782313) showed higher values for BMI (CC/CT: N = 174; 24.70 ± 0.33 kg/m2 vs. TT: N = 278; 23.41 ± 0.26 kg/m2; P = 0.003) and this accounted for 1.9% of the BMI phenotype (P = 0.002).
Males with a copy of the rare allele (T) for rs6548238 (TMEM18 gene) had lower amounts of subcutaneous fat pretraining (CT/TT: n = 65; 156,534 ± 7,415 mm3, CC: n = 136; 177,825 ± 5,139 mm3, P = 0.019). This SNP explained 0.12% of phenotype.
Two of the SNPs that we examined were associated with changes in subcutaneous fat with resistance training. Females with a copy of the G allele for a missense variant in the SH2B1 gene (rs7498665) that changes a Thr to Ala at position 484 was associated with less of a difference in subcutaneous fat volume using a dominant model after the 12-week training period (AG/GG: n = 191; 9,813 ± 2,250 mm3 vs. AA: n = 126; 770 ± 2,772 mm3; P = 0.011). This SNP explained 2.0% of the phenotype in women. The FTO rs9939609SNP was associated with difference in subcutaneous fat volume in men using a dominant model (AT/AA: n = 83; −798.35 ± 2,624.30 mm3 vs. TT: n = 47; 9,435.23 ± 3,494.44 mm3; P = 0.02). This SNP explained 4.1% of the phenotype in men. However, adjustment for multiple testing showed only the baseline BMI in females associated with allele (C) for rs17782313 (MC4R) to remain statistically significant.
Discussion
The current obesity epidemic is related to decreased habitual physical activity levels, changes in dietary intake, and genetic predisposition to obesity (11). In this study, we hypothesized that GWAS loci associated with BMI in older populations may have a stronger genetic effect on BMI in a younger, healthy cohort. However, our results show that only one of the eight GWAS-identified SNPs was associated with BMI in our young population. Specifically, we ascertained that a variant near the MC4R gene (rs17782313) is associated with BMI values in young women. This work confirms one of the GWAS results from Willer et al. (4) for BMI and extends the findings to a younger population. The gender specificity observed for this variant warrants further investigation.
We found the rare allele for rs6548238 in the TMEM18 gene to be associated with lower values of subcutaneous fat volume in the upper arm before the 12-week training period in males. This same allele (T) shows an association with lower values of BMI in 91,469 individuals as part of a large GWAS (4,5). The TMEM18 gene is involved in the modulation of neural stem cell migration to glimoa cells (12). In addition, this gene is expressed at high levels in the brain and hypothalamus and points to a neuronal component as part the development of obesity (4).
The young adult cohort studied completed a 12-week supervised unilateral resistance-training program of the upper nondominant arm. After completion of the resistance training, both males and females showed an increase in BMI and subcutaneous fat volume (see Table 1). We hypothesized that the alleles associated with higher BMI values in the GWAS from Willer et al. (4) would be associated with stunted changes in BMI and subcutaneous fat volume in response to resistance training. The response of subcutaneous fat to resistance training has been varied, with multiple studies showing a reduction in subcutaneous fat as measured by magnetic resonance imaging (13–15) and one study showing no change in subcutaneous fat (16) with resistance training. We discovered that males with a copy of the rare allele (A) for the FTO variant (rs9939609) lost a significant amount of subcutaneous fat in the upper arm with resistance training. This result is in agreement with a article by Mitchell et al. (17) that showed women with two copies of the rare allele for the rs8050136 SNP (in complete linkage disequilibrium with rs9939609) lost significantly more weight than carriers of the common allele with aerobic training (cycle ergometers or treadmill). However, another study showed that carriers of the common allele for rs8050136 lost three times greater percent body fat with aerobic exercise (18). Both of these studies used a different form of exercise (aerobic exercise) than the FAMUSS study (resistance training). In addition, both studies used different durations for their aerobic exercise program, with the Mitchell et al. (17) study using 6 months (3–4 sessions a week) and the Rankinen et al. (18) study using a 20-week program (3 sessions a week). Our data show that there is an interaction between the FTO rs9939609 SNP and resistance training in men. However, this association needs to be explored further and tested in other cohorts using different exercise modalities.
In addition, we discovered an interaction between a missense mutation in the SH2B1 gene (rs7498665; Thr484Ala) with resistance training in women. Women with two copies of the common allele (A allele; Thr) did not gain as much subcutaneous fat as women with a copy of the rare risk allele (G) after the unilateral- resistance training. The common allele has been shown to be associated with a lower value of BMI in 91,469 individuals as part of a large GWAS (4,5). Mice with a disruption of the Sh2b1 gene resulted in metabolic disorders including hyperlipidemia, leptin resistance, hyperphagia, obesity, hyperglycemia, insulin resistance, and glucose intolerance (19). This gene is thought to regulate energy balance (19) and may interact with resistance training to promote weight homeostasis (20).
There are some limitations to our study. Only one arm was trained for the 12-week time period; therefore, individuals did not see any loss in weight or BMI, which has been shown for whole body resistance training (13–15). In addition, we do not have a replication group that provides insurance against errors and bias that can affect an individual study’s results (21). Finally, three of the associations are not significant after correction for multiple testing but the results have biological relevance because of previous associations for BMI values (4,5). In addition, the statistical community is divided on correction methods for multiple testing as the Bonferonni correction may be too harsh and thus increasing false negatives (22). The results would need to be validated in additional populations (21).
