Abstract
Creutzfeldt–Jakob disease (CJD) is a rare but invariably fatal neurodegenerative disease caused by misfolding of an endogenous protein into an alternative pathogenic conformation. The details of protein misfolding and aggregation are not well understood nor are the mechanism(s) by which the aggregated protein confers cellular toxicity. While there is as yet no clear consensus about how best to intervene therapeutically in CJD, prion infections can be propagated in cell culture and in experimental animals, affording both in vitro and in vivo models of disease. Here we review recent lead discovery efforts for CJD, with a focus on our own efforts to optimize 2-aminothiazole analogues for anti-prion potency in cells and for brain exposure in mice. The compounds that emerged from this effort were found to be efficacious in multiple animal models of prion disease even as they revealed new challenges for the field, including the emergence of resistant prion strains.
Introduction
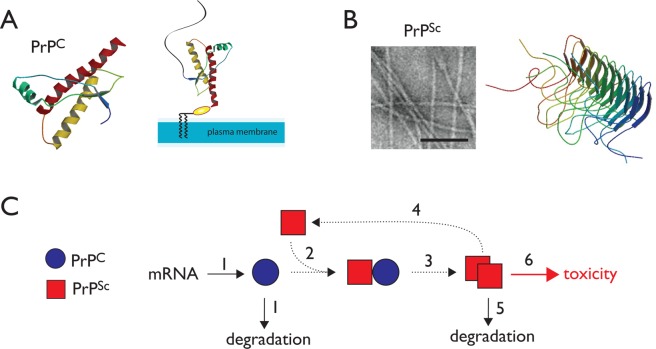
Prion diseases are a group of fatal neurodegenerative disorders characterized by neuronal loss, vacuolation, and the accumulation of amyloid protein aggregates in the central nervous system.1−3 They include Creutzfeldt–Jakob disease (CJD) in humans, scrapie in sheep, bovine spongiform encephalopathy (BSE) in cattle, and chronic wasting disease (CWD) in cervids. In humans, prion diseases such as CJD result in widespread neurological abnormalities including dementia and ataxia.4,5 The symptomatic phase of the disease, characterized by rapidly progressive neurodegeneration, typically occurs after a long latent incubation period.6 The initiation and progression of disease are thought to occur through the conformational conversion of an endogenous membrane-bound protein, PrPC, to an aggregated conformation termed PrPSc.1,2,7 Endogenous PrPC is localized to the plasma membrane via an glycophosphatidylinositol anchor in the unstructured N-terminal domain (Figure 1A). The structure of human PrPC has been determined by NMR,8 and X-ray structures of antibody-bound mouse PrPC have appeared recently.9,10 Pathogenic PrPSc on the other hand forms a fibrillar aggregate (Figure 1B) that has so far thwarted attempts to determine atomic-resolution structures. However, solid-state NMR has revealed the β solenoid structure of the Het-S prion from the filamentous fungus Podospora anserine, and this fold provides one possible model for PrPSc (Figure 1B).11 Once formed, PrPSc can template its own formation in a chain reaction that utilizes the cellular pool of PrPC as substrate (Figure 1C). PrPC appears to be a nonessential protein, and although some neuroprotective functions have been ascribed to it, the deletion of PrPC does not cause any gross developmental and behavioral phenotypes in mice, goats, or cattle.12−14 Thus, CJD and other prion disorders are thought to be gain-of-function diseases. Although the accumulation of PrPSc in the central nervous system is believed to be the basis of neurodegeneration, the exact cellular mechanisms of its toxicity are poorly understood.15
Figure 1.
Mechanism of prion propagation and potential points of therapeutic intervention. (A) The NMR structure of the folded domain of human PrPC is shown. (B) PrPSc forms fibrillar aggregates as shown in the electron micrograph (bar indicates 1000 Å). The β solenoid structure of the fungal Het-S prion provides one possible model of the pathogenic fold. (C) A simplified model for the propagation of PrPSc suggests several potential sites for therapeutic intervention.
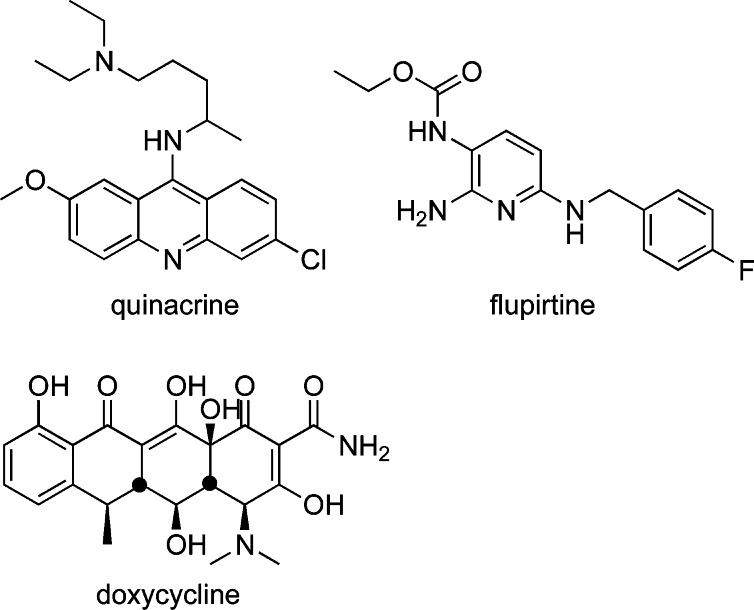
The self-replicating nature of PrPSc accounts for its ability to be horizontally transmitted in the infectious forms of the disease. Given the potential public health implications of transmissible latent prions in the food and blood supply, there is significant interest in the development of effective drugs for prion diseases.16 However, there are presently no effective therapeutics available. A number of approved drugs have been found to have anti-prion effects in cell culture experiments, and this has spurred efforts to repurpose these agents to treat CJD patients. Hence, the approved drugs flupirtine maleate,17 quinacrine,18 and doxycycline19 (Figure 2) have been studied in placebo-controlled human clinical trials for CJD. Unfortunately, none of these trials could establish any improvement in survival compared to placebo, highlighting the need to develop new molecules designed specifically to treat CJD.
Figure 2.
Approved drugs studied in human clinical trials for CJD.
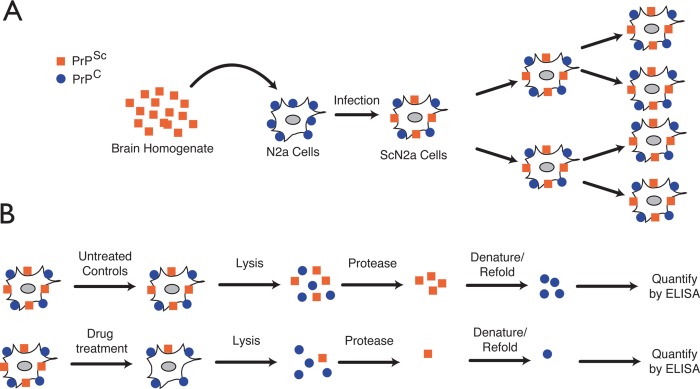
Historically, drug discovery efforts against prion diseases have been facilitated by the availability of robust cellular models that take advantage of the infectious nature of prion aggregates.20 Exposure of cultured cells to prion-containing extracts results in intracellular accumulation of PrPSc.21 These cells persistently maintain high levels of infectious prions upon subpassage (Figure 3A). Prion levels in infected cells can be readily quantified by fairly simple immunoassays. These assays commonly exploit the differential susceptibility of PrPC and PrPSc to proteolysis. In a typical assay, cell lysates are treated with proteinase K to degrade PrPC and the remaining PrPSc is quantified with anti-PrP antibodies (Figure 3B). By comparing PrPSc levels between treated and untreated cells, compounds that reduce prion load can be readily identified. These relatively convenient cell-based assays have been used extensively for identification of anti-prion compounds. However, it should be noted that protease-sensitive forms of PrPSc have been described22 and the presence of these forms may be overlooked by protease-based assays.
Figure 3.
Cell-based assay for identification of anti-prion compounds. (A) Neuroblastoma cell lines are exposed to PrPSc to initiate the infection process. Once infected, cells continuously transmit PrPSc to daughter cells upon passage. (B) Infected cells are exposed to test compounds at various concentrations and time durations. The effect of a compound on PrPSc levels is quantified by treating cell lysates with proteinase K and quantifying the level of remaining PrPSc by ELISA.
