Abstract
When exposed to far-red light followed by darkness, wild-type Arabidopsis thaliana seeds fail to germinate or germinate very poorly. We have previously shown that the ethylene receptor ETR1 (ETHYLENE RESPONSE1) inhibits and ETR2 stimulates seed germination of Arabidopsis during salt stress. This function of ETR1 requires the full-length receptor. These roles are independent of ethylene levels and sensitivity and are mainly mediated by a change in abscisic acid (ABA) sensitivity. In the current study we find that etr1-6 and etr1-7 loss-of-function mutant seeds germinate better than wild-type seeds after illumination with far-red light or when germinated in the dark indicating an inhibitory role for ETR1. Surprisingly, this function of ETR1 does not require the receiver domain. No differences between these mutants and wild-type are seen when germination proceeds after treatment with white, blue, green, or red light. Loss of any of the other four ethylene receptor isoforms has no measurable effect on germination after far-red light treatment. An analysis of the transcript abundance for genes encoding ABA and gibberellic acid (GA) metabolic enzymes indicates that etr1-6 mutants may produce more GA and less ABA than wild-type seeds after illumination with far-red light which correlates with the better germination of the mutants. Epistasis analysis suggests that ETR1 may genetically interact with the phytochromes (phy), PHYA and PHYB to control germination and growth. This study shows that of the five ethylene receptor isoforms in Arabidopsis, ETR1 has a unique role in modulating the effects of red and far-red light on plant growth and development.
Keywords: ethylene receptors, ethylene, phytochrome, light signaling, cross-talk, seed germination
INTRODUCTION
Ethylene is a gaseous plant hormone that is involved in many developmental and physiological processes in higher plants (Mattoo and Suttle, 1991; Abeles et al., 1992). In Arabidopsis thaliana, there are five receptor isoforms that mediate responses to ethylene called ETHYLENE RESPONSE1 (ETR1), ETR2, ETHYLENE INSENSITIVE4 (EIN4), ETHYLENE RESPONSE SENSOR1 (ERS1), and ERS2 (Chang et al., 1993; Hua and Meyerowitz, 1998; Hua et al., 1998; Sakai et al., 1998). Genetic studies have shown that in the absence of ethylene, the receptors positively regulate CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) which acts as a negative regulator of the pathway. The role of CTR1 is to inhibit downstream components of the pathway and prevent ethylene responses (Kieber et al., 1993). According to current models, ethylene binding to the receptors reduces the activity of the receptors. This leads to reduced activity of the CTR1 kinase resulting in reduced phosphorylation of EIN2 protein (Chen et al., 2011; Ju et al., 2012; Qiao et al., 2012). This reduction in EIN2 phosphorylation leads to a decrease in ubiquitination of EIN2 causing a rise in EIN2 protein levels and proteolytic separation of the C-terminal portion of the protein from the N-terminal portion (Qiao et al., 2009, 2012; Ju et al., 2012; Wen et al., 2012). The C-terminal region of EIN2 causes a rise in the levels of the EIN3 and EIL1 transcription factors (Guo and Ecker, 2003; Yanagisawa et al., 2003; Gagne et al., 2004) which leads to most ethylene responses.
The plant ethylene receptors fall into two subfamilies with subfamily 1 comprising ETR1 and ERS1 and subfamily 2 comprising ETR2, EIN4, and ERS2. These receptors have homology to bacterial two-component receptors that signal via histidine autophosphorylation followed by phosphotransfer to an aspartate on a receiver domain. However, only subfamily 1 receptors have histidine kinase activity in vitro with the subfamily 2 receptors having serine/threonine kinase activity in vitro (Gamble et al., 1998; Moussatche and Klee, 2004). ETR1 is unique in that it is the only ethylene receptor in Arabidopsis with both histidine kinase activity and a receiver domain and it may be the only receptor with histidine kinase activity in vivo (Moussatche and Klee, 2004). ETR1 histidine kinase activity is not required for ethylene signaling in plants (Wang et al., 2003; Xie et al., 2006; Qu et al., 2007) but does modulate signal output via an unknown mechanism (Binder et al., 2004; Kim et al., 2011; Hall et al., 2012). All of the Arabidopsis ethylene receptor isoforms are involved in ethylene signaling (Hua et al., 1995, 1998; Hall and Bleecker, 2003). However, even though the receptors have overlapping roles for certain traits, it is also clear that they have non-redundant roles (Hall and Bleecker, 2003; Binder et al., 2004, 2006; Seifert et al., 2004; O’Malley et al., 2005; Xie et al., 2006; Qu et al., 2007; Plett et al., 2009a,b; Liu et al., 2010; Kim et al., 2011). In some cases, individual isoforms have roles that are opposite from the other isoforms in the control of certain phenotypes (Binder et al., 2006; Liu et al., 2010; Wilson et al., 2014). This is likely to be a common attribute of ethylene signaling in land plants since certain receptors have a prominent role in controlling specific phenotypes in other species including both eudicots and monocots (Tieman et al., 2000; Kevany et al., 2007; Chen et al., 2009; Wuriyanghan et al., 2009). The mechanistic basis for these unique roles is unknown.
Seed germination is a critical developmental stage of the plant that is controlled by both hormones such as abscisic acid (ABA), gibberellic acid (GA), and ethylene and environmental factors such as light quality and quantity (reviewed by Bentsink and Koorneef, 2008; Linkies and Leubner-Metzger, 2012). Ethylene stimulates germination of Arabidopsis seeds (Bleecker et al., 1988; Wilson et al., 2014) and ethylene insensitive etr1-1 mutants have diminished germination (Chiwocha et al., 2005). Ethylene appears to function in this process by antagonizing the inhibitory effects of ABA on germination (Chiwocha et al., 2005; Linkies et al., 2009). However, we recently showed that loss of ETR1 enhances and loss of ETR2 diminishes seed germination during salt stress (Wilson et al., 2014). These contrasting roles for ETR1 and ETR2 are not predicted by current models of ethylene signaling and appear to largely be independent of ethylene. Rather, ETR1 and ETR2 are affecting seed germination during salt stress predominantly via regulation of ABA synthesis or signaling (Wilson et al., 2014). These prior observations suggested that there might be other traits regulated by the ethylene receptors that are unpredicted for by current models of ethylene signaling.
Light intensity and quality affect Arabidopsis seed germination and the phytochrome (phy) family of photoreceptors has a critical role in this regulation (Shinomura et al., 1994, 1996). The phy are evolutionarily related to the ethylene receptors with both receptors having homology to bacterial two-component receptors and are thought to have been acquired from cyanobacteria (Bleecker, 1999; Mount and Chang, 2002). It is noteworthy that even though the phy are evolutionarily related to the ethylene receptors and also contain putative histidine kinase domains, plant phy have serine/threonine protein kinase activity (Yeh and Lagarias, 1998; Fankhauser et al., 1999) much like the subfamily two ethylene receptors. The phy are red/far-red photoreversible receptors that affect many developmental and growth processes in plants including germination (Neff et al., 2000). Illumination with red light drives phy to the active form of the receptor that is far-red absorbing (PFR) and illumination with far-red light drives it toward the red absorbing form (PR) that is inactive. There are complex interactions between light and phytohormones in the control of germination (reviewed by Bae and Choi, 2008; Finkelstein et al., 2008; Jaillais and Chory, 2010; Lau and Deng, 2010; Leivar and Quail, 2010). Here we report that loss of ETR1 reduces the effect of far-red light on germination and is affecting the transcript abundance of genes encoding metabolic enzymes for both GA and ABA. Epistasis analysis shows that ETR1 genetically interacts with PHYA and PHYB. However, the effects of ETR1 on seed germination may be independent of light suggesting a model where ETR1 acts in parallel with the photoreceptors to control seed germination.
MATERIALS AND METHODS
PLANT MATERIALS AND CHEMICALS
The etr1-6, etr1-7, etr2-3, and ein4-4 mutants are lab stocks that have been previously characterized (Hua and Meyerowitz, 1998). The phya-t (SALK 014575C) and phyb-9 (CS6217) mutants were obtained from the Arabidopsis Biological Resource Center and have previously been described (Rösler et al., 2007; Sung et al., 2007). The receptor combinatorial mutants and transformants used in this study have previously been described (Hua and Meyerowitz, 1998; Kim et al., 2011; Wilson et al., 2014). All mutants are in the Columbia background. GA was from ACROS Organics (Belgium) and the ABA biosynthesis inhibitor norflurazon was from Fluka (Switzerland). L-α-(2-amino ethoxyvinyl)-glycine (AVG) was a gift from Rohm Haas (Philadelphia).
The etr1-6;phya-t and etr1-6;phyb-9 mutants were obtained by crossing phya-t to etr1-6 and phyb-9 to etr1-6. The double mutants were selected for in the F2 generation by first growing seedlings in constant far-red light to identify homozygous phya-t mutants or constant red light to identify homozygous phyb-9 mutants. Tall seedlings were transferred to soil and subsequently genotyped with dCAPs primers to identify homozygous etr1-6 plants. The dCAPs primers used for genotyping etr1-6 were 5′-ACTCGTTGAAGTCGTCCCTGATC-3′ (forward) and 5′-ATGTGAGAGAGCTACAGCCAC-3′ (reverse). These primers yield an expected polymerase chain reaction (PCR) product of 121 bp. Digestion of this PCR product with BslI results in a 23 and 98 bp fragment for wild-type ETR1 and does not digest product from etr1-6, while digestion with Hpy188I results in a 24 and 97 bp product from etr1-6 and does not digest PCR product from wild-type ETR1. Homozygous F3 or F4 seeds were used for germination assays and analysis of hypocotyl lengths.
