Abstract
Myofibroblasts secrete matrix during chronic injury, and their ablation ameliorates fibrosis. Development of new biomarkers and therapies for CKD will be aided by a detailed analysis of myofibroblast gene expression during the early stages of fibrosis. However, dissociating myofibroblasts from fibrotic kidney is challenging. We therefore adapted translational ribosome affinity purification (TRAP) to isolate and profile mRNA from myofibroblasts and their precursors during kidney fibrosis. We generated and characterized a transgenic mouse expressing an enhanced green fluorescent protein (eGFP)–tagged L10a ribosomal subunit protein under control of the collagen1α1 promoter. We developed a one-step procedure for isolation of polysomal RNA from collagen1α1-eGFPL10a mice subject to unilateral ureteral obstruction and analyzed and validated the resulting transcriptional profiles. Pathway analysis revealed strong gene signatures for cell proliferation, migration, and shape change. Numerous novel genes and candidate biomarkers were upregulated during fibrosis, specifically in myofibroblasts, and we validated these results by quantitative PCR, in situ, and Western blot analysis. This study provides a comprehensive analysis of early myofibroblast gene expression during kidney fibrosis and introduces a new technique for cell-specific polysomal mRNA isolation in kidney injury models that is suited for RNA-sequencing technologies.
The kidney interstitium, composed of vasculature, stroma, and immune cells, plays a central role in regulating kidney function, blood flow, immune surveillance, recovery from kidney injury, and progression of CKD. There is general agreement that kidney stroma, which includes pericytes, resident fibroblasts, and perivascular fibroblasts, contains the predominant myofibroblast progenitor population in fibrotic kidney disease. A precise definition of the transcriptional responses of kidney stroma to injury would aid in the identification of new therapeutic targets and potential novel circulating biomarkers of kidney fibrosis.
Over the last decade, functional genomics has been used successfully to generate comprehensive datasets, including transcriptional profiles during the course of disease. Gene profiling yields systematic, nonbiased information describing the complete transcriptional profile for kidney in health and disease. The maturity of the technology, its low cost, the availability of data analysis programs (such as gene ontology analysis1 or gene set enrichment analysis2), and the presence of institutional microarray core facilities have made such analyses relatively straightforward. This technology is increasingly used in kidney research, for example to identify the transcriptional circuitry of polycystic kidney disease modifiers,3 to distinguish polycystic kidney disease profiles with that of allograft nephropathy,4 and to analyze the transcriptome in ischemia-reperfusion injury5–7 and in renal fibrosis.8,9 The Genitourinary Database Molecular Anatomy Project (GudMap) serves as a repository for gene expression datasets in developing and adult kidneys. In one successful effort by this consortium, laser capture microdissection coupled with gene profiling delineated the transcriptional profiles of the major structures during kidney development.10
A critical limitation to transcriptomic approaches to the study of kidney biology and disease is the kidney’s cellular complexity: gene arrays from whole tissue reflect the averaged expression of >26 different cell types rather than a pure cell population. This limitation is exacerbated in disease states where inflammatory cells invade and distort normal architecture. Isolation of single cell populations by FACS is one approach to solve this problem, but it is limited by the availability of suitable antibodies and the process of kidney disaggregation itself, which rapidly induces transcriptional stress responses. Laser capture microdissection of frozen sections offers an alternative strategy, but it cannot resolve interstitial cell types, which are intermixed and closely apposed to other cell types within the interstitium.
Here we describe an approach to circumvent these problems using a method for the isolation of cell-specific RNA from genetically defined cell types in the mammalian kidney. Following the recent success in obtaining cell type–specific gene expression signatures in adult neurons,11,12 we created a transgenic mouse with heterologous expression of the ribosomal subunit protein L10a tagged with enhanced green fluorescent protein (eGFP-L10a), under control of the col1α1 promoter. Col1α1-eGFP-L10a mice express the fusion protein in podocytes in kidney cortex and in pericytes in kidney medulla.13,14 During fibrosis, eGFP-L10a is expressed specifically in α-smooth muscle actin (SMA)–positive interstitial myofibroblasts. We have adapted a one-step affinity purification protocol for the kidney in order to isolate polysomal mRNA from myofibroblasts or their precursors in healthy and fibrotic kidneys.12 We performed microarray of this cell-specific, actively translated mRNA and generated a comprehensive pericyte and myofibroblast transcriptional profile during the early stages of renal fibrosis. We have validated the approach using quantitative PCR (qPCR) and in situ analysis, as well as bioinformatics analysis. This dataset will serve as a valuable resource for the CKD community, and the approach should open new avenues into investigation of kidney stromal biology.
Results
Creation and Characterization of a Peri/FibroTRAP Transgenic Mouse
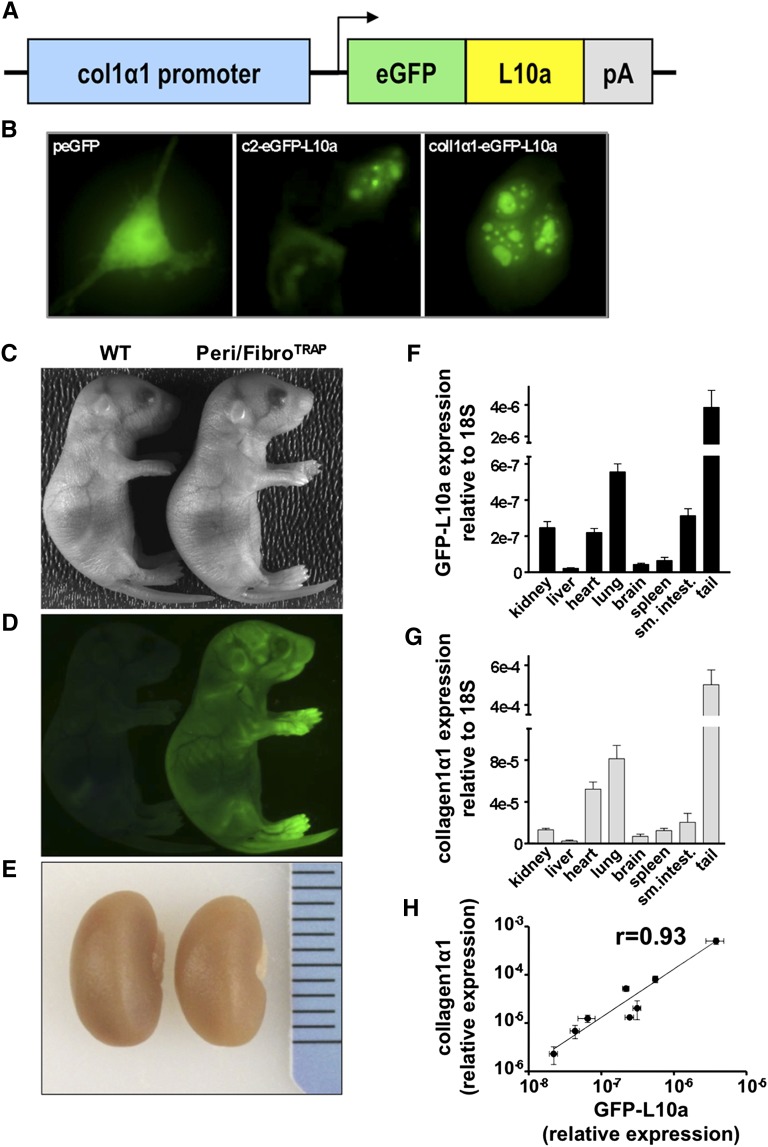
An eGFP-L10a cDNA was inserted downstream of the well characterized collagen1α1 promoter fragment to create the Col1α1-eGFP-L10a allele (Figure 1A). To validate this allele, it was transfected into 3T3 fibroblasts along with plasmids driving expression of eGFP or eGFP-L10a under control of the strong cytomegalovirus enhancer. Both eGFP-L10a constructs demonstrated a nucleolar subcellular distribution, as expected for ribosomes (Figure 1B). We also verified that we could efficiently immunoprecipitate eGFP from transfected human embryonic kidney 293 cells (Supplemental Figure 1). Transgenic founders were generated by standard techniques, and two independent founder lines were evaluated, each giving equivalent results. The resulting line, hereafter referred to as Peri/FibroTRAP, was characterized by intense eGFP fluorescence in positive pups compared with littermate controls, reflecting skin expression of Col1α1-eGFP-L10a, and normal kidney sizes (Figure 1, C–E). It was important to verify that eGFP-L10a expression in Peri/FibroTRAP mice faithfully recapitulated endogenous COL1α1 expression, so RNA from a variety of tissues was harvested, and levels of eGFP-L10a and endogenous Col1α1 were compared. This analysis revealed an excellent correlation, with a calculated Spearman correlation coefficient (r) of 0.93 (Figure 1, F–H).
Figure 1.