Once a compound with anti-prion activity has been identified and optimized, its in vivo efficacy can be evaluated in mouse models of disease.20,23 Inoculation of mice with prion isolates results in neurologic disease and recapitulates typical symptoms and histopathologies associated with the specific prion strain. Prion infected mice have well-defined and predictable incubation periods that can range from several weeks to several months, depending on the prion strain. Thus, monitoring survival times of prion-infected mice in the presence or absence of a lead compound provides a convenient, if time-consuming, approach for evaluating drug efficacy in vivo. Alternatively, recent studies have indirectly measured neurological damage as an indicator of disease progression and treatment effectiveness. In these models, up-regulation of glial fibrillary acidic protein (GFAP), a widely used marker of neuronal damage, is detected in Tg(Gfap-luc) mice using bioluminescent imaging (BLI).24 Importantly, this new BLI model can provide an indication of compound efficacy weeks before clinical symptoms of disease emerge.
It is important to note that the above assays are inherently phenotypic in nature and do not provide information about the pathways or molecular targets that might be involved in anti-prion action of small molecules (Figure 1C). A wide variety of possible molecular mechanisms can be envisioned for small molecule anti-prion agents. Thus, an anti-prion therapeutic might exert effects on disease via (1) influencing the expression or degradation of PrPC, (2) inhibiting the interaction of PrPC with PrPSc, (3) inhibiting the conversion of PrPC to PrPSc via binding to PrPC and/or to cellular factors involved in misfolding, (4) capping or promoting fragmentation of the growing PrPSc fiber, (5) directly interacting with PrPSc aggregates leading to their degradation, or (6) modulating other cellular pathways such as those involved in the clearance of misfolded protein.
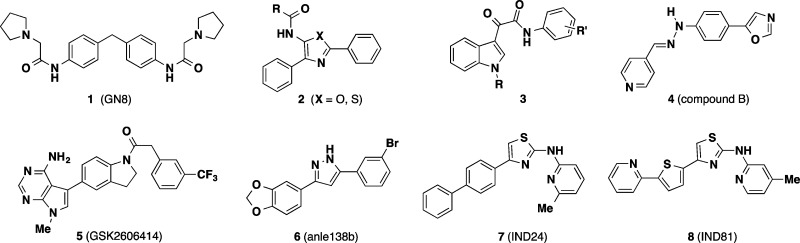
By 2010, a number of research groups had utilized cell-based assays to identify a striking variety of compounds with apparent anti-prion effects (Chart 1). These included the symmetrical biphenylmethylene 1 (GN8),25 phenothiazine antipsychotics, statins, tetracycline antibiotics, diaryl oxazoles and thiazoles (2), indole-3-glyoxylamides (3), and polyanionic or cationic dendrimers, among many other chemotypes described in recent comprehensive reviews.26,27 In an effort to explore anti-prion mechanisms, Kuwata and co-workers studied the binding of several distinct classes of anti-prion compounds to recombinant PrPC using SPR and NMR.28 The anti-prion compounds studied were classified by the nature of their interaction with PrPC as (1) stoichiometric binders or “medical chaperones”, (2) nonspecific, superstoichiometric binders, (3) compounds that cause aggregation/precipitation of PrPC, or (4) noninteracting compounds that likely have a different target. One caveat with this important work is that the binding studies were conducted with a PrPC construct (PrP121–231) lacking N-terminal residues that have since been found to be important for the interaction of the tricyclic antipsychotic promazine with a cryptic small molecule binding site on PrPC.10 This recent finding that promazine binding induces the organization of otherwise unstructured N-terminal residues in PrPC must be taken into account when designing protein constructs for future biophysical studies of small molecule binding to PrPC.
Chart 1. Selected Small Molecules with Anti-Prion Activitya.
a Compound 1 and 4–8 have demonstrated efficacy in animal models of prion disease.
Despite considerable success in identifying new chemotypes, relatively few compounds have been evaluated in animal models, and those that have performed poorly. Quinacrine for example fails to significantly prolong survival in mice despite initial favorable effects on prion load in vivo. This failure has been attributed to a combination of low brain exposure due to P-glycoprotein (P-gp) mediated drug efflux and to the formation of quinacrine-resistant prion strains.29 The antibiotics tetracycline and doxycycline were shown to extend survival in Syrian hamsters; however, in these studies drug was co-incubated with the infectious brain homogenate prior to the inoculation of animals.30 The relevance of this study design to true therapeutic intervention is unclear, and the ultimate failure of doxycycline in human trials19 casts further doubt on the experimental model. In another study, mice treated subcutaneously with analogue 1 lived ∼20 days longer than untreated controls, amounting to a ∼12% extension of survival.25 Among in vivo efficacy studies published prior to 2013, the arylhydrazone 4 (compound B) is the only agent that produced a ≥50% extension of survival in treated animals compared to controls.31
Notably, the year 2013 saw the disclosure of several new small molecules reported to exhibit promising effects in prion animal models. Among these was the compound 5 (GSK2606414),32,33 an inhibitor of the kinase PERK, a key mediator of one branch of the unfolded protein response (UPR). The UPR is initiated in response to the accumulation of misfolded protein and PERK functions both to detect misfolded protein in the ER and to phosphorylate the translation initiation factor eIF2α, leading to repression of translation. In the context of neurodegenerative disease, repression of translation may contribute to a loss of synaptic function and neuronal death. Previously, genetic manipulation of the PERK pathway had been shown to be neuroprotective in mice, and in the more recent study similar effects were observed in mice treated with 5.32,33 While the effects of 5 on survival were not examined in this work, modulation of the PERK pathway appears to merit further investigation, as this approach could conceivably be effective in multiple misfolding diseases.
The other notable anti-prion small molecules disclosed in 2013 include the diarylpyrazole 6 (anle138b)34 and the 2-aminothiazole analogues 7 (IND24) and 8 (IND81).35 These compounds were identified in phenotypic assays of the variety described above, and optimized analogues were subsequently found to extend the survival of infected mice ∼2-fold (∼100%) compared to untreated controls. The molecular targets of these agents remain to be identified. In this review, we will detail the discovery, optimization, and in vivo evaluation of the 2-aminothiazole chemotype represented by compounds 7 and 8. This work involved the efforts of a multidisciplinary team of scientists from multiple laboratories at University of California, San Francisco, the contributions of which are explicitly acknowledged here and in the primary references cited herein.
Screening and Hit Profiling
Most anti-prion compounds have been discovered by ad hoc screening of small collections of known bioactive compounds. In fact, the only moderate-throughput cell-based screening effort published prior to 2010 was a screen of 2000 compounds utilizing a dot blot assay.36 This screen identified 17 anti-prion compounds that included naturally occurring polyphenols, phenothiazines, antihistamines, statins, and antimalarial compounds including quinacrine. Another screen of 10 000 compounds involved a cell-free assay to identify compounds that interfered with the interaction of PrPC and PrPSc.37 A number of these compounds were shown to be active in cell culture. However, the activity of the hits identified in these two screens was not recapitulated in vivo.
In 2010, the Prusiner group sought to extend these studies by initiating a larger cell-based screen of 10 000 diverse leadlike compounds utilizing an ELISA-based assay.38 In this screen, N2a cells infected with scrapie prions (ScN2a) were incubated with 5 μM test compound for 5 days (quinacrine was used as a positive control). Following proteolysis of cell lysates, the resulting change in protease-resistant prion levels was quantified by sandwich ELISA. Importantly, the cytotoxic effects of each compound were also analyzed in a parallel screen. Although the 96-well format prion assay was inherently heterogeneous in nature (drug incubation and signal readout were conducted in different plates), the screen was shown to have a high level of quantitative precision, reproducibility, and signal linearity.
The ELISA screen identified 121 compounds that reduced PrPSc levels by >50% at 5 μM without significant cellular toxicity (1.2% hit rate). The hit set was evaluated for nascent SAR and synthetic tractability. Four structural scaffolds were selected for follow-up studies, including quinazolines, 8-hydroxyquinolines, 2-arylbenzoxazoles, and 2-aminothiazoles.38 The first three scaffolds were eventually deprioritized for reasons ranging from chemical instability to inscrutable SAR. This left the 2-aminothiazole series (henceforth denoted AMT), which demonstrated tractable SAR and was amenable to parallel synthesis using the classical Hantzsch thiazole synthesis. Of potential concern was that the parent 2-amino-4-arylthiazole ring system is known to be subject to P450-mediated oxidation leading to the production of electrophilic metabolites.39 We judged this risk was somewhat mitigated in our leads, which possessed electron-withdrawing heteroaryl substitution on the 2-amino function and thus would be less prone to oxidation. Although the potential oxidative liability of AMTs was never explicitly investigated, we found empirically that 2-pyridyl AMTs like 7 and 8 were generally well tolerated in mice over the long courses of therapy employed in the mouse infection models (see below).
