INTRODUCTION
During the past few decades, numerous studies described the microbiota of endodontic infections (Sundqvist, 1992, Brito et al., 2007, Siqueira & Roças, 2009, Tavares et al., 2011). In particular, the use of molecular biology techniques has expanded the breadth of knowledge of this topic. They have allowed a better characterization of the microbial composition of the root canal system and have confirmed that anaerobic species predominate in endodontic infections (Brito et al., 2007, Siqueira & Roças, 2009, Tavares et al., 2011).
Despite the advances in the field, little is known regarding the microbial composition of the root canal infections in HIV positive (HIV+) patients. The HIV/acquired immunodeficiency syndrome (AIDS) affects 33.4 million individuals worldwide and remains a global pandemic disease (UNAIDS, 2011). HIV infected individuals are considered a high-risk group to develop opportunistic infections (Ramirez-Amador et al., 2003) including lesions of the oral cavity (Coogan et al., 2005). In addition, the mouth may represent a microbial reservoir that can harbor medically important microorganisms (Scannapieco et al., 2003, Didilescu et al., 2005, de Souza Gonçalves et al., 2009).
Checkerboard DNA-DNA hybridization is a high-throughput molecular method that allows the identification and quantification of a wide range of bacterial species in multiple samples simultaneously. This technique has been employed in the study of the microbiota in saliva (Sachdeo et al., 2008), supragingival plaque (Haffajee et al., 2008, Teles et al., 2011), subgingival plaque (Teles et al., 2008, Uzel et al., 2011), on oral soft tissues (Mager et al., 2003), on dentures (Sachdeo et al., 2008), from dental implants (Gerber et al., 2006) and in samples from root canals (Brito et al., 2007, Tavares et al., 2011).
The level of detection of the checkerboard DNA-DNA hybridization technique is between 104 and 107 bacterial cells of a given species in each sample. Because the microbial content of samples from root canals may be below this level, a DNA amplification step has been used to enhance detection limits (Brito et al., 2007, Teles et al., 2007, Tavares et al., 2011). Multiple Displacement Amplification (MDA) enables whole genomic amplification of DNA targets with minimal bias (Dean et al., 2002). The template is replicated again and again by a “hyperbranching” mechanism of strand displacement synthesis (Lizardi et al., 1998), with the polymerase laying down a new copy as it displaces previously made copies. Samples as small as 1 ng can be amplified 1000–10,000 fold (Mai et al., 2004). This method allows the uniform amplification of the whole genomes present in a sample and has been effectively used as an aid in Checkerboard DNA-DNA Hybridization (Brito et al., 2007, Tavares et al., 2011).
The aim of the present study was to compare the microbial profile of endodontic infections in HIV− and HIV+ subjects and to seek intra-oral and medically important microbial taxa in those root canals.
MATERIALS AND METHODS
Subject population and sample collection
Forty HIV− subjects and twenty HIV+ subjects were enrolled in the study. HIV+ individuals were referred from the Reference Center of Infectious and Parasitary Diseases Orestes Diniz, Belo Horizonte, Minas Gerais, Brazil a center that specializes in HIV/Aids treatment (de Brito et al., 2009). HIV− subjects were recruited at the Department of Endodontics, Federal University of Minas Gerais (UFMG), Belo Horizonte, Brazil, where all study participants were examined and sample collection was performed.
To be included in the study, HIV− and HIV+ study participants had to present at least one tooth with endodontic infection and radiographically detected periradicular tissue alteration. Individuals who underwent antibiotic treatment within three months prior to the beginning of the endodontic therapy were not eligible to participate. This study was approved by the Ethics Committee of the Federal University of Minas Gerais (ETIC 122/08).
All selected teeth had clinical crowns that permitted effective rubber dam isolation. There was no history of trauma associated with the selected teeth, periodontal involvement or previous root canal treatment. Samples from multi-rooted teeth were taken from the largest root canal always associated with the periapical lesion.
After patients signed an informed consent, microbial samples from selected root canals were collected. The selection and preparation of the teeth was performed as previously described (Brito et al., 2007). In brief the 60 selected teeth were isolated using a rubber dam. Complete asepsis was employed, using the methodology proposed by Moller, 1966. Hydrogen peroxide (30%) was applied on the isolated crown, followed by 5% iodine that was inactivated by a 5% sodium thiosulfate solution. The samples were taken by scraping or filing the root canal walls with a #10 K-type hand file (Maillefer, Ballaigues, Switzerland). The file was introduced into the canal to a level approximately 1 mm short of the tooth apex. The tooth length was defined using an apex locator (Root ZX® II- J.Morita-USA). After removal from the canal, the final 4 mm of the file was removed using a sterile pair of surgical scissors and placed in a microcentrifuge tube containing 20 μl of alkaline lysis buffer (400 mM KOH, 100 mM dithiothreitol, 10 mM EDTA). After 10 min of incubation on ice, 20 μl of neutralization solution (400 mM HCl, 600 mM Tris-HCl, pH 0.6) was added. Samples were kept at 4°C until analysis.
Multiple displacement amplification (MDA) of root canal samples
Multiple displacement amplification was performed as previously described (Brito et al., 2007, Teles et al., 2007, Tavares et al., 2011). The Illustra™ GenomiPhi™ V2 DNA Amplification Kit (GE Healthcare, USA) was used for whole genomic amplification as described by the manufacturer. In brief, 1 μl of each of the DNA templates (i.e. endodontic samples) was added to 9 μl of sample buffer (50 mM Tris-HCl pH 8.2, 0.5 mM EDTA containing random hexamer primers) in 200 μl microcentrifuge tubes (Stratagene, La Jolla, CA, USA). Templates in sample buffer were heat denatured at 95°C for 3 min in a Perkin-Elmer Thermocycler and cooled to 4°C. One μl of phi 29 DNA polymerase mix including additional random hexamers was mixed on ice with 9 μl of reaction buffer containing dNTPs. The mixture was then added to the denatured sample to make a final volume of 20 μl and incubated at 30°C for 2 hours. Ten ng of Lambda DNA (contained in 1 μl) was used as a control. The amplification reaction was terminated by incubation of the samples at 65°C for 10 min. The amplified material was either immediately used, stored short-term at 4°C or at −20°C for longer storage.