The Col1a1-eGFP-L10a transgenic mouse faithfully recapitulates endogenous COL1α1 expression. (A) Transgene structure. A cDNA encoding the eGFP-L10a fusion protein was inserted downstream of the col1α1 promoter/enhancer element. (B) 3T3 cells were transiently transfected with peGFP (control), c2-eGFP-L10a (positive control), or the col1α1-eGFP-L10a plasmid. C2-eGFP-L10a (middle) and col1α1-eGFP-L10a (right) transfected cells show the expected ribosomal localization pattern (nucleoli, perinuclear region). eGFP (left) transfected cells show a homogenously distributed fluorescence throughout the cell body. (C–E) Positive founders (right) exhibit prominent green fluorescence reflecting eGFP-L10a expression in skin fibroblasts. Gross kidney morphology is indistinguishable between mice. (F and G) qPCR comparison of eGFP-L10a and endogenous col1α1 expression in Peri/FibroTRAP animals. Expression levels of eGFP-L10a and endogenous col1α1 transcripts show similar patterns across various organs. Mean±SEM, n=4 for each data point. (H) Robust positive correlation between eGFP-L10a and native col1α1 expression. Data points reflect mean relative expression values±SEM of eGFP-L10a and col1α1 mRNA expression.
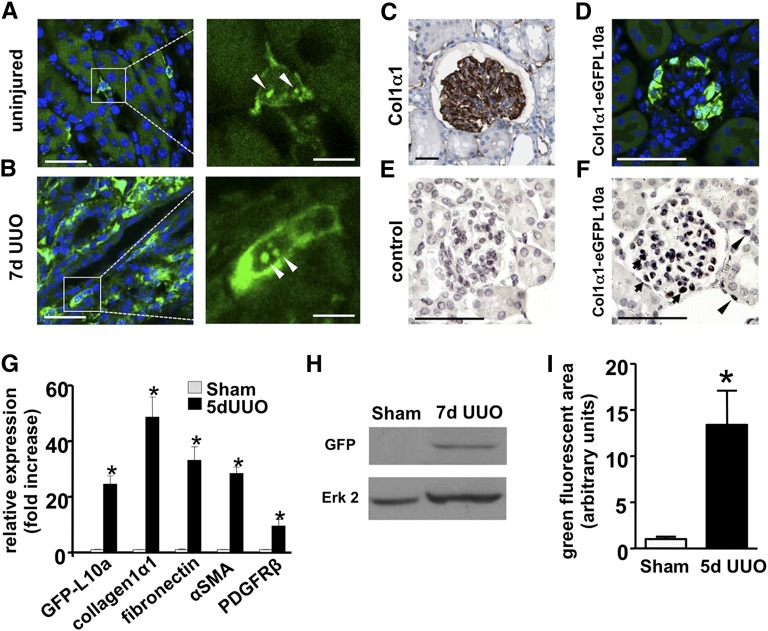
In uninjured adult kidney of Peri/FibroTRAP mice, eGFP-L10a was expressed in interstitial cells in the kidney medulla (Figure 2A). After 7 days of unilateral ureteral obstruction (UUO), there was a substantial expansion of eGFP-L10a expression exclusively in the interstitium (Figure 2B). Positive cells demonstrated the typical ribosomal nucleolar expression pattern on high magnification (Figure 2, A and B). Although some reports have failed to detect COL1α1 protein in podocytes,15–17 other studies have detected Col1α1 mRNA in podocytes.18 COL1α1 protein is expressed in human podocytes (Figure 2C), and the eGFPL10a transgene was also expressed in podocytes of Peri/FibroTRAP kidneys whether assessed by epifluorescence (Figure 2D) or by immunohistochemistry (Figure 2, E and F). There was much weaker expression of the transgene in cortical pericytes, detectable only by anti-GFP immunohistochemistry, compared with the strong medullary pericyte expression (Figure 2F, Supplemental Figure 2). These results are the same as those that we obtained with an independently created Col1α1-eGFP-Cre-ERt2 transgenic mouse line, ruling out position-dependent transgene effects.13
Figure 2.
Enhanced eGFP-L10a expression during fibrosis in Peri/FibroTRAP mice. (A) Representative micrographs of uninjured kidney medulla. eGFP-L10a+ cells (green) are readily detected by epifluorescence and are found in the interstitium and not the tubular compartment. Note perinuclear and nucleolar (arrow heads) expression (right), a pattern consistent with ribosomal location. (B) Kidney medulla after 7 days of UUO. The population of eGFP-L10a+ cells shows vast expansion but remains confined to the tubulointerstitial space. Note accentuated perinuclear and nucleolar (arrow heads) fluorescence on higher magnification (right). Scale bars, left: 20 µm; right: 5 µm. (C) Immunohistochemical analysis of a normal, 16-year-old human kidney shows strong podocyte expression of col1α1, image from The Human Protein Atlas.50 (D) A similar podocyte eGFP-L10a expression pattern is observed in glomeruli from a normal adult Peri/FibroTRAP kidney. (E and F) By enhancing immunohistochemical analysis (antibody against GFP), glomerular expression and weak interstitial expression is also observed (arrow heads). (G) qPCR analysis shows a robust, approximately 25-fold induction of renal eGFP-L10a mRNA expression as well as other fibrotic readouts including col1α1, fibronectin, αSMA, and PDGFRβ during UUO. Data represent mean values±SEM, n=5–6 for each data point, *P<0.001. (H) Immunoblot showing induction of eGFP-L10a protein (about 50 kDa in size) in fibrotic kidney (7 days UUO). Expression is so low in uninjured medulla that protein is not detected at same exposure time. (I) Quantification of green fluorescent area in Peri/FibroTRAP kidney sections confirms a significant increase in signal at 5 days of UUO compared with uninjured controls. Data are shown as mean±SEM, n=3 for each data point; *P<0.05.
The eGFP-L10a transgene was upregulated by about 25-fold after 5 days of UUO, as reflected by qPCR, and the fusion protein could be detected in lysates from 7 day fibrotic kidney medulla (Figure 2, G and H). Expression of eGFP-L10a was much lower in uninjured kidney medulla, explaining why the protein was undetectable in control kidney lysates. As expected, the relative GFP-positive area expanded during fibrosis as well (Figure 2I). EGFP-L10a in the Peri/FibroTRAP mouse was strongly expressed in other collagen1α1-positive tissues, such as skin and heart, suggesting the ability to use this mouse model for translational ribosome affinity purification (TRAP) in extrarenal tissues (data not shown).
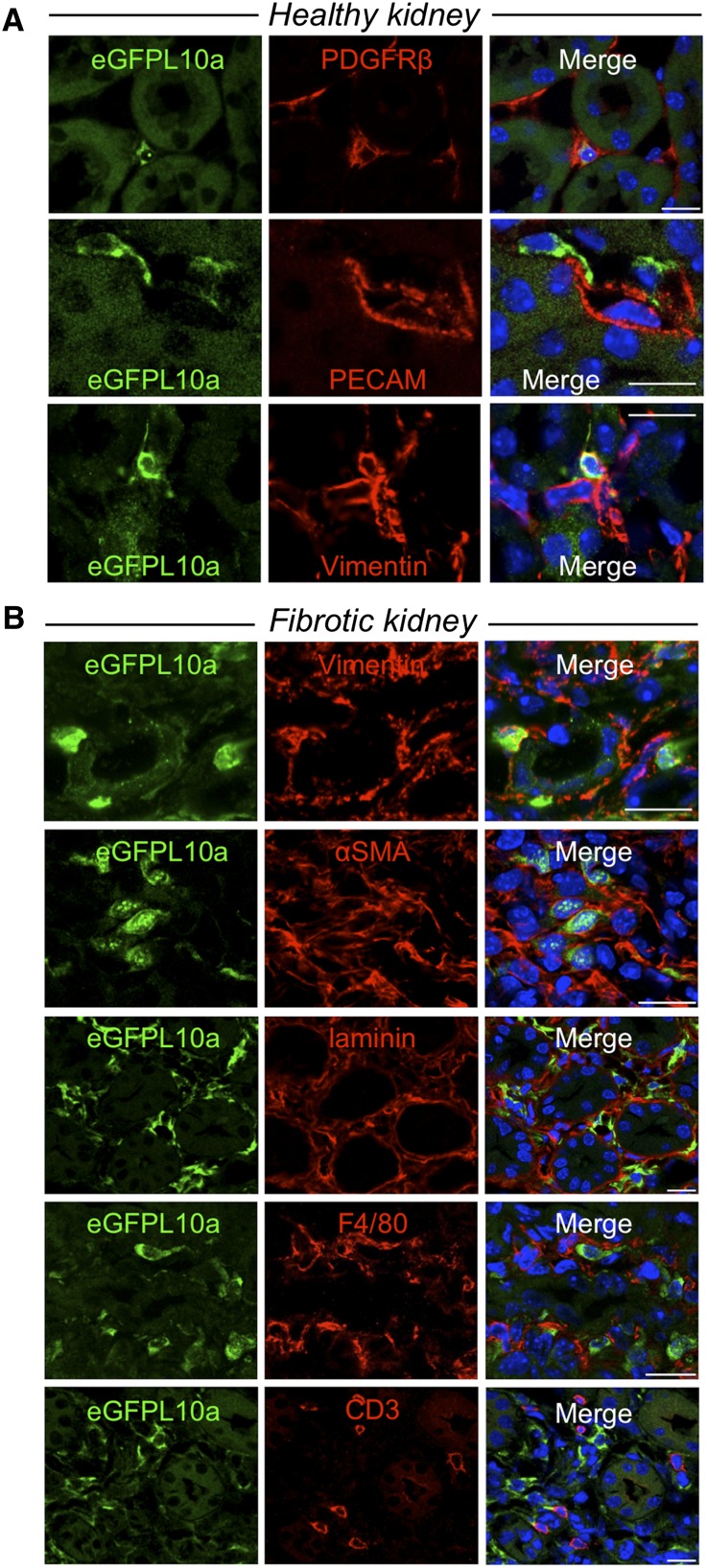
To better define the interstitial cell population that expresses eGFP-L10a in Peri/FibroTRAP mice, we next costained for several markers of interstitial cell types in healthy tissue and in fibrosis. All medullary eGFP-L10a–positive cells were positive for the pericyte/fibroblast markers vimentin19 and platelet-derived growth factor receptor-β (PDGFR-β) and negative for the endothelial cell marker platelet endothelial cell adhesion molecule (Figure 3A). Whether PDGFR-β also marks resident fibroblasts is an unresolved controversy, hence our term “Peri/FibroTRAP” mouse.20 At day 5 of UUO, eGFP-L10a–positive cells were bounded by laminin-positive basement membrane, confirming that there is no transgene expression in tubule epithelia (Figure 3B). They remained positive for vimentin. Although every medullary eGFP-L10a–positive cell was αSMA positive, roughly 50% of αSMA-positive interstitial cells were eGFP-L10a negative (Figure 3B). This could reflect mosaic expression of the transgene or an extrarenal source of kidney myofibroblasts that do not express collagen1α1, as recently suggested.20 EGFP-L10a was not expressed in myeloid lineage cells because they were negative for both F4/80 and CD3 (Figure 3B). Taken together, these experiments demonstrate that the Peri/FibroTRAP mouse faithfully recapitulates endogenous collagen1α1 expression with eGFP-L10a expressed in medullary pericytes and that these cells differentiate into activated myofibroblasts during fibrosis.