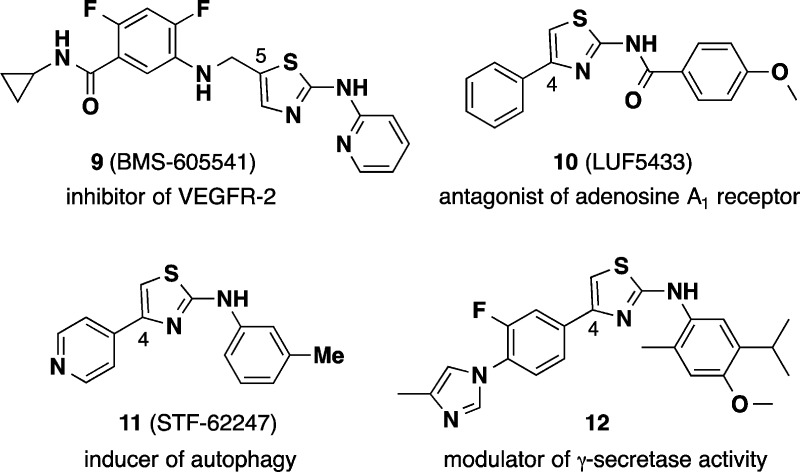
The early AMT analogues were further profiled in orthogonal assays. Thus, using an assay that exploits the propensity of PrPSc to precipitate in the presence of phosphotungstic acid,22 we demonstrated that early AMTs were able to clear both protease-sensitive and protease-resistant forms of PrPSc aggregates. We also found that the compounds did not disaggregate PrPSc in cell-free assays, nor did they alter the expression level of endogenous PrPC.38 This suggested that AMTs might act by interfering with the formation of new PrPSc in the cell, either directly by interference in the misfolding/assembly process or indirectly by modulating endogenous cellular clearance mechanisms. A review of the literature raised other mechanistic hypotheses. For example, the 2-arylaminothiazole ring system is found in a number of pharmacological agents, including kinase inhibitors (e.g., dasatanib, VEGFR inhibitor 9),40,41 and in adenosine A1 receptor antagonists (e.g., 10).42,43 However, as the SAR of anti-prion AMTs became better defined, action at kinases or adenosine receptors appeared increasingly unlikely. For example, AMTs bearing benzamide substituents as in compound 10 lacked anti-prion effect. Similarly, modification of the putative hinge-binding motif to ablate kinase inhibition did not diminish anti-prion activity.
During the course of SAR studies, AMT analogues more closely related to our leads appeared in the literature (Chart 2), including compounds reported to induce autophagy (e.g., 11)44 and modulate γ-secretase activity (12).45,46 Despite close structural resemblance, anti-prion AMTs do not appear to induce autophagy (S. Ghaemmaghami, unpublished data) nor do they inhibit γ-secretase activity (S. L. Wagner, personal communication). The molecular target(s) and mechanism of action of AMTs thus remain under active investigation. The recent finding10 that flexible, unstructured regions of PrPC contribute to forming a cryptic small molecule binding site has encouraged further study of direct interactions between AMTs and PrPC.
Chart 2. Pharmacologic Agents Bearing the 2-Aminothiazole Ring System.
Structure–Activity Trends
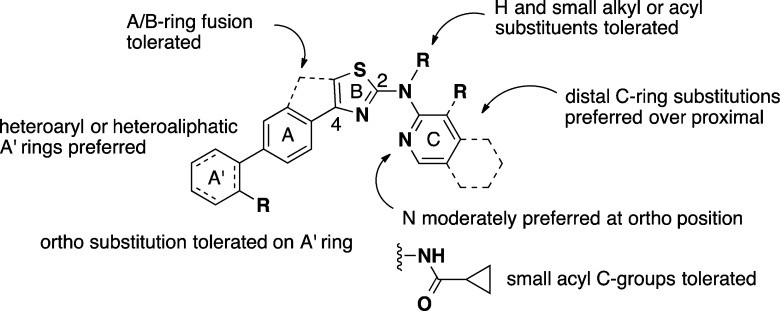
Our structure–activity studies of the AMT chemotype have been described elsewhere47,48 and are summarized briefly here (Figure 4). One confounding issue in our early SAR studies was the nonlinear dependence of EC50 values on PrPC expression levels in different N2a cell lines. Thus, early SAR work using the original ScN2a assay led quickly to a number of analogues with low nanomolar potency. While encouraging, the high sensitivity of this assay to AMTs made it challenging to discern clear SAR trends. Concurrent with the early chemistry effort, the Prusiner lab had developed additional N2a cell lines for screening purposes, including the so-called “clone-3” cell line that overexpresses PrPC.49 Interestingly, we found that AMT analogues were generally much less potent (10- to 100-fold higher EC50 values) when the clone-3 line was employed in the ScN2a assay. This effect was not limited to AMTs but was true of other anti-prion classes as well, including hydrazone 4. Further improvements to the ELISA assay using the clone-3 cell line resulted in a very robust assay with good dynamic range and excellent precision.48 The “ScN2a-cl3” assay thus supplanted the earlier assay and allowed for more subtle SAR trends to be discerned, at the cost of less impressive EC50 values.
Figure 4.
Summary of anti-prion SAR for aminothiazole analogues.
Anti-prion AMTs comprise an 2-aminothiazole “B-ring” substituted on the 2-amino function with a small acyl “C-group” or aryl “C-ring” and at C-4 of the aminothiazole ring with an aryl or heteroaryl “A-ring” (Figure 4). Replacement of the aminothiazole ring with other heterocycles was examined briefly but with unsatisfactory results. With regard to the C-ring, we found that only small acyl C-groups (e.g., acetamide, cyclopropylamide) were tolerated, while a wider variety of C-rings could be employed, pyridine and quinoline being favored. The 2-amino function could be further alkylated or acylated, indicating that a hydrogen-bond donor was not required at this position. With regard to the A-ring, both five- and six-membered heteroaryl rings were tolerated, and these could be appended to a second aromatic or aliphatic “A′-ring”. The dihedral angle of the A- to B-ring connection proved to be important, with coplanar (including fused) A/B-ring analogues generally active, while those with enforced orthogonal A/B-rings lacked the anti-prion effect.47
Given the coplanar and highly conjugated pharmacophore, it was unsurprising that poor aqueous solubility became an issue when formulating AMTs for oral dosing. This issue was addressed to some degree by the introduction of heteroaliphatic A′-rings (morpholine, piperazine) or by the introduction of ortho substituents in the A′-ring to enforce orthogonal disposition of the A′ and A-ring. Unlike with the A/B-ring connection, staggered A′-ring/A-ring dihedrals were accommodated without significant loss of anti-prion activity. Although notable improvements were made in terms of potency and druglike properties, the majority of efficacy studies performed to date have involved the relatively early analogues 7 and 8, for which suitable oral formulations were eventually developed.50
Structure–Brain Exposure Trends
The team recognized early on that new anti-prion chemotypes should be evaluated as soon as feasible for brain exposure in animals. Indeed, the fact that many early AMT analogues exhibited good brain exposure in mice encouraged further work on the series. To maximize the number of AMTs that could be evaluated, pharmacokinetic studies were focused on determining brain exposure (AUC) in mice on oral dosing. It was expected that the most promising compounds would later be evaluated in full PK studies with both iv and po dosing to derive additional PK parameters. Bioanalysis was performed on whole brain homogenate, and the bound and unbound fractions were estimated later for select analogues using in vitro brain tissue binding assays.50 By use of this approach, the fraction unbound in mouse brain tissue was estimated at ∼8–9% for compounds 7 and 8.
Among the first ∼100 AMT analogues synthesized, nearly a quarter were evaluated in PK studies. We selected analogues that covered a breadth of A- and C-ring chemotypes, intentionally including analogues with only modest potency in the ScN2a-cl3 assay. The goal was to derive structure–brain exposure relationships, and it was understood that these relationships would be unrelated to in vitro anti-prion SAR. In fact the first 27 AMTs evaluated exhibited brain exposure (AUC) values distributed over 4 orders of magnitude. While the presence of more than one hydrogen bond donor was associated with poor brain exposure, we unfortunately could not derive correlations with other properties such as molecular weight or polar surface area (PSA). It should be noted, however, that most of the compounds in this set already fall within recommended51 PSA and MW ranges for brain-penetrant small molecules. Also, it is clear that the brain exposure values obtained vary as a result of differences in oral absorption, clearance, and metabolism, as well as from intrinsic differences in brain penetrance. Nevertheless, by prioritizing PK studies early in the discovery process, we identified a number of promising analogues.
