Abstract
Sequencing technology is increasingly demonstrating the impact of genomic copy number variation (CNV) on phenotypes. Opposing variation in growth, head size, cognition and behaviour is known to result from deletions and reciprocal duplications of some genomic regions. We propose normative inversion of face shape, opposing difference from a matched norm, as a basis for investigating the effects of gene dosage on craniofacial development. We use dense surface modelling techniques to match any face (or part of a face) to a facial norm of unaffected individuals of matched age, sex and ethnicity and then we reverse the individual’s face shape differences from the matched norm to produce the normative inversion. We demonstrate for five genomic regions, 4p16.3, 7q11.23, 11p15, 16p13.3 and 17p11.2, that such inversion for individuals with a duplication or (epi)-mutation produces facial forms remarkably similar to those associated with a deletion or opposite (epi-)mutation of the same region, and vice versa. The ability to visualise and quantify face shape effects of gene dosage is of major benefit for determining whether a CNV is the cause of the phenotype of an individual and for predicting reciprocal consequences. It enables face shape to be used as a relatively simple and inexpensive functional analysis of the gene(s) involved.
Electronic supplementary material
The online version of this article (doi:10.1007/s00439-014-1455-z) contains supplementary material, which is available to authorized users.
Introduction
In recent years, genome-wide screening technologies have helped identify large numbers of genomic structural variants (GSVs). Duplications and deletions, usually indicated as copy number variants (CNVs), are the most prevalent GSVs and have been shown to make an important contribution to development and disease (Valsesia et al. 2013; Weischenfeldt et al. 2013). CNVs have been associated with mirrored or opposing phenotypes at several loci. For example, CNVs of the 7q11.23 Williams-Beuren syndrome (OMIM: #194050) region cause neuro developmental disorders with a multi-faceted phenotype and variable expressivity. Typically, individuals with 7q11.23 micro deletions have specific facial dysmorphism, supravalvular aortic stenosis, hypercalcaemia and a distinctive cognitive profile including heightened sociability and relative strength of language over visuo-spatial processing. In contrast, the 7q11.23 reciprocal duplication results in different facial dysmorphism, low sociability and prominent speech delay (Schubert 2009; Merla et al. 2010). This deletion-duplication opposing nature of a phenotype also occurs for 17p11.2. Individuals with Smith-Magenis syndrome (OMIM: #182290), caused by a deletion in 17p11.2 or point mutation in RAI at 17p11.2, exhibit attention-seeking and overt friendliness (Potocki et al. 2000, 2007; Lupski and Stankiewicz 2005). In contrast, the reciprocal duplication causes Potocki-Lupski syndrome where behaviour is characterized by autism spectrum disorders. Opposing over/undergrowth effects result from deletions (Sotos syndrome (OMIM: #117550)) and duplications of NSD1 at 5q35 (Zhang et al. 2011; Rosenfeld et al. 2013; Žilina et al. 2013; Franco et al. 2010; Dikow et al. 2013). Opposing extreme BMI phenotypes have also been associated with gene dosage at 16p11.2 (Jacquemont et al. 2011). In the two latter regions, there is also opposing microcephaly and macrocephaly. Hypomethylation of imprinting control region 1 at 11p15 and maternal duplication of 11p15 have been described as major (epi) genetic disturbances in Silver-Russell syndrome (OMIM: # 180860) resulting in severe undergrowth. Opposite (epi)-mutations involved in Beckwith-Wiedemann syndrome (OMIM: # 130650) cause overgrowth, suggesting that Silver-Russell and Beckwith-Wiedemann syndromes are genetically and clinically opposite (Eggermann 2009).
Based on these observations, we hypothesised that in opposite CNV pairings, there could be quantitatively opposite facial phenotypes. Facial morphology is determined in part by a large number of genes and enhancers acting in concert, and a decrease in dosage in some genes will lead to abnormal morphology at various parts of the face. An increase in dosage may therefore lead to a related abnormal morphology at the same parts of the face. To test this hypothesis, we developed a transformation, normative inversion, for reversing the differences of an individual’s 3D face shape from a facial norm, the average of an age-sex-ethnicity matched control group.