Figure 3.
Immunostaining demonstrates cell type–specific expression of eGFP-L10a in Peri/FibroTRAP kidney. (A) In uninjured kidney, eGFP-L10a+ cells (green) of kidney medulla stain positive for pericyte/fibroblast markers PDGFR-β and vimentin and are closely associated with endothelial cell marker platelet endothelial cell adhesion molecule (PECAM)+peritubular capillaries, consistent with a pericyte/fibroblast identity. (B) In fibrotic kidney (5 days UUO), eGFP-L10a+ cells remain strictly confined to the tubulointerstitial compartment, as documented by antilaminin staining, continue to express vimentin but also express the myofibroblast marker αSMA, suggesting a pericyte/fibroblast to myofibroblast transformation. Although spatially close, eGFP-L10a+ cells do not costain with macrophage marker F4/80 or T-cell marker CD3. Scale bars: 10 µm.
Myofibroblast Transcriptome Analysis during UUO by Translational Ribosome Affinity Purification
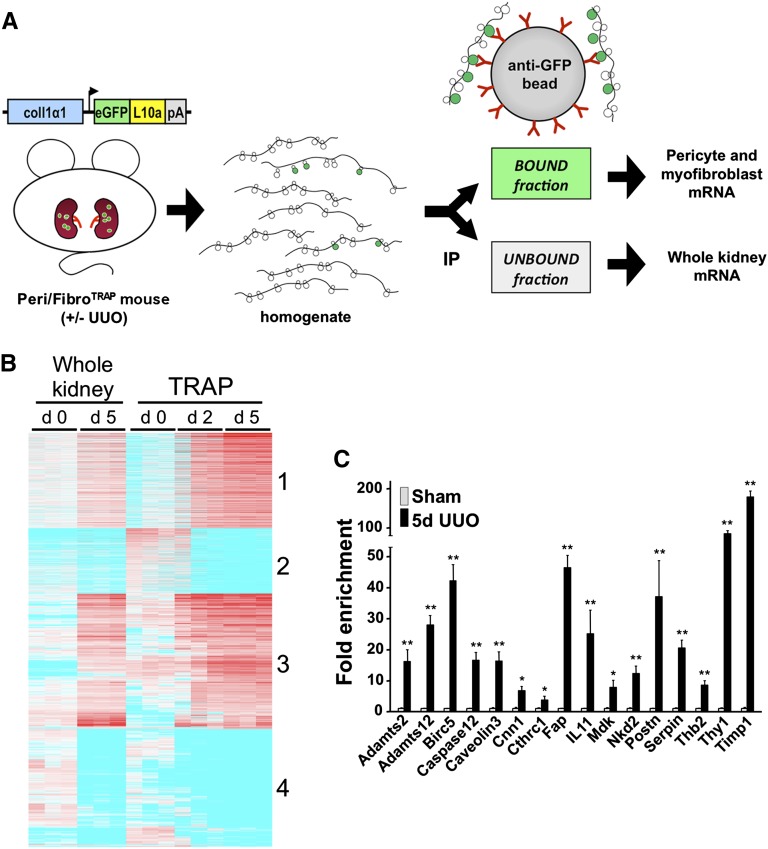
Mice were subjected to sham or UUO surgery, and polysomal RNA was isolated from kidney medulla at day 2 or day 5 using the TRAP procedure (Figure 4A). We excluded kidney cortex from these preparations to eliminate podocyte RNA from the analysis. RNA quality was excellent as assessed by bioanalyzer (Supplemental Figure 3). RNA yields were lowest in the sham samples (10–20 ng per kidney), as expected, with higher yields from UUO kidneys (50–100 ng per kidney). We analyzed gene expression by microarray analysis in both the bound fraction, which reflects pericyte/myofibroblast-specific RNA, and the unbound fraction, which reflects total kidney medulla RNA. After hybridization to Mouse Genome 430 2.0 chips data were successfully normalized by the Robust Multichip Average method as reflected by overlapping density curves (Supplemental Figure 4A).
Figure 4.
Microarray analysis of TRAP-isolated RNA and candidate validation by qPCR in experimental fibrosis. (A) Schematic illustrating the principles and workflow of TRAP. Kidney medulla from Peri/FibroTRAP mice is harvested, immediately homogenized, and immunoprecipitated (IP) with anti-GFP–coated magnetic beads. Polysomes containing the tagged ribosomal protein eGFP-L10a bind to the beads, thus representing the pericyte-specific mRNA fraction; the unbound fraction represents the whole medulla. The mRNA is isolated and processed for downstream analysis. (B) Heat map of microarray results (Affymetrix Mouse Genome 430 2.0 chips) showing differential gene expression in bound, pericyte/myofibroblast-specific mRNA fractions extracted from sham, UUO day 2 and UUO day 5 kidneys from Peri/FibroTRAP mice. Unbound RNA, which reflects total kidney medulla, is also shown between sham and UUO day 5. Note clustering into 4 distinct groups. (C) Expression of 16 candidate genes identified as upregulated by TRAP was validated by qPCR of whole kidney RNA samples. Data are shown as mean±SEM, n=4 for each data point; *P<0.05; **P<0.01.
All four groups from day 0 and day 5 showed excellent separation by multidimensional scaling plot (Supplemental Figure 4B), so we focused on these four groups for further comparative analysis. We filtered the 14,425 genes with differential expression by four-way ANOVA among day 0, day 5, bound, or unbound samples. This yielded 8325 genes that were then subject to filtering by post hoc testing, where we compared bound day 5 to bound day 0 samples, yielding 4803 genes. We further divided these genes into four groups: genes that were upregulated or downregulated, and within these two groups, genes that were TRAP-specific (defined as genes with a difference in fold-change expression of at least 2-fold higher or lower in the bound compared with the unbound fraction) or genes that were not TRAP-specific (genes whose fold-change in expression was similar in the bound and unbound fractions). In this fashion, we identified genes whose expression is specific to kidney myofibroblasts (a gene whose expression primarily changed in the bound but not the unbound fraction) versus genes that are expressed in myofibroblasts and other kidney cell types or a myofibroblast-specific gene whose expression change is so dramatic that it is detected in whole kidney lysate (a gene whose expression changed in both bound and unbound forms).
Among TRAP-specific upregulated genes, group 1 consisted of 1107 genes that were upregulated at day 5 in myofibroblasts but not in the whole kidney (Table 1). Group 2 consisted of 753 genes that were downregulated in myofibroblasts but were unchanged in the whole kidney (Table 2). Among genes whose expression changed in both TRAP and unbound samples, group 3 had 1575 genes that were upregulated and group 4 consisted of 1366 downregulated genes. A heat map summarizing these four clusters is shown in Figure 4B. Because a theoretical advantage of TRAP is the ability to detect gene changes that would be missed in whole kidney arrays, we next asked how many unique genes were identified by TRAP that would have been missed in whole kidney arrays. Among genes whose expression changed during UUO, whole kidney and TRAP identified 897 genes in common. However, TRAP identified an additional 1042 genes that were not identified by whole kidney analysis, indicating a substantial enrichment of myofibroblast genes that would not have been detected by standard whole kidney arrays.