SEED GERMINATION EXPERIMENTS
To minimize biological variation, seeds were harvested on the same day from plants grown together under similar conditions (Hensel et al., 1993), stored in a desiccator at room temperature, and allowed to after-ripen for at least 3 weeks. These were then mechanically sorted as previously described (Ellwell et al., 2011). Seeds between 250 and 300 μm in size were surface sterilized in 70% ethanol for 30 s, dried on filter paper and placed on agar plates containing half-strength Murashige and Skoog (MS) basal medium with Gamborg’s vitamins (Sigma, St. Louis, MO, USA), pH 5.7, 0.8% (w/v) agar, with no added sugar. Plates were sealed with micropore surgical tape (3M, St. Paul, MN, USA) so that ethylene did not accumulate (Buer et al., 2003). Twenty seeds of one genotype were plated per plate with a spacing of 5 mm between seeds and three plates per genotype per condition were used in each experiment.
Where used, GA and the ABA synthesis inhibitor, norflurazon were prepared as 10,000× stocks in ethanol, filter sterilized, and added to the media at the indicated concentration after autoclaving the media. Solvent control plates contained 0.01% ethanol. In some cases, 5 μM AVG was added to block ethylene biosynthesis. Ethylene treatments were conducted by placing the plates of seeds in sealed containers and injecting ethylene to yield the indicated concentrations.
Unless otherwise specified, seeds were treated with 45–55 μmol m-2 s-1 white light for 4 h. This was followed by a 3 h treatment with 12 μmol m-2 s-1 blue (λmax = 470 nm), green (λmax = 525 nm), red (λmax = 672 nm), or far-red (λmax = 732 nm) light. Monochromatic light was provided by LED arrays (Quantum Devices, Inc., Barneveld, WI, USA). For positive controls, seeds were treated for 3 h with 45–55 μmol m-2 s-1 white light. After light treatment, seeds were allowed to germinate for 7 days in darkness at which time germination was evaluated. For dark controls, seeds were treated for 5 min with far-red light immediately after sowing without a pre-treatment with white light (Oh et al., 2007) and then allowed to germinate in darkness for 7 days. In some experiments, seeds were germinated on 150 mM NaCl under continuous white light.
Germination was scored as the visible rupture of the testa (seed coat). After each experiment, plates containing seeds that did not germinate were transferred to white light conditions (16 h of light/8 h of dark photoperiod) and germination evaluated after 7 days. In all cases, seeds reached at least 95% germination showing that the seeds were viable.
ETHYLENE MEASUREMENTS
To measure ethylene production, 22 mg of seeds were placed on 1 mL of half-strength MS media in sealed 6 mL glass vials, treated with white or far-red light, and ethylene levels in the headspace measured every 6 h over several days using an ETD-300 photoacoustic ethylene detector (Sensor Sense, The Netherlands) as previously described (Wilson et al., 2014).
HYPOCOTYL GROWTH EXPERIMENTS
To examine the growth of hypocotyls, seeds were placed on agar plates and illuminated with 45–55 μmol m-2 s-1 white light for 24 h. The plates of seeds were then grown vertically for 6 days under continuous red or far-red light. At that time, the plates were scanned with a flat-bed scanner and seedling length measured using ImageJ (ver. 1.43u).
RNA ISOLATION AND QUANTITATIVE REAL-TIME REVERSE TRANSCRIPTASE (qRT)-PCR
The transcript abundance of several Arabidopsis genes that encode GA and ABA metabolic enzymes was examined using quantitative real-time reverse transcriptase (qRT)-PCR. This included gene transcripts for GA3 oxidase 1 and 2 (GA3ox1, GA3ox2) encoding for two GA biosynthesis enzymes, GA2ox2 encoding for a GA degradation enzyme, zeaxanthin epoxidase (ZEP), 9-cis-epoxycarotenoid dioxygenase6 and 9 (NCED6, NCED9) encoding for three ABA biosynthesis enzymes, and ABA 8′-hydroxylase (CYP707A2) encoding for an ABA degradation enzyme. For this, total RNA was isolated from either 25 mg dry seeds or 25 mg (dry weight) of seeds that were imbibed in half strength MS and light treated as specified for the indicated times. RNA was isolated according to methods modified from Meng and Feldman (2010). Instead of resuspending the pellet in Trizol, the RNA was further purified using the Spectrum Plant Total RNA Kit (Sigma, St. Louis, MO, USA). Total RNA was treated with DNase I (Invitrogen, Carlsbad, CA, USA) and 800 ng of the RNA was used for cDNA synthesis with the ImProm-II Reverse Transcription System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Each qPCR reaction consisted of 5 μL of SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, USA), 0.5 μL each of the forward and Reverse primers (10 μM) and 4 μL of cDNA diluted 1:8. The qPCR reactions were run on a Bio-Rad iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with the following conditions: an initial denaturation step of 95∘C for 1 min followed by 45 cycles of 15 s at 95°C, 30 s at 58°C, and 10 s at 72°C.
Transcript data were normalized to At3g12210 (Dekkers et al., 2011) using the method of Livak and Schmittgen (2001) for each seed line for each condition to obtain the relative amounts of target gene transcripts between plant backgrounds for each treatment. The primers used for GA3ox2 were 5′-GTTCTTTAATAAGAAGATGTGGTCCG-3′ (forward) and 5′CATCAACTTGGCTGCCAACTTT-3′ (reverse). Other primers used have been previously described (Seo et al., 2004; Kim et al., 2012; Shu et al., 2013).
STATISTICS
Data were analyzed with Student’s t-tests and considered statistically different with a P value <0.05.
RESULTS
ETR1 INHIBITS SEED GERMINATION AFTER EXPOSURE TO FAR-RED LIGHT
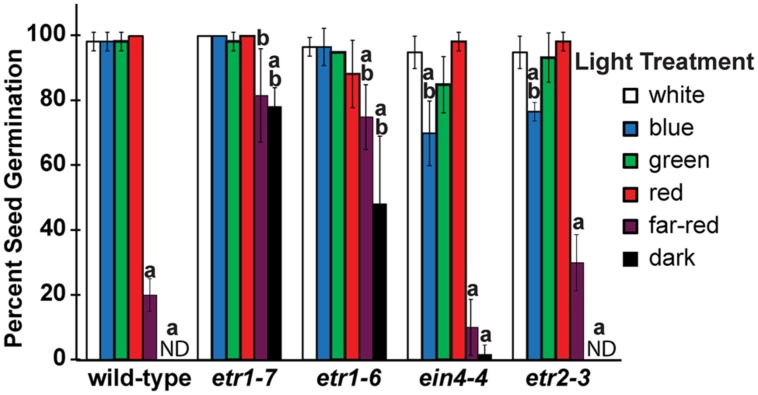
We recently reported that loss of ETR1 led to better seed germination during salt stress when seeds were germinated in white light (Wilson et al., 2014). During the course of those experiments we noted that loss of ETR1 also caused better seed germination during salt treatment when the seeds were germinated in darkness (data not shown). This led us to determine the role of individual ethylene receptor isoforms on seed germination under different light conditions. To do this, we measured the percent of seed germination for receptor loss-of-function mutants 7 days following a 3 h treatment with white, blue, green, red, or far-red light as described in Section “Materials and Methods.” Following white light treatment, all seed lines reached at least 95% germination (Figure 1). The extent of wild-type seed germination was not altered by blue, green, or red light. However, germination of wild-type seeds was drastically reduced by treatment with far-red light. This is consistent with prior research (Shinomura et al., 1994). By contrast, the effect of far-red light on the germination of etr1-6 and etr1-7 loss-of-function mutants was significantly reduced even though responses to blue, green, and red light were not altered by the loss of ETR1. This effect of etr1 mutants was observed with multiple seed batches. Loss of ETR2 or EIN4 did not affect responses to far-red light, but did have a small and statistically significant effect on germination in blue light (Figure 1). Loss of ERS1 or ERS2 had no measurable effect on germination in any of the light conditions tested (data not shown). Interestingly, when germination occurred in darkness without a white light pre-treatment, wild-type, etr2-3, and ein4-4 seeds failed to germinate or germinated very poorly, whereas etr1 loss-of-function mutants germinated to an extent that was similar to germination after far-red illumination (Figure 1). These data suggest that ETR2 and EIN4 promote seed germination after exposure to blue light and ETR1 inhibits germination after exposure to far-red light and in darkness.
FIGURE 1.
Ethylene receptors affect germination in response to different wavelengths of light. Seed germination of wild-type seeds was compared to ethylene receptor loss-of-function mutants. Seeds were treated with 4 h of white light, followed by 3 h of different colors of monochromatic light or white light as indicated. For dark controls, seeds were treated for 5 min with far-red light immediately after sowing without a pre-treatment with white light. The seeds were then placed in darkness for 7 days. At that time seed germination was measured and the percent of seed germination calculated. Plots show the average ± SD from at least three biological replicates. ND denotes no germination detected. Data were analyzed by t-tests and differences considered statistically significant with P < 0.05. aSignificant difference from white light for that ecotype; bSignificant difference of mutant from wild-type after the same light treatment.
ETR1 FUNCTIONS OPPOSITELY TO ETR2 TO INFLUENCE SEED GERMINATION AFTER EXPOSURE TO FAR-RED LIGHT
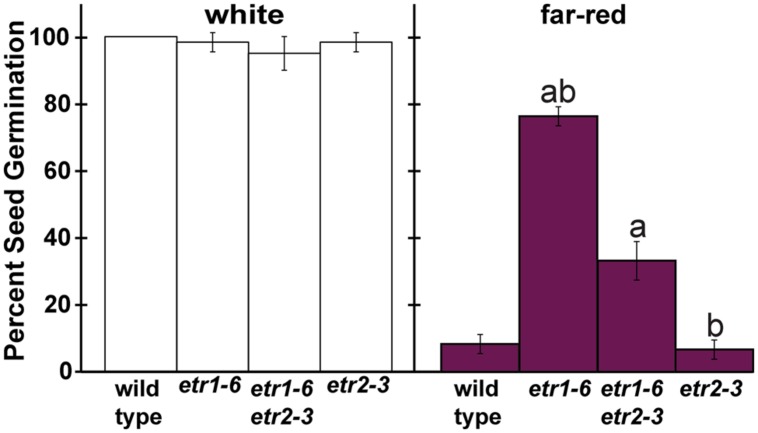
In seed germination under salt stress, etr2-3 loss-of-function mutants were shown to have reduced germination compared to wild-type seeds and ETR1 and ETR2 were found to act additively (Wilson et al., 2014). In contrast, under the far-red light conditions used, the germination of etr2-3 mutants was indistinguishable from wild-type seeds (Figure 1). However, an effect of ETR2 on seed germination in these conditions might have been masked by the fact that the wild-type seeds germinated poorly and a further reduction might have been obscured. To determine whether or not ETR1 and ETR2 act additively to control seed germination after far-red light, we compared seed germination of etr1-6 and etr2-3 to germination of etr1-6;etr2-3 double mutants. Following a 3 h white light treatment, the seed germination of the single and double mutants were not significantly different and reached at least 95% germination (Figure 2). A different pattern emerged after a 3 h far-red light treatment. Here, the etr1-6 seeds germinated significantly better and the etr2-3 seeds germinated significantly worse than the etr1-6;etr2-3 double mutants. These results show that ETR1 and ETR2 have opposite effects on seed germination after far-red light treatment.