The DNA content of the samples was measured prior to and after amplification using the Picogreen™ dsDNA quantification assay (Invitrogen, Carlsbad, CA, USA). Picogreen™ is a fluorescent nucleic acid stain that allows the quantification of as little as 25 pg/mL of double stranded DNA in samples. The microbiological content of the amplified samples was analyzed using checkerboard DNA-DNA hybridization.
Bacterial strains and growth conditions, DNA isolation, preparation of DNA probes and Checkerboard DNA-DNA hybridization
The 107 reference strains used for the preparation of DNA probes are listed in Table 1. The grown conditions of the selected bacterial strains have been described earlier (Socransky et al., 2004, Brito et al., 2007, Teles et al., 2007, Tavares et al., 2011).
Table 1.
Strains of microbial species used to prepare DNA probes and standards
| Bacterial strains (a) | |
|---|---|
| Acinetobacter baumannii (19606) | Legionella pneumophila (33152) |
| Actinomyces georgiae (49285) | Leptotrichia buccalis (14201) |
| Actinomyces gerencseriae (23860) | Mobiluncus mulieris (35243) |
| Actinomyces israelii (12102) | Mogibacterium timidum (33093) |
| Actinomyces meyeri (35568) | Neisseria gonorrhea (21823) |
| Actinomyces naeslundii (12104) | Neisseria meningitidis (13077) |
| Actinomyces odontolyticus (17929) | Neisseria mucosa (19696) |
| Actinomyces viscosus (43146) | Olsenella uli (49627) |
| Aggregatibacter actinomycetemcomitans (b) | Peptostreptococcus anaerobius (27337) |
| Atopobium parvulum (33793) | Parvimonas micra (33270) |
| Bacteroides fragilis (25285) | Porphyromonas endodontalis (35406) |
| Bacteroides ureolyticus (33387) | Porphyromonas gingivalis (33277) |
| Campylobacter gracilis (33236) | Prevotella denticola (35308) |
| Campylobacter rectus (33238) | Prevotella heparinolytica (35895) |
| Campylobacter showae (51146) | Prevotella intermedia (25611) |
| Capnocytophaga gingivalis (33624) | Prevotella loescheii (15930) |
| Capnocytophaga ochracea (33596) | Prevotella melaninogenica (25845) |
| Capnocytophaga sputigena (33612) | Prevotella nigrescens (33563) |
| Clostridium difficile (9689) | Prevotella oris (33573) |
| Corynebactehum diphtheriae (13812) | Prevotella tannerae (51259) |
| Corynebactehum matruchotii (14266) | Propionibacterium acnes (c) |
| Dialister pneumosintes (GBA27) | Propionibactehum propionicum (14157) |
| Eikenella corrodens (23834) | Rothia dentocariosa (17931) |
| Enterococcus faecalis (10100) | Salmonella enterica (27870) |
| Enterobacter aerogenes (13048) | Selenomonas artemidis (43528) |
| Enterobacter agglomerans (27155) | Selenomonas noxia (43541) |
| Enterobacter cloacae (10699) | Selenomonas sputigena (35185) |
| Enterobacter gergoviae (33028) | Serratia liquifasciens (11367) |
| Enterobacter sakazakii (12868) | Slackia exigua (700122) |
| Escherichia coli (10798) | Staphylococcus aureus (14458) |
| Eubacterium limosum (8486) | Staphylococcus epidermidis (14990) |
| Eubacterium nodatum (33099) | Staphylococcus warneri (27836) |
| Eubacterium saburreum (33271) | Stenotrophomonas maltophilia (13637) |
| Eubacterium saphenum (49989) | Streptococcus anginosus (33397) |
| Filifactor alocis (35896) | Streptococcus constellatus (27823) |
| Fusobacterium naviforme (25832) | Streptococcus gordonii (10558) |
| Fusobacterium necrophorum (25286) | Streptococcus intermedius (27335) |
| Fusobacterium nucleatum ss. nucleatum (25586) | Streptococcus mitis (49456) |
| Fusobacterium nucleatum ss. polymorphum (10953) | Streptococcus mutans (25175) |
| Fusobacterium nucleatum ss. vincentii (49256) | Streptococcus oralis (35037) |
| Fusobacterium periodonticum (33693) | Streptococcus parasanguinis (15912) |
| Gardnerella vaginalis (49145) | Streptococcus pneumoniae (49619) |
| Gemella hemolysans (10379) | Streptococcus salivarius (27945) |
| Gemella morbillorum (27824) | Streptococcus sanguinis (10556) |
| Granulicatella adiacens (49175) | Streptococcus sobrinus (33478) |
| Haemophilus aphrophilus (33389) | Streptococcus vestibularis (49124) |
| Haemophilus influenzae (33533) | Tannerella forsythia (43037) |
| Haemophilus paraphrophilus (29242) | Treponema denticola (B1) |
| Haemophilus segnis (33393) | Treponema socranskii (S1) |
| Hafnia alvei (13337) | Veillonella dispar (17748) |
| Helicobacter pylori (43504) | Veillonella parvula (10790) |
| Klebsiella oxytoca (12833) | Fungal strains (a) |
| Lactobacillus acidophilus (4356) | Candida albicans (10231) |
| Lactobacillus casei (393) | Candida tropicalis (750) |
All strains were obtained from the American Type Culture Collection (ATCC number in parentheses) except for Treponema denticola B1 and Treponema socranskii S1, which were obtained from The Forsyth Institute.