Table 1.
Top 25 Myofibroblast-Specific Upregulated genes
| Symbol | Name | Fold-Change | P Value |
|---|---|---|---|
| Bub1 | Budding uninhibited by benzimidazoles 1 homolog | 69.18 | <0.001 |
| Cdca5 | Cell division cycle associated 5 | 67.04 | <0.001 |
| Tubb3 | Tubulin, β3 class III | 63.87 | <0.001 |
| Fap | Fibroblast activation protein | 60.78 | <0.001 |
| Gm15428 | Predicted pseudogene 15428 | 58.83 | <0.001 |
| Dmp1 | Dentin matrix protein 1 | 57.46 | 0.001 |
| Aldh1a2 | Aldehyde dehydrogenase family 1, subfamily A2 | 52.60 | 0.001 |
| Cdkn3 | Cyclin-dependent kinase inhibitor 3 | 50.87 | <0.001 |
| Hmmr | Hyaluronan mediated motility receptor | 47.76 | <0.001 |
| Ccnb2 | Cyclin B2 | 43.63 | <0.001 |
| Crlf1 | Cytokine receptor-like factor 1 | 42.65 | <0.001 |
| Nuf2 | NDC80 kinetochore complex component, homolog | 40.90 | <0.001 |
| Fbln2 | Fibulin 2 | 38.00 | <0.001 |
| Birc5 | Baculoviral IAP repeat-containing 5 | 37.66 | <0.001 |
| Lgi2 | Leucine-rich repeat LGI family, member 2 | 37.47 | <0.001 |
| Actg2 | Actin, γ2, smooth muscle, enteric | 28.51 | 0.004 |
| Rgs16 | Regulator of G protein signaling 16 | 26.83 | <0.001 |
| Lrrc15 | Leucine rich repeat containing 15 | 26.68 | <0.001 |
| E2f8 | E2F transcription factor 8 | 26.35 | <0.001 |
| Gda | Guanine deaminase | 26.32 | 0.002 |
| Thbs2 | Thrombospondin 2 | 26.24 | <0.001 |
| Col5a3 | Collagen, type V, α 3 | 25.61 | <0.001 |
| Actc1 | Actin, α, cardiac muscle 1 | 25.34 | 0.001 |
| 5730559C18Rik | RIKEN cDNA 5730559C18 gene | 24.70 | <0.001 |
| Thbs4 | Thrombospondin 4 | 24.60 | 0.005 |
Fold-change is the ratio of day 5 compared with day 0 expression. Genes were excluded if raw expression at day 5 was≤500.
Table 2.
Top 25 myofibroblast-specific downregulated Genes
| Symbol | Name | Fold Change | P Value |
|---|---|---|---|
| Spnb3 | Spectrin β3 | 0.03 | <0.03 |
| Limch1 | LIM and calponin homology domains-containing protein 1 | 0.04 | <0.01 |
| Lmo7 | LIM domain 7 | 0.05 | <0.01 |
| Lmtk2 | Lemur tyrosine kinase 2 | 0.05 | <0.001 |
| Syne2 | Spectrin repeat-containing, nuclear envelope 2 | 0.06 | <0.01 |
| Ppp1r9a | Protein phosphatase 1, regulatory subunit 9A | 0.07 | <0.02 |
| Cgn | Cingulin | 0.07 | 0.03 |
| Misp | Mitotic spindle-positioning | 0.07 | 0.012 |
| C130021I20Rik | Riken cDNA C130021I20 | 0.07 | <0.03 |
| Myom2 | Myomesin 2 | 0.08 | <0.04 |
| Myo6 | Myosin VI | 0.08 | 0.01 |
| Zfp185 | Zinc finger protein 185 | 0.08 | <0.02 |
| Tmod2 | Tropomodulin 2 | 0.08 | <0.01 |
| Cgnl1 | Cingulin-like 1 | 0.09 | 0.02 |
| Dbn1 | Drebrin 1 | 0.09 | <0.01 |
| Myo15b | Myosin XVB | 0.09 | 0.03 |
| Aim1 | Absent in melanoma 1 | 0.09 | 0.01 |
| Mprip | Myosin phosphatase Rho-interacting protein | 0.10 | 0.001 |
| Dab1 | Disabled 1 | 0.10 | 0.02 |
| Fgd4 | FYVE, RhoGEF and PH domain containing 4 | 0.10 | 0.001 |
| Scel | Sciellin | 0.10 | 0.01 |
| Sorl1 | Sortilin-related receptor, LDLR class A repeats-containing | 0.11 | <0.01 |
| Zbtb20 | Zinc finger and BTB domain-containing 20 | 0.11 | <0.01 |
| Slc5a3 | Solute carrier family 5 (inositol transporters), member 3 | 0.11 | 0.002 |
| B4galt6 | UDP-gal:βGlcNAcβ 1,4-galactosyltransferase, polypeptide 6 | 0.11 | <0.01 |
Fold-change is the ratio of day 5 compared with day 0 expression. Genes were excluded if raw expression at day 0 was ≤500.
Within the TRAP-specific upregulated gene signature group 1, eight collagen genes were identified including Col5a3, Col6a4, Col7a1, Col8a1, Col10a1, Col11a1, Col12a1, and Col16a1. Message for Col1a1 itself, used to identify pericytes/myofibroblasts in this transgenic system, was enriched by 7-fold in bound versus unbound samples, confirming enrichment by TRAP. Wnt4, which we have recently identified as a gene specifically upregulated in medullary myofibroblasts during kidney fibrosis, was also present in this group.21 Together, these results provide internal validation that the Peri/FibroTRAP mouse identifies collagens and other known genes expressed in medullary myofibroblasts.
The four subsets were then individually analyzed by gene ontology (GO) analysis to identify biologic functions within each group. Eighteen separate functional categories were computed within each group, including “molecular function,” “biological process” “cellular component,” “pathway,” relevant PubMed articles, microRNA targets, and others. These results are presented in an annotated, searchable spreadsheet (Supplemental Material). As an example of the information contained within this rich dataset, the top “biological process” GO terms in group 1 (strongly upregulated TRAP-specific genes) reflect cell proliferation, morphogenesis, and matrix binding (“anaphase-promoting complex binding,” “microtubule motor activity,” and “extracellular matrix binding”). Cell cycle is a remarkably strong theme within this group of genes: the top 5 terms for GO terms “biological process,” “cellular component,” and “pathway” are all cell cycle–related categories. In an analysis of downregulated genes from group 2, the top three GO terms in the same category relate cell shape (“actin binding,” “cytoskeletal protein binding,” and “actin filament binding”). Similarly, under the “cellular component” term and among TRAP-specific downregulated genes, categories include “cell junction” and “anchoring junction.” These GO terms strongly suggest that pericyte to myofibroblast transition is associated with loss of cell–cell connections, cytoskeletal rearrangement, and cell proliferation.
Analysis of individual genes with high expression and large changes in expression reveal genes with no previous connection to kidney but intriguing roles in fibrosis and cell proliferation in other tissues. Lemur tyrosine kinase 2 (Lmtk2) is a membrane anchored kinase expressed at very high levels in pericytes at day 0 but downregulated to just 5% of this expression by day 5. In the brain, Lmtk2 promotes TGF-β–dependent Smad2 translocation to the nucleus by inactivating GSK3β.22 Its downregulation during early UUO may therefore reflect a compensatory and adaptive antifibrotic response to chronic injury. Similarly, Thrombospondin-4 (Thbs4) is an extracellular matrix glycoprotein now recognized to induce the endoplasmic reticulum stress response after cardiac injury.23 Knockout mice are more susceptible to fibrosis and transgenic mice are protected from fibrosis, so by extension Thbs4 upregulation in kidney myofibroblasts may also reflect an adaptive, antifibrotic endoplasmic reticulum stress response to injury. The top 25 myofibroblast-specific upregulated or downregulated genes are presented in Tables 1 and 2.