FIGURE 2.
ETR1 functions oppositely to ETR2 to control seed germination after treatment with far-red light. The percent of wild-type, etr1-6, etr2-3, and etr1-6;etr2-3 seed germination was determined 7 days after a far-red light treatment. For comparison, germination after a white light treatment is included. Plots show the average ± SD from at least three biological replicates. Data were analyzed by t-tests and differences considered statistically significant with P < 0.05.aStatistical difference from wild-type in that light condition; bSignificant difference of the single mutant from the double.
THE ETR1 RECEIVER DOMAIN IS NOT REQUIRED FOR ETR1 FUNCTION IN SEED GERMINATION AFTER EXPOSURE TO FAR-RED LIGHT
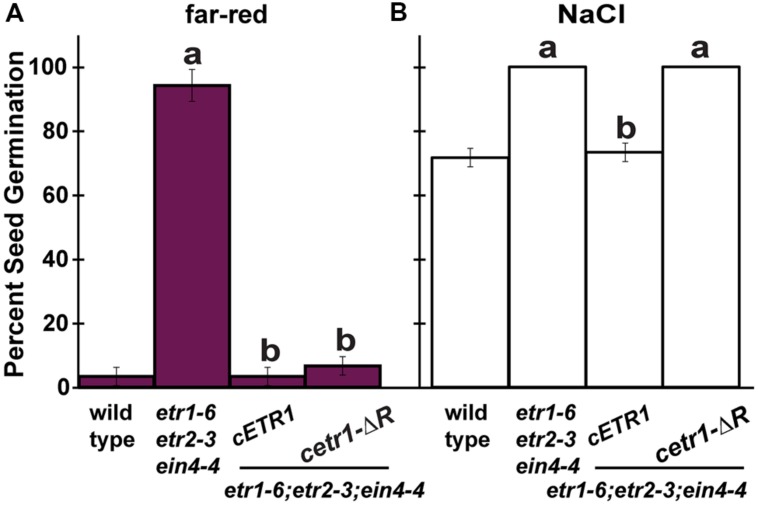
We previously showed that etr1-6;etr2-3;ein4-4 triple mutants germinate better than wild-type seeds during salt stress under white light (Wilson et al., 2014). Interestingly, transformation of this triple mutant with a cDNA construct for full-length ETR1 (cETR1) resulted in a reduction in germination, whereas transformation with a truncated ETR1 transgene lacking the receiver domain (cetr1-ΔR) failed to alter germination (Wilson et al., 2014). Both constructs are under the control of the ETR1 promoter and have been shown to be expressed and functional (Kim et al., 2011) supporting the idea that the full-length ETR1 receptor is required to affect seed germination during salt stress under white light. To determine whether or not the ETR1 receiver domain is also required to modulate seed germination after treatment with far-red light, we examined the seed germination of these transformants. Comparable to our results with germination on salt, the etr1-6;etr2-3;ein4-4 triple mutants germinated better than wild-type seeds after illumination with far-red light (Figure 3). However, in contrast to germination on salt, both the full-length and truncated ETR1 transgenes reduced seed germination to wild-type levels after far-red illumination indicating that the receiver domain of ETR1 is not required to function in this trait. When transferred to white light conditions for 7 days, all seed lines reached at least 98% germination (data not shown). This pattern of rescue is different from what we obtained when studying the role of ETR1 in the control of germination during salt stress in white light (Wilson et al., 2014). To confirm this difference, we germinated seeds in white light under salt stress. Under salt stress, wild-type seeds reached approximately 72% germination while the etr1-6;etr2-3;ein4-4 triple mutants reached 100% germination (Figure 3). The full-length cETR1, but not the truncated cetr1-ΔR transgene, reduced germination to wild-type levels as previously reported (Wilson et al., 2014). These results reveal that the function of ETR1 varies depending upon environmental conditions.
FIGURE 3.
After far-red illumination, the better germination of etr1;etr2;ein4 triple mutants is reversed by both a full-length and truncated ETR1 transgene lacking the receiver domain. (A) The percent of germination 7 days after treatment with far-red light for wild-type, etr1-6;etr2-3;ein4-4 triple mutants and these triple mutants transformed with cDNA for full-length ETR1 (cETR1) or a truncated ETR1 lacking the receiver domain (cetr1-ΔR) was determined. (B) For comparison, the percent germination of these seed lines 7 days after sowing on 150 mM NaCl in the presence of continuous white light was determined. Data were analyzed by t-tests and differences considered statistically significant with P < 0.05. aSignificant difference from wild-type germinated under the same conditions.bSignificant rescue of germination by the transgene.
ETHYLENE MAY HAVE A ROLE IN MEDIATING THE EFFECTS OF ETR1 ON GERMINATION AFTER EXPOSURE TO FAR-RED ILLUMINATION
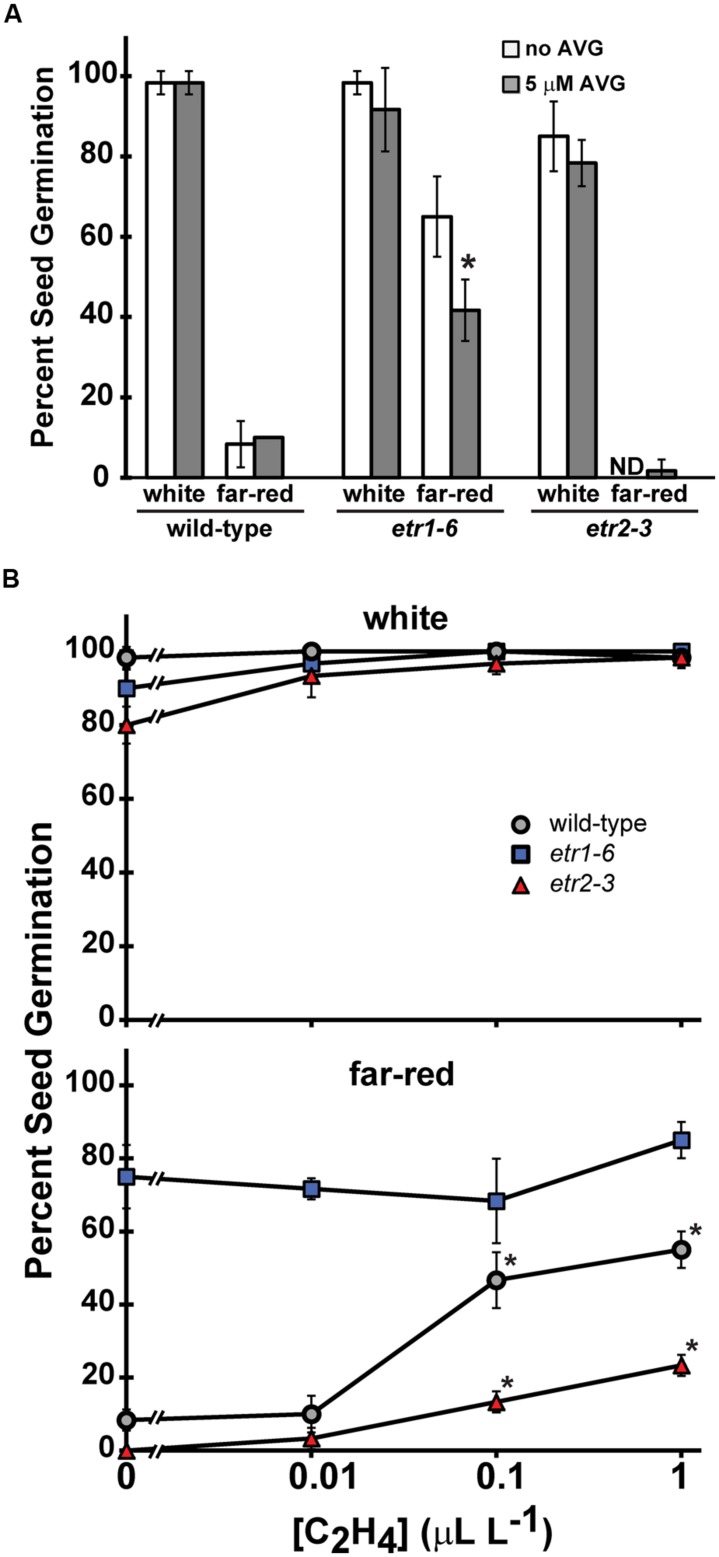
In order to determine whether differences in ethylene production or sensitivity are responsible for the difference between the etr1 loss-of-function mutants and wild-type in germination following far-red treatment, we examined the effect of ethylene and the ethylene biosynthesis inhibitor, AVG, on germination. Following white light treatment, AVG did not have a statistically significant effect on the germination of wild-type, etr1-6 or etr2-3 seeds (Figure 4A). AVG also did not affect wild-type or etr2-3 germination following far-red treatment. However, germination of etr1-6 was statistically reduced by 20% by AVG after far-red light (Figure 4A). Although AVG reduced germination of etr1-6 seeds in far-red, this mutant still had better seed germination than wild-type. These observations suggest that part of the difference in germination between wild-type and the etr1-6 mutants may be due to alterations in ethylene levels. To more directly determine this, we measured ethylene levels of germinating seeds after white or far-red light treatment. We found that ethylene levels were indistinguishable between etr1-6 and wild-type seeds during germination in both conditions (data not shown) suggesting that higher ethylene levels are not responsible for the enhanced germination of etr1-6 seeds after far-red illumination.
