Abstract
Improved technology for reconstructing cryo-electron microscopy (cryo-EM) images has now made it possible to determine secondary structural features of membrane proteins in enveloped viruses. The structure of mature dengue virus particles was determined to a resolution of 9.5 Å by cryo-EM and image reconstruction techniques, establishing the secondary structural disposition of the 180 envelope (E) and 180 membrane (M) proteins in the lipid envelope. The α-helical ‘stem’ regions of the E molecules, as well as part of the N-terminal section of the M proteins, are buried in the outer leaflet of the viral membrane. The ‘anchor’ regions of E and the M proteins each form antiparallel E-E and M-M transmembrane α-helices, leaving their C termini on the exterior of the viral membrane, consistent with the predicted topology of the unprocessed polyprotein. This is one of only a few determinations of the disposition of transmembrane proteins in situ and shows that the nucleocapsid core and envelope proteins do not have a direct interaction in the mature virus.
Knowledge of protein structures within cellular membranes is limited to only a few examples of membrane proteins whose structures have been determined in situ1–3. Although X-ray crystallography has succeeded in the analysis of the PRD1 membrane-containing bacteriophage4, in general it is difficult to crystallize integral membrane proteins or enveloped viruses. Improved technology for reconstructing cryo-EM images has now made it possible to determine secondary structural features of membrane proteins in enveloped viruses. We report here the structure and disposition of the membrane proteins in mature dengue virus.
Dengue viruses belong to the Flavivirus genus of the family Flaviviridae. The flaviviruses are insect-transmitted, icosahedral, enveloped RNA viruses that infect vertebrates and frequently cause serious, sometimes fatal, infections in humans5,6. Other viruses belonging to the same genus are West Nile, yellow fever, Japanese encephalitis and tick-borne encephalitis virus (TBEV). During the last half of the twentieth century, instances of dengue hemorrhagic fever, which usually results from the sequential infection by more than one of the four dengue virus serotypes, have spread from southeast Asia to most tropical and semitropical regions on Earth. As there is no effective dengue virus vaccine or antiviral agent, the spread of dengue virus infection has become a major health concern and a subject of special interest to the World Health Organization. Similarly, the spread of the closely related West Nile virus into North America has become a prominent public health issue in the United States7.
The positive-sense, 10.2-kb RNA genome of dengue virus has a single, long open reading frame that is translated into a polyprotein5,8. Signal sequences direct the translocation of the polyprotein several times across the endoplasmic reticulum membrane to be subsequently cleaved by cellular and virally encoded proteinases. Located at the N terminus of the polyprotein is the capsid protein (C), followed by the premembrane (prM) and envelope (E) glycoproteins. The prM protein is cleaved by the cellular protease furin9, releasing the N-terminal 91 amino acids and leaving 180 copies of the 75-residue M protein along with 180 E proteins anchored in the viral membrane5.
The structure of the 500–Å diameter dengue virus (Fig. 1) has been studied by cryo-EM10. The previous low-resolution cryo-EM map, in conjunction with an X-ray crystallographic structure of the trypsin-cleaved, homologous, TBEV E glycoprotein11,12, had shown that the mature virus has 90 E dimers arranged in a herringbone pattern10. In contrast to the smooth surface of the mature virus, immature virions, in which the prM protein has not yet been cleaved, have a rough surface characterized by 60 spikes that reach to an external diameter of 600 Å (ref. 13). Neither the herringbone organization of the mature virus nor the pattern of spikes in the immature particles obey the T = 3 quasi-symmetry that would be predicted for icosahedral particles with 180 identical protein subunits14.
Figure 1.