Methods
Study participants
3D facial images were available for 387 Caucasian controls and individuals with Williams-Beuren syndrome, Smith-Magenis syndrome and Wolf-Hirschhorn syndrome (OMIM: # 194190) from previous studies (Hammond et al., 2004, 2005, 2012). Individuals with Rubinstein-Taybi syndrome were recruited during attendance at national family support group meetings in the Netherlands, Norway and UK. Through clinical co-authors, we also recruited individuals specifically for this study with confirmed mutations causing Beckwith-Wiedemann (n = 1) and Silver-Russell syndromes (n = 17) as well as individuals with associated duplications at 4p16.3 (n = 1), 7q11.23 (n = 4), 16p13.3 (n = 1) and 17p11.2 (n = 1). All subjects were of Caucasian origin except for one duplication 7q11.23 case which was of North African (Moroccan) origin.
Normative inversion of facial morphology
Initially, we constructed a dense surface model (DSM) of the 3D facial images of individuals with an identified syndrome or CNV and the 387 controls (Hammond and Suttie 2012). In such models, we retain sufficient principle component analysis (PCA) modes to cover 99 % of all shape variation. The inclusion of a large number of controls and a high proportion of PCA modes enables accurate synthesis of faces in the model.
The subset of controls of the same ethnicity and gender were ordered by age and a series of running mean faces of 50 contiguously aged individuals were computed. This enabled every face to be matched to a same ethnicity-gender facial norm of closely matched age. Within the multi-dimensional DSM representation, all faces were projected onto a hyperline from the face being inverted based at (1, 0) to the matched norm based at (−1, 0). The line was extended to a point the same distance away at (−3, 0) where the face shape corresponds to an inverted facial form whose shape differences from the norm are exactly opposite to those of the original face. Such a normative inversion can be applied to any face.
A similar notion of facial inversion, termed anti-face, was previously used to investigate psychological aspects of the perception of face shape differences (Blanz et al. 2000). In this earlier study, various mean faces were employed to compute an opposite face shape, sometimes even based on a mixed set of male and female individuals. Therefore, we retain the use of “normative inversion” in order to emphasize the age-sex-ethnicity matched nature of the mean used in our inversion process.
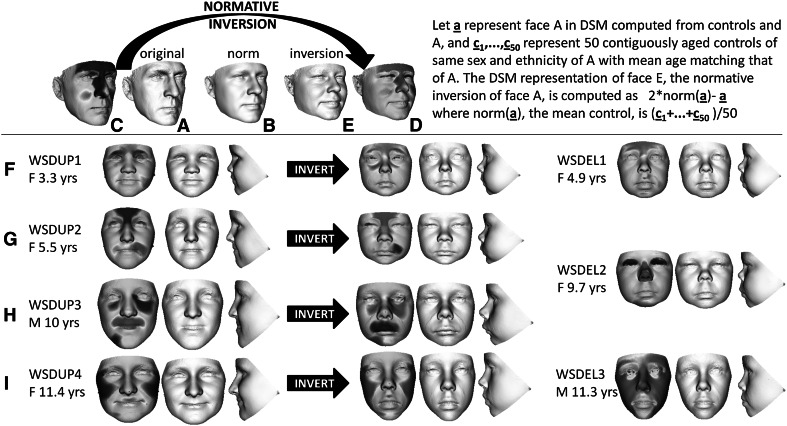
The face signature of an individual is the term used for its 3D shape difference from its matched norm (Hammond and Suttie 2012). It can be visualised as a heat map reflecting the normalized differences at each of 25,000 + surface points captured by commercial 3D imaging devices. Figure 1a, c show the face surface and signature heat map of one of the authors where the red–green–blue spectrum highlights regional differences (contraction-coincidence-expansion) orthogonal to the face surface compared to the matched norm. The extremes of red and blue reflect normalised differences of ± 2 SDs or more. Figure 1b shows the matched facial norm and Fig. 1d, e the surface and heat map of the normative inversion of A with respect to B. We apply an analogous transformation to all faces of interest.