FIGURE 4.
The effects of L-α-(2-amino ethoxyvinyl)-glycine (AVG) and ethylene on seed germination after treatment with far-red light. The number of seeds that germinated after far-red or white illumination was counted and the percent of seeds that germinated was calculated. (A) Seeds on plates containing the ethylene biosynthesis inhibitor, AVG, were treated with far-red light and then germinated for 7 days in the dark. The average ± SD from at least three biological replicates is plotted. ND denotes no germination detected. Data were analyzed by t-tests and differences considered statistically significant with P < 0.05. *Denotes that AVG had a significant effect compared to the non-treated control in that light condition. (B) The effects of increasing concentrations of ethylene as indicated were measured after treatment with white light or far-red light. The average ± SD from at least three biological replicates is plotted. Data were analyzed by t-tests and differences considered statistically significant with P < 0.05. *Denotes that ethylene significantly increased the percent of germination after-far red light over the ethylene untreated control.
Ethylene can stimulate seed germination of lettuce and cocklebur after far-red illumination (Abeles and Lonski, 1969; Burdett and Vidaver, 1971; Esashi et al., 1987). We wished to determine if this was the case for Arabidopsis too and to see if the etr1-6 and etr2-3 mutants had altered ethylene sensitivity when assayed in these conditions. One explanation for etr1 loss-of-function mutants germinating better than wild-type is that they are more sensitive to ethylene. This has been observed when assaying growth of dark-grown Arabidopsis seedlings (Hua and Meyerowitz, 1998; Cancel and Larsen, 2002), but is not true for seeds germinating on salt (Wilson et al., 2014). After white light, 0.01 ppm ethylene increased the germination of etr2-3 seeds slightly, but significantly (P < 0.05), and higher concentrations of ethylene caused no measurable additional increase in germination. By contrast, ethylene had no measurable effect on the germination of wild-type or etr1-6 seeds after white light illumination (Figure 4B). After treatment with far-red light, ethylene increased seed germination of wild-type seeds (Figure 4B). By contrast, ethylene had no measurable effect on germination of the etr1-6 mutants indicating that these mutants are largely un-responsive to ethylene for this trait. We also examined etr2-3 mutants and found that ethylene enhanced germination of these seeds after far-red illumination, but the effect was smaller than seen with wild-type seeds. The highest concentration of ethylene used (1 ppm) failed to completely eliminate the differences in germination between etr1-6, etr2-3, and wild-type seeds. This is at least 1000-fold higher than the concentration of ethylene released by the germinating seeds under these conditions. Thus, ethylene may have a role in mediating the differences in germination between wild-type and etr1 mutants, but other factors are likely to be more important.
ABA AND GA HAVE A ROLE IN MEDIATING THE EFFECTS OF ETR1 ON GERMINATION AFTER EXPOSURE TO FAR-RED ILLUMINATION
There are complex interactions between hormones and light to control germination (reviewed by Finkelstein et al., 2008; Lau and Deng, 2010; Leivar and Quail, 2010). We previously found that ETR1 predominantly affects ABA sensitivity to alter seed germination during salt stress (Wilson et al., 2014). We therefore were curious to know if ETR1 is also predominantly affecting seed germination after treatment with far-red light in the same way.
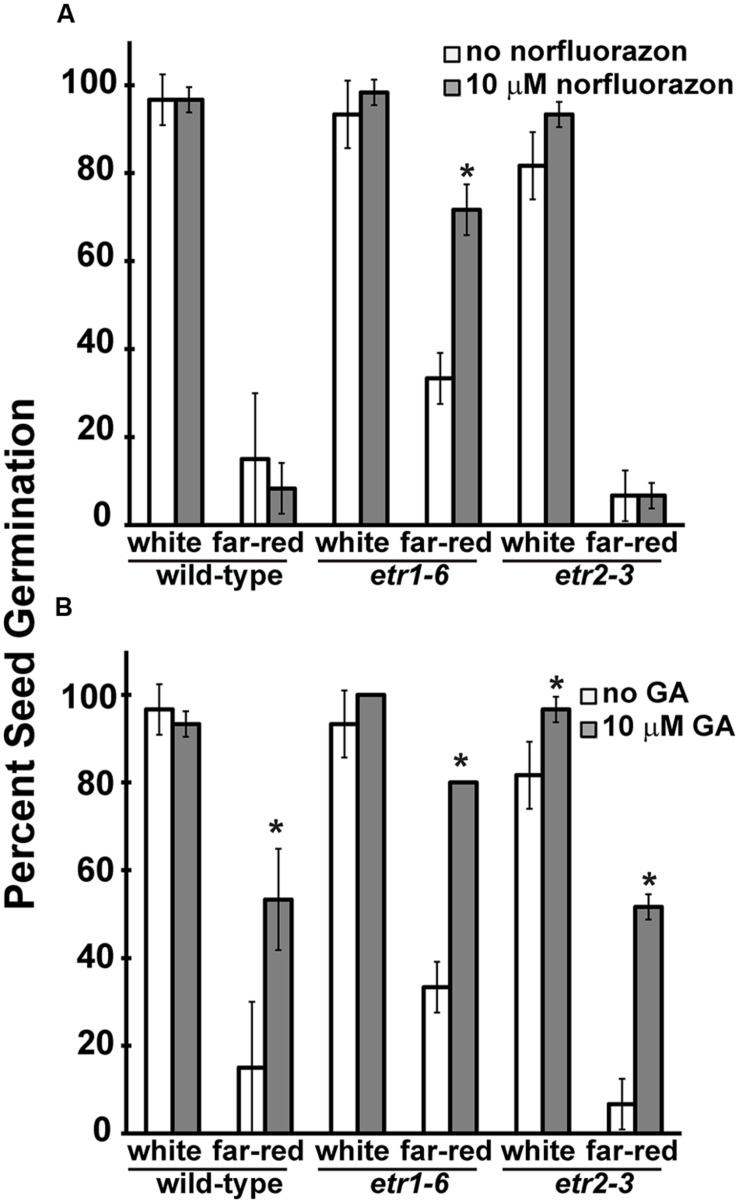
Abscisic acid is well known for inhibiting germination and GA for stimulating Arabidopsis seed germination (Hilhorst and Karssen, 1992; Garciarrubio et al., 1997). In order to test whether differences in ABA or GA or both were responsible for the differences in germination following far-red light treatment, we treated the seeds with an ABA biosynthesis inhibitor, norflurazon, or with GA. Initially, these chemicals were dissolved in ethanol to yield a final concentration of 0.1% (v/v) ethanol. However, this concentration of ethanol significantly (P < 0.05) reduced germination of etr1-6 and etr2-3 seeds after white light treatment to 55 ± 22 and 45 ± 9%, respectively. As previously observed, this concentration of ethanol had no measureable effect on the germination of Columbia seeds (Hirayama et al., 2004). Even though norflurazon and GA affected germination as expected, this added effect of the ethanol made interpretation of the data difficult. Because of this, we used stocks that yielded a final ethanol concentration of 0.01% (v/v). At this concentration of ethanol, there was no statistical difference (P < 0.05) between wild-type and the mutants following white light treatment (Figure 5). However, following far-red treatment, this concentration of ethanol significantly (P < 0.05) reduced the germination of etr1-6 (Figure 1 vs. Figure 5). In Figure 1, that has no added ethanol, etr1-6 reached 75 ± 10% germination, but when ethanol was added to the plates (Figure 5) etr1-6 only reached 33 ± 6% germination following far-red treatment. These experiments were conducted at the same time and with the same seed batches. We found that addition of 10 μM norflurazon, resulted in a significant improvement of etr1-6 germination following far-red treatment, but surprisingly had no effect on wild-type or etr2-3 seed germination (Figure 5A). Treatment with 10 μM GA significantly increased the germination following far-red treatment of all three seed lines tested (P < 0.05), but was not able to improve the germination of wild-type and etr2-3 to that seen by etr1-6 (Figure 5B). These data suggest that ETR1 is having a complex effect on ABA and GA to control seed germination after far-red illumination. This is somewhat different from the role of ETR1 in controlling seed germination during salt stress (Wilson et al., 2014).
FIGURE 5.
The effects of gibberellic acid (GA) and norflurazon on seed germination after treatment with far-red light. Wild-type, etr1-6, and etr2-3 seeds were germinated after treatment with white or far-red light. Seed germination was then measured 7 days later. The effects of (A) norflurazon and (B) GA were assessed. All experiments were performed in triplicate. The average percent seed germination ± SD is plotted. Data were analyzed by t-tests and differences considered statistically significant with P < 0.05. *Denotes that the compound had a significant effect compared to the non-treated control in that light condition.