Validation of Novel Myofibroblast Gene Expression Identified by TRAP
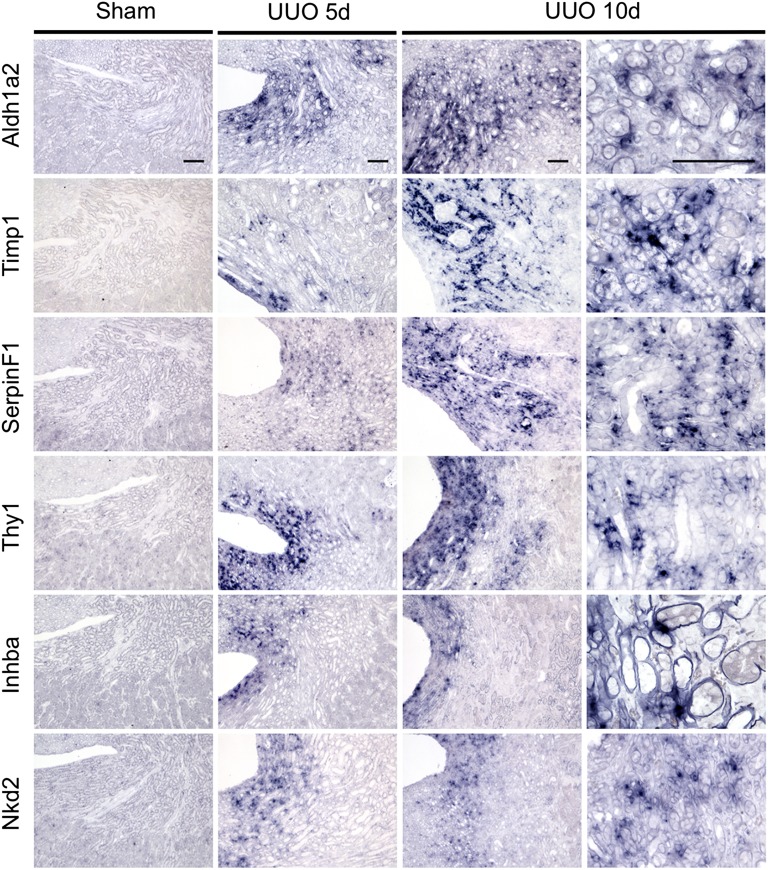
To further confirm that genes identified by this analysis reflect transcriptional changes in myofibroblasts and their progenitors in vivo, we next selected candidate genes that were novel and were substantially upregulated. We performed qPCR amplification from RNA isolated from whole kidney lysate in sham or UUO kidneys to validate mRNA induction. This confirmed strong upregulation of all 16 genes (Figure 4C). To confirm that upregulation of candidate genes occurred in interstitial myofibroblasts, we next performed RNA in situ hybridization analysis on sections prepared from sham or UUO mouse kidneys. We focused on genes from both groups 1 and 3, choosing three examples from each group with the highest absolute expression values as well as fold-change from baseline. We hypothesized that genes in group 3 were upregulated in both bound and unbound fractions because of high absolute expression of these genes in myofibroblasts, although it could also be explained by high expression in both myofibroblasts and other cells, such as tubular epithelium. The six genes selected include Aldh1a2, Timp1, SerpinF1, Thy1, Inhba, and Nkd2.
Aldh1a2, upregulated 52-fold in myofibroblasts, is the rate-limiting enzyme in retinoic acid synthesis and is expressed in nephrogenic mesenchyme during development. Retinoids play important roles in nephrogenesis.24,25 Timp1 is an inhibitor of matrix metalloproteases and has known profibrotic roles in some contexts by inhibition of extracellular matrix degradation.26,27 It was upregulated 57-fold by day 5 of UUO. The SerpinF1 gene produces the pigment epithelium–derived factor (PEDF), a secreted multifunctional protein that regulates stem cell fate.28,29 In kidneys, PEDF serves as a marker for diabetic nephropathy, and exogenous PEDF ameliorates glomerular disease.30,31 Thy1, a heavily glycosylated cell surface protein, regulates cell–cell and cell–matrix interactions, is expressed in certain fibroblast and mesenchymal cell populations, and plays roles in fibrosis.32 Inhba encodes the secreted protein inhibin-βA, a subunit of both activin and inhibin that acts as both a growth and differentiation factor.33 Interestingly, activin A is strongly upregulated in activated hepatic stellate cells, the myofibroblast progenitors in the liver.34 Nkd2 is a poorly characterized gene with no known roles in kidney. It antagonizes both canonical and noncanonical Wnt/β-catenin signaling, and it also appears to regulate trafficking of the epidermal growth factor receptor ligand, TGF-α.35,36 All six of these genes were undetectable in sham kidneys but showed strong upregulation specifically in the medullary interstitium at both 5 and 10 days after UUO (Figure 5).
Figure 5.
RNA in situ hybridization validates interstitial expression of novel myofibroblast genes during fibrosis. In situ hybridization comparing sham surgery versus UUO day 5 and day 10 for six novel myofibroblast genes strongly induced during fibrosis. All genes show an interstitial and not tubular expression pattern with strongest expression in medulla and extending to cortical interstitium. Scale bars: 100 µm.
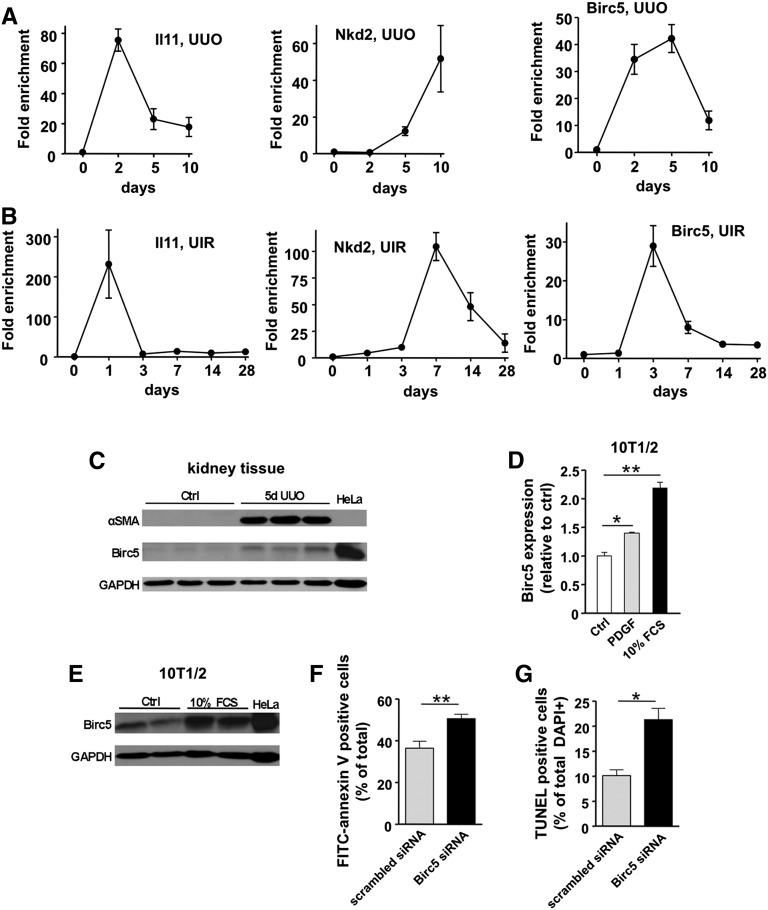
We further analyzed the time course for expression of three of the genes identified by TRAP. IL11 encodes IL-11, a hematopoietic cytokine with antiapoptotic and antinecrotic properties, ameliorates renal injury when administered exogenously.37 Endogenous IL11 transcripts rose at early time points in two separate kidney fibrosis models, UUO and unilateral ischemia-reperfusion injury, and fell back toward normal expression levels later during the disease course (Figure 6, A and B). NKD2 mRNA was strongly upregulated at later time points in fibrosis. Birc5, also known as survivin, is a negative regulator of apoptosis and is under development as a drug target in cancer therapy.38 Birc5 was strongly upregulated in group 1 by TRAP analysis and showed upregulation in both fibrotic models as well (Figure 6, A and B). BIRC5 protein could be detected in lysates from whole kidney, and Birc5 mRNA and protein were induced by growth factor or serum treatment of the pericyte-like cell line 10T1/2 (Figure 6, C–E). Small interfering RNA (siRNA)–mediated Birc5 knockdown enhanced 10T1/2 cell apoptosis, consistent with a prosurvival role for Birc5 in these mesenchymal cells (Figure 6G).
Figure 6.
Differential expression kinetics of genes identified by TRAP in two fibrosis models and functional validation of Birc5 as a potential target. (A) Time course analysis of IL11, Nkd2, and Birc5 (survivin) expression during UUO. (B) Expression of the same genes was analyzed in a second fibrotic model, unilateral ischemia-reperfusion (UIR). Note similar expression patterns. Mean±SEM, n=3–5 for each time point. (C) Analysis of renal Birc5 protein expression during UUO. Note concomitant induction of myofibroblast marker αSMA. HeLa cell lysate was used as positive control for Birc5. (D) In vitro stimulation of pericyte-like 10T1/2 cells with PDGF (20 ng/ml) or FCS (10%) for 24 hours causes a significant increase in Birc5 expression. Mean±SEM, n=3 for each data point; *P<0.01; **P<0.001. (E) Birc5 protein is also increased in 10T1/2 cells upon stimulation with 10% FCS compared to unstimulated control (Ctrl). (F) Analysis of cell apoptosis by FITC-annexin V staining of 10T1/2 cells. Knockdown of Birc5 by siRNA results in significant increase of apoptotic rate compared with scrambled siRNA-transfected control cells. Serum starvation was used to induce apoptosis. Mean±SEM, n=13–17 for each data point. (G) Analysis of TUNEL labeling confirms enhanced cell apoptosis upon siRNA mediated knockdown of Birc5. Mean±SEM, n=3–4; *P<0.01; **P<0.001. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
We describe here a new transgenic line that allows profiling of translated RNAs from kidney myofibroblasts during injury. This line, and the associated TRAP protocol adapted to kidney, will allow sensitive detection of pericyte, fibroblast, and myofibroblast gene expression analysis in a simple, single-step purification procedure. It will also enable gene expression analysis from other cell types where collagen1α1 is expressed, including mesenchymal cells from heart and skin, and podocytes in kidney cortex. Why collagen1α1 protein has not been consistently detected in mouse glomeruli is unclear, but one possibility is a podocyte splice variant lacking a relevant epitope. Regardless, the eGFP-L10a transgene is strongly expressed in podocytes, and this will be an advantage for isolating podocyte-specific RNA.