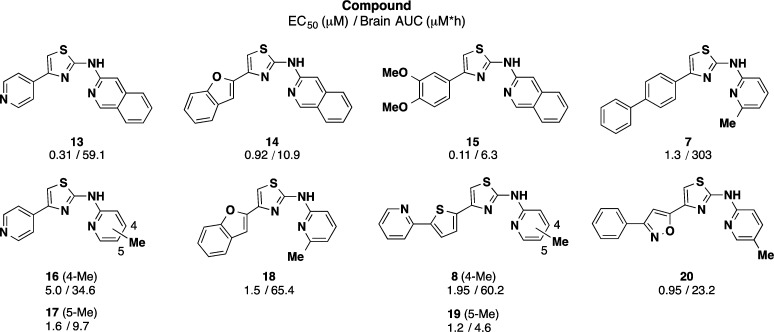
Some of the more promising AMT analogues from the first round of PK studies are presented above (Chart 3). Of three quinoline C-ring analogues (13, 14, and 15) with submicromolar potency, analogue 13 bearing a pyridyl A-ring exhibited the highest brain AUC value. Also apparent is that seemingly small structural changes can have significant effects on brain exposure. Thus, in the case of regioisomeric analogues 16/17 and 8/19, the 4-methyl congeners exhibited significantly higher brain exposure in vivo than the 5-methyl comparators. Other compounds exhibiting high brain exposure included the benzofuran 18, phenylisoxazole 20, and especially the biphenyl A-ring analogue 7, which exhibited the highest brain AUC among the initial 27 AMTs evaluated. The ratio of brain AUC/EC50 was used as a criterion for advancement into additional in vivo PK studies, and by this measure compounds 7, 8, 13, 15, 18, and 20 merited further evaluation.
Chart 3. Select Early AMT Analogues Evaluated for Brain Exposure in Micea.
a The EC50 values shown are for the ScN2a-cl3 assay, while brain AUC values reflect analysis of total brain homogenate following a single oral dose of 40 mg/kg in mice.50
Animal models of prion disease typically require 100–200 days to evaluate efficacy, depending on the incubation time of the prion strain employed. Previous studies with quinacrine had established that the test article could be conveniently administered daily as part of a liquid rodent diet.29 We used this formulation to evaluate AMT analogues 7, 8, 13, 15, 18, and 20 in dose escalation studies. Mice received test article in their feed at approximate doses of 40, 80, 130, or 210 mg kg–1 day–1 for 3 days (Table 1). Plasma and brain samples were collected at the end of the last dosing cycle and analyzed as in the single-dose PK studies. Since only a single time point was evaluated in these dosing studies, the resulting concentrations must be regarded as pseudo-steady-state concentrations. Compounds 7 and 8 emerged from these studies as the most suitable for efficacy trials, both compounds achieving pseudo-steady-state brain concentrations in the micromolar range at the 40 mg kg–1 day–1 dose and showing linear exposure with escalating dose.
Table 1. Pseudo Steady-State Brain Concentrations of Selected AMTs after 3-Day Dosing in a Liquid Rodent Diet at Various Daily Doses50,a.
| brain concentration (μM) after 3 days at the indicated dose |
||||
|---|---|---|---|---|
| compd | 40 mg kg–1 day–1 | 80 mg kg–1 day–1 | 130 mg kg–1 day–1 | 210 mg kg–1 day–1 |
| 7 | 8.70 ± 1.46 | 19.3 ± 2.28 | 31.8 ± 5.46 | 37.4 ± 9.06 |
| 8 | 3.00 ± 0.52 | 7.45 ± 1.00 | 13.0 ± 2.81 | 19.3 ± 3.24 |
| 13 | 0.02 ± 0.03 | 0.08 ± 0.03 | 0.04 ± 0.03 | 0.10 ± 0.07 |
| 15 | 0.03 ± 0.03 | 0.06 ± 0.07 | 0.14 ± 0.04 | 0.32 ± 0.11 |
| 18 | 1.31 ± 0.39 | 2.70 ± 0.52 | 3.74 ± 0.41 | 3.23 ± 2.62 |
| 20 | 0.88 ± 0.68 | 5.39 ± 2.93 | 23.3 ± 13.2 | 12.6 ± 8.62 |
Drug concentrations are for total brain homogenate.
Among other analogues, compounds 20 and 18 exhibited nonlinear exposure profiles while compounds 13 and 15 exhibited surprisingly poor exposure when dosed in the liquid diet. In general AMT analogues exhibited higher exposure in brain than in plasma, suggesting that the class is not subject to P-gp mediated efflux. Assuming a free fraction of ∼8%, compounds 7 and 8 were thus predicted to achieve free, steady-state concentrations in excess of their EC50 values at doses of 80 mg kg–1 day–1 and higher (for 7) or 130 mg kg–1 day–1 and higher (for 8). Even at the highest dose of 210 mg kg–1 day–1, mice receiving 7 or 8 exhibited no adverse clinical or behavioral effects, suggesting that the compounds would be well tolerated on prolonged dosing in an animal model of prion disease. Accordingly, compounds 7 and 8 became the focus of further PK and efficacy studies, as described below.
Having identified 7 and 8 as candidates for study in animal models of disease, we sought to determine their full PK profile compared to that of hydrazone 4 (Table 2). The three compounds were evaluated head-to-head with iv and po dosing in female FVB mice. In nearly every respect, 7 appeared to be the superior compound. Compared to 4, compound 7 achieved nearly 30-fold higher brain exposure following oral dosing and also exhibited a much longer half-life, lower clearance, a superior brain/plasma ratio, and greater bioavailability. Compound 8 was also superior to 4 in most respects and exhibited the highest volume of distribution of the three compounds. Although full toxicology studies were not conducted for any AMT analogues, we observed over the course of dozens of efficacy studies that compounds 7 and 8 produced no overt signs of toxicity in mice, even on prolonged dosing over many weeks at doses of 210 mg kg–1 day–1. In contrast we found that hydrazone 4 exhibited lethal toxicity at a dose of 150 mg kg–1 day–1 and could only be employed in efficacy trials at doses of ≤110 mg kg–1 day–1.52
Table 2. Pharmacokinetic Parameters for AMT Analogues and Compound B after a Single Dose of 1 mg/kg (iv) or 10 mg/kg (po)50.
| compd | route/matrixa | Vss (L/kg)b | CL (L h–1 kg–1)c | t1/2 (h)d | Cmax (μM)e | AUC (μM·h)f | B/P g | F (%)h |
|---|---|---|---|---|---|---|---|---|
| 7 | iv/pl | 2.46 | 0.92 | 2.16 | 2.49 ± 2.65 | 2.78 ± 1.09 | 2.60 | 40.3 |
| po/pl | 4.65 | 1.66 ± 0.11 | 11.2 ± 1.31 | |||||
| po/br | nd | 2.45 ± 0.74 | 29.1 ± 1.24 | |||||
| 8 | iv/pl | 12.6 | 9.05 | 1.18 | 0.52 ± 0.28 | 0.30 ± 0.04 | 5.53 | 27.3 |
| po/pl | 0.98 | 0.43 ± 0.03 | 0.82 ± 0.39 | |||||
| po/br | nd | 1.62 ± 0.54 | 4.54 ± 2.28 | |||||
| 4 | iv/pl | 1.53 | 4.56 | 0.22 | 1.76 ± 0.14 | 0.83 ± 0.12 | 0.51 | 24.8 |
| po/pl | 1.01 | 0.83 ± 0.38 | 2.06 ± 0.06 | |||||
| po/br | nd | 0.46 ± 0.16 | 1.06 ± 0.10 |
Route of administration and matrix analyzed (brain, br; plasma, pl).
Volume of distribution at steady state.
Clearance.
Half-life.
Maximal concentration.
Brain exposure expressed as area under the drug concentration–time curve.
Brain/plasma AUC ratio.
Bioavailability.