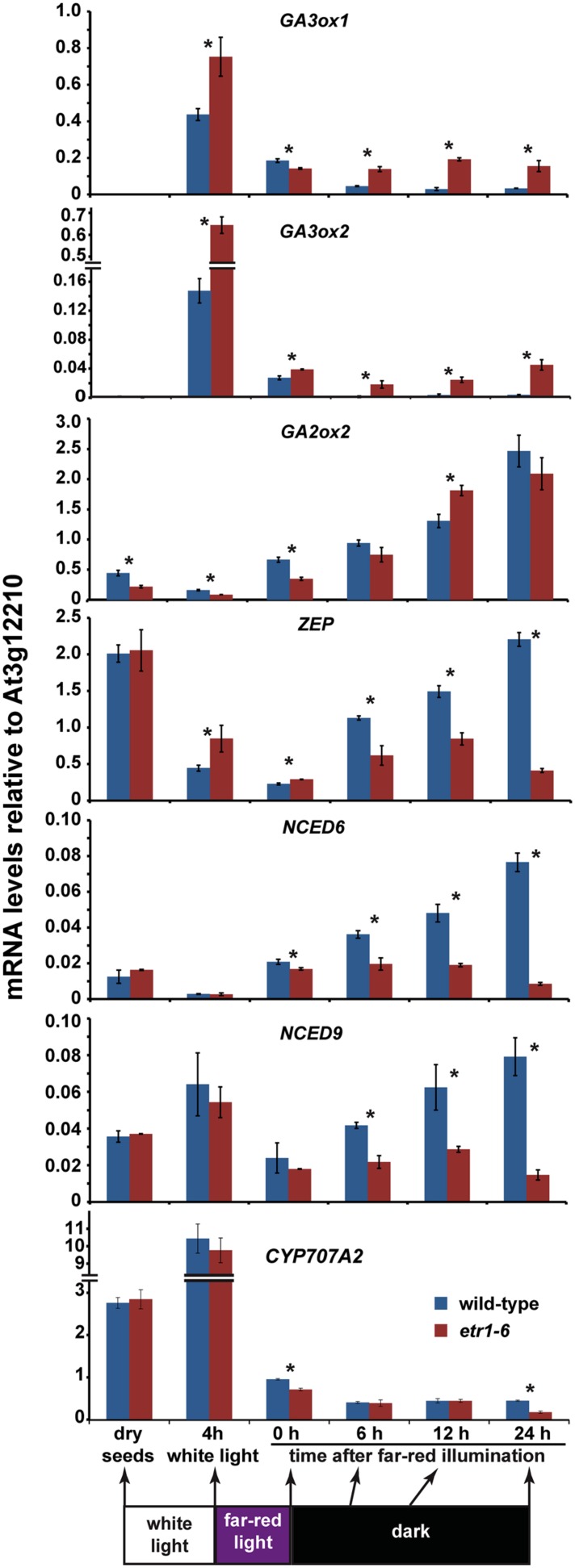
To explore this complex interaction more, we used qRT-PCR to examine the time-course of changes in transcript levels of selected genes during germination under the conditions used above. We chose genes encoding for enzymes for GA biosynthesis (GA3ox1, GA3ox2), GA degradation (GA2ox2), ABA biosynthesis (ZEP, NCED6, NCED9), and ABA degradation (CYP707A2). These genes are important for seed development and germination (Yamauchi et al., 2004, 2007; Lefebvre et al., 2006; Okamoto et al., 2006). Generally consistent with prior studies (Yamauchi et al., 2004; Seo et al., 2006; Finch-Savage et al., 2007), imbibing wild-type seeds in white light led to large increases in the transcript abundance of GA3ox1, GA3ox2, and CYP707A2 and significant (P < 0.05) decreases in GA2ox2, ZEP, and NCED6 (Figure 6). The transcript abundance of NCED9 increased slightly, but overall these results are predicted to lead to increased GA levels and decreased ABA levels. Immediately following treatment with far-red light, the transcript abundance of GA3ox1, GA3ox2, ZEP, NCED9, and CYP707A2 decreased and the levels of GA2ox2 and NCED6 increased in wild-type seeds. Overall, these transcriptional changes are predicted to cause ABA levels to rise and GA levels to decrease compared to white light and lead to inhibited germination. In the wild-type seeds, the transcript levels of each gene showed different patterns of change over the 24 h period following far-red illumination. GA3ox1 and GA3ox2 showed a small decrease 6 h following far-red light but then plateaued at a very low level and CYP707A2 showed very little change in transcript abundance after far-red illumination. By contrast, the transcript levels of ZEP, NCED6, and NCED9 increased with time after far-red light treatment.
FIGURE 6.
Transcript levels for select GA and ABA metabolism genes are affected by loss of ETR1. The levels of transcript for GA3ox1, GA3ox2, GA2ox2, ZEP, NCED6, NCED9, and CYP707A2 in wild-type and etr1-6 mutants at various times during germination were measured using qRT-PCR. Data were normalized to the levels of At3g12210 in each seed line at each stage of germination. Transcript levels of dry seeds and imbibed seeds treated with light as indicated were compared. Data were analyzed by t-tests and differences considered statistically significant with P < 0.05. *Denotes that the levels of transcript at that time-point are statistically different between wild-type and etr1-6 seeds.
The effect of the etr1-6 mutation on the transcript levels of these genes was evident at various times during germination. After white-light treatment, the etr1-6 mutants had higher levels of GA3ox1, GA3ox2, and ZEP and slightly lower levels of GA2ox2 (P < 0.05) compared to the wild-type seeds (Figure 6). Immediately following treatment with far-red light, the levels of GA3ox1, GA2ox2, NCED6, and CYP707A2 were slightly but significantly (P < 0.05) lower in etr1-6 mutants compared to wild-type and GA3ox2 and ZEP were slightly, but significantly higher in the mutants compared to wild-type. More differences between the etr1-6 seeds and wild-type became apparent with increasing time after treatment with far-red light with a trend toward higher levels of GA3ox1 and GA3ox2 and lower levels of ZEP, NCED6, and NCED9 in the etr1-6 mutants compared to wild-type. The differences in the levels of ZEP, NCED6, and NCED9 appear to arise because the levels of these gene transcripts increased in the wild-type seeds, but remained fairly constant in the etr1-6 mutants. Since the transcript levels of GA and ABA metabolic genes generally correlate with the levels of GA and ABA, respectively (Qin and Zeevaart, 1999; Iuchi et al., 2001; Reid et al., 2002; Seo et al., 2004; Lefebvre et al., 2006; Reinecke et al., 2013), our results suggest that the etr1-6 seeds are likely to have higher levels of GA and lower levels of ABA than wild-type seeds following treatment with far-red light. These alterations in GA and ABA levels are predicted to cause better germination of the mutant seeds compared to the wild-type seeds after far-red illumination.
ETR1 GENETICALLY INTERACTS WITH PHYA AND PHYB TO CONTROL GERMINATION
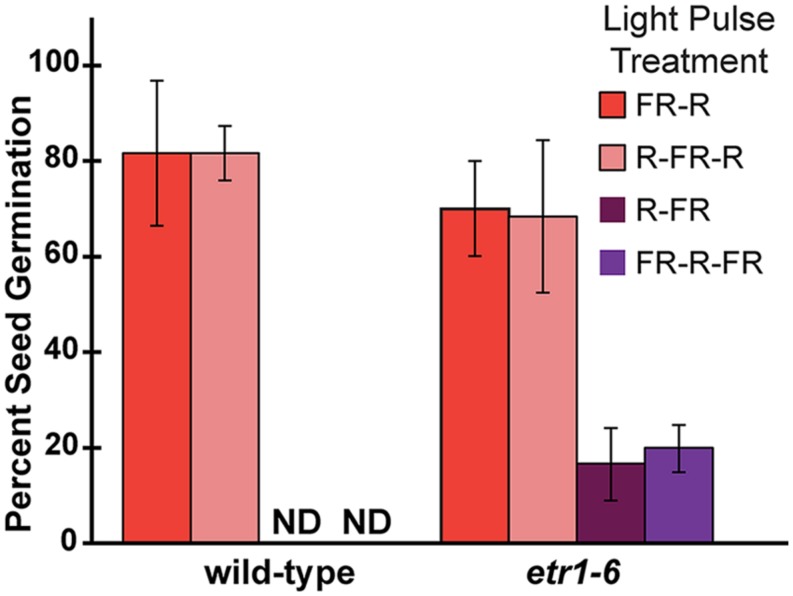
The phy have an important role in seed germination and responses to far-red light are mediated by the phy (Shinomura et al., 1994, 1996; Poppe and Schäfer, 1997; Hennig et al., 2002; Rösler et al., 2007) suggesting that ETR1 is affecting events influenced by the phy. The phy act as photoreversible switches driven by red and far-red light to control germination of seeds including germination of Arabidopsis (Shinomura et al., 1994; Casal and Sánchez, 1998). To confirm that loss of ETR1 is affecting phy-influenced germination, we treated seedlings with a series of 5 min pulses of red and far-red light followed by darkness (Figure 7). From this we found that there was no measurable difference in wild-type and etr1-6 germination rates in seeds treated with a pulse series that ended with red light. By contrast, if the pulse series ended with far-red light, wild-type seedlings failed to germinate, but approximately 20% of the etr1-6 seeds germinated.
FIGURE 7.
The effect of ETR1 on germination after far-red light treatment is reversed by red light. Seeds were treated with white light for 3 h, followed by a series of 5 min pulses of red (R) and far-red (FR) light as shown. The percent of seeds that germinated was determined 7 days later. The average ± SD from at least three biological replicates is plotted. ND denotes no germination detected.