TRAP was first developed to profile molecularly distinct neuron subgroups in mice.11,12 Recently, TRAP has been applied to heart regeneration and expanded to Drosophila, zebrafish, and Xenopus.39–41 The TRAP approach has many advantages compared with standard RNA isolation protocols. Most important, TRAP obviates the need for cell purification procedures, such as FACS, before RNA profiling. These procedures introduce cell stresses that alter RNA expression profiles, especially in tissue injury models characterized by inflammation and fibrosis, where cell dissociation may require extensive proteolytic digestion and mechanical force. The single-step purification method is easily within the capacity of any scientific laboratory, is inexpensive, and does not require FACS machinery or expertise. Our results in uninjured kidney medulla, where Col1α1-eGFP-L10a+ pericytes make up <0.5% of the cellular content, demonstrate that this approach can be used successfully in rare populations as well. Finally, RNA isolated by TRAP should be well suited as a substrate for next-generation sequencing technologies.
The precise origin of myofibroblasts during kidney fibrosis remains controversial, with the bulk of evidence favoring resident mesenchymal progenitors as the primary source.20,42 It is not yet clear whether these progenitors are resident fibroblasts, pericytes, or as-yet-undefined subsets of these cells, as recently reviewed.43 The searchable dataset that accompanies this paper should provide a substantial source of novel gene markers that might help further define various interstitial stromal cell subsets in kidney. Our validation of the TRAP dataset identified many genes that may play important roles in regulating myofibroblast biology during fibrosis. Future research will be required to better define the roles of the product of Aldh1a2, retinoic acid, as well as PEDF and inhibin-βA in fibrosis.
There is a strong clinical need to develop novel biomarkers of CKD that identify early stages of disease and correlate with prognosis or response to therapy. Genes in group 1, whose expression is upregulated primarily in myofibroblasts during injury, should serve as a resource for discovering novel CKD biomarker candidates. For example, screening among these genes identifies the secreted protein cytokine receptor–like factor 1 (Crlf1),44 which has very low expression in healthy kidney, but is upregulated 43-fold in myofibroblasts by day 5 of UUO. Thrombospondin-4 (Thbs4) is also secreted and has known roles in cardiac fibrosis,22 and its expression increases 25-fold in myofibroblasts with nearly undetectable expression in whole medulla. Insulin-like growth factor binding protein-6 (Igfbp6) is another candidate secreted biomarker, upregulated 4.4-fold and a known target of hedgehog signaling,45 which is activated in kidney myofibroblasts.46 This group also contains numerous cell surface proteins that might serve as tissue biomarkers, such as the Wnt antagonist frizzled-related protein (Frzb), and killer cell lectin–like receptor subfamily B member 1A (Klrb1a).
Pathway analyses of pericyte to myofibroblast transition reflect a strong gene signature corresponding to cell proliferation, cytoskeletal changes, loss of cell–cell contacts, and motility genes. Altogether, these changes support the notion that cell shape change and acquisition of a motile phenotype are critical responses of kidney stromal cells during chronic injury. The signaling pathways most notably upregulated include integrin and platelet-derived growth factor signaling, and upregulated gene families include snail homologs as well as the Wnt modulating gene family, secreted frizzled-related proteins. These pathways comprise known important regulators of myofibroblast function and kidney fibrosis, providing further internal validation of this approach.
In summary, we have generated and characterized a novel tool for translational profiling of cells that express col1α1. The resulting atlas of pericyte and myofibroblast gene expression in kidney medulla will serve as a rich resource for investigators interested in CKD and will guide further investigation of these critical cells in fibrosis.
Concise Methods
Mice
All mouse studies were performed according to the animal experimental guidelines issued by the Animal Care and Use Committee at Harvard University. EGFP and L10a sequences were separately PCR amplified from the C2-eGFP-L10a plasmid12 and sequentially inserted into the EcoRI and SalI sites of the pGL3-Col1α1-Cre plasmid, replacing Cre downstream of the collagen 1α1 promoter/enhancer element.47 After sequencing and in vitro validation, the linearized Col1α1-eGFP-L10a transgene was introduced into F1-hybrid embryos (Gene Modification Facility, Harvard University) by pronuclear injection to generate transgenic mice. Genomic DNA was obtained from tail biopsies and two independent Col1α1-eGFP-L10a founders identified by genotyping using the following primers: (product 497 bp) 5′-GGCATCGACTTCAAGGAGGA-3′ (F), 5′-GGTCGTAGTTCTTCAGGCTGA-3′ (R). Transgenic mice were maintained on a C57BL/6J x DBA/2 mixed background.
UUO
For the induction of kidney fibrosis, the UUO model was used as previously described.46 Briefly, animals were anesthetized with pentobarbital sodium (60 mg/kg body wt intraperitoneally) before surgery and put in a prone position. The left kidney was accessed by the retroperitoneal approach and the ureter ligated twice with a 4–0 silk suture.
Unilateral Ischemia-Reperfusion Injury
Animals were anesthetized with pentobarbital sodium (60 mg/kg body wt intraperitoneally) before surgery. Body temperatures were controlled at 36.5–37.5°C throughout the procedure. The left kidney was exposed through flank incision and subjected to ischemia by clamping the renal pedicle with nontraumatic microaneurysm clamps (Roboz, Rockville, MD) for 35 minutes. After clamp removal, return of blood flow was confirmed and the incision closed with surgical staples. Saline, 0.5 ml, was given intraperitoneally 2 hours after surgery.
TRAP
Purification of polysomally bound mRNA from whole organ lysate was performed as described, with modifications.12 A detailed step-by-step protocol is available upon request. Briefly, mice were perfused with ice-cold PBS under deep anesthesia, kidneys were removed and decapsulated, and the medulla was rapidly microdissected and minced on ice and then transferred to 1 ml of ice-cold polysome extraction buffer. Dynabeads (MyOne T1 Dynabeads; Invitrogen, Carlsbad, CA) coated with monoclonal anti-GFP antibodies (clones 19C8 and 19F7; Rockefeller University Monoclonal Antibody Core Facility) were added to the postmitochondrial supernatant of medulla extract and together incubated at 4°C with end-over-end rotation for 4 hours. Following incubation, beads were collected on a magnetic rack, repeatedly washed with high-salt wash buffer and immediately resuspended in Qiagen RNeasy lysis buffer. RNA was purified using an RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA) with in-column DNase digestion (RNase-Free DNase Set; Qiagen).
Quantitation and Amplification of mRNA and Microarray Analysis
Purified mRNA samples were analyzed by chip-based capillary electrophoresis using an Agilent RNA 6000 PicoChip kit and the Agilent 2100 Bioanalyzer System (Agilent Technologies, Santa Clara, CA) to assess RNA quantity and quality. The Ovation RNA Amplification System V2 (NuGEN Technologies, Inc., San Carlos, CA) was used to process and amplify samples of satisfactory quality for subsequent microarray analysis. Expression analysis was carried out on Affymetrix GeneChip 430 v2.0 arrays, and scanning was performed with an Affymetrix 3000 GeneChip scanner.
Bioinformatic Analysis
Microarray data were normalized by Robust Multichip Average using GeneSpring software (Agilent Technologies). Multidimensional scaling plots, heat map, and hierarchical clustering were also performed using GeneSpring. Gene Ontology was performed using the ToppGene Suite (Cincinnati Children’s Hospital Medical Center). Microarrays have been deposited in the GEO database (GEO ID GSM1219324 through GSM1219338) and the GUDMAP database (GUDMAP ID 21179 through 21193).
In Situ Hybridization
In situ hybridization was performed using the InsituPro VSi robot (Intavis) according to a previously published protocol, unless stated otherwise.48 Briefly, formalin-fixed, paraffin-embedded tissue sections were dewaxed, permeabilized with proteinase K (10 µg/ml; 2×10 minutes), postfixed, and incubated in acetylation solution before loading into the machine. Slides were incubated with hybridization buffer (50% formamide, 5× saline-sodium citrate, 1% SDS, 50 µg/ml yeast tRNA, 50 µg/ml heparin) containing 0.5–1 µg/ml of digoxigenin (DIG)-labeled riboprobe twice for 6 hours at 68°C. For each gene, a DNA template of approximately 750 bp was generated from plasmid clones by PCR and transcribed to produce a DIG-labeled antisense riboprobe (for details, see www.gudmap.org). Hybridization was followed by stringency washes, RNase treatment (2 µg/ml), blocking, and anti–DIG-AP antibody incubation (Roche; 11093274910; 2×3 hours, 1:1000). After disassembly of the hybridization chambers, slides were washed and incubated with chromogenic substrate (BM Purple; Roche) for a maximum of 7 days. Once a sufficient staining intensity was reached, sections were postfixed and mounted in Glycergel (Dako).