Fig. 1.
Normative inversion of face shape. a The face surface of an adult Caucasian male control. b The average face of 50 adult male Caucasians whose mean age matches that of a. c A heat map of the face signature of a normalised against the 50 individuals whose average is b. d The inverted heat map of c. e. The face surface whose face signature has heat map d. f Left to right a triptych of face signature, portrait and profile of individual 1 with a duplication of 7q11.23; then the normative inversion of the duplication case; and finally an individual with a confirmed deletion of 7q11.23 whose face closely resembles the inversion. g Left to right a triptych of face signature, portrait and profile of individual 2 with a duplication of 7q11.23; then the normative inversion of the duplication case; and finally an individual with a confirmed deletion of 7q11.23 whose face closely resembles the inversion. h Left to right a triptych of face signature, portrait and profile of individual 3 with a duplication of 7q11.23; then the normative inversion of the duplication case. i Left to right a triptych of face signature, portrait and profile of individual 4 with a duplication of 7q11.23; then the normative inversion of the duplication case; and finally an individual with a confirmed deletion of 7q11.23 whose face closely resembles the inversion. Note that in the original and inverted face signatures of rows f–h, the red–green–blue of the heat maps are opposite with red and blue regions interchanged. The red–green–blue spectrum in all images represents regions of contraction-coincidence-expansion relative orthogonal to the face surface of the matched norm with extreme red-blue indicating difference beyond 2SD (color figure online)
Results
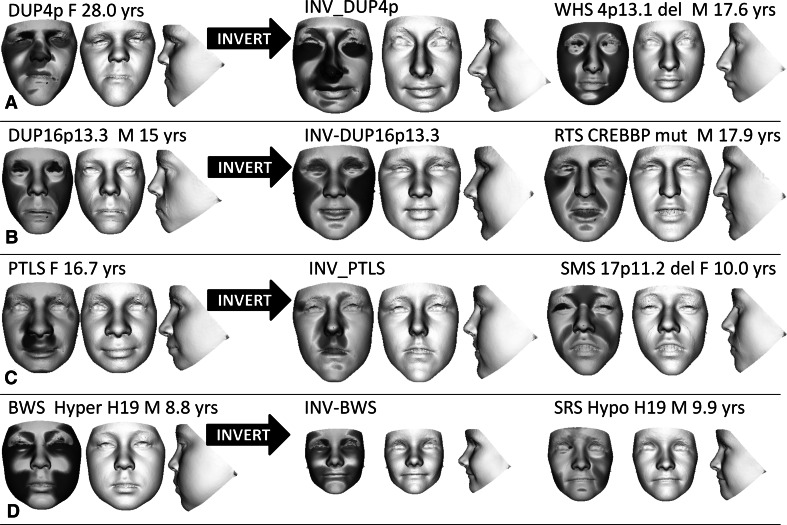
In Fig. 1f–i, we demonstrate the effect of inverting the face shape of three unpublished individuals and one previously published individual with a duplication of 7q11.23 (Malenfant et al. 2012). Clinically, the features of the normative inversions of their faces strongly resemble those of Williams-Beuren syndrome (flat nasal bridge, short upturned nose, long philtrum, full lips, malar flattening, and micro/retrognathia). This similarity is further emphasised by their comparison with three adjacent faces of individuals with a confirmed 7q11.23 Williams-Beuren syndrome deletion. To test normative face inversion further, the procedure was repeated for individuals with duplications of 4p16.3, 16p13.3 and 17p11.1, and with Beckwith-Wiedemann syndrome caused by H19 hypermethylation. Clinical evaluation of the normative inversions of facial features of these cases establishes their similarity to facial characteristics of individuals with Wolf-Hirschhorn (Fig. 2a), Rubinstein-Taybi (Fig. 2b), Smith–Magenis (Fig. 2c) and Silver–Russell syndromes due to H19 hypomethylation (Fig. 2d), respectively.