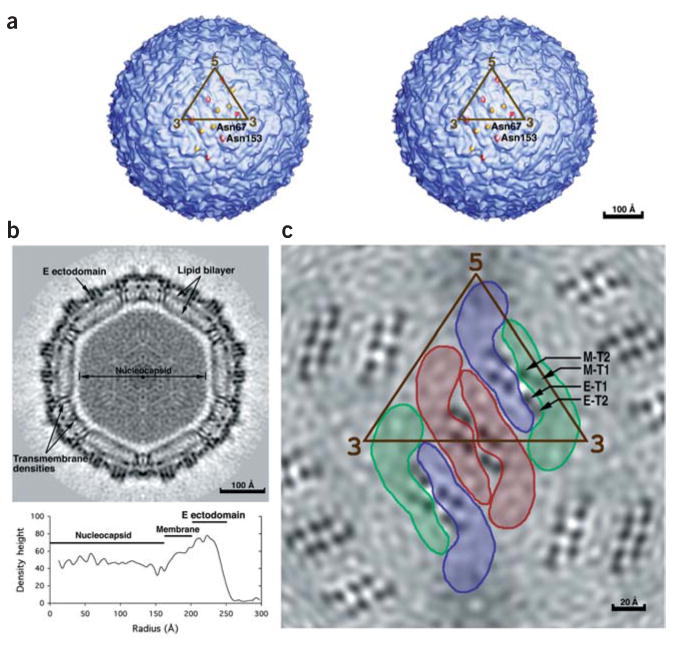
The dengue virus structure. (a) Stereo view of the viral surface at a resolution of 12.0 Å. The brown triangle demarcates the limits of one icosahedral asymmetric unit as defined by the five- and three-fold axes. Note the two protrusions per monomer corresponding to the glycosylation sites at Asn67 (yellow) and Asn153 (red). (b) A central cross section looking down an icosahedral three-fold axis, showing the polygonal shape of the membrane. The darkness of the shading is proportional to the magnitude of the cryo-EM density. Viral components are labeled. Maximum density heights are plotted below on a relative scale as a function of radius. (c) A radial cryo-EM density section at a radius of 185 Å, corresponding to the center of the lipid membrane, highlighting the herringbone arrangement of the three E dimers. The density is indicated in gray scale, with the highest density being the blackest. Shown also in brown is the limit of one icosahedral asymmetric unit. The boundaries of the E glycoprotein dimers are also indicated. The E dimer on the icosahedral twofold axis is red, whereas the monomers of the general-position dimer are blue and green. The transmembrane helices are viewed in cross section and marked for the green monomer according to the nomenclature of Figure 2.
The 395-residue TBEV E fragment11 crystallized as a dimer12, consistent with the expectation that maturation results in the formation of homodimers15. Each monomer consists of three domains: the structurally central, N-terminal domain I, followed by the dimerization domain II and finally the C-terminal, Ig-like domain III. Domain II contains the hydrophobic ‘fusion’ peptide (residues 98–110) essential for virus-cell fusion16,17. Domain III has been proposed to function as the binding site for cellular receptors and has been recognized as the receptor attachment site in competition experiments with monoclonal antibodies12,18–20. The 101-residue C terminus of the TBEV E glycoprotein, which was not part of the X-ray structure, is called the ‘stem-anchor’ region. The stem is composed of residues 396 to about 449, and the hydrophobic transmembrane anchor region is composed of residues 450–496. The stem and transmembrane anchor regions of flaviviruses have been predicted to each consist of two α-helices15,21 (Fig. 2).
Figure 2.
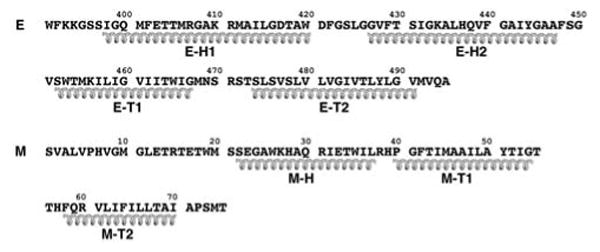
Secondary structural predictions based on the primary sequences of the E and M stem-anchor regions. Helical coils represent the E stem (E-H1, E-H2) and transmembrane anchor (E-T1, E-T2) and the M stem (M-H) and transmembrane anchor (M-T1, M-T2) regions.
A 9.5-Å resolution cryo-EM structure of mature dengue virus now shows that the two α-helices in the stem region of E are buried in the outer leaflet of the viral membrane, and the anchor region of E is arranged in an antiparallel, coiled coil transmembrane structure. Similarly, an α-helix near the N terminus of M is partially buried in the outer lipid bilayer, and the C-terminal region of M consists of a pair of antiparallel helices traversing the membrane. In contrast to alphaviruses, both the E and M proteins have their C termini in the outer leaflet of the lipid bilayer, suggesting that the deposition of the polyprotein after post-translational cleavage may be independent of the formation of the nucleocapsid and influence both budding and fusion.