ATCC strains 43718 and 29523
ATCC strains 11827 and 11828
Preparation of probes and standards for quantification
Checkerboard DNA-DNA Hybridization was performed as previously described (Socransky et al., 2004). To prepare probes and standards, each species listed in Table 1 was grown on agar plates (except the two spirochetes, which were grown in broth) for 3–7 days. The cells were harvested and placed in 1.5 mL microcentrifuge tubes containing 1 mL of TE buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 7.6). Cells were washed twice by centrifugation in TE buffer at 1300×g for 10 min. The cells were resuspended and lysed with either 10% SDS and Proteinase K (20 mg/mL) for Gram-negative strains or in 150 μl of an enzyme mixture containing 15 mg/mL lysozyme (Sigma) and 5 mg/mL achromopeptidase (Sigma) in TE buffer (pH 8.0) for gram-positive strains. The pelleted cells were resuspended by 15 s of sonication and incubated at 37°C for 1 h. DNA was isolated and purified using the method of Smith et al. (1989). The concentration of the purified DNA was determined by spectrophotometric measurement of the absorbance at 260 nm. The purity of the preparations was assessed by the ratio of the absorbances at 260 and 280 nm. Whole genomic DNA probes were prepared from each of the 107 test strains by labeling 1 – 3 μg of DNA with digoxigenin (Boehringer Mannheim, Indianapolis, IN, USA) using a random primer technique (Feinberg & Vogelstein, 1983).
Sample preparation and microbial analysis
Following amplification and quantification, the amplified endodontic samples were boiled for 10 min. Approximately 1500 ng of DNA (5 μl) of the amplified sample were placed in a microcentrifuge tube containing 1 mL of TE buffer prior to boiling. The samples were placed into the extended slots of a Minislot 30 apparatus (Immunetics, Cambridge, MA, USA), concentrated onto a nylon membrane (Boehringer Mannheim) by vacuum and fixed onto the membrane by cross-linking using ultraviolet light (Stratalinker 1800, La Jolla, CA, USA) followed by baking at 120°C for 20 min. The Minislot device permitted the deposition of 28 different samples in individual lanes on a single membrane, as well as two control lanes containing the standards for quantification: 1 and 10 ng of DNA of each bacterial species tested, equivalent to 105 and 106 cells, respectively.
Checkerboard DNA-DNA hybridization was performed as previously described by Socransky et al., (2004). The membrane with fixed DNA was placed in a Miniblotter 45 apparatus (Immunetics) with the lanes of DNA at 90° to the channels of the device. A 30 × 45 “checkerboard” pattern was produced. Each channel was used as an individual hybridization chamber for separate DNA probes. Bound probes were detected by anti-digoxigenin antibody conjugated with alkaline phosphatase and a chemifluorescent substrate. Signal intensities of the endodontic samples and the standards (containing 105 and 106 cells of each species) on the same membrane were measured using a Storm FluorImager (Molecular Dynamics, Sunnyvale, CA, USA). Signals were converted to absolute counts by comparison with standards on the membrane (Socransky et al., 2004). Failure to detect a signal was recorded as zero.
Three membranes were run for each sample: one containing the “standard” 40 DNA probes routinely used to examine periodontal samples as well as a probe to detect Streptococcus mutans. A second membrane employed 42 probes to species thought to be implicated in endodontic infections. A third membrane was used to assess levels of medically important microbial taxa. Sensitivity and specificity tests were performed for all probes before performing the checkerboard DNA-DNA hybridization analysis, using a protocol similar to that described by Socransky et al, (2004).
Data analysis
The microbial data were expressed in two ways. The prevalence of each species, reflected by presence/absence data, indicated the proportion of samples in which the species were detected at >105 cells in amplified samples. Since the sample DNA was amplified, absolute numbers could not be determined. Thus, proportions of the total DNA probe count for each species comprised were computed for each sample and then averaged across subjects in each group separately.
Significance of differences between the prevalence of individual taxa in samples from HIV− and HIV+ subjects was sought using a Chi-square analysis. Significance of differences between the proportions of test species in samples from HIV− and HIV+ subjects was sought using the Mann Whitney test.
RESULTS
Patients characteristics
Among the 60 samples included in this study, 28 were from single rooted and 32 from multi-rooted teeth. In HIV− group 57.5% of samples were from females (n= 23) and the mean age of the subjects was 30.7 (±10.9) (range: 11– 64 years). In HIV+ group, 65% of samples were from females (n=13) and the mean age of the subjects was 42.3 (± 8.6) (range: 20–60 years). Additionally, 80.0% of the HIV+ individuals had CD4 + T-cell counts below 500 cell/mm3, while 70.0% had viral loads below 10,000 copies/mm3.
Quantification of DNA before and after MDA of endodontic samples
DNA from each root canal sample was amplified using MDA. The amount of DNA (± Standard Deviation - SD) present in the samples before the amplification averaged 6.1 (± 1.8) ng and 7.6 (± 1.2) μg after amplification, an approximately 1,000 - fold amplification.
Microbial species in root canal samples
The mean number of species (±standard error of the mean; SEM) detected in amplified root canal samples from HIV− and HIV+ subjects at a detection threshold of 104 cells was 53.3 (± 5.8) and 36.5 (± 6.2), respectively.