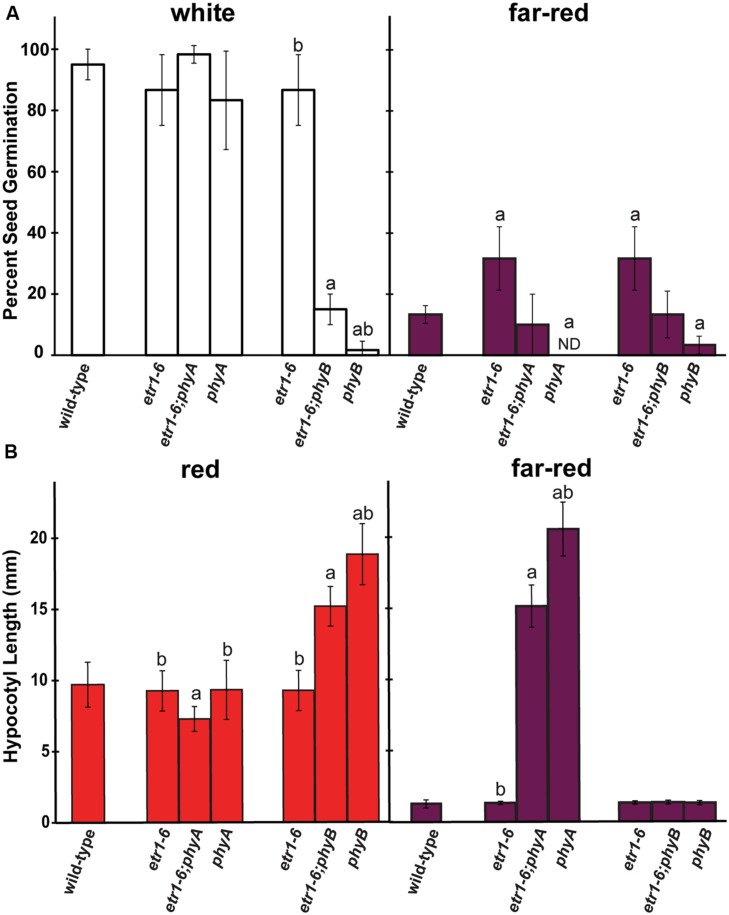
Of the phy, phyA and phyB have the predominant roles in controlling Arabidopsis seed germination (Shinomura et al., 1994, 1996; Rösler et al., 2007). To further explore the interaction between ETR1 and the phy, we generated etr1-6;phya-t and etr1-6;phyb-9 double mutants. After treatment with white light, the germination rates of etr1-6, etr1-6;phya-t, and phya-t were indistinguishable from wild-type seeds (Figure 8A). By contrast, the phyb-9 seeds had severely diminished germination after white light treatment, consistent with previous reports (Reed et al., 1994; Shinomura et al., 1994). The etr1-6;phyb-9 double mutants had a percent germination worse than the etr1-6 single mutants and slightly better than the phyb-9 mutants following white light treatment. After treatment with far-red light, the phya-t and phyb-9 mutants had significantly worse germination than wild-type seeds and the etr1-6 mutants significantly better germination (Figure 8A). Both the etr1-6;phya-t and etr1-6;phyb-9 double mutants had a percent germination intermediate between the single mutants. However, these were not statistically different.
FIGURE 8.
Epistasis analysis of ETR1 and the phytochromes (phy), PHYA and PHYB after light treatment. (A) Seeds were treated with white light for 3 h, followed by either 4 h of white or far-red light as shown. The percent of seed germination was then determined 7 days later. The average ± SD from at least three biological replicates is plotted. ND denotes no germination detected. (B) Seeds were treated with white light for 24 h followed by continuous illumination with red or far-red light for 6 days as shown. The lengths of the hypocotyls were then measured. The average ± SD is plotted. In both panels, the data were analyzed by t-tests and differences considered statistically significant with P < 0.05. aStatistical difference from wild-type in that light condition; bSignificant difference of the single mutant from the double.
We also examined hypocotyl growth in red and far-red light (Figure 8B). Consistent with previous studies, the phyB mutants grew taller than wild-type in red light and the phyA mutants grew taller than wild-type in far-red light (Reed et al., 1994; Quail et al., 1995; Smith et al., 1997). The growth of the etr1-6 seedlings was similar to wild-type in both light conditions. In red light, the etr1-6;phyb-9 double mutants were shorter than the phyb-9 mutants and taller than the etr1-6 mutants. Similarly, in far-red light the etr1-6;phya-t double mutants were shorter than the phya-t seedlings and taller than the etr1-6 seedlings. The etr1-6;phya-t double mutants were also slightly, but significantly, shorter than either single mutant in red light. Together, these results on seed germination and hypocotyl growth show that ETR1 genetically interacts with PHYA and PHYB.
ETR1 AFFECTS GERMINATION INDEPENDENTLY OF LIGHT
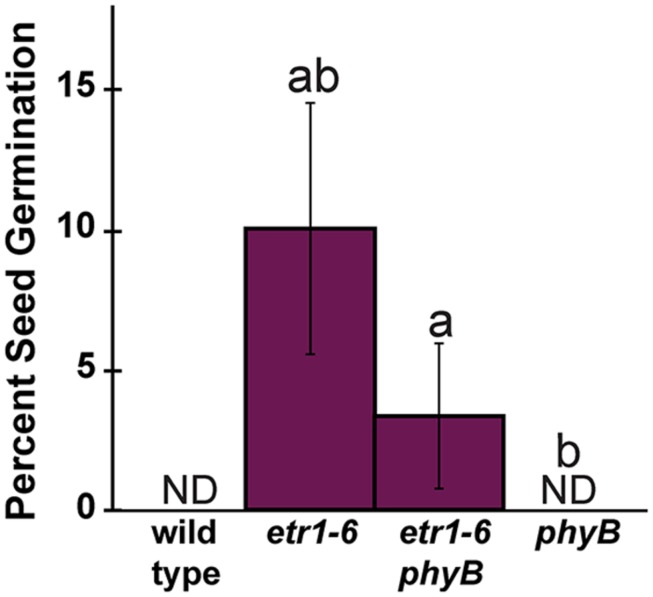
From the above experiments, it is unclear whether or not ETR1 is affecting responses to light by affecting signaling from phyA or phyB. We have previously shown that etr1-6 loss-of-function mutants are more sensitive to ABA when measuring seed germination under white light (Wilson et al., 2014) indicating that the effects of ETR1 could be independent of these photoreceptors. Since phyB has the major role in controlling germination in the dark (Bentsink and Koorneef, 2008), we examined the effect of ETR1 on the germination of phyb-9 mutants in the dark. In dark conditions, wild-type and phyb-9 seeds failed to germinate, whereas etr1-6 mutants did germinate (Figure 9). The etr1-6;phyb-9 also germinated under these conditions but had a percent germination lower than the etr1-6 single mutant seeds. This indicates that ETR1 can affect germination independently of light.
FIGURE 9.
Epistasis analysis of ETR1 and PHYB in the dark. Seeds were germinated in the dark for 7 days and the percent of seed germination was then determined. The average ± SD from at least three biological replicates is plotted. ND denotes no germination detected. The data were analyzed by t-tests and differences considered statistically significant with P < 0.05. aStatistical difference from wild-type in that light condition; bSignificant difference of the single mutant from the double mutant.
DISCUSSION
It has been suggested that there is a network of two-component receptors in plants that fine-tune signaling (Hwang et al., 2002; Hass et al., 2004; Wilson et al., 2014). In this study, we found that ETR1, a two-component-like receptor for ethylene, inhibits germination after far-red light treatment and in the dark. This is similar to results we obtained when examining germination during salt stress (Wilson et al., 2014). However, in the prior study, the effect of ETR1 was to mainly affect ABA signaling or synthesis. In the current study, ETR1 was found to affect ABA, GA, and perhaps ethylene. It has previously been shown that in Arabidopsis phyA and B affect the transcript abundance of ETR2 and ERS1 (Tepperman et al., 2004), and phy interacting factor1 (PIF1), which is important for phy signaling, affects the transcript abundance of ETR2 and EIN4 (Oh et al., 2009). Additionally, phyB is involved in the circadian control of ethylene synthesis in sorghum (Finlayson et al., 1998) showing that there is likely to be signaling cross-talk from the phy to ethylene signaling and synthesis. Results presented in the current study show that there is also signaling from ETR1, but not the other four ethylene receptor isoforms, that affects responses to red and far-red light.
Previous research has shown that the receiver domain of ETR1 affects receptor function in the control of growth of dark-grown seedlings (Binder et al., 2004, 2006; Qu and Schaller, 2004; Kim et al., 2011) and germination during salt stress under white light (Wilson et al., 2014). In the current study, we found that transformation of etr1-6;etr2-3;ein4-4 triple mutants with either cETR1 or cetr1-ΔR rescued germination after far-red light treatment to wild-type levels showing that the receiver domain of ETR1 is not required in the control of this trait. This is intriguing because the receiver domain is required for ETR1 function in the control of germination during salt stress under white light (Wilson et al., 2014; this study). These results indicate that environmental conditions affect the function of the ETR1 ethylene receptor to regulate seed germination. The mechanism for this is not known but one possibility is that a downstream target unique to ETR1 is altered upon illumination. There is precedence for this since light regulates the levels of one downstream target of EIN3, PIF3, to alter the effects of ethylene on plant growth in the dark versus light (Zhong et al., 2012).
A major question is whether or not the effect of ETR1 on seed germination is ethylene-dependent or not. We found that the ethylene biosynthesis inhibitor, AVG, partially reversed the effect of etr1-6 on germination after treatment far-red light, but addition of ethylene up to 1 ppm had no effect on germination of the mutant after far-red illumination. It is possible that the etr1 loss-of-function mutants are producing high levels of ethylene and are saturated for ethylene responses. However, this seems unlikely since etr1-6 produced very low levels of ethylene that were indistinguishable from the germinating wild-type seeds after illumination with far-red light. Transfer of these seeds to white light conditions resulted in an increase in germination showing that the seeds were still capable of better germination. Additionally, AVG has effects that are independent of its effect on ethylene biosynthesis (Larsen and Chang, 2001; Soeno et al., 2010) confounding the results with this compound. Thus, it is seems unlikely that ethylene is the major factor contributing to the differences in germination between wild-type and etr1 loss-of-function mutants observed in this study.
Results presented here indicate that ETR1 is affecting germination by affecting the levels of GA and ABA. We observed that the transcript abundance of genes encoding certain GA and ABA metabolic genes are altered in etr1-6 mutants consistent with higher GA levels and lower ABA levels after far-red light treatment in the etr1 loss-of-function mutants compared to wild-type. Additionally, we have previously shown that etr1-6 mutants are less sensitive to ABA than wild-type (Wilson et al., 2014). Together, these results would explain the better germination of the etr1 loss-of-function mutants. Consistent with these results, application of GA improved the germination of all seed lines and norflurazon improved seed germination of etr1-6 mutants.