Fig. 2.
Normative inversion of duplications of 4p16.3, 16p13.3, 17p11.2 and H19 hypermethylation Beckwith-Wiedemann cases. Using the same format as Fig. 1, each row includes a face triptych comprising signature, portrait and profile of a duplication (4p16.3, 16p13.3, 17p11.2) or hypermethylation (Beckwith-Wiedemann syndrome) case; the normative inversion of the original; and, finally a triptych for an individual with a deletion or mutation (Wolf–Hirschhorn syndrome, Rubinstein–Taybi syndrome, Smith–Magenis syndrome) or opposite methylation (Silver–Russell syndrome) whose face shape closely resembles that of the original case. The inversions of the dup 4p16.3 and Beckwith–Wiedemann cases display strong similarity respectively with features of Wolf–Hirschhorn and Silver–Russell syndromes. The inversion of the dup 16p13.3 case displays Rubinstein–Taybi features such as hooded eyes, significant convexity of the zygomatic arch and exposure of the columella. Although somewhat narrower than is usual in Smith–Magenis syndrome, the inversion of the Potocki–Lupski case displays the typical upward curve to the upward lip, the hidden columella and mid-facial flatness
A convincing objective confirmation of these clinical observations would be to demonstrate the effect of normative inversion along a duplication-deletion axis—for example, classifying each duplication case and its inverse relative to proximity to the mean duplication and mean deletion cases (taking care to omit the test individual from the calculation of the mean duplication). But there are an insufficient number of identified duplication cases available and so a compromise was to measure the effect of inversion of the face of an individual with a duplication along a control-deletion axis. A moment’s reflection confirms that as a result of normative inversion there would be much stronger movement of closest mean classification along a duplication-deletion axis than along a control-deletion axis, as the duplication group mean would be more “repulsing” to the inverted duplication. But until more duplication cases can be identified, the control-deletion axial comparison will have to substitute for the duplication-deletion axial comparison.
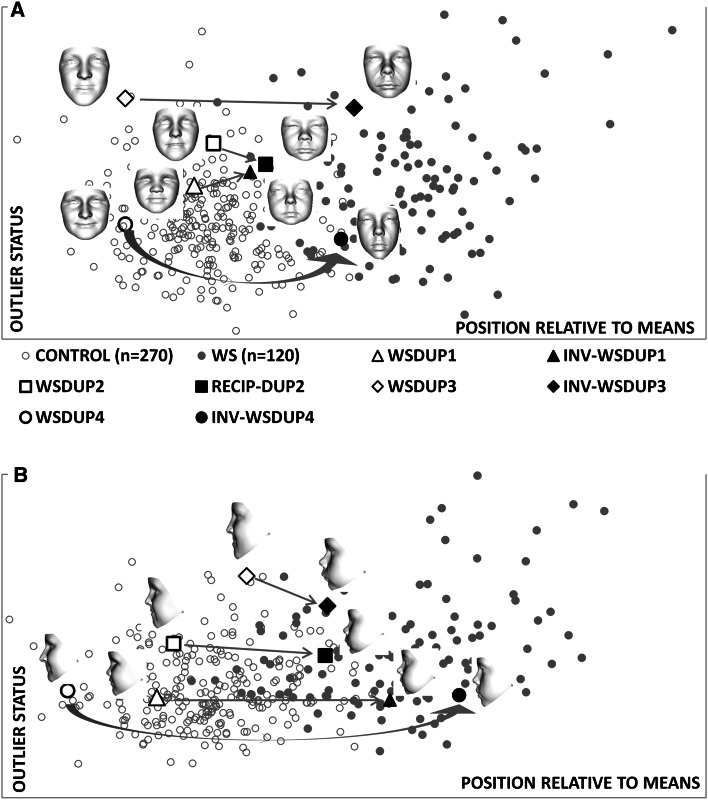
We computed the change in relative similarity to the average faces of controls and individuals with Williams–Beuren syndrome resulting from normative inversion of the faces of each duplication 7q11.23 case. Using the full face without ears, the inverted facial forms are classified at the periphery of, or within, the Williams-Beuren syndrome cluster (Fig. 3a). Thus, normative inversion produces considerable position change from proximity to the average control towards the average Williams–Beuren syndrome face. For a thin ribbon-like surface along the mid-line facial profile (see Fig. S1 in supplementary material), normative inversion of all three duplication 7q11.23 cases results in even stronger classification within the Williams-Beuren syndrome cluster (Fig. 3b). These quantitative results confirm the clinical interpretation of the inverted face signatures as being Williams–Beuren syndrome-like, especially in the facial mid-line.