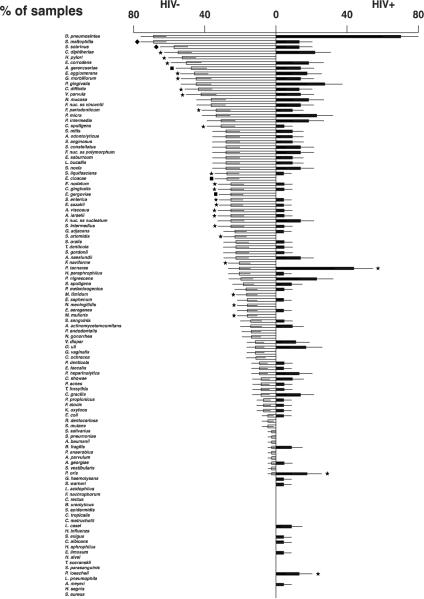
Figure 1 demonstrates the mean percentage of samples exhibiting counts of each of the 107 microbial species detected at the level of > 105 bacterial cells. The most prevalent taxa detected in the HIV− group were Dialister pneumosintes (68%), Stenotrophomonas maltophilia (68%), Streptococcus sobrinus (57%), Corynebacterium diphtheriae (55%) and Helicobacter pylori (52%). Among HIV+ individuals D. pneumosintes (70%), Prevotella tannerae (43%), Porphyromonas gingivalis (27%), Parvimonas micra (23%), Prevotella nigrescens (23%), and Corynebacterium diphtheriae (22%) were the most frequently detected taxa. Significant differences in prevalence between the two groups were observed. S. maltophilia, S. sobrinus, C. diphtheriae and H. pylori were more commonly detected in the HIV− group, whereas P. tannerae, Prevotella oris and Prevotella loescheii were found more frequently in samples from HIV+ subjects (p<0.05).
Figure 1.
Bilateral bar chart of the frequency of detection (prevalence ± SEM) of 107 microbial taxa in root canal samples taken from 40 HIV negative patients (white bars) and 20 HIV+ patients (black bars). A detection level threshold of 105 cells was employed. The significance of differences between groups was determined using the Chi-square analysis. The data are ordered in descending order of prevalence in HIV negative samples. ss, subsp. ◆ p< 0.001; ∎ p< 0.01; ★ p< 0.05.
The mean proportions of the 107 microbial species in amplified root canal samples from HIV− and HIV+ patients are presented in Fig. 2. Among HIV− subjects, D. pneumosintes, C. diphtheriae and Candida albicans were the most abundant taxa, representing 7.4% (± 1.0), 4.8 % (± 0.6) and 4.2% (± 0.8) of the total DNA probe counts (±SEM), respectively. Among HIV+ individuals, P. tannerae (11.1% ± 4.0) and D. pneumosintes (9.4% ± 1.6) were the bacterial species detected in highest mean proportions.
Figure 2.
Bilateral bar chart of the mean percentages of the DNA probe counts (± SEM) for 107 microbial species in MDA-amplified root canal samples taken from 40 HIV negative patients (white bars) and 20 HIV+ patients (black bars). The proportion of each species was averaged across subjects in the two clinical groups separately. The significance of differences between groups was determined using the Mann-Whitney test. The data are ordered in descending order of mean percentages of DNA probe counts detected in HIV negative samples. ss, subsp. ◆ p< 0.001; ∎ p< 0.01; ★ p< 0.05. Note: only taxa that comprised at least 1% of the total microbiota were eligible to be considered statistically significantly different.
There were significant differences in the proportions of the test taxa between the two groups. The mean proportions of P. tannerae, Olsenella uli, Veilonella dispar, Bacteroides fragilis and Actinomyces meyeri were higher in HIV+ positive than in HIV− patients (p<0.05). C. diphtheria, H. pylori, Clostridium difficile, Fusobacterium naviforme and Streptococcus mutans, were detected in higher mean proportions in HIV− than in HIV + patients (p<0.05).
DISCUSSION
Even though HIV/AIDS remains a global health priority (UNAIDS, 2011) and infected individuals are at high-risk for opportunistic infections, there is limited information regarding endodontic infections in HIV+ patients. There are a few reports on the effects of endodontic treatment (Quesnell et al., 2005, Shetty et al., 2006, Suchina et al., 2006, Alley et al., 2008), but virtually nothing is known regarding the root canal microbiota of HIV+ individuals. Thus, the goal of the current investigation was to compare the microbial profile of endodontic infections in HIV− and HIV+ subjects and to seek intra-oral and medically important microbial taxa in those root canals using MDA and checkerboard DNA-DNA hybridization techniques.
The presence and proportions of 107 different taxa were evaluated in samples recovered from root canal infections from HIV + and HIV − individuals. The mean number of species per sample was 53.3 (± 5.8) and 36.5 (± 6.2) in HIV− and HIV+ subjects, respectively. These values are in accord with previous findings from HIV− subjects, using a similar approach (Brito et al., 2007). Samples from HIV+ patients exhibited lower mean number of taxa than HIV− subjects. These results are in contrast with those from studies of the periodontal microbiota of HIV+ and HIV− patients (Tsang & Samaranayake, 2001, Patel et al., 2003). Those differences can be attributed to many factors, including the method of microbial identification, the number and types of samples collected, the microbial species analyzed, and the environmental pressures to which the microbiota under study was exposed (Socransky & Haffajee, 2005). Conversely, our results are similar to those reported by others (Tenenbaum et al., 1997, Gonçalves L de et al., 2007). Gonçalves et al (2007) investigated the periodontal microbiota of HIV − and HIV+ Brazilian patients under HAART and observed higher mean prevalence (and counts) of bacterial species in seronegative individuals. The authors suggest that HAART might have provided a protective effect and suppressed the local microbiota. Also in line with their results, we observed significantly higher prevalence of Eikenella corrodens, Gemella morbillorum, Veillonella parvula, Eubacterium nodatum, Capnocytophaga gingivalis and Actinomyces viscosus in HIV − individuals.
D. pneumosintes, P. tannerae and P. gingivalis were the most frequently detected taxa in HIV+ subjects. D. pneumosintes is frequently found in endodontic infections and seems to be an important pathogen (Siqueira & Roças, 2002, Brito et al., 2007, Tavares et al., 2011), particularly in persistent root canal infections (Siqueira et al., 2005). Periodontal pockets appear to be a reservoir for D. pneumosintes, as it has been is commonly detected in subgingival biofilms (Contreras et al., 2000, Shchipkova et al., 2010), particularly in refractory periodontitis (Colombo et al., 2009) and in HIV+ individuals (Aas et al., 2007).