Another major question is whether ETR1 is affecting germination via the phy signaling pathway, via a phy-independent pathway, or both. Previous studies provide a tentative link between ETR1 and phyB (Urao et al., 2000; Sweere et al., 2001; To et al., 2004; Salomé et al., 2006; Scharein et al., 2008). However, our data suggest that ETR1 is affecting responses to light that are independent of the phy. Support for this is our observation that etr1 loss-of-function mutants are more sensitive to ABA in white light (Wilson et al., 2014) and germinate better than wild-type seeds in the dark (this study). Additionally, etr1;phyb double mutants germinated in the dark, whereas phyb single mutants did not. Similarly, after treatment with far-red light the phyA mutants did not germinate but the etr1;phyA double mutants did germinate. Thus, the effects of ETR1 on germination are occurring in the absence of actively signaling phy. These observations coupled with our observation that ETR1 affects the transcript levels of genes encoding for ABA and GA metabolic enzymes support a model where ETR1 is affecting germination by altering ABA and GA levels and ABA sensitivity in parallel with the effects of phy signaling on these parameters.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the technical assistance of Abigail Hall, Joonyup Kim, and Daniel Rose and thank Kenneth McFarland and Albrecht von Arnim for the use of equipment. We also thank Albrecht von Arnim for feedback on this manuscript. This research was supported by a NSF fellowship to Rebecca L. Wilson and a NSF grant (MCB-0918430) to Brad M. Binder.
REFERENCES
- Abeles F., Morgan P., Saltveit M. J. (1992). Ethylene in Plant Biology. San Diego, CA: Academic Press [Google Scholar]
- Abeles F. B., Lonski J. (1969). Stimulation of lettuce seed germination by ethylene. Plant Physiol. 44 277–280 10.1104/pp.44.2.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G., Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59 281–311 10.1146/annurev.arplant.59.032607.092859 [DOI] [PubMed] [Google Scholar]
- Bentsink L., Koorneef M. (2008). Seed dormancy and germination. Arabidopsis Book 6:e0119 10.1199/tab.0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder B. M., O’Malley R. C., Wang W., Moore J. M., Parks B. M., Spalding E. P., et al. (2004). Arabidopsis seedling growth response and recovery to ethylene. A kinetic analysis. Plant Physiol. 136 2913–2920 10.1104/pp.104.050369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder B. M., O’Malley R. C., Wang W., Zutz T. C., Bleecker A. B. (2006). Ethylene stimulates nutations that are dependent on the ETR1 receptor. Plant Physiol. 142 1690–1700 10.1104/pp.106.087858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker A. B. (1999). Ethylene perception and signaling: an evolutionary perspective. Trends Plant Sci. 4 269–274 10.1016/S1360-1385(99)01427-2 [DOI] [PubMed] [Google Scholar]
- Bleecker A. B., Estelle M. A., Somerville C., Kende H. (1988). Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241 1086–1089 10.1126/science.241.4869.1086 [DOI] [PubMed] [Google Scholar]
- Buer C. S., Wasteneys G. O., Masle J. (2003). Ethylene modulates root-wave responses in Arabidopsis. Plant Physiol. 132 1085–1096 10.1104/pp.102.019182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett A. N., Vidaver W. E. (1971). Synergistic action of ethylene with gibberellin or red light in germinating lettuce seeds. Plant Physiol. 48 656–657 10.1104/pp.48.5.656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancel J. D., Larsen P. B. (2002). Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol. 129 1557–1567 10.1104/pp.003780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J. J., Sánchez R. A. (1998). Phytochromes and seed germination. Seed Sci. Res. 8 317–329 10.1017/S0960258500004256 [DOI] [Google Scholar]
- Chang C., Kwok S. F., Bleecker A. B., Meyerowitz E. M. (1993). Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262 539–544 10.1126/science.8211181 [DOI] [PubMed] [Google Scholar]
- Chen R., Binder B. M., Garrett W. M., Tucker M. L., Cooper B., Chang C. (2011). Proteomic responses in Arabidopsis thaliana seedlings treated with ethylene. Mol. Biosyst. 7 2637–2650 10.1039/c1mb05159h [DOI] [PubMed] [Google Scholar]
- Chen T., Liu J., Lei G., Liu Y.-F., Li Z.-G., Tao J.-J., et al. (2009). Effects of tobacco ethylene receptor mutations on receptor kinase activity, plant growth and stress responses. Plant Cell Physiol. 50 1636–1650 10.1093/pcp/pcp107 [DOI] [PubMed] [Google Scholar]
- Chiwocha S., Cutler A., Abrams S. R., Ambrose S., Yang J., Ross A. R., et al. (2005). The etr1-2 mutation in Arabidopsis thaliana affects the abscisice acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 42 35–48 10.1111/j.1365-313X.2005.02359.x [DOI] [PubMed] [Google Scholar]
- Dekkers B. J. W., Willems L., Bassel G. W., Van Bolderen-Veldkamp R. P., Ligterink W., Hilhorst H. W. M., et al. (2011). Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 53 28–37 10.1093/pcp/pcr113 [DOI] [PubMed] [Google Scholar]
- Ellwell A. L., Gronwald D. S., Miller N. D., Spalding E. P., Brooks T. L. D. (2011). Separating parental environment from seed size effects on next generation growth and development in Arabidopsis. Plant Cell Environ. 34 291–301 10.1111/j.1365-3040.2010.02243.x [DOI] [PubMed] [Google Scholar]
- Esashi Y., Hase S., Kojima K. (1987). Light actions in the germination of cocklebur seeds. J. Exp. Bot. 189 702–710 10.1093/jxb/38.4.702 [DOI] [Google Scholar]
- Fankhauser C., Yeh K.-C., Lagarias C. J., Zhang H., Elich T. D., Chory J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284 1539–1541 10.1126/science.284.5419.1539 [DOI] [PubMed] [Google Scholar]
- Finch-Savage W. E., Cadman C. S. C., Toorop P. E., Lynn J. R., Hilhorst H. W. M. (2007). Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 51 60–78 10.1111/j.1365-313X.2007.03118.x [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59 387–415 10.1146/annurev.arplant.59.032607.092740 [DOI] [PubMed] [Google Scholar]
- Finlayson S. A., Lee I.-J., Morgan P. W. (1998). Phytochrome B and the regulation of circadian ethylene production in Sorghum. Plant Physiol. 116 17–25 10.1104/pp.116.1.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J. M., Smalle J., Gingerich D. J., Walker J. M., Yoo S. D., Yanagisawa S., et al. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. U.S.A. 101 6803–6808 10.1073/pnas.0401698101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble R. L., Coonfield M. L., Schaller G. E. (1998). Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95 7825–7829 10.1073/pnas.95.13.7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciarrubio A., Legaria J. P., Covarrubias A. A. (1997). Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 203 182–187 10.1007/s004250050180 [DOI] [PubMed] [Google Scholar]
- Guo H. W., Ecker J. R. (2003). Plant responses to ethylene gas are mediated by SCF (EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115 667–677 10.1016/S0092-8674(03)00969-3 [DOI] [PubMed] [Google Scholar]
- Hall A. E., Bleecker A. B. (2003). Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell 15 2032–2041 10.1105/tpc.013060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. P., Shakeel S. N., Amir M., Haq N. U., Qu X., Schaller G. E. (2012). Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol. 159 682–695 10.1104/pp.112.196790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C., Lohrmann J., Albrecht V., Sweere U., Hummel F., Yoo S., et al. (2004). The response regulator 2 mediates ethylene signalling and hormone integration in Arabidopsis. EMBO J. 23 3290–3302 10.1038/sj.emboj.7600337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Stoddart W. M., Dieterle M., Whitelam G. C., Schäfer E. (2002). Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol. 128 194–200 10.1104/pp.010559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel L. L., Grbic V., Baumgarten D. A., Bleecker A. B. (1993). Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5 553–564 10.1105/tpc.5.5.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst H. W. M., Karssen C. M. (1992). Seed dormancy and germination: the role of abscisic acid and gibberellins and the importance of hormone mutants. Plant Growth Regul. 11 225–238 10.1007/BF00024561 [DOI] [Google Scholar]
- Hirayama T., Fujishige N., Kunii T., Nishimura N., Iuchi S., Shinozaki K. (2004). A novel ethanol-hypersensitive mutant of Arabidopsis. Plant Cell Physiol. 45 703–711 10.1093/pcp/pch078 [DOI] [PubMed] [Google Scholar]
- Hua J., Chang C., Sun Q., Meyerowitz E. M. (1995). Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269 1712–1714 10.1126/science.7569898 [DOI] [PubMed] [Google Scholar]
- Hua J., Meyerowitz E. M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94 261–271 10.1016/S0092-8674(00)81425-7 [DOI] [PubMed] [Google Scholar]
- Hua J., Sakai H., Nourizadeh S., Chen Q. H. G., Bleecker A. B., Ecker J. R., et al. (1998). EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10 1321–1332 10.1105/tpc.10.8.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D., Chen H. C., Sheen J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129 500–515 10.1104/pp.005504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27 325–333 10.1046/j.1365-313x.2001.01096.x [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Chory J. (2010). Unraveling the paradoxes of plant hormone signaling integration. Nat. Struct. Mol. Biol. 17 642–645 10.1038/nsmb0610-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C., Yoon G. M., Shemansky J. M., Lin D., Yin I., Chang J., et al. (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109 19486–19491 10.1073/pnas.1214848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevany B. M., Tieman D. M., Taylor M. G., Cin V. D., Klee H. J. (2007). Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 51 458–467 10.1111/j.1365-313X.2007.03170.x [DOI] [PubMed] [Google Scholar]
- Kieber J. J., Rothenberg M., Roman G., Feldman K. A., Ecker J. R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72 427–441 10.1016/0092-8674(93)90119-B [DOI] [PubMed] [Google Scholar]
- Kim H., Helmbrecht E. E., Stalans M. B., Schmitt C., Patel N., Wen C.-K., et al. (2011). Ethylene receptor ETR1 domain requirements for ethylene responses in Arabidopsis seedlings. Plant Physiol. 156 417–429 10.1104/pp.110.170621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Wilson R. L., Case J. B., Binder B. (2012). A comparative study of ethylene growth response kinetics in eudicots and monocots reveals a role for gibberellin in growth inhibition and recovery. Plant Physiol. 160 1567–1580 10.1104/pp.112.205799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P. B., Chang C. (2001). The Arabidopsis eer1 mutant has enhanced ethylene responses in the hypocotyl and stem. Plant Physiol. 125 1061–1073 10.1104/pp.125.2.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. S., Deng X. W. (2010). Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13 571–577 10.1016/j.pbi.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., et al. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45 309–319 10.1111/j.1365-313X.2005.02622.x [DOI] [PubMed] [Google Scholar]