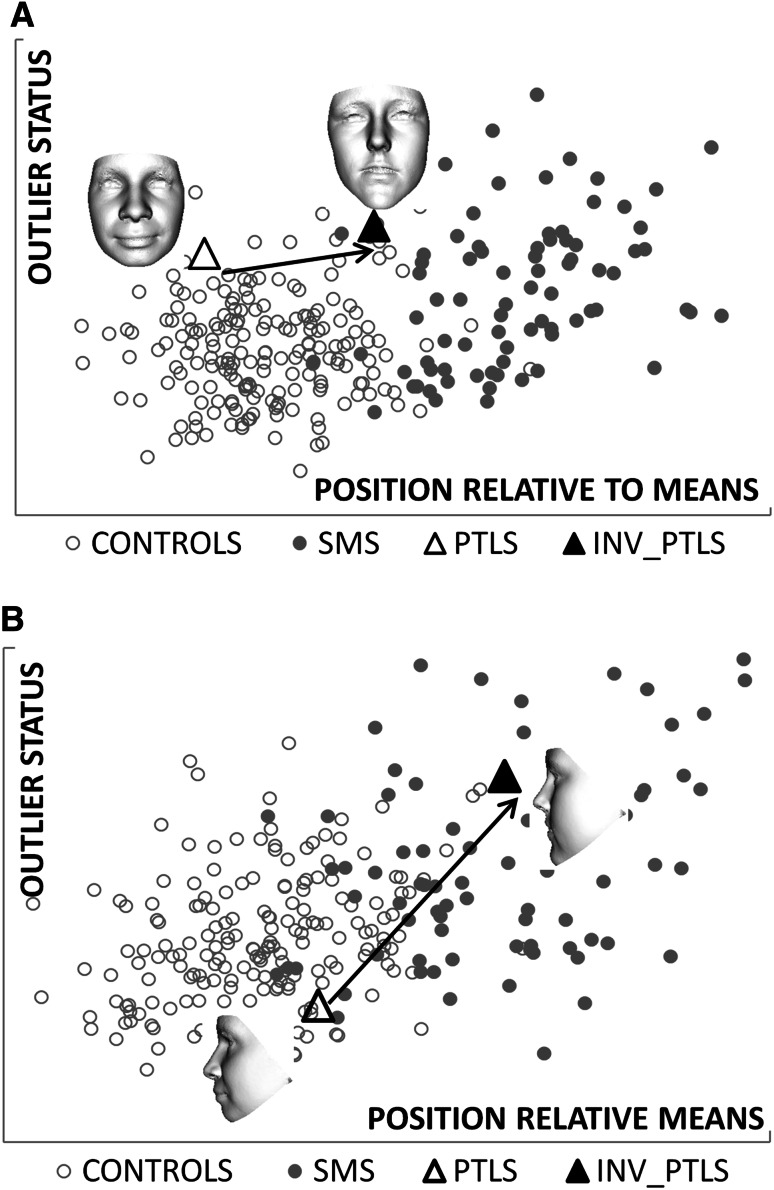
Fig. 3.