Tissue Preparation, Histology, Immunofluorescence, and Immunohistochemistry
Mice were euthanized and immediately perfused via left ventricle with ice-cold PBS for 1 minute. Kidneys were removed, cut sagittally or transversely, fixed in 4% paraformaldehyde on ice for 1–2 hours and transferred to sucrose solution (30%) overnight for cryoprotection. Immunohistological staining of kidney sections was performed as previously described. Briefly, rehydrated paraffin-embedded (3–4 μm) or cryopreserved (7 μm) sections were labeled with primary antibodies, including chicken anti-GFP (Aves Labs; 1:500), rabbit antilaminin (Sigma-Aldrich; 1:100), rat anti-PDGFRβ (eBioscience; 1:200), Cy3-conjugated anti-αSMA (Sigma-Aldrich; 1:500), rat anti-platelet endothelial cell adhesion molecule (eBioscience; 1:100), rat anti-F4/80 (Abcam, Inc.; 1:100), and rabbit anti-CD3 (Vector Laboratories; 1:200). Slides were subsequently exposed to corresponding FITC- or Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and mounted with 4′,6-diamidino-2-phenylindole-containing Vectashield mounting medium (Vector Laboratories). Staining was examined by fluorescence microscopy (Nikon C1 confocal and Nikon eclipse 90i), and semiautomated quantitation was performed using ImageJ software (http://rsbweb.nih.gov/ij/). Immunohistochemical staining for eGFP was performed as described49 on formalin fixed, paraffin-embedded 3–4-μm sections. Sections were rehydrated and antigens retrieved using heated citrate. Primary antibody against eGFP (Abcam, Inc.; #6556; 1:200) was incubated at room temperature for 3 hours. Staining was visualized using horseradish peroxidase–coupled secondary antibodies (Vectastain Elite; Vector Laboratories).
Cell Culture and Immunoblotting
NIH/3T3 fibroblasts, human embryonic kidney 293, and 10T1/2 pericyte-like cells were cultured using standard cell culture techniques. Lipofectamine 2000 (Invitrogen) was used for vector transfection and DharmaFECT (Thermo Fisher Scientific, Waltham, MA) for Birc5 siRNA knockdown experiments. All siRNAs were obtained from Dharmacon (Thermo Fisher Scientific). For Western analysis, radioimmunoprecipitation assay lysis buffer containing a protease inhibitor cocktail (Roche, Indianapolis, IN) was used to generate protein lysates from both renal tissue and cultured cells. For protein detection, polyvinylidene fluoride membranes were incubated with primary antibodies, including chicken anti-GFP (Aves Labs; 1:2000), rabbit anti-αSMA (Abcam, Inc.; 1:2000), and rabbit anti-Birc5 (Novus Biologicals; 1:1000). Antibodies directed against Erk2 (Santa Cruz Biotechnology; 1:1000) and glyceraldehyde 3-phosphate dehydrogenase (Bethyl Labs; 1:4000) were used as loading controls. Membranes were subsequently probed with corresponding horseradish peroxidase–conjugated secondary antibodies.
Real-Time PCR
RNA was extracted from snap-frozen tissue stored at −80°C using standard techniques (RNAeasy kit; Qiagen) and cDNA produced using iScript reverse transcription (Bio-Rad). Real-time PCR was performed using the iQ-SYBR Green Supermix (Bio-Rad) and the iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad) for detection of mRNA levels, including eGFP-L10a, Col1α1, Fn1, αSMA, PDGFRβ, Il11, Nkd2 and Birc5. Housekeeping genes 18S and GAPDH were used as internal controls.
Apoptosis Assays
Cell apoptosis was studied using TUNEL staining (Roche) followed by quatification of the ratio of TUNEL+/total 4′,6-diamidino-2-phenylindole+ cell nuclei as well as Annexin-V labeling (Roche). Experiments were conducted according to the manufacturer’s recommendations.
Statistical Analyses
Data are given as mean±SEM. Statistical analyses were performed using the unpaired t test to determine differences between two groups and ANOVA to compare data among groups. P values <0.05 were considered to represent statistically significant differences.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors thank Nat Heintz for the eGFP-L10a plasmids and Oliver Hofmann and Shannon Sui of the Harvard School of Public Health Bioinformatics Core for an initial analysis of the microarray data.
This work was supported by a grant from the National Institutes of Health (DK088923), the Harvard Stem Cell Institute, the Genzyme Renal Innovations Program, and an Established Investigator Award from the American Heart Association (all to B.D.H.) and by ARRA-DK87389 from the National Institutes of Health (to J.S.D.). Work in A.P.M.’s laboratory was supported by a grant from California Institute for Regenerative Medicine (LA1-06536). I.G. was supported by a fellowship from the Deutsche Forschungsgemeinschaft (GR 3301/4-1) and grants from the Philipps-University Marburg and the Von Behring-Röntgen-Foundation.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013101143/-/DCSupplemental.
References
- 1.Huntley RP, Binns D, Dimmer E, Barrell D, O’Donovan C, Apweiler R: QuickGO: A user tutorial for the web-based Gene Ontology browser. Database (Oxford) 2009: bap010, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP: Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong Y, Ma Z, Patel V, Fischer E, Hiesberger T, Pontoglio M, Igarashi P: HNF-1beta regulates transcription of the PKD modifier gene Kif12. J Am Soc Nephrol 20: 41–47, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schieren G, Rumberger B, Klein M, Kreutz C, Wilpert J, Geyer M, Faller D, Timmer J, Quack I, Rump LC, Walz G, Donauer J: Gene profiling of polycystic kidneys. Nephrol Dial Transplant 21: 1816–1824, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Yuen PS, Jo SK, Holly MK, Hu X, Star RA: Ischemic and nephrotoxic acute renal failure are distinguished by their broad transcriptomic responses. Physiol Genomics 25: 375–386, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devarajan P, Mishra J, Supavekin S, Patterson LT, Steven Potter S: Gene expression in early ischemic renal injury: Clues towards pathogenesis, biomarker discovery, and novel therapeutics. Mol Genet Metab 80: 365–376, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Supavekin S, Zhang W, Kucherlapati R, Kaskel FJ, Moore LC, Devarajan P: Differential gene expression following early renal ischemia/reperfusion. Kidney Int 63: 1714–1724, 2003 [DOI] [PubMed] [Google Scholar]
- 8.Maluf DG, Mas VR, Archer KJ, Yanek K, Gibney EM, King AL, Cotterell A, Fisher RA, Posner MP: Molecular pathways involved in loss of kidney graft function with tubular atrophy and interstitial fibrosis. Mol Med 14: 276–285, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ju W, Eichinger F, Bitzer M, Oh J, McWeeney S, Berthier CC, Shedden K, Cohen CD, Henger A, Krick S, Kopp JB, Stoeckert CJ, Jr, Dikman S, Schröppel B, Thomas DB, Schlondorff D, Kretzler M, Böttinger EP: Renal gene and protein expression signatures for prediction of kidney disease progression. Am J Pathol 174: 2073–2085, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, Zhang P, GUDMAP project : GUDMAP: The genitourinary developmental molecular anatomy project. J Am Soc Nephrol 19: 667–671, 2008 [DOI] [PubMed] [Google Scholar]
- 11.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N: Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135: 749–762, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N: A translational profiling approach for the molecular characterization of CNS cell types. Cell 135: 738–748, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grgic I, Brooks CR, Hofmeister AF, Bijol V, Bonventre JV, Humphreys BD: Imaging of podocyte foot processes by fluorescence microscopy. J Am Soc Nephrol 23: 785–791, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin SL, Kisseleva T, Brenner DA, Duffield JS: Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am J Pathol 173: 1617–1627, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung GS, Kim MK, Choe MS, Lee KM, Kim HS, Park YJ, Choi HS, Lee KU, Park KG, Lee IK: The orphan nuclear receptor SHP attenuates renal fibrosis. J Am Soc Nephrol 20: 2162–2170, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng J, Truong LD, Wu X, Kuhl D, Lang F, Du J: Serum- and glucocorticoid-regulated kinase 1 is upregulated following unilateral ureteral obstruction causing epithelial-mesenchymal transition. Kidney Int 78: 668–678, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doi S, Zou Y, Togao O, Pastor JV, John GB, Wang L, Shiizaki K, Gotschall R, Schiavi S, Yorioka N, Takahashi M, Boothman DA, Kuro-o M: Klotho inhibits transforming growth factor-beta1 (TGF-beta1) signaling and suppresses renal fibrosis and cancer metastasis in mice. J Biol Chem 286: 8655–8665, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minto AW, Fogel MA, Natori Y, O’Meara YM, Abrahamson DR, Smith B, Salant DJ: Expression of type I collagen mRNA in glomeruli of rats with passive Heymann nephritis. Kidney Int 43: 121–127, 1993 [DOI] [PubMed] [Google Scholar]