Results
The E ectodomain
The amino acid sequence of the dengue virus E glycoprotein is 37% identical to that of TBEV E glycoprotein. An alignment of flavivirus E proteins and the known crystal structure of the TBEV E dimer12 (PDB entry 1SVB) were used to build a homology model of the dengue virus E glycoprotein. The density outside the viral membrane was fit with the dengue dimer homology model using the program EMfit22 (Table 1). A difference map, in which the density corresponding to the fitted E dimers was set to zero, showed density peaks close to Asn67 (with heights of 4.4, 5.3 and 4.2 standard deviations for the red, green and blue outlined molecules in Fig. 1c, respectively) and Asn153 (with heights of 5.7, 5.8 and 5.8 standard deviations for the red, green and blue outlined molecules in Fig. 1c, respectively) for each of the three E glycoproteins in an icosahedral asymmetric unit. These two peaks were within 10 Å of the potentially glycosylated asparagine residues in Asn-X-Ser/Thr sequence motifs and, thus, were assigned to represent carbohydrate modifications. Previous studies presented conflicting data as to whether there were one or two carbohydrate sites23,24.
Table 1. Results obtained by fitting crystallographic structural components into the cryo-EM map.
| Molecule 1 | Molecule 2 | Molecule 3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Model | Sumfa | Clasha | -Dena | D1 | D2 | D3 | D1 | D2 | D3 | D1 | D2 | D3 |
| DEN E ectob (All)c | 38.3 | 3.6 | 3.0 | 36.2 | 39.4 | 36.0 | 38.9 | 40.3 | 37.3 | 38.5 | 39.3 | 35.8 |
| TBEV E ectob (All)c | 37.2 | 4.4 | 3.8 | 35.8 | 38.2 | 34.9 | 38.2 | 39.1 | 35.3 | 37.9 | 38.2 | 33.5 |
| DEN E stem-trb (Cα)c | 38.8 | 0.0 | 0.0 | 45.9 | 42.3 | 44.6 | 41.6 | 38.4 | 34.8 | 42.0 | 36.1 | 41.5 |
| DEN Mb (Cα)c | 36.5 | 0.0 | 0.0 | 44.2 | 37.6 | 44.6 | 31.6 | 39.1 | 32.0 | |||
Molecules 1, 2 and 3 are the three molecules in one icosahedral asymmetric unit corresponding to the red, green and blue outlines in Figure 1c. D1, D2 and D3 in rows 1 and 2 refer to the three domains of the E protein's ectodomain; D1, D2 and D3 in row 3 refer to E-H1 and E-H2 in stem and E-T1 plus E-T2 in the transmembrane region (tr); D1 and D3 in row 4 refer to M-H and M-T1 plus M-T2.
Sumf, mean density height averaged over all atoms where the maximum density is scaled to have a value of 100; clash, percentage of atoms in one molecule that approach another symmetry-related molecule closer than 6 Å (if Cα atoms are used for fitting) or 3.2 Å (if all atoms are used for fitting); -den, percentage of atoms in negative density.
ecto, ectodomain of the E homology model (residues 1–394) or TBEV E (residues 1–395) proteins; stem-tr, model of the dengue E (DEN E) stem-anchor region (residues 400–495); DEN M, stem-anchor region of the dengue M protein (residues 20–75).
All indicates that main chain and side chain atoms were used for fitting; Cα indicates that only Cα atoms were used.
The lipid bilayer
The characteristic lipid bilayer is readily recognizable between 165-Å and 205-Å radii, consistent with the lipid radii of the immature virus13, but is a revision of the previously reported membrane location10. However, the envelope is markedly polygonal (Fig. 1b) rather than spherical, with constrictions where proteins cross the membrane forming the vertices of the polygon. A similar, but less pronounced, deviation from a spherical membrane envelope was associated with the transmembrane regions in the structure of immature dengue virus particles13, suggesting that the protein assembly influences the overall shape of the membrane.