- Leivar P., Quail P. H. (2010). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16 19–28 10.1016/j.tplants.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A., Leubner-Metzger G. (2012). Beyond gibberellins and abscisic acid: how ethylene and jasmonates control seed germination. Plant Cell Rep. 31 253–270 10.1007/s00299-011-1180-1 [DOI] [PubMed] [Google Scholar]
- Linkies A., Müller K., Morris K., Turečková V., Wenk M., Cadman C. S. C., et al. (2009). Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21 3803–3822 10.1105/tpc.109.070201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Xu C., Wen C.-K. (2010). Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Biol. 10:60 10.1186/1471-2229-10-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mattoo A. K., Suttle J. C.(eds) (1991). The Plant Hormone Ethylene. Boca Raton: CRC Press, Inc [Google Scholar]
- Meng L., Feldman L. (2010). A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol. J. 5 183–186 10.1002/biot.200900211 [DOI] [PubMed] [Google Scholar]
- Mount S. M., Chang C. (2002). Evidence for a plastid origin of plant ethylene receptor genes. Plant Physiol. 130 10–14 10.1104/pp.005397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussatche P., Klee H. J. (2004). Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J. Biol. Chem. 279 48734–48741 10.1074/jbc.M403100200 [DOI] [PubMed] [Google Scholar]
- Neff M. M., Fankhauser C., Chory J. (2000). Light: an indicator of time and place. Genes Dev. 14 257–271 [PubMed] [Google Scholar]
- O’Malley R. C., Rodriguez F. I., Esch J. J., Binder B. M., O’Donnell P., Klee H. J., et al. (2005). Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J. 41 651–659 10.1111/j.1365-313X.2004.02331.x [DOI] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., et al. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21 403–419 10.1105/tpc.108.064691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., et al. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19 1192–1208 10.1105/tpc.107.050153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Kuwahara A., Seo M., Kushiro T., Asami T., Hirai N., et al. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141 97–107 10.1104/pp.106.079475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plett J. M., Cvetkovska M., Makenson P., Xing T., Regan S. (2009a). Arabidopsis ethylene receptors have different roles in Fumonisin B1-induced cell death. Physiol. Mol. Plant Pathol. 74 18–26 10.1016/j.pmpp.2009.08.004 [DOI] [Google Scholar]
- Plett J. M., Mathur J., Regan S. (2009b). Ethylene receptor ETR2 controls trichome branching by regulating microtubule assembly in Arabidopsis thaliana. J. Exp. Bot. 60 3923–3933 10.1093/jxb/erp228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe C., Schäfer E. (1997). Seed germination of Arabidopsis thaliana phyA/phyB double mutants is under phytochrome control. Plant Physiol. 114 1487–1492 10.1104/pp.114.4.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Chang K. N., Yazaki J., Ecker J. R. (2009). Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 23 512–521 10.1101/gad.1765709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Shen Z., Huang S.-S. C., Schmitz R. J., Urich M. A., Briggs S. P., et al. (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338 390–393 10.1126/science.1225974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Zeevaart J. A. D. (1999). The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. U.S.A. 96 15354–15361 10.1073/pnas.96.26.15354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Hall B., Gao Z., Schaller G. E. (2007). A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 7:3 10.1186/1471-2229-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Schaller G. E. (2004). Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol. 136 2961–2970 10.1104/pp.104.047126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P. H., Boylan M. T., Parks B. M., Short T. W., Xu Y., Wagner D. (1995). Phytochromes: photosensory perception and signal transduction. Science 268 675–680 10.1126/science.7732376 [DOI] [PubMed] [Google Scholar]
- Reed J. W., Nagatani A., Elich T. D., Fagan M., Chory J. (1994). Phytochrome A and Phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J. B., Botwright N. A., Smith J. J., O’Neill D. P., Kerckhoffs L. H. J. (2002). Control of gibberellin levels and gene expression during de-etiolation in pea. Plant Physiol. 128 734–741 10.1104/pp.010607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke D. M., Wickramarathna A. D., Ozga J. A., Kurepin L. V., Jin A. L., Good A. G., et al. (2013). Gibberellin 3-oxidase gene expression patterns influence gibberellin biosynthesis, growth, and development in pea. Plant Physiol. 163 929–945 10.1104/pp.113.225987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösler J., Klein I., Zeidler M. (2007). Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc. Natl. Acad. Sci. U.S.A. 104 10737–10742 10.1073/pnas.0703855104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Hua J., Chen Q. H. G., Chang C., Medrano L. J., Bleecker A. B., et al. (1998). ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95 5812–5817 10.1073/pnas.95.10.5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P. A., To J. P. C., Kieber J. J., Mcclung C. R. (2006). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18 55–69 10.1105/tpc.105.037994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharein B., Voet-Van-Vormizeele J., Harter K., Groth G. (2008). Ethylene signaling: identification of a putative ETR1-AHP1 phosphorelay complex by fluorescence spectroscopy. Anal. Biochem. 377 72–76 10.1016/j.ab.2008.03.015 [DOI] [PubMed] [Google Scholar]
- Seifert G. J., Barber C., Wells B., Roberts K. (2004). Growth regulators and the control of nucleotide sugar flux. Plant Cell 16 723–730 10.1105/tpc.019661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Aoki H., Koiwai H., Kamiya Y., Nambara E., Koshiba T. (2004). Comparative studies on the Arabidopsis Aldehyde Oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 45 1694–1703 10.1093/pcp/pch198 [DOI] [PubMed] [Google Scholar]
- Seo M., Hanada A., Kuwahara A., Endo A., Okamoto M., Yamauchi Y., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48 354–366 10.1111/j.1365-313X.2006.02881.x [DOI] [PubMed] [Google Scholar]
- Shinomura T., Nagatani A., Chory J., Furuya M. (1994). The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol. 104363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T., Nagatani A., Hanzawa H., Kubota M., Watanabe M., Furuya M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 93 8129–8133 10.1073/pnas.93.15.8129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K., Zhang H., Wang S., Chen M., Wu Y., Tang S., et al. (2013). ABI4 Regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9:e1003577 10.1371/journal.pgen.1003577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., Xu Y., Quail P. H. (1997). Antagonistic but complementary actions of phytochromes A and B allow optimum seedling de-etiolation. Plant Physiol. 114 637–641 10.1104/pp.114.2.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno K., Goda H., Ishii T., Ogura T., Tachikawa T., Sasaki E., et al. (2010). Auxin biosynthesis inhibitors, identified by a genomics-based approach, provide insights into auxin biosynthesis. Plant Cell Physiol. 51 524–536 10.1093/pcp/pcq032 [DOI] [PubMed] [Google Scholar]
- Sung D.-Y., Lee D., Harris H., Raab A., Feldmann J., Meharg A., et al. (2007). Identification of an arsenic tolerant double mutant with a thiol-mediated component and increased arsenic tolerance in phyA mutants. Plant J. 49 1064–1075 10.1111/j.1365-313X.2006.03018.x [DOI] [PubMed] [Google Scholar]
- Sweere U., Eichenberg K., Lohrmann J., Mira-Rodado V., Bäurle I., Kudla J., et al. (2001). Interaction of the response regulator ARR4 with Phytochrome B in modulating red light signaling. Science 294 1108–1111 10.1126/science.1065022 [DOI] [PubMed] [Google Scholar]
- Tepperman J. M., Hudson M. E., Khanna R., Zhu T., Chang S. H., Wang X., et al. (2004). Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38 725–739 10.1111/j.1365-313X.2004.02084.x [DOI] [PubMed] [Google Scholar]
- Tieman D. V., Taylor M. G., Ciardi J. A., Klee H. J. (2000). The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc. Natl. Acad. Sci. U.S.A. 97 5663–5668 10.1073/pnas.090550597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To J. P. C., Haberer G., Ferreira F. J., Deruère J., Mason M. G., Schaller G. E., et al. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 658–671 10.1105/tpc.018978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T., Miyata S., Yamaguchi-Shinozaki K., Shinozaki K. (2000). Possible his to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 478 227–232 10.1016/S0014-5793(00)01860-3 [DOI] [PubMed] [Google Scholar]
- Wang W., Hall A. E., O’Malley R., Bleecker A. B. (2003). Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc. Natl. Acad. Sci. U.S.A. 100 352–357 10.1073/pnas.0237085100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Zhang C., Ji Y., Zhao Q., He W., An F., et al. (2012). Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 22 1613–1616 10.1038/cr.2012.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. L., Kim H., Bakshi A., Binder B. M. (2014). The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol. 165 1353–1366 10.1104/pp.114.241695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuriyanghan H., Zhang B., Cao W.-H., Ma B., Lei G., Liu Y.-F., et al. (2009). The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 21 1473–1494 10.1105/tpc.108.065391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Liu Q., Wen C.-K. (2006). Receptor signal output mediated by the ETR1 N-terminus is primarily subfamily I receptor dependent. Plant Physiol. 142 492–508 10.1104/pp.106.082628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Ogawa M., Kuwahara A., Hanada A., Kamiya Y., Yamaguchi S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 367–378 10.1105/tpc.018143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Takeda-Kamiya N., Hanada A., Ogawa M., Kuwahara A., Seo M., et al. (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 48 555–561 10.1093/pcp/pcm023 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S., Yoo S. D., Sheen J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425 521–525 10.1038/nature01984 [DOI] [PubMed] [Google Scholar]
- Yeh K.-C., Lagarias J. C. (1998). Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. U.S.A. 95 13976–13981 10.1073/pnas.95.23.13976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wang L., Xi Y., Li J., et al. (2012). A molecular framework of light-controlled phytochrome action in Arabidopsis. Curr. Biol. 22 1530–1535 10.1016/j.cub.2012.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]