- 19.Marxer-Meier A, Hegyi I, Loffing J, Kaissling B: Postnatal maturation of renal cortical peritubular fibroblasts in the rat. Anat Embryol (Berl) 197: 143–153, 1998 [DOI] [PubMed] [Google Scholar]
- 20.LeBleu VS, Taduri G, O’Connell J, Teng Y, Cooke VG, Woda C, Sugimoto H, Kalluri R: Origin and function of myofibroblasts in kidney fibrosis. Nat Med 19: 1047–1053, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiRocco DP, Kobayashi A, Taketo MM, McMahon AP, Humphreys BD: Wnt4/β-catenin signaling in medullary kidney myofibroblasts. J Am Soc Nephrol 24: 1399–1412, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manser C, Guillot F, Vagnoni A, Davies J, Lau KF, McLoughlin DM, De Vos KJ, Miller CC: Lemur tyrosine kinase-2 signalling regulates kinesin-1 light chain-2 phosphorylation and binding of Smad2 cargo. Oncogene 31: 2773–2782, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch JM, Maillet M, Vanhoutte D, Schloemer A, Sargent MA, Blair NS, Lynch KA, Okada T, Aronow BJ, Osinska H, Prywes R, Lorenz JN, Mori K, Lawler J, Robbins J, Molkentin JD: A thrombospondin-dependent pathway for a protective ER stress response. Cell 149: 1257–1268, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niederreither K, Fraulob V, Garnier JM, Chambon P, Dollé P: Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech Dev 110: 165–171, 2002 [DOI] [PubMed] [Google Scholar]
- 25.Wingert RA, Selleck R, Yu J, Song HD, Chen Z, Song A, Zhou Y, Thisse B, Thisse C, McMahon AP, Davidson AJ: The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet 3: 1922–1938, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshiji H, Kuriyama S, Miyamoto Y, Thorgeirsson UP, Gomez DE, Kawata M, Yoshii J, Ikenaka Y, Noguchi R, Tsujinoue H, Nakatani T, Thorgeirsson SS, Fukui H: Tissue inhibitor of metalloproteinases-1 promotes liver fibrosis development in a transgenic mouse model. Hepatology 32: 1248–1254, 2000 [DOI] [PubMed] [Google Scholar]
- 27.Kim H, Oda T, López-Guisa J, Wing D, Edwards DR, Soloway PD, Eddy AA: TIMP-1 deficiency does not attenuate interstitial fibrosis in obstructive nephropathy. J Am Soc Nephrol 12: 736–748, 2001 [DOI] [PubMed] [Google Scholar]
- 28.Ramírez-Castillejo C, Sánchez-Sánchez F, Andreu-Agulló C, Ferrón SR, Aroca-Aguilar JD, Sánchez P, Mira H, Escribano J, Fariñas I: Pigment epithelium-derived factor is a niche signal for neural stem cell renewal. Nat Neurosci 9: 331–339, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez R, Jennings LL, Knuth M, Orth AP, Klock HE, Ou W, Feuerhelm J, Hull MV, Koesema E, Wang Y, Zhang J, Wu C, Cho CY, Su AI, Batalov S, Chen H, Johnson K, Laffitte B, Nguyen DG, Snyder EY, Schultz PG, Harris JL, Lesley SA: Screening the mammalian extracellular proteome for regulators of embryonic human stem cell pluripotency. Proc Natl Acad Sci U S A 107: 3552–3557, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Zheng Z, Li R, Lu J, Bao Y, Ying X, Zeng R, Jia W: Urinary pigment epithelium-derived factor as a marker of diabetic nephropathy. Am J Nephrol 32: 47–56, 2010 [DOI] [PubMed] [Google Scholar]
- 31.Fujimura T, Yamagishi S, Ueda S, Fukami K, Shibata R, Matsumoto Y, Kaida Y, Hayashida A, Koike K, Matsui T, Nakamura K, Okuda S: Administration of pigment epithelium-derived factor (PEDF) reduces proteinuria by suppressing decreased nephrin and increased VEGF expression in the glomeruli of adriamycin-injected rats. Nephrol Dial Transplant 24: 1397–1406, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Hagood JS, Prabhakaran P, Kumbla P, Salazar L, MacEwen MW, Barker TH, Ortiz LA, Schoeb T, Siegal GP, Alexander CB, Pardo A, Selman M: Loss of fibroblast Thy-1 expression correlates with lung fibrogenesis. Am J Pathol 167: 365–379, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia Y, Schneyer AL: The biology of activin: Recent advances in structure, regulation and function. J Endocrinol 202: 1–12, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patella S, Phillips DJ, Tchongue J, de Kretser DM, Sievert W: Follistatin attenuates early liver fibrosis: Effects on hepatic stellate cell activation and hepatocyte apoptosis. Am J Physiol Gastrointest Liver Physiol 290: G137–G144, 2006 [DOI] [PubMed] [Google Scholar]
- 35.Van Raay TJ, Coffey RJ, Solnica-Krezel L: Zebrafish Naked1 and Naked2 antagonize both canonical and non-canonical Wnt signaling. Dev Biol 309: 151–168, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Hao M, Cao Z, Ding W, Graves-Deal R, Hu J, Piston DW, Coffey RJ: Naked2 acts as a cargo recognition and targeting protein to ensure proper delivery and fusion of TGF-alpha containing exocytic vesicles at the lower lateral membrane of polarized MDCK cells. Mol Biol Cell 18: 3081–3093, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HT, Park SW, Kim M, Ham A, Anderson LJ, Brown KM, D’Agati VD, Cox GN: Interleukin-11 protects against renal ischemia and reperfusion injury. Am J Physiol Renal Physiol 303: F1216–F1224, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horowitz JC, Ajayi IO, Kulasekaran P, Rogers DS, White JB, Townsend SK, White ES, Nho RS, Higgins PD, Huang SK, Sisson TH: Survivin expression induced by endothelin-1 promotes myofibroblast resistance to apoptosis. Int J Biochem Cell Biol 44: 158–169, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson FL, Mills EA, Wang X, Guo C, Chen DF, Marsh-Armstrong N: Cell type-specific translational profiling in the Xenopus laevis retina. Dev Dyn 241: 1960–1972, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang Y, Gupta V, Karra R, Holdway JE, Kikuchi K, Poss KD: Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc Natl Acad Sci U S A 110: 13416–13421, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas A, Lee PJ, Dalton JE, Nomie KJ, Stoica L, Costa-Mattioli M, Chang P, Nuzhdin S, Arbeitman MN, Dierick HA: A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLoS ONE 7: e40276, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphreys BD, Lin SL, Kobayashi A, Hudson TE, Nowlin BT, Bonventre JV, Valerius MT, McMahon AP, Duffield JS: Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol 176: 85–97, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kramann R, DiRocco DP, Humphreys BD: Understanding the origin, activation and regulation of matrix-producing myofibroblasts for treatment of fibrotic disease. J Pathol 231: 273–289, 2013 [DOI] [PubMed] [Google Scholar]
- 44.Kass DJ, Yu G, Loh KS, Savir A, Borczuk A, Kahloon R, Juan-Guardela B, Deiuliis G, Tedrow J, Choi J, Richards T, Kaminski N, Greenberg SM: Cytokine-like factor 1 gene expression is enriched in idiopathic pulmonary fibrosis and drives the accumulation of CD4+ T cells in murine lungs: Evidence for an antifibrotic role in bleomycin injury. Am J Pathol 180: 1963–1978, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu XF, Guo CY, Liu J, Yang WJ, Xia YJ, Xu L, Yu YC, Wang XP: Gli1 maintains cell survival by up-regulating IGFBP6 and Bcl-2 through promoter regions in parallel manner in pancreatic cancer cells. J Carcinog 8: 13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabian SL, Penchev RR, St-Jacques B, Rao AN, Sipilä P, West KA, McMahon AP, Humphreys BD: Hedgehog-Gli pathway activation during kidney fibrosis. Am J Pathol 180: 1441–1453, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krempen K, Grotkopp D, Hall K, Bache A, Gillan A, Rippe RA, Brenner DA, Breindl M: Far upstream regulatory elements enhance position-independent and uterus-specific expression of the murine alpha1(I) collagen promoter in transgenic mice. Gene Expr 8: 151–163, 1999 [PMC free article] [PubMed] [Google Scholar]
- 48.Rumballe B, Georgas K, Little MH: High-throughput paraffin section in situ hybridization and dual immunohistochemistry on mouse tissues. Cold Spring Harb Protoc doi:10.1101/pdb.prot5030, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Humphreys BD, Xu F, Sabbisetti V, Grgic I, Naini SM, Wang N, Chen G, Xiao S, Patel D, Henderson JM, Ichimura T, Mou S, Soeung S, McMahon AP, Kuchroo VK, Bonventre JV: Chronic epithelial kidney injury molecule-1 expression causes murine kidney fibrosis. J Clin Invest 123: 4023–4035, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F: Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28: 1248–1250, 2010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.