The center of the transmembrane region, corresponding to the lower density of the aliphatic chains, is traversed by six pairs of higher density regions per icosahedral asymmetric unit (Fig. 1c). Each pair represents two α-helices joined by density in the internal lipid leaflet. A variety of different secondary structural predictions (Fig. 2) of the E and M transmembrane regions show two α-helical regions linked by four to six amino acids rich in polar residues15. The polar linker residues are unlikely to be buried within the central hydrophobic region of the lipid bilayer, but are presumably embedded within the inner lipid leaflet. The linking residues between the transmembrane helices do not seem to penetrate into the space around the nucleocapsid core, but remain associated with the inner phospholipid polar head groups. Thus, it is apparent that the transmembrane structures of these proteins are antiparallel helices, implying that each polypeptide traverses the membrane from the external to the internal and back again to the external lipid leaflet as anticipated based on sequence analyses21.
Each of the three monomers in an icosahedral asymmetric unit is associated with two pairs of transmembrane helices, corresponding to four pairs per dimer. The four pairs of transmembrane helices associated with the E dimer on the icosahedral two-fold axis can be transformed onto the four associated with the E dimer in the general position (Fig. 1c) by the same operations as pertain to the final fit of the E glycoproteins. This shows that the same organization of transmembrane densities is associated with each type of dimer (Fig. 1c). Presumably, one pair of E antiparallel transmembrane helices belongs to each E dimer and one pair of M antiparallel transmembrane helices belongs to the two M proteins associated with each E dimer. The lengths of the second transmembrane helices (E-T2 and M-T2) are slightly different and can be matched with the lengths of the E (~18 residues) and M (~14 residues) transmembrane α-helix predictions. This assignment was subsequently verified by observing the connectivity of the E ectodomain with the E-H1 and E-H2 stem helices and with the E-T1 and E-T2 transmembrane helices. The E transmembrane helices form an antiparallel coiled coil and could be fit with the dimeric, antiparallel coiled coil found in colicin E3 (see Methods), whereas each of the M transmembrane helices had to be fit independently (Table 1, Fig. 3). Similar sets of E and M transmembrane helices were found in immature dengue virus particles, although the relationship of the E-M heterodimer to the E and M transmembrane regions is different13.
Figure 3.
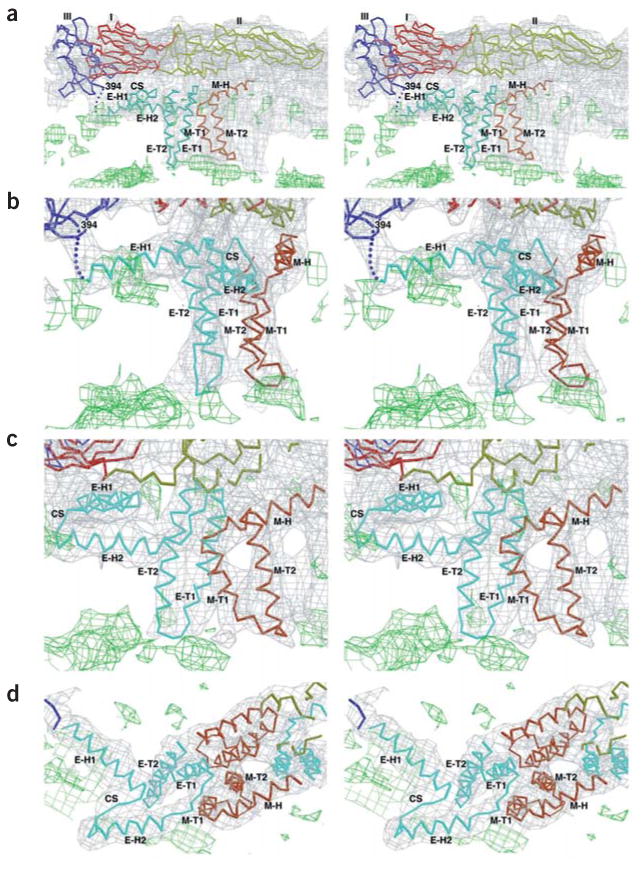
Stereoscopic diagrams showing the fit of the Cα backbones for the E and M regions into the cryo-EM density (gray) of the outer lipid (green) leaflet associated with the E dimer on the icosahedral two-fold axis. E ectodomains I, II and III are red, yellow and blue, respectively; stem-anchor region of E, cyan; M protein, orange; cryoEM density of the lipid bilayers, green. The stem and transmembrane helices are labeled with the nomenclature shown in Figure 2. Contour levels are chosen arbitrarily. The contour level for the lipid (green) is lower than that for the protein (gray). (a) Side view showing E and M monomers. (b) Enlarged view of a with a +50° rotation about the vertical axis to more clearly show the fit of E-H1 into the density. (c) Enlarged view of a with a –20° rotation about the vertical axis to more clearly show the fit of E-H2 and M-H into the density. (d) Top view of helices E-H1, E-H2 and M-H.