Prevotella and Porphyromonas species are among the taxa that form black pigmented colonies. These include P. tannerae, P. denticola, Prevotella intermedia, Prevotella nigrescens, Prevotella loeschii, Porphyromonas endodontalis and P. gingivalis. Such taxa have been found in high frequency in endodontic infections (Xia et al., 2000, Vianna et al., 2005, Brito et al., 2007, Sassone et al., 2007, Tavares et al., 2011). In the present study, P. tannerae, P. oris and P. loescheii were significantly more prevalent in HIV+ individuals. Using the same methodology as the one employed in this study, a previous study from our group, found that P. tannerae and P. oris were the most prominent taxa in endodontic infections in HIV− subjects (Brito et al. (2007). P. gingivalis, a periodontal pathogen member of the “red complex” (Socransky et al., 1998), has been detected in root canal infections (Brito et al., 2007, Sassone et al., 2007, Tavares et al., 2011). In this study it was more frequently detected in HIV+ than in HIV− subjects, similarly to the results demonstrated by others in periodontal diseases (Chattin et al., 1999, Scully et al., 1999, Gonçalves L de et al., 2007). Taken together, these outcomes suggest potential role for those taxa in the pathogenesis of root canal infections.
An earlier investigation from our group focusing on HIV− population found a somewhat different microbial profile to the one described herein. Brito et al (2007) found that P. tannerae, Acinetobacter baumanni and Actinomyces meyerii were the most prevalent endodontic taxa, whereas in the present study D. pneumosintes, S. maltophilia and S. sobrinus were, overall, the most frequently detected bacterial species. There are a number of reasons for this apparent discrepancy. First, in the present manuscript, a more comprehensive probe panel was employed, as thirty additional probes were included. Therefore, certain taxa were not evaluated in the earlier paper, including S. maltophilia, S. sobrinus, C. diphtheria, and H. pylori which are some of the most prevalent microbial species in this study (Fig.1). Further, a few species that were evaluated in the 2007 study were not assessed in the present evaluation. In addition, in the present study the frequency of detection (prevalence) of each taxon was plotted in Figure 1 based on its presence at a level of at least 105 cells in a given sample, instead of the 104 criterium used in the previous paper. Finally, both studies are cross-sectional investigations of the endodontic microbiota and one of the limitations of cross-sectional studies is that they provide a snapshot view of a dynamic process.
When samples from HIV+ and HIV− subjects were compared regarding the mean proportions of the tested taxa, we observed that P. tannerae, Olsenella uli, Veillonella dispar, Bacteroides fragilis, and Actinomyces meyeri were significantly more abundant in HIV + than in HIV− individuals. P. tannerae (Xia et al., 2000, Brito et al., 2007, Tavares et al., 2011) and O. uli (Chavez de Paz et al., 2004, Roças & Siqueira, 2005) have been commonly found in endodontic infections. B. fragilis is more frequently isolated from intra-abdominal infections, and it is not typically associated with dentoalveolar infections. However, using cultivation techniques, Rams et al (1991)(Rams et al., 1991) could recover B. fragilis from subgingival plaque samples collected from 8 HIV-infected periodontitis patients. It is noteworthy that this species is resistant to virtually all classes of antimicrobials (Rasmussen et al., 1997).
C. albicans is a oral commensal yeast that, in the presence of predisposing conditions, can rise in levels and lead to disease states (Cannon & Chaffin, 1999). It is usually involved in opportunistic infections in patients AIDS (Back-Brito et al., 2009). In this study, the presence C. albicans and Candida tropicalis was assessed. C. albicans was the most predominant yeast in both groups, as demonstrated elsewhere (Miranda et al., 2009). Its low prevalence and proportion in HIV+ individuals in the present study is in contrast with studies that analyzed fungal species in oral lesions in HIV-infected patients (Baradkar & Kumar, 2009, Domaneschi et al., 2011), but it is in accord with other studies that evaluated the presence of fungal species in subgingival plaque samples from HIV-infected periodontitis subjects (Rams et al., 1991).
It has been shown that bacterial communities within the oral cavity, may be reservoirs of respiratory pathogens (Scannapieco & Rethman, 2003, Didilescu et al., 2005). Few researchers have examined endodontic infections as possible sources of those microorganisms (Chaudhry et al., 1997, Nandakumar et al., 2008). In this investigation, we sought the presence of important lung pathogens, including D. pneumosintes, Enterobacter aerogenes, Streptococcus pneumoniae and Legionella pneumophila in the root canal microbiota. Because HIV/AIDS patients are known to have an elevated risk of contracting serious respiratory diseases (Perello et al., 2010), we compared samples from HIV-negative as well as HIV-positive patients. D. pneumosintes was abundant in both groups (Fig.1 and 2), but all other pathogens were found in much lower levels and there no significant differences between groups regarding their presence or proportions.
H. pylori is primarily recovered from the stomach, is responsible for certain forms of gastritis or peptic ulcers. Because it has been found in endodontic infections and periodontal diseases, it has been postulated that the oral cavity might be a reservoir for H. pylori (de Souza Goncalves et al., 2009, Gao et al., 2011, Tavares et al., 2011) possibly leading to the reinfection of patients after treatment for those diseases (Zou & Li, 2011). Several studies have indicated that H. pylori infections were common in HIV-infected patients (Romanelli et al., 2007, de Souza Goncalves et al., 2009), although many others described a low prevalence of H. pylori infection in such individuals (Lv et al., 2007, Panos et al., 2007) as observed in this study.
The present study provides a comprehensive analysis of the root canal microbiota in HIV positive individuals. The lack of similar studies poses a challenge regarding the inferences that can be made on the present dataset. Thus, one can only cautiously elaborate on what the microbial described above represents. It is possible that the observed microbiota reflects the impact of the HIV infection on the root canal microbiota. Alternatively, the observed microbial community might also represent the effect of the HAART regimen, which may have kept the endodontic microbiota under control, despite the compromised immune state (Gonçalves L de et al., 2007). Finally, it is also possible that the observed microbial profile represents a microbial shift in the oral microbiota of HIV+ individuals. Because the oral environment is the source of endodontic bacteria, if it changes after HIV infection, subsequent root canal infections are likely to reflect that change. A similar event has been demonstrated by others in the salivary microbiota (Navazesh et al., 2005) and in periodontal biofilms (de Souza Gonçalves et al., 2009).