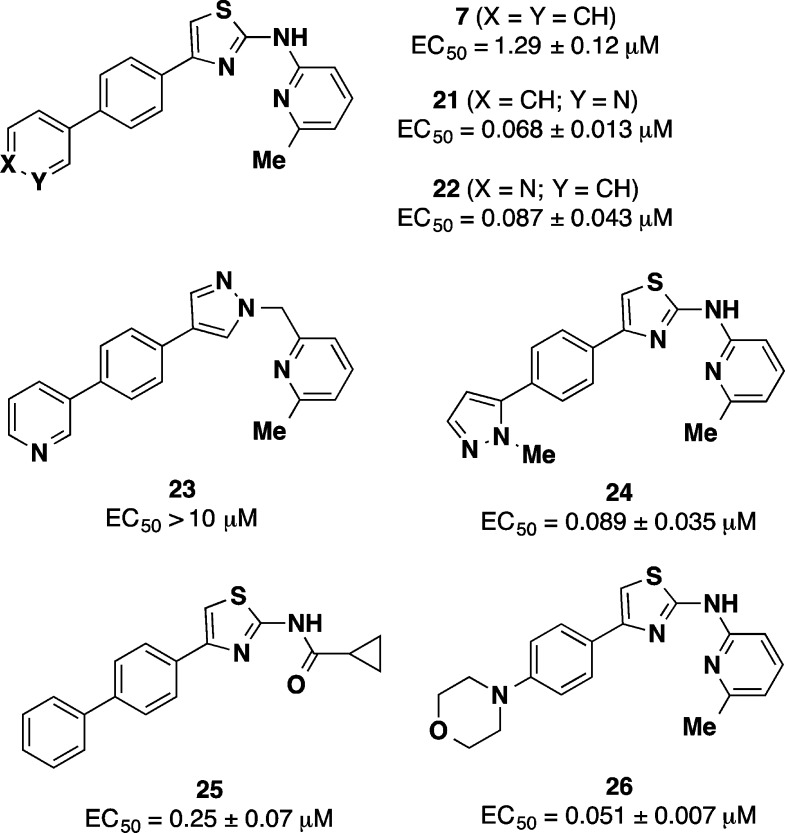
Even as compound 7 became the focus of extensive in vivo evaluation (see below), further optimization of this promising lead continued.48 Interestingly, we found that replacement of the terminal phenyl ring in 7 with heteroaromatic or heteroaliphatic rings produced analogues with clearly superior potency in the ScN2a-cl3 assay (Chart 4, Table 3). Thus, pyridyl (21 and 22), pyrazole (24), and morpholine (26) A′ ring congeners of 7 all exhibited EC50 values below 100 nM, a greater than 10-fold improvement in potency compared to 7. Compound 21 also exhibited excellent brain exposure in animals (Table 3), yielding a greater than 20-fold improvement in brain AUC/EC50 ratio for 21 compared to 7. An attempted bioisosteric replacement of the aminothiazole ring in 21 with pyrazolylmethyl (analogue 23) unfortunately led to an utter loss of anti-prion activity. Whether this effect is due to the pyrazole ring, the sp3 linkage between B-ring and C-ring, or some combination of the two is unclear. More promising results were had with a series of cyclopropylamide C-group analogues, including compound 25, the direct analogue of 7. Analogue 25 exhibited potency and in vivo brain exposure superior to 7, with a brain AUC/EC50 ratio 10-fold higher than for 7. The new generation of AMT analogues exemplified by 21 and 25 are only now being evaluated in animal efficacy models.
Chart 4. Anti-Prion Activities (ScN2a-cl3 Assay) for Structural Analogues of 7a.
a Additional PK data for some of these analogues is provided in Table 3.48
Table 3. Anti-Prion Activity and Select PK Parameters for AMT Analogues Related to 7a.
| compd | ScN2a-cl3 EC50 (μM) | brain Cmax (μM) | brain AUC (μM·h) | plasma AUC (μM·h) |
|---|---|---|---|---|
| 7 | 1.29 ± 0.12 | 2.45 ± 0.74 | 9.78 ± 2.07 | 6.99 ± 0.73 |
| 21 | 0.068 ± 0.013 | 2.46 ± 0.85 | 10.90 ± 1.75 | 15.60 ± 0.06 |
| 22 | 0.087 ± 0.043 | 0.50 ± 0.005 | 1.68 ± 0.10 | nd |
| 25 | 0.248 ± 0.067 | 4.34 ± 0.53 | 18.60 ± 3.75 | 12.70 ± 1.11 |
The PK study involved a single oral dose of 10 mg/kg.48
Efficacy in Animal Models
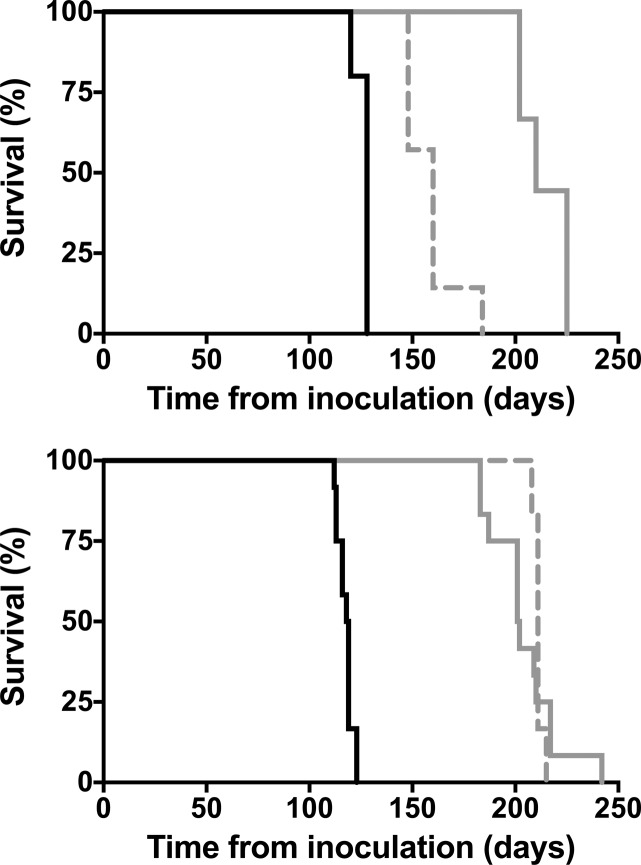
The effects of compounds 7 and 8 in prion infected mice have been extensively studied, as recently reported.35 Oral administration of either compound is found to significantly extend the survival of mice infected with various prion strains (Figure 5). For example, mice infected with the Rocky Mountain Laboratories (RML) scrapie strain and treated with 7 at 210 mg kg–1 day–1 survived for 204 ± 5 days, compared to 118 ± 1 days for vehicle-treated controls. Similarly, treatment with compound 7 under the same protocol extended survival from 126 ± 2 days to 214 ± 4 days in mice inoculated with the ME7 scrapie strain. In a chronic wasting disease (CWD) model involving transgenic mice expressing elk PrPC, compound 7 extended survival from 108 ± 3 days to 237 ± 0 days. When treatment with 7 was initiated 60 days postinoculation (rather than 1 dpi), significant lifespan extension was still observed in mice inoculated with RML or ME7 strains (Figure 5). This finding is significant given that BLI imaging of these mice reveals that brain disease is already progressing by day 60. However, despite significant and reproducible extension of lifespan in scrapie and CWD animal models, AMT-treated mice eventually showed accumulation of PrPSc in their brains and ultimately succumbed to disease. Most disappointingly, compound 7 proved to be entirely ineffective against human CJD prions in susceptible transgenic mice expressing human PrPC.
Figure 5.
Kaplan–Meier survival curves for mice infected with ME7 (top) and RML prions (bottom) . The vehicle-treated cohort is shown as a solid black line. Treated cohorts received compound 7 from day 1 postinoculation (solid gray line) or from day 60 postinoculation (dashed gray line).
The animal studies summarized above clearly demonstrate that the in vivo efficacy of AMTs is dependent on the specific prion strain employed and that even infections involving susceptible prion strains cannot be fully cured with the compounds. In a number of elegant experiments, the Prusiner lab demonstrated that the eventual failure of AMTs can be accounted for by treatment-induced selection of AMT-resistant prion strains.35 For example, prions isolated from the brains of terminal AMT-treated mice were resistant to AMTs, both in cell culture and in vivo. This result also helped to explain the initially surprising finding that a dose of 50 mg kg–1 day–1 of compound 7 provided comparable benefit as the higher dose of 210 mg kg–1 day–1 (K. Giles, manuscript in preparation). Efficacy is limited in both cases by the emergence of resistant prion strains, so greater drug concentrations at the higher dose provided no additional benefit.