Secondary structural predictions of the 56-residue stem region of E (residues 395–450) (Fig. 2) suggest that there are two consecutive mostly amphipathic helices (E-H1 and E-H2) that would join the C-terminal residue 394 of the fitted E tryptic fragment to the transmembrane region15,21,25. The fitted E tryptic fragment and the N terminus of the E transmembrane region are joined consecutively by two cylindrical densities in the outer lipid bilayer. These densities could be readily fit (Table 1) with model amphipathic α-helices, tentatively placing the hydrophobic sides of the helices facing the hydrophobic center of the membrane (Fig. 3).
With the identification of the secondary structural elements belonging to E, it was then possible to assign the remaining larger uninterpreted density features to the M protein. Secondary structural predictions of the M protein show a weakly amphipathic α-helix (M-H) followed by the two transmembrane α-helices (M-T1, M-T2)25. The cryo-EM density shows that the M-H helix is partially buried at its C terminus in the outer lipid leaflet and makes an angle of ~20° with the membrane surface (Fig. 3a). The N terminus of helix M-H is close to the ‘hole’ between the two monomers that form the homodimer of the E ectodomain (Fig. 4). Some uninterpreted density between helix M-H and the i-j loop12 in domain II of the E protein might correspond to the first ~20 amino acids of the M protein.
Figure 4.
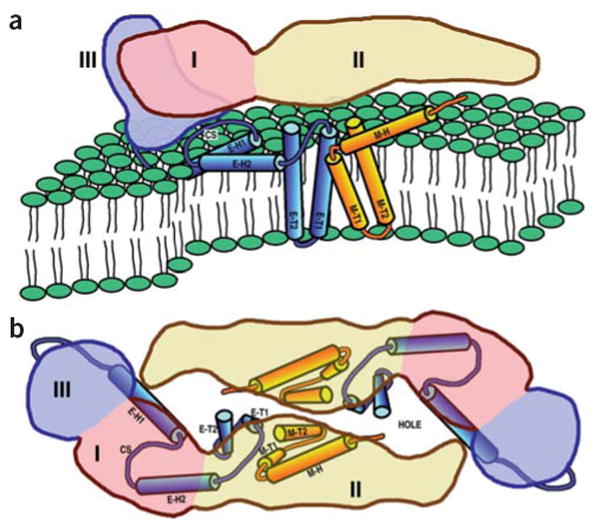
Diagrams of the dengue virus ectodomain and transmembrane domain proteins. The volume occupied by the ectodomain of an E monomer is pink (domain I), yellow (domain II) and lilac (domain III). The stem and anchor helices of E and M are blue and orange, respectively. Helices are identified by the nomenclature shown in Figure 2. CS represents the conserved sequence between E-H1 and E-H2. (a) View as in Figure 3a. (b) View as in Figure 3d with the superimposed E ectodomain homodimer.
The considerable separation of the helical components in the lipid made it possible to establish the position and connectivity of the secondary structural elements even at a resolution of 9.5 Å. Nevertheless, the precise orientation of the helices around their long axes and the amino acid residues that start and end the helices are uncertain.
The nucleocapsid core
The outer and inner limits of the membrane envelope and the outer limits of the nucleocapsid (150 Å) are at almost the same radii in the mature virus as in immature dengue particles13 (Fig. 1b). The large 15-Å gap of low, but not zero, density between the polygonal membrane and the core suggests that there probably is a unique structure that is in random contact with the membrane.