In conclusion, the present study showed that endodontic infections in HIV+ patients present a complex microbiota. There were significant differences in prevalence and mean proportions of microbial taxa between HIV− and HIV+ subjects. In addition, many oral and non-oral pathogens were detected in both groups. Collectively, these data suggest that treatment and prevention of endodontic infections might have a potential positive impact in clinical outcomes in HIV+ individuals.
ACKNOWLEDGMENTS
This work was supported in part by NIH/NIDCR grants R03 DE021742 (F.T.), U01DE021127 (R.T.), the Eleanor and Miles Shore Fellowship Program for Scholars in Medicine (The Forsyth Institute/Harvard Medical School) (F.T.). The authors wish to thank the postgraduate program at the School of Dentistry of UFMG. LCNB is a CAPES fellow; APRS and LQV are CNPq fellows.
REFERENCES
- Aas JA, Barbuto SM, Alpagot T, Olsen I, Dewhirst FE, Paster BJ. Subgingival plaque microbiota in HIV positive patients. J Clin Periodontol. 2007;34:189–95. doi: 10.1111/j.1600-051X.2006.01034.x. [DOI] [PubMed] [Google Scholar]
- Alley BS, Buchanan TH, Eleazer PD. Comparison of the success of root canal therapy in HIV/AIDS patients and non-infected controls. Gen Dent. 2008;56:155–7. [PubMed] [Google Scholar]
- Back-Brito GN, Mota AJ, Vasconcellos TC, Querido SM, Jorge AO, Reis AS, Balducci I, Koga-Ito CY. Frequency of Candida spp. in the oral cavity of Brazilian HIV-positive patients and correlation with CD4 cell counts and viral load. Mycopathologia. 2009;167:81–7. doi: 10.1007/s11046-008-9153-9. [DOI] [PubMed] [Google Scholar]
- Baradkar VP, Kumar S. Species identification of Candida isolates obtained from oral lesions of HIV infected patients. Indian J Dermatol. 2009;54:385–6. doi: 10.4103/0019-5154.57622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito LC, Teles FR, Teles RP, Franca EC, Ribeiro-Sobrinho AP, Haffajee AD, Socransky SS. Use of multiple-displacement amplification and checkerboard DNA-DNA hybridization to examine the microbiota of endodontic infections. J Clin Microbiol. 2007;45:3039–49. doi: 10.1128/JCM.02618-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon RD, Chaffin WL. Oral colonization by Candida albicans. Crit Rev Oral Biol Med. 1999;10:359–83. doi: 10.1177/10454411990100030701. [DOI] [PubMed] [Google Scholar]
- Chattin BR, Ishihara K, Okuda K, Hirai Y, Ishikawa T. Specific microbial colonizations in the periodontal sites of HIV-infected subjects. Microbiol Immunol. 1999;43:847–52. doi: 10.1111/j.1348-0421.1999.tb01219.x. [DOI] [PubMed] [Google Scholar]
- Chaudhry R, Kalra N, Talwar V, Thakur R. Anaerobic flora in endodontic infections. Indian J Med Res. 1997;105:262–5. [PubMed] [Google Scholar]
- Chavez de Paz LE, Molander A, Dahlen G. Gram-positive rods prevailing in teeth with apical periodontitis undergoing root canal treatment. Int Endod J. 2004;37:579–87. doi: 10.1111/j.1365-2591.2004.00845.x. [DOI] [PubMed] [Google Scholar]
- Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–32. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras A, Doan N, Chen C, Rusitanonta T, Flynn MJ, Slots J. Importance of Dialister pneumosintes in human periodontitis. Oral Microbiol Immunol. 2000;15:269–72. doi: 10.1034/j.1399-302x.2000.150410.x. [DOI] [PubMed] [Google Scholar]
- Coogan MM, Greenspan J, Challacombe SJ. Oral lesions in infection with human immunodeficiency virus. Bull World Health Organ. 2005;83:700–6. [PMC free article] [PubMed] [Google Scholar]
- de Brito LC, da Rosa MA, Lopes VS, e Ferreira EF, Vieira LQ, Sobrinho AP. Brazilian HIV-infected population: assessment of the needs of endodontic treatment in the post-highly active antiretroviral therapy era. Journal of endodontics. 2009;35:1178–81. doi: 10.1016/j.joen.2009.05.004. [DOI] [PubMed] [Google Scholar]
- de Souza Gonçalves L, Souto R, Colombo AP. Detection of Helicobacter pylori, Enterococcus faecalis, and Pseudomonas aeruginosa in the subgingival biofilm of HIV-infected subjects undergoing HAART with chronic periodontitis. Eur J Clin Microbiol Infect Dis. 2009;28:1335–42. doi: 10.1007/s10096-009-0786-5. [DOI] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci U S A. 2002;99:5261–6. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didilescu AC, Skaug N, Marica C, Didilescu C. Respiratory pathogens in dental plaque of hospitalized patients with chronic lung diseases. Clin Oral Investig. 2005;9:141–7. doi: 10.1007/s00784-005-0315-6. [DOI] [PubMed] [Google Scholar]
- Domaneschi C, Massarente DB, de Freitas RS, de Sousa Marques HH, Paula CR, Migliari DA, Antunes JL. Oral colonization by Candida species in AIDS pediatric patients. Oral Dis. 2011;17:393–8. doi: 10.1111/j.1601-0825.2010.01765.x. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gao J, Li Y, Wang Q, Qi C, Zhu S. Correlation between distribution of Helicobacter pylori in oral cavity and chronic stomach conditions. J Huazhong Univ Sci Technolog Med Sci. 2011;31:409–12. doi: 10.1007/s11596-011-0391-6. [DOI] [PubMed] [Google Scholar]