In summary, we found that a variant near the MC4R gene (rs17782313) is associated with BMI in young females and a variant in the TMEM18 gene (rs6548238) to be associated with subcutaneous fat volume in males. In addition, we found that the FTO SNP that has been associated with exercise response (17,18,23) has an effect on subcutaneous fat loss following resistance training in males and, a missense variant in the SH2B1 gene was associated with a greater gain in subcutaneous fat following resistance training in women. These findings emphasize that genetics can be utilized in combination with resistance training to lower subcutaneous fat levels and may ultimately provide the public with genetic information that can influence exercise choice.
Supplementary Material
Acknowledgments
This study was supported by a grant from the National Institutes of Health 3R01AR055100-06. F.E.O.-S. designed the study, genotyped the samples, analyzed data, and drafted the manuscript. B.T.H. assisted in genotyping. H.G.D. ran the statistics, analyzed and interpreted data and drafted the manuscript. P.M.C., M.J.H., P.D.T., T.J.A., P.M.G., N.M.M., L.S.P., P.S.V., and R.F.Z. recruited the subjects, collected the clinical data, and supervised exercise trainings. J.M.D. and E.P.H. designed the study and drafted the manuscript.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/oby
All authors contributed to the final critical revision of the manuscript.
Disclosure
The authors declared no conflict of interest.
REFERENCES
- 1.Obesity: preventing and managing the global epidemic. Report of a WHO consultation. World Health Organ Tech Rep Ser. 2000;894:i–xii. 1–253. [PubMed] [Google Scholar]
- 2.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 6.Herbert A, Gerry NP, McQueen MB, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 7.Orkunoglu-Suer FE, Gordish-Dressman H, Clarkson PM, et al. INSIG2 gene polymorphism is associated with increased subcutaneous fat in women and poor response to resistance training in men. BMC Med Genet. 2008;9:117. doi: 10.1186/1471-2350-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson PD, Moyna N, Seip R, et al. Functional polymorphisms associated with human muscle size and strength. Med Sci Sports Exerc. 2004;36:1132–1139. doi: 10.1249/01.mss.0000132274.26612.23. [DOI] [PubMed] [Google Scholar]
- 9.Kostek MA, Pescatello LS, Seip RL, et al. Subcutaneous fat alterations resulting from an upper-body resistance training program. Med Sci Sports Exerc. 2007;39:1177–1185. doi: 10.1249/mss.0b0138058a5cb. [DOI] [PubMed] [Google Scholar]
- 10.Cluskey M, Grobe D. College weight gain and behavior transitions: male and female differences. J Am Diet Assoc. 2009;109:325–329. doi: 10.1016/j.jada.2008.10.045. [DOI] [PubMed] [Google Scholar]
- 11.Campbell WW, Crim MC, Young VR, Evans WJ. Increased energy requirements and changes in body composition with resistance training in older adults. Am J Clin Nutr. 1994;60:167–175. doi: 10.1093/ajcn/60.2.167. [DOI] [PubMed] [Google Scholar]
- 12.Jurvansuu J, Zhao Y, Leung DS, et al. Transmembrane protein 18 enhances the tropism of neural stem cells for glioma cells. Cancer Res. 2008;68:4614–4622. doi: 10.1158/0008-5472.CAN-07-5291. [DOI] [PubMed] [Google Scholar]
- 13.Wilmore JH. Alterations in strength, body composition and anthropometric measurements consequent to a 10-week weight training program. Med Sci Sports. 1974;6:133–138. [PubMed] [Google Scholar]
- 14.Treuth MS, Ryan AS, Pratley RE, et al. Effects of strength training on total and regional body composition in older men. J Appl Physiol. 1994;77:614–620. doi: 10.1152/jappl.1994.77.2.614. [DOI] [PubMed] [Google Scholar]
- 15.Staron RS, Leonardi MJ, Karapondo DL, et al. Strength and skeletal muscle adaptations in heavy-resistance-trained women after detraining and retraining. J Appl Physiol. 1991;70:631–640. doi: 10.1152/jappl.1991.70.2.631. [DOI] [PubMed] [Google Scholar]
- 16.Häkkinen K, Pakarinen A, Kraemer WJ, Newton RU, Alen M. Basal concentrations and acute responses of serum hormones and strength development during heavy resistance training in middle-aged and elderly men and women. J Gerontol A Biol Sci Med Sci. 2000;55:B95–B105. doi: 10.1093/gerona/55.2.b95. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell JA, Church TS, Rankinen T, et al. FTO genotype and the weight loss benefits of moderate intensity exercise. Obesity (Silver Spring) 2010;18:641–643. doi: 10.1038/oby.2009.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rankinen T, Rice T, Teran-Garcia M, Rao DC, Bouchard C. FTO genotype is associated with exercise training-induced changes in body composition. Obesity (Silver Spring) 2010;18:322–326. doi: 10.1038/oby.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren D, Zhou Y, Morris D, et al. Neuronal SH2B1 is essential for controlling energy and glucose homeostasis. J Clin Invest. 2007;117:397–406. doi: 10.1172/JCI29417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirier P, Després JP. Exercise in weight management of obesity. Cardiol Clin. 2001;19:459–470. doi: 10.1016/s0733-8651(05)70229-0. [DOI] [PubMed] [Google Scholar]
- 21.Hattersley AT, McCarthy MI. What makes a good genetic association study? Lancet. 2005;366:1315–1323. doi: 10.1016/S0140-6736(05)67531-9. [DOI] [PubMed] [Google Scholar]
- 22.Rice TK, Schork NJ, Rao DC. Methods for handling multiple testing. Adv Genet. 2008;60:293–308. doi: 10.1016/S0065-2660(07)00412-9. [DOI] [PubMed] [Google Scholar]
- 23.Andreasen CH, Stender-Petersen KL, Mogensen MS, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57:95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.