Unlike the nucleocapsid shell of alphaviruses26 and, indeed, of most viruses, there is no clear distinction between the presumed capsid protein shell and the RNA. The maximum height of the nucleocapsid density is 60% of that observed for the outer glycoprotein shell (Fig. 1b), as is also the case for the immature particles13. This, together with the short amino acid sequence of the capsid (101 residues in the dengue virus capsid protein compared with 264 residues in Sindbis virus), gives rise to the hypotheses10,27 that the core of flaviviruses might not have a unique structure, or that its icosahedral orientation is not synchronized with the larger and dominant external structure, or that the core has an asymmetric structure with a random orientation within the icosahedral envelope. As neither the E nor M proteins appear to extend beyond the limits of the cytoplasmic side of the lipid membrane, the capsid is probably randomly oriented relative to the outer glycoprotein shell.
Discussion
The results reported here represent a ‘quasi’-atomic description of a mature flavivirus. The only other enveloped virus with an equivalent amount of structural information for the virion is that of an alphavirus26,28. Although the E1 and E proteins in alpha- and flaviviruses have a similar structure29 and have been shown to form heterodimers with PE2 (the E2 precursor30) and prM, respectively31, there are also some major differences that are reflected in the biological properties of these viruses.
Contrary to previous expectations15,21, the stem region of E is a monotopic membrane protein component present only in the outer leaflet of the viral lipid bilayer. In contrast, the E1 glycoprotein in alphaviruses is well separated from the membrane surface, with no indication of buried sequences in the outer membrane leaflet26. The stem regions of E and M in flaviviruses are amphipathic α-helices half buried in the outer lipid leaflet. Their interactions with the lipid phosphate head groups likely will be influenced by pH changes, thereby contributing to the forces that induce the conformational rearrangements known to occur on fusion10.
The topology of the polyprotein in viral membrane envelopes has generally been deduced from amino acid sequences and limited biochemical studies. Visualization of the transmembrane domains in dengue virus particles (Fig. 4) confirms the predicted antiparallel orientations of the transmembrane helices and shows that the stem region of the E protein is a membrane component of the outer lipid leaflet. However, in alphaviruses the E1 and E2 proteins associate with each other to form parallel coiled coils across the viral membrane with their C termini emerging from the inner lipid leaflet26. This requires that, in alphaviruses, the C termini of the E2 glycoproteins are withdrawn from the membrane after post-translational cleavage of the membrane-bound polyprotein. This difference between flavi- and alphaviruses may be related, in part, to differences in their assembly pathways in which the association of the nucleocapsid with an envelope lipid bilayer and particle budding is completed earlier in the endoplasmic reticulum for flaviviruses, as opposed to later at the plasma membrane for alphaviruses.
Differences in viral morphogenesis are expected to have an impact on the properties of the nucleocapsids. In alphaviruses, the 33 cytoplasmic residues of each of the 240 copies of the E2 protein bind to a corresponding capsid protein, thus assuring that the icosahedral symmetry of the external glycoprotein assembly is synchronized with that of the internal core structure32,33. In contrast, the E and M proteins of flaviviruses are not exposed on the cytoplasmic side of the viral membrane. Therefore, they do not provide an anchor to synchronize the orientation of the nucleocapsid with respect to the external glycoprotein shell, and, hence, leave the nucleocapsid separated from the inner lipid bilayer. It would seem probable that the different relationship of the nucleocapsid to the external shell may be reflected both in the budding and fusion processes. For example, alphaviruses normally mature when preformed nucleocapsids bud through the plasma membrane, as can readily be seen in electron micrographs of infected cells34. This is the result of the interactions between the cytoplasmic domain of E2 and the nucleocapsid, described above, which provides one of the driving forces for virus assembly. However, preformed nucleocapsids are almost never seen in flavivirus-infected cells35,36. Thus, different mechanisms are at work in the assembly of alphaviruses and flaviviruses.
Methods
Electron microscopy and image processing
Dengue virus was prepared and cryo-EM image data were recorded as described10. A total of 1,691 dengue virus images were selected from 78 micrographs recorded with defocus settings ranging between 0.8 and 4.8 μm under focus and used for the final cryo-EM reconstruction. Particle processing was as described26 with the exception that orientations were refined by a procedure37 based on a reciprocal space refinement38. Phase agreement (<50°) and Fourier shell correlation coefficients (>0.5) indicated that the resolution of the final map was 9.5 Å. The pixel separation in the map representing the reconstruction was 2.80 Å.