Another intriguing finding is that selective drug pressure must be continuously maintained in vivo in order to preserve the AMT-resistant strain. Thus, passage of AMT-resistant prions in naive, untreated mice returned prions that were once again susceptible to AMTs. This result may suggest that AMT treatment leads to the selection and proliferation of a resistant prion strain that is otherwise uncompetitive with AMT-sensitive strains. The ability of AMTs to induce the formation of drug resistant strains is reminiscent of similar effects observed with quinacrine and swainsonine.29,53 Although quinacrine does not extend survival in animal models of disease, it does rapidly select for quinacrine resistant prions in vivo.19 In cell culture experiments, swainsonine exposure leads to selection for resistant prion strains as well. The passage of these resistant prions in untreated mice produces prions that are once again susceptible to the compound. Thus, it appears that drug resistance may be a general confounding issue for anti-prion therapeutics as it is in other therapeutic areas. What is remarkable about this particular form of drug resistance is that it apparently results from “conformational mutagenesis” of PrPSc rather than from mutations in the nucleic acid sequence of a gene.
The fascinating relationship between PrPSc conformational strains, prion species barriers, and drug resistance has been reviewed recently.54 The consensus that is emerging from various studies is that strain adaptation results from the interplay between two related phenomena: (1) conformational transformation of a replicating prion to create a heterogeneous pool of novel conformations (mutation) and (2) enrichment of conformations with high proliferative capacity within this pool (selection). This “conformational drift” continuously produces pools of structurally distinct self-replicating prions that can be acted upon by natural selection. Thus, strains that are resistant to drugs can be spontaneously generated and selected upon continuous drug treatment.
While the native fold of PrPC appears to be a rather poor target for small molecules, the conformational mutation of replicating prions described above may well involve the transient formation of cryptic small molecule binding sites, and these sites may well differ between strains. The first clear evidence in support of this hypothesis comes from the recently published X-ray structure of promazine bound to PrPC.10 Remarkably, the binding site revealed in this structure is formed in part from N-terminal residues that are unstructured in all existing X-ray and NMR structures of PrPC. Thus, promazine binding induces the formation of a new antiparallel β-strand and two tandem β-turns in the normally unstructured region PrP(117–124). The authors further demonstrate that promazine binding allosterically stabilizes more distal regions of the protein via the formation of new cation−π and hydrogen bonding interactions in the β2−α2 loop and enhanced hydrophobic interactions at the interface of the α2−α3 helices, among other changes. One can therefore propose a plausible hypothesis of small-molecule anti-prion action that involves binding to and stabilizing specific PrPC conformations. In this scenario, strain specific action can be readily understood by positing alternative misfolding pathways that avoid the conformation(s) trapped by specific therapeutic compounds.
Drug discovery and development for a rare and as yet poorly understood disease like CJD are highly challenging. The studies of anti-prion AMTs described herein provide a number of valuable lessons for future discovery efforts targeting these diseases. First, it is clear that the anti-prion effects of small molecule leads can be strain-specific. Thus, compounds identified in phenotypic screens utilizing prion strains from non-human organisms (such as RML) may have limited efficacy against CJD in humans. This fact highlights the urgent need for CJD-infected cell models (as yet unknown), as these would presumably be more congruent with prion infection of differentiated neurons in the brain. Second, a more target-oriented drug discovery effort could help circumvent the problem of drug resistance. For example, targeting cellular pathways that are broadly involved in protein quality control may prove to be an effective strategy to find more widely effective compounds. Third, more sophisticated dosing regiments involving cocktails of diverse drugs and/or intermittent dosing may be required to mitigate the issue of drug resistance. Finally, as with any challenging problem in drug discovery, ultimate success will depend on the robust, sustained, and collaborative efforts of a multidisciplinary team of committed clinicians and scientists.
The insights gathered from anti-prion AMT studies not only are relevant to prion medicine but also may have important implications for related neurological conditions. A number of neurodegenerative diseases such as Alzheimer’s disease (AD), amylotrophic lateral schlerosis (ALS), and Parkinson’s disease share many features in common with prion disease.55,56 These prion-like diseases are associated with seeded aggregation pathways analogous to the misfolding of PrPC to PrPSC, but they lack the robust infectiousness that has enabled an expanded repertoire of in vivo and in vitro prion disease models. Thus, prions are a highly tractable prototype of protein misfolding disorders and provide an invaluable source of insight into similar diseases. Not only can successes in prion drug discovery offer research leads for these related conditions, but the challenges and lessons learned could have a broad impact as well. For example, accumulating evidence suggests that other prion-like proteins such as tau (associated with Alzheimer’s disease and other taupathies) and α-synuclein (associated with Parkinson’s disease and some forms of Alzheimer’s disease) may occur in distinct forms with different disease phenotypes.57,58 While it remains to be seen if these different forms behave like prion strains, the emerging similarities pose the question of whether strain specificity and drug resistance will affect treatment discovery for other neurodegenerative diseases as well.
Acknowledgments
The authors acknowledge the contributions of a large team of researchers whose sustained efforts produced the results detailed in this review and in the primary publications cited herein. The work reviewed in this manuscript was funded by grants from the National Institutes of Health as well as from several private foundations. Detailed funding information is provided in the primary publications cited herein.
Glossary
Abbreviations Used
- CJD
Creutzfeldt–Jakob disease
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- BSE
bovine spongiform encephapathy
- SPR
surface plasmon resonance
- PrPC
endogenous prion protein
- PrPSc
misfolded scrapie prion protein
- ScN2a
scrapie-infected murine neuroblastoma cell
- BLI
bioluminescent imaging
- GFAP
glial fibrillary acidic protein
- Tg(Gfap-luc)
transgenic mice with the luciferase gene under control of the gfap promoter
- ELISA
enzyme-linked immunosorbent assay
- AMT
2-aminothiazole
- VEGFR
vascular endothelial growth factor receptor
- AUC
area under the plasma (or brain) exposure curve
- PK
pharmacokinetic
- PSA
polar surface area
- RML
Rocky Mountain Laboratories
- CWD
chronic wasting disease
- dpi
days postinoculation
Biographies
Sina Ghaemmaghami is an Assistant Professor of Biology at the University of Rochester, NY. He received his Ph.D. from Duke University, NC, and conducted his postdoctoral studies at the University of California, San Francisco. His laborotary studies the regulation of protein folding and degradation and their role in the pathogenesis of neurodegenerative diseases.
Miranda Russo is an undergraduate at the University of Rochester, NY, with an interest in prion biology. She is conducting research on the anti-prion mechanism of 2-aminothiazoles.