- Gerber J, Wenaweser D, Heitz-Mayfield L, Lang NP, Persson GR. Comparison of bacterial plaque samples from titanium implant and tooth surfaces by different methods. Clin Oral Implants Res. 2006;17:1–7. doi: 10.1111/j.1600-0501.2005.01197.x. [DOI] [PubMed] [Google Scholar]
- Gonçalves L de S, Soares Ferreira SM, Souza CO, Souto R, Colombo AP. Clinical and microbiological profiles of human immunodeficiency virus (HIV)-seropositive Brazilians undergoing highly active antiretroviral therapy and HIV-seronegative Brazilians with chronic periodontitis. J Periodontol. 2007;78:87–96. doi: 10.1902/jop.2007.060040. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS, Patel MR, Song X. Microbial complexes in supragingival plaque. Oral Microbiol Immunol. 2008;23:196–205. doi: 10.1111/j.1399-302X.2007.00411.x. [DOI] [PubMed] [Google Scholar]
- Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet. 1998;19:225–32. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Lv FJ, Luo XL, Meng X, Jin R, Ding HG, Zhang ST. A low prevalence of H. pylori and endoscopic findings in HIV-positive Chinese patients with gastrointestinal symptoms. World J Gastroenterol. 2007;13:5492–6. doi: 10.3748/wjg.v13.i41.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–54. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- Mai M, Hoyer JD, McClure RF. Use of multiple displacement amplification to amplify genomic DNA before sequencing of the alpha and beta haemoglobin genes. J Clin Pathol. 2004;57:637–40. doi: 10.1136/jcp.2003.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TT, Vianna CR, Rodrigues L, Monteiro AS, Rosa CA, Correa A., Jr. Diversity and frequency of yeasts from the dorsum of the tongue and necrotic root canals associated with primary apical periodontitis. Int Endod J. 2009;42:839–44. doi: 10.1111/j.1365-2591.2009.01601.x. [DOI] [PubMed] [Google Scholar]
- Nandakumar R, Whiting J, Fouad AF. Identification of selected respiratory pathogens in endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2008;106:145–51. doi: 10.1016/j.tripleo.2008.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navazesh M, Mulligan R, Pogoda J, Greenspan D, Alves M, Phelan J, Greenspan J, Slots J. The effect of HAART on salivary microbiota in the Women's Interagency HIV Study (WIHS) Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2005;100:701–8. doi: 10.1016/j.tripleo.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Panos GZ, Xirouchakis E, Tzias V, Charatsis G, Bliziotis IA, Doulgeroglou V, Margetis N, Falagas ME. Helicobacter pylori infection in symptomatic HIV-seropositive and -seronegative patients: a case-control study. AIDS Res Hum Retroviruses. 2007;23:709–12. doi: 10.1089/aid.2006.0174. [DOI] [PubMed] [Google Scholar]
- Patel M, Coogan M, Galpin JS. Periodontal pathogens in subgingival plaque of HIV-positive subjects with chronic periodontitis. Oral Microbiol Immunol. 2003;18:199–201. doi: 10.1034/j.1399-302x.2003.00064.x. [DOI] [PubMed] [Google Scholar]
- Perello R, Miro O, Marcos MA, Almela M, Bragulat E, Sanchez M, Agusti C, Miro JM, Moreno A. Predicting bacteremic pneumonia in HIV-1-infected patients consulting the ED. Am J Emerg Med. 2010;28:454–9. doi: 10.1016/j.ajem.2009.01.024. [DOI] [PubMed] [Google Scholar]
- Quesnell BT, Alves M, Hawkinson RW, Jr., Johnson BR, Wenckus CS, BeGole EA. The effect of human immunodeficiency virus on endodontic treatment outcome. J Endod. 2005;31:633–6. doi: 10.1097/01.don.0000157985.88883.81. [DOI] [PubMed] [Google Scholar]
- Ramirez-Amador V, Esquivel-Pedraza L, Sierra-Madero J, Anaya-Saavedra G, Gonzalez-Ramirez I, Ponce-de-Leon S. The changing clinical spectrum of human immunodeficiency virus (HIV)-related oral lesions in 1,000 consecutive patients: A 12-year study in a referral center in Mexico. Medicine (Baltimore) 2003;82:39–50. doi: 10.1097/00005792-200301000-00004. [DOI] [PubMed] [Google Scholar]
- Rams TE, Andriolo M, Jr., Feik D, Abel SN, McGivern TM, Slots J. Microbiological study of HIV-related periodontitis. J Periodontol. 1991;62:74–81. doi: 10.1902/jop.1991.62.1.74. [DOI] [PubMed] [Google Scholar]
- Rasmussen BA, Bush K, Tally FP. Antimicrobial resistance in anaerobes. Clin Infect Dis. 1997;24(Suppl 1):S110–20. doi: 10.1093/clinids/24.supplement_1.s110. [DOI] [PubMed] [Google Scholar]
- Roças IN, Siqueira JF., Jr. Species-directed 16S rRNA gene nested PCR detection of Olsenella species in association with endodontic diseases. Lett Appl Microbiol. 2005;41:12–6. doi: 10.1111/j.1472-765X.2005.01723.x. [DOI] [PubMed] [Google Scholar]
- Romanelli F, Smith KM, Murphy BS. Does HIV infection alter the incidence or pathology of Helicobacter pylori infection? AIDS Patient Care STDS. 2007;21:908–19. doi: 10.1089/apc.2006.0215. [DOI] [PubMed] [Google Scholar]
- Sachdeo A, Haffajee AD, Socransky SS. Biofilms in the edentulous oral cavity. J Prosthodont. 2008;17:348–56. doi: 10.1111/j.1532-849X.2008.00301.x. [DOI] [PubMed] [Google Scholar]
