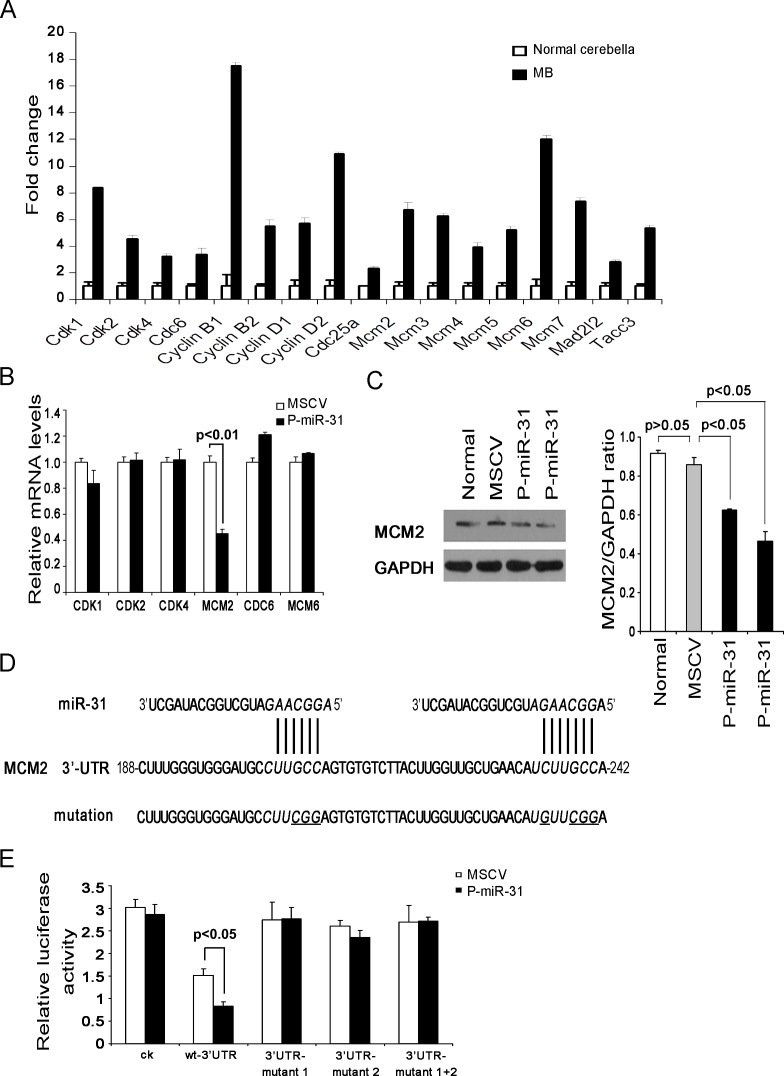
Figure 3. MCM2 is a direct target gene of miR-31.
(A) A selected group of cell-cycle associated genes differentially expressed in normal cerebellar and medulloblastoma tissues as determined by microarray analysis. (B) Quantitative RT-PCR analysis of putative miR-31 targets, CDK1, CDK2, CDK4, MCM2, CDC6 and MCM6, in vector control and miR-31 expressing DAOY cells. Data were analyzed according to the comparative Ct method, with GAPDH as a reference. The expression level in vector control cells was set to 1. (C) Immunoblot analysis of MCM2 protein levels in vector control and two independent pools of miR-31 expressing DAOY cells. GAPDH served as an internal control. The bar graph represents the relative MCM2 band intensity (on right panel). It was calculated as a ratio of MCM2 and GAPDH. (D) Predicted miR-31 target recognition sites in the 3'-UTR of human MCM2. The wild type luciferase reporter was generated with a DNA fragment covering two putative miR-31 binding sites. Mutations in the two miR-31 target sites are underlined. (E) Luciferase assays for the effect of re-expressing miR-31 response on the MCM2 3'-UTR reporters and its mutant variants in DAOY cells. The relative luciferase activity, defined as the ratio of the activity of MCM2 3'-UTR reporter (firefly) to that of the internal control (Renilla), was determined 48 h after transfection, and data representing the average of three independent experiments were shown.