Adam R. Renslo is an Associate Professor of Pharmaceutical Chemistry at the University of California, San Francisco. He received his Ph.D. from Massachusetts Institute of Technology, MA, and conducted postdoctoral research at The Scripps Research Institute, CA. He began his career in the pharmaceutical industry where he contributed to the discovery of multiple antibacterial drug candidates from the oxazolidinone class. His laboratory at University of California, San Francisco, applies fragment-based lead discovery to challenging drug targets and is exploring new drug delivery approaches for infectious disease and cancer.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Colby D. W.; Prusiner S. B. Prions. Cold Spring Harbor Perspect Biol. 2011, 3, a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B. Novel proteinaceous infectious particles cause scrapie. Science 1982, 216, 136–144. [DOI] [PubMed] [Google Scholar]
- Prusiner S. B. Shattuck lecture—neurodegenerative diseases and prions. N. Engl. J. Med. 2001, 344, 1516–1526. [DOI] [PubMed] [Google Scholar]
- Brown P.; Cathala F.; Castaigne P.; Gajdusek D. C. Creutzfeldt–Jakob disease: clinical analysis of a consecutive series of 230 neuropathologically verified cases. Ann. Neurol. 1986, 20, 597–602. [DOI] [PubMed] [Google Scholar]
- Roos R.; Gajdusek D. C.; Gibbs C. J. Jr. The clinical characteristics of transmissible Creutzfeldt–Jakob disease. Brain 1973, 96, 1–20. [DOI] [PubMed] [Google Scholar]
- Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci. 2001, 24, 519–550. [DOI] [PubMed] [Google Scholar]
- Weissmann C. The state of the prion. Nat. Rev. Microbiol. 2004, 2, 861–871. [DOI] [PubMed] [Google Scholar]
- Zahn R.; Liu A.; Luhrs T.; Riek R.; von Schroetter C.; Lopez Garcia F.; Billeter M.; Calzolai L.; Wider G.; Wuthrich K. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonati T.; Reimann R. R.; Falsig J.; Baral P. K.; O’Connor T.; Hornemann S.; Yaganoglu S.; Li B.; Herrmann U. S.; Wieland B.; Swayampakula M.; Rahman M. H.; Das D.; Kav N.; Riek R.; Liberski P. P.; James M. N.; Aguzzi A. The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein. Nature 2013, 501, 102–106. [DOI] [PubMed] [Google Scholar]
- Baral P. K.; Swayampakula M.; Rout M. K.; Kav N. N.; Spyracopoulos L.; Aguzzi A.; James M. N. Structural basis of prion inhibition by phenothiazine compounds. Structure 2014, 22, 291–303. [DOI] [PubMed] [Google Scholar]
- Wasmer C.; Lange A.; Van Melckebeke H.; Siemer A. B.; Riek R.; Meier B. H. Amyloid fibrils of the het-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science 2008, 319, 1523–1526. [DOI] [PubMed] [Google Scholar]
- Bueler H.; Fischer M.; Lang Y.; Bluethmann H.; Lipp H. P.; DeArmond S. J.; Prusiner S. B.; Aguet M.; Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface prp protein. Nature 1992, 356, 577–582. [DOI] [PubMed] [Google Scholar]
- Richt J. A.; Kasinathan P.; Hamir A. N.; Castilla J.; Sathiyaseelan T.; Vargas F.; Sathiyaseelan J.; Wu H.; Matsushita H.; Koster J.; Kato S.; Ishida I.; Soto C.; Robl J. M.; Kuroiwa Y. Production of cattle lacking prion protein. Nat. Biotechnol. 2007, 25, 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G.; Chen J.; Xu Y.; Zhu C.; Yu H.; Liu S.; Sha H.; Chen J.; Xu X.; Wu Y.; Zhang A.; Ma J.; Cheng G. Generation of goats lacking prion protein. Mol. Reprod. Dev. 2009, 76, 3. [DOI] [PubMed] [Google Scholar]
- Aguzzi A.; Falsig J. Prion propagation, toxicity and degradation. Nat. Neurosci. 2012, 15, 936–939. [DOI] [PubMed] [Google Scholar]
- Hewitt P. E.; Llewelyn C. A.; Mackenzie J.; Will R. G. Creutzfeldt–Jakob disease and blood transfusion: results of the UK transfusion medicine epidemiological review study. Vox Sang. 2006, 91, 221–230. [DOI] [PubMed] [Google Scholar]
- Otto M.; Cepek L.; Ratzka P.; Doehlinger S.; Boekhoff I.; Wiltfang J.; Irle E.; Pergande G.; Ellers-Lenz B.; Windl O.; Kretzschmar H. A.; Poser S.; Prange H. Efficacy of flupirtine on cognitive function in patients with CJD: a double-blind study. Neurology 2004, 62, 714–718. [DOI] [PubMed] [Google Scholar]
- Geschwind M. D.; Kuo A. L.; Wong K. S.; Haman A.; Devereux G.; Raudabaugh B. J.; Johnson D. Y.; Torres-Chae C. C.; Finley R.; Garcia P.; Thai J. N.; Cheng H. Q.; Neuhaus J. M.; Forner S. A.; Duncan J. L.; Possin K. L.; Dearmond S. J.; Prusiner S. B.; Miller B. L. Quinacrine treatment trial for sporadic Creutzfeldt–Jakob disease. Neurology 2013, 81, 2015–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haik S.; Marcon G.; Mallet A.; Tettamanti M.; Welaratne A.; Giaccone G.; Azimi S.; Pietrini V.; Fabreguettes J. R.; Imperiale D.; Cesaro P.; Buffa C.; Aucan C.; Lucca U.; Peckeu L.; Suardi S.; Tranchant C.; Zerr I.; Houillier C.; Redaelli V.; Vespignani H.; Campanella A.; Sellal F.; Krasnianski A.; Seilhean D.; Heinemann U.; Sedel F.; Canovi M.; Gobbi M.; Di Fede G.; Laplanche J. L.; Pocchiari M.; Salmona M.; Forloni G.; Brandel J. P.; Tagliavini F. Doxycycline in Creutzfeldt–Jakob disease: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2014, 13, 150–158. [DOI] [PubMed] [Google Scholar]
- Geissen M.; Leidel F.; Eiden M.; Hirschberger T.; Fast C.; Bertsch U.; Tavan P.; Giese A.; Kretzschmar H.; Schatzl H. M.; Groschup M. H. From high-throughput cell culture screening to mouse model: identification of new inhibitor classes against prion disease. ChemMedChem 2011, 6, 1928–1937. [DOI] [PubMed] [Google Scholar]
- Vilette D. Cell models of prion infection. Vet. Res. 2008, 39, 10. [DOI] [PubMed] [Google Scholar]
- Safar J. G.; Geschwind M. D.; Deering C.; Didorenko S.; Sattavat M.; Sanchez H.; Serban A.; Vey M.; Baron H.; Giles K.; Miller B. L.; Dearmond S. J.; Prusiner S. B. Diagnosis of human prion disease. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 3501–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner S.; Klein M. A.; Frigg R.; Pekarik V.; Parizek P.; Raeber A.; Glatzel M.; Schwarz P.; Rulicke T.; Weissmann C.; Aguzzi A. Neuroinvasion of prions: insights from mouse models. Exp. Physiol. 2000, 85, 705–712. [DOI] [PubMed] [Google Scholar]
- Tamguney G.; Francis K. P.; Giles K.; Lemus A.; DeArmond S. J.; Prusiner S. B. Measuring prions by bioluminescence imaging. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 15002–15006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwata K.; Nishida N.; Matsumoto T.; Kamatari Y. O.; Hosokawa-Muto J.; Kodama K.; Nakamura H. K.; Kimura K.; Kawasaki M.; Takakura Y.; Shirabe S.; Takata J.; Kataoka Y.; Katamine S. Hot spots in prion protein for pathogenic conversion. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 11921–11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevitt C. R.; Collinge J. A systematic review of prion therapeutics in experimental models. Brain 2006, 129, 2241–2265. [DOI] [PubMed] [Google Scholar]
- Sim V. L. Prion disease: chemotherapeutic strategies. Infect. Disord.: Drug Targets 2012, 12, 144–160. [DOI] [PubMed] [Google Scholar]
- Kamatari Y. O.; Hayano Y.; Yamaguchi K.; Hosokawa-Muto J.; Kuwata K. Characterizing antiprion compounds based on their binding properties to prion proteins: implications as medical chaperones. Protein Sci. 2013, 22, 22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S.; Ahn M.; Lessard P.; Giles K.; Legname G.; DeArmond S. J.; Prusiner S. B. Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 2009, 5, e1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forloni G.; Iussich S.; Awan T.; Colombo L.; Angeretti N.; Girola L.; Bertani I.; Poli G.; Caramelli M.; Grazia Bruzzone M.; Farina L.; Limido L.; Rossi G.; Giaccone G.; Ironside J. W.; Bugiani O.; Salmona M.; Tagliavini F. Tetracyclines affect prion infectivity. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 10849–10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teruya K.; Kawagoe K.; Kimura T.; Chen C. J.; Sakasegawa Y.; Doh-ura K. Amyloidophilic compounds for prion diseases. Infect. Disord.: Drug Targets 2009, 9, 15–22. [DOI] [PubMed] [Google Scholar]