Fitting atomic structures to the cryo-EM density
The cryo-EM density outside the viral membrane was fit with the homology model of the dengue E glycoprotein dimer using EMfit22 in the same two-step process as described10. However, instead of using only the Cα atoms, all nonhydrogen atoms were used (Table 1). The fit of the model to the density showed that the angle between domains I and III of the homology model required a consistent 4.5° adjustment in each of the three independent E monomers. In addition, the loop between residues 240 to 250 in domain II was outside density and needed repositioning. As the absolute hand of the cryo-EM map was unknown, the fitting procedure was attempted for both possible enantiomorphs. However, only one enantiomorph gave satisfactory results.
The stem regions of the E and M proteins (helices E-H1, E-H2 and M-H) were initially fit by manually placing regularized polyalanine α-helices into the cryo-EM density corresponding to one monomer in the dimer on the icosahedral two-fold axis. The transmembrane helices of E (E-T1 and E-T2) were fit simultaneously by using the colicin E3 Cα coordinates as the best available model structure of an antiparallel coiled coil (PDB entry 1JCH). The transmembrane helices of M (M-T1 and M-T2) were independently fit with regularized polyalanine α-helices as these were not associated with each other as in a coiled coil. The resultant fitted components of the stem and anchor regions of E in one monomer were then associated with each other as a rigid body and used as a model to fit simultaneously into all three E monomers in the icosahedral asymmetric unit using the program EMfit22 (Table 1). The stem-anchor region of the M protein was also fit similarly (Table 1).
Coordinates
The Cα atom coordinates of the fitted E, M and TM proteins have been deposited with the Protein Data Bank (accession code 1P58).
Acknowledgments
We thank E. Strauss for helpful discussions and R. Ashmore, C.R. Xiao, Y. Ji and D. Marinescu for the use of their various computer programs that were essential for calculating the image reconstruction. We are grateful for an equipment grant from the Keck Foundation. This work was supported by a US National Institutes of Health (NIH) Program Project grant to M.G.R., R.J.K. and T.S.B., by NIH grants to T.S.B. and J.H.S. and by a US National Science Foundation grant to T.S.B.
Footnotes
Note added in proof: The structure of the dengue virus E protein was published39 subsequent to the submission of our manuscript. The r.m.s. deviation between Cα atoms with respect to the homology model used in the above manuscript is 3.5 Å.
Competing Interests Statement: The authors declare that they have no competing financial interests.
References
- 1.Werten PJL, et al. Progress in the analysis of membrane protein structure and function. FEBS Lett. 2002;529:65–72. doi: 10.1016/s0014-5793(02)03290-8. [DOI] [PubMed] [Google Scholar]
- 2.Unger VM, Kumar NM, Gilula NB, Yeager M. Projection structure of a gap junction membrane channel at 7 Å resolution. Nat Struct Biol. 1997;4:39–43. doi: 10.1038/nsb0197-39. [DOI] [PubMed] [Google Scholar]
- 3.Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electroncrystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 4.Cockburn JJB, Bamford JKH, Grimes JM, Bamford DH, Stuart DI. Crystallization of the membrane-containing bacteriophage PRD1 in quartz capillaries by vapour diffusion. Acta Crystallogr D. 2003;59:538–540. doi: 10.1107/s0907444902023491. [DOI] [PubMed] [Google Scholar]
- 5.Lindenbach BD, Rice CM. Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, Pennsylvania, USA: 2001. pp. 991–1041. [Google Scholar]
- 6.Burke DS, Monath TP. Flaviviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, Pennsylvania, USA: 2001. pp. 1043–1125. [Google Scholar]
- 7.Solomon T, Mallewa M. Dengue and other emerging flaviviruses. J Infect. 2001;42:104–115. doi: 10.1053/jinf.2001.0802. [DOI] [PubMed] [Google Scholar]
- 8.Hahn YS, et al. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology. 1988;162:167–180. doi: 10.1016/0042-6822(88)90406-0. [DOI] [PubMed] [Google Scholar]