- Sassone L, Fidel R, Figueiredo L, Fidel S, Faveri M, Feres M. Evaluation of the microbiota of primary endodontic infections using checkerboard DNA-DNA hybridization. Oral Microbiol Immunol. 2007;22:390–7. doi: 10.1111/j.1399-302X.2007.00376.x. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA, Bush RB, Paju S. Associations between periodontal disease and risk for nosocomial bacterial pneumonia and chronic obstructive pulmonary disease. A systematic review. Ann Periodontol. 2003;8:54–69. doi: 10.1902/annals.2003.8.1.54. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA, Rethman MP. The relationship between periodontal diseases and respiratory diseases. Dent Today. 2003;22:79–83. [PubMed] [Google Scholar]
- Scully C, Porter SR, Mutlu S, Epstein JB, Glover S, Kumar N. Periodontopathic bacteria in English HIV-seropositive persons. AIDS Patient Care STDS. 1999;13:369–74. doi: 10.1089/apc.1999.13.369. [DOI] [PubMed] [Google Scholar]
- Shchipkova AY, Nagaraja HN, Kumar PS. Subgingival microbial profiles of smokers with periodontitis. J Dent Res. 2010;89:1247–53. doi: 10.1177/0022034510377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty K, Garcia J, Leigh J. Success of root canal therapy in HIV-positive patients. Gen Dent. 2006;54:397–402. [PubMed] [Google Scholar]
- Siqueira JF, Jr., Roças IN. Dialister pneumosintes can be a suspected endodontic pathogen. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2002;94:494–8. doi: 10.1067/moe.2002.125202. [DOI] [PubMed] [Google Scholar]
- Siqueira JF, Jr., Roças IN. Diversity of endodontic microbiota revisited. J Dent Res. 2009;88:969–81. doi: 10.1177/0022034509346549. [DOI] [PubMed] [Google Scholar]
- Siqueira JF, Jr., Roças IN, Cunha CD, Rosado AS. Novel bacterial phylotypes in endodontic infections. J Dent Res. 2005;84:565–9. doi: 10.1177/154405910508400615. [DOI] [PubMed] [Google Scholar]
- Smith GL, Socransky SS, Smith CM. Rapid method for the purification of DNA from subgingival microorganisms. Oral Microbiol Immunol. 1989;4:47–51. doi: 10.1111/j.1399-302x.1989.tb00406.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–87. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Smith C, Martin L, Haffajee JA, Uzel NG, Goodson JM. Use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiol Immunol. 2004;19:352–62. doi: 10.1111/j.1399-302x.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- Suchina JA, Levine D, Flaitz CM, Nichols CM, Hicks MJ. Retrospective clinical and radiologic evaluation of nonsurgical endodontic treatment in human immunodeficiency virus (HIV) infection. J Contemp Dent Pract. 2006;7:1–8. [PubMed] [Google Scholar]
- Sundqvist G. Associations between microbial species in dental root canal infections. Oral Microbiol Immunol. 1992;7:257–62. doi: 10.1111/j.1399-302x.1992.tb00584.x. [DOI] [PubMed] [Google Scholar]
- Tavares WL, Neves de Brito LC, Teles RP, Massara ML, Ribeiro Sobrinho AP, Haffajee AD, Socransky SS, Teles FR. Microbiota of deciduous endodontic infections analysed by MDA and Checkerboard DNA-DNA hybridization. Int Endod J. 2011;44:225–35. doi: 10.1111/j.1365-2591.2010.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles F, Haffajee AD, Socransky SS. Multiple displacement amplification as an aid in checkerboard DNA-DNA hybridization. Oral Microbiol Immunol. 2007;22:118–25. doi: 10.1111/j.1399-302X.2007.00333.x. [DOI] [PubMed] [Google Scholar]
- Teles FR, Teles RP, Uzel NG, Song XQ, Torresyap G, Socransky SS, Haffajee AD. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. J Periodontal Res. 2011 doi: 10.1111/j.1600-0765.2011.01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles RP, Patel M, Socransky SS, Haffajee AD. Disease progression in periodontally healthy and maintenance subjects. J Periodontol. 2008;79:784–94. doi: 10.1902/jop.2008.070485. [DOI] [PubMed] [Google Scholar]
- Tenenbaum H, Elkaim R, Cuisinier F, Dahan M, Zamanian P, Lang JM. Prevalence of six periodontal pathogens detected by DNA probe method in HIV vs non-HIV periodontitis. Oral Dis. 1997;3(Suppl 1):S153–5. doi: 10.1111/j.1601-0825.1997.tb00350.x. [DOI] [PubMed] [Google Scholar]
- Tsang CS, Samaranayake LP. Predominant cultivable subgingival microbiota of healthy and HIV-infected ethnic Chinese. Apmis. 2001;109:117–26. doi: 10.1034/j.1600-0463.2001.d01-113.x. [DOI] [PubMed] [Google Scholar]
- UNAIDS . Report on the global AIDS epidemic. Geneva, Switzerland: 2011. [Google Scholar]
- Uzel NG, Teles FR, Teles RP, Song XQ, Torresyap G, Socransky SS, Haffajee AD. Microbial shifts during dental biofilm re-development in the absence of oral hygiene in periodontal health and disease. J Clin Periodontol. 2011;38:612–20. doi: 10.1111/j.1600-051X.2011.01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianna ME, Horz HP, Gomes BP, Conrads G. Microarrays complement culture methods for identification of bacteria in endodontic infections. Oral Microbiol Immunol. 2005;20:253–8. doi: 10.1111/j.1399-302X.2005.00221.x. [DOI] [PubMed] [Google Scholar]
- Xia T, Baumgartner JC, David LL. Isolation and identification of Prevotella tannerae from endodontic infections. Oral Microbiol Immunol. 2000;15:273–5. doi: 10.1034/j.1399-302x.2000.150411.x. [DOI] [PubMed] [Google Scholar]
- Zou QH, Li RQ. Helicobacter pylori in the oral cavity and gastric mucosa: a meta-analysis. J Oral Pathol Med. 2011;40:317–24. doi: 10.1111/j.1600-0714.2011.01006.x. [DOI] [PubMed] [Google Scholar]