- Moreno J. A.; Halliday M.; Molloy C.; Radford H.; Verity N.; Axten J. M.; Ortori C. A.; Willis A. E.; Fischer P. M.; Barrett D. A.; Mallucci G. R. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med. 2013, 5, 206ra138. [DOI] [PubMed] [Google Scholar]
- Axten J. M.; Medina J. R.; Feng Y.; Shu A.; Romeril S. P.; Grant S. W.; Li W. H.; Heerding D. A.; Minthorn E.; Mencken T.; Atkins C.; Liu Q.; Rabindran S.; Kumar R.; Hong X.; Goetz A.; Stanley T.; Taylor J. D.; Sigethy S. D.; Tomberlin G. H.; Hassell A. M.; Kahler K. M.; Shewchuk L. M.; Gampe R. T. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J. Med. Chem. 2012, 55, 7193–7207. [DOI] [PubMed] [Google Scholar]
- Wagner J.; Ryazanov S.; Leonov A.; Levin J.; Shi S.; Schmidt F.; Prix C.; Pan-Montojo F.; Bertsch U.; Mitteregger-Kretzschmar G.; Geissen M.; Eiden M.; Leidel F.; Hirschberger T.; Deeg A.; Krauth J.; Zinth W.; Tavan P.; Pilger J.; Zweckstetter M.; Frank T.; Bähr M.; Weishaupt J.; Uhr M.; Urlaub H.; Teichmann U.; Samwer M.; Bötzel K.; Groschup M.; Kretzschmar H.; Griesinger C.; Giese A. Anle138b: a novel oligomer modulator for disease-modifying therapy of neurodegenerative diseases such as prion and Parkinson’s disease. Acta Neuropathol. 2013, 125, 795–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D. B.; Lu D.; Geva M.; Watts J. C.; Bhardwaj S.; Oehler A.; Renslo A. R.; DeArmond S. J.; Prusiner S. B.; Giles K. Drug resistance confounding prion therapeutics. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, E4160–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocisko D. A.; Baron G. S.; Rubenstein R.; Chen J.; Kuizon S.; Caughey B. New inhibitors of scrapie-associated prion protein formation in a library of 2000 drugs and natural products. J. Virol. 2003, 77, 10288–10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsch U.; Winklhofer K. F.; Hirschberger T.; Bieschke J.; Weber P.; Hartl F. U.; Tavan P.; Tatzelt J.; Kretzschmar H. A.; Giese A. Systematic identification of antiprion drugs by high-throughput screening based on scanning for intensely fluorescent targets. J. Virol. 2005, 79, 7785–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S.; May B. C.; Renslo A. R.; Prusiner S. B. Discovery of 2-aminothiazoles as potent antiprion compounds. J. Virol. 2010, 84, 3408–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalgutkar A. S.; Driscoll J.; Zhao S. X.; Walker G. S.; Shepard R. M.; Soglia J. R.; Atherton J.; Yu L.; Mutlib A. E.; Munchhof M. J.; Reiter L. A.; Jones C. S.; Doty J. L.; Trevena K. A.; Shaffer C. L.; Ripp S. L. A rational chemical intervention strategy to circumvent bioactivation liabilities associated with a nonpeptidyl thrombopoietin receptor agonist containing a 2-amino-4-arylthiazole motif. Chem. Res. Toxicol. 2007, 20, 1954–1965. [DOI] [PubMed] [Google Scholar]
- Borzilleri R. M.; Bhide R. S.; Barrish J. C.; D’Arienzo C. J.; Derbin G. M.; Fargnoli J.; Hunt J. T.; Jeyaseelan R. Sr.; Kamath A.; Kukral D. W.; Marathe P.; Mortillo S.; Qian L.; Tokarski J. S.; Wautlet B. S.; Zheng X.; Lombardo L. J. Discovery and evaluation of N-cyclopropyl-2,4-difluoro-5-((2-(pyridin-2-ylamino)thiazol-5-ylmethyl)amino)benzamide (BMS-605541), a selective and orally efficacious inhibitor of vascular endothelial growth factor receptor-2. J. Med. Chem. 2006, 49, 3766–3769. [DOI] [PubMed] [Google Scholar]
- Das J.; Chen P.; Norris D.; Padmanabha R.; Lin J.; Moquin R. V.; Shen Z.; Cook L. S.; Doweyko A. M.; Pitt S.; Pang S.; Shen D. R.; Fang Q.; de Fex H. F.; McIntyre K. W.; Shuster D. J.; Gillooly K. M.; Behnia K.; Schieven G. L.; Wityak J.; Barrish J. C. 2-Aminothiazole as a novel kinase inhibitor template. Structure–activity relationship studies toward the discovery of N-(2-chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide (dasatinib, BMS-354825) as a potent pan-Src kinase inhibitor. J. Med. Chem. 2006, 49, 6819–6832. [DOI] [PubMed] [Google Scholar]
- Press N. J.; Taylor R. J.; Fullerton J. D.; Tranter P.; McCarthy C.; Keller T. H.; Brown L.; Cheung R.; Christie J.; Haberthuer S.; Hatto J. D.; Keenan M.; Mercer M. K.; Press N. E.; Sahri H.; Tuffnell A. R.; Tweed M.; Fozard J. R. A new orally bioavailable dual adenosine a2b/a3 receptor antagonist with therapeutic potential. Bioorg. Med. Chem. Lett. 2005, 15, 3081–3085. [DOI] [PubMed] [Google Scholar]
- van Muijlwijk-Koezen J. E.; Timmerman H.; Vollinga R. C.; Frijtag von Drabbe Kunzel J.; de Groote M.; Visser S.; Ijzerman A. P. Thiazole and thiadiazole analogues as a novel class of adenosine receptor antagonists. J. Med. Chem. 2001, 44, 749–762. [DOI] [PubMed] [Google Scholar]
- Turcotte S.; Chan D. A.; Sutphin P. D.; Hay M. P.; Denny W. A.; Giaccia A. J. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell 2008, 14, 90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbers T.; Flohr A.; Jolidon S.; David-Pierson P.; Jacobsen H.; Ozmen L.; Baumann K. Aminothiazoles as gamma-secretase modulators. Bioorg. Med. Chem. Lett. 2011, 21, 6554–6558. [DOI] [PubMed] [Google Scholar]
- Kounnas M. Z.; Danks A. M.; Cheng S.; Tyree C.; Ackerman E.; Zhang X.; Ahn K.; Nguyen P.; Comer D.; Mao L.; Yu C.; Pleynet D.; Digregorio P. J.; Velicelebi G.; Stauderman K. A.; Comer W. T.; Mobley W. C.; Li Y. M.; Sisodia S. S.; Tanzi R. E.; Wagner S. L. Modulation of gamma-secretase reduces beta-amyloid deposition in a transgenic mouse model of Alzheimer’s disease. Neuron 2010, 67, 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo-Godoy A.; Gever J.; Fife K. L.; Silber B. M.; Prusiner S. B.; Renslo A. R. 2-Aminothiazoles as therapeutic leads for prion diseases. J. Med. Chem. 2011, 54, 1010–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Silber B. M.; Rao S.; Gever J. R.; Bryant C.; Gallardo-Godoy A.; Dolghih E.; Widjaja K.; Elepano M.; Jacobson M. P.; Prusiner S. B.; Renslo A. R. 2-Aminothiazoles with improved pharmacotherapeutic properties for treatment of prion disease. ChemMedChem 2013, 8, 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S.; Ullman J.; Ahn M.; St Martin S.; Prusiner S. B. Chemical induction of misfolded prion protein conformers in cell culture. J. Biol. Chem. 2010, 285, 10415–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber B. M.; Rao S.; Fife K. L.; Gallardo-Godoy A.; Renslo A. R.; Dalvie D. K.; Giles K.; Freyman Y.; Elepano M.; Gever J. R.; Li Z.; Jacobson M. P.; Huang Y.; Benet L. Z.; Prusiner S. B. Pharmacokinetics and metabolism of 2-aminothiazoles with antiprion activity in mice. Pharm. Res. 2013, 30, 932–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock S. A.; Pennington L. D. Structure–brain exposure relationships. J. Med. Chem. 2006, 49, 7559–7583. [DOI] [PubMed] [Google Scholar]
- Lu D.; Giles K.; Li Z.; Rao S.; Dolghih E.; Gever J. R.; Geva M.; Elepano M. L.; Oehler A.; Bryant C.; Renslo A. R.; Jacobson M. P.; Dearmond S. J.; Silber B. M.; Prusiner S. B. Biaryl amides and hydrazones as therapeutics for prion disease in transgenic mice. J. Pharmacol. Exp. Ther. 2013, 347, 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Browning S.; Mahal S. P.; Oelschlegel A. M.; Weissmann C. Darwinian evolution of prions in cell culture. Science 2010, 327, 869–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Vedam L.; Ghaemmaghami S. Strain specificity and drug resistance in anti-prion therapy. Curr. Top. Med. Chem. 2013, 13, 2397–2406. [DOI] [PubMed] [Google Scholar]
- Prusiner S. B. Cell biology. A unifying role for prions in neurodegenerative diseases. Science 2012, 336, 1511–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker M.; Walker L. C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 2013, 501, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. L.; Covell D. J.; Daniels J. P.; Iba M.; Stieber A.; Zhang B.; Riddle D. M.; Kwong L. K.; Xu Y.; Trojanowski J. Q.; Lee V. M. Distinct alpha-synuclein strains differentially promote tau inclusions in neurons. Cell 2013, 154, 103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavaguera F.; Akatsu H.; Fraser G.; Crowther R. A.; Frank S.; Hench J.; Probst A.; Winkler D. T.; Reichwald J.; Staufenbiel M.; Ghetti B.; Goedert M.; Tolnay M. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 9535–9540. [DOI] [PMC free article] [PubMed] [Google Scholar]