- 9.Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J Gen Virol. 2003;84:183–191. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- 10.Kuhn RJ, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinz FX, et al. The flavivirus envelope protein E: isolation of a soluble form from tick-borne encephalitis virus and its crystallization. J Virol. 1991;65:5579–5583. doi: 10.1128/jvi.65.10.5579-5583.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 15.Stiasny K, Allison SL, Marchler-Bauer A, Kunz C, Heinz FX. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J Virol. 1996;70:8142–8147. doi: 10.1128/jvi.70.11.8142-8147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001;75:4268–4275. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roehrig JT, Johnson AJ, Hunt AR, Bolin RA, Chu MC. Antibodies to dengue 2 virus E-glycoprotein syntheti(c peptides identify antigenic conformation. Virology. 1990;177:668–675. doi: 10.1016/0042-6822(90)90532-v. [DOI] [PubMed] [Google Scholar]
- 18.Bhardwaj S, Holbrook M, Shope RE, Barrett ADT, Watowich SJ. Biophysical characterization and vector-specific antagonist activity of domain III of the tick-borne flavivirus envelope protein. J Virol. 2001;75:4002–4007. doi: 10.1128/JVI.75.8.4002-4007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol. 2001;75:7769–7773. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beasley DWC, Barrett ADT. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J Virol. 2002;76:13097–13100. doi: 10.1128/JVI.76.24.13097-13100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allison SL, Stiasny K, Stadler K, Mandl CW, Heinz FX. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J Virol. 1999;73:5605–5612. doi: 10.1128/jvi.73.7.5605-5612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with X-ray crystallographic structures. J Struct Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 23.Johnson AJ, Guirakhoo F, Roehrig JT. The envelope glycoproteins of dengue 1 and dengue 2 viruses grown in mosquito cells differ in their utilization of potential glycosylation sites. Virology. 1994;203:241–249. doi: 10.1006/viro.1994.1481. [DOI] [PubMed] [Google Scholar]
- 24.Smith GW, Wright PJ. Synthesis of proteins and glycoproteins in dengue type 2 virus-infected vero and Aedes albopictus cells. J Gen Virol. 1985;66:559–571. doi: 10.1099/0022-1317-66-3-559. [DOI] [PubMed] [Google Scholar]
- 25.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W, et al. Placement of the structural proteins in Sindbis virus. J Virol. 2002;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones CT, et al. Flavivirus capsid protein is a dimeric α-helical protein. J Virol. 2003;77:7147–7179. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mancini EJ, Clarke M, Gowen B, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional anatomy of an enveloped virus, Semliki Forest virus. Mol Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- 29.Lescar J, et al. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 30.Barth BU, Wahlberg JM, Garoff H. The oligomerization reaction of the Semliki Forest virus membrane protein subunits. J Cell Biol. 1995;128:283–291. doi: 10.1083/jcb.128.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenz IC, Allison SL, Heinz FX, Helenius A. Folding and dimerization of tickborne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J Virol. 2002;76:5480–5491. doi: 10.1128/JVI.76.11.5480-5491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S, et al. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 33.Skoging U, Vihinen M, Nilsson L, Liljeström P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 34.Strauss JH, Strauss EG, Kuhn RJ. Budding of alphaviruses. Trends Microbiol. 1995;3:346–350. doi: 10.1016/s0966-842x(00)88973-8. [DOI] [PubMed] [Google Scholar]
- 35.Ng ML, Tan SH, Chu JJ. Transport and budding at two distinct sites of visible nucleocapsids of West Nile (Sarafend) virus. J Med Virol. 2001;65:758–764. doi: 10.1002/jmv.2101. [DOI] [PubMed] [Google Scholar]
- 36.Strauss JH, Strauss EG. Current advances in yellow fever research. In: Koprowski H, Oldstone MBA, editors. More About Microbe Hunters—Then and Now. Medi-Ed Press; Bloomington, Illinois, USA: 1996. pp. 113–137. [Google Scholar]
- 37.Ji Y, Marinescu DC, Zhang W, Baker TS. Orientation Refinement of Virus Structures with Unknown Symmetry. IEEE Computer Society; Los Alamitos, California, USA: 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossmann MG, Tao Y. Cryo-electron microscopy reconstruction of partially symmetric objects. J Struct Biol. 1999;125:196–208. doi: 10.1006/jsbi.1999.4083. [DOI] [PubMed] [Google Scholar]
- 39.Modis Y, Ogata S, Clements D, Harrison SCA. ligand–binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]