Closest mean classification of inversions of dup7q11.23 cases. The arrows emphasise position change in closest mean classification for the faces of the duplication cases in Fig. 1 f–h and their normative inversions. The horizontal axis determines relative similarity to the mean of the control group compared to the mean of the affected group. The vertical axis reflects the outlier status in terms of distance from the hyperline linking the means of the two groups under comparison. In a, the inversions of two duplication cases are classified at the periphery of the Williams–Beuren syndrome cluster. A third duplication inverts to well within the Williams–Beuren syndrome cluster. In b, when only the curvilinear mid-line facial profile is considered, all inversions are within the Williams–Beuren syndrome cluster. This is consistent with clinical evaluation suggesting the inverted faces to be somewhat wider than the typical Williams-Beuren syndrome facies but very Williams–Beuren syndrome-like in nose, lips and mid-line profile
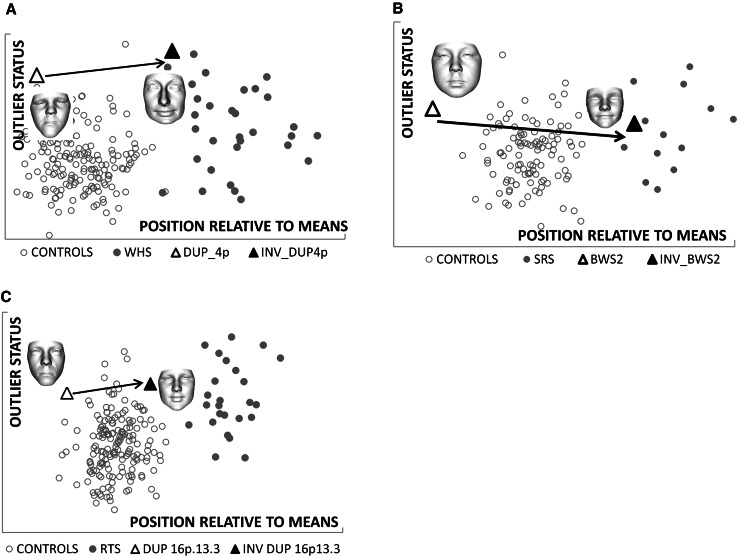
Classification of face shape using the closest mean algorithm also produced positive recognition of the relevant facial characteristics for the other four genomic regions (Figs. 4, 5). For the Smith–Magenis and Potocki–Lupski syndromes comparison at 17p11.2, an additional shape classification for the mid-line facial profile was undertaken to demonstrate once again that shape inversion of a more localised region can produce more convincing objective results (Fig. 5).
Fig. 4.
Closest mean classification of face for individuals with duplications of 4p16.3, 16p13.3 and H19 hypermethylation, and their normative inversions. Scatter plots a–c depict the results of closest mean classification of paired groups: controls and affected individuals with Wolf–Hirschhorn syndrome, Silver–Russell syndrome or Rubinstein–Taybi syndrome. The arrows indicate the change in closest mean classification position of an individual and their normative inversion. In each case, the result of the normative inversion is to alter the original face to be more like individuals in the affected group. Each scatter provides quantitative corroboration of the clinical evaluation of the face shape change resulting from the inversion
Fig. 5.
Closest mean classification of face and mid-line profile for individual with duplication of 17p11.2 and their normative inversions. Scatter plots a and b depict the result of closest mean classification of controls and individuals with Smith–Magenis syndrome. The arrows indicate the change in closest mean classification position of an individual and their normative inversion. In each case, the result of the normative inversion is to alter the original face or mid-line profile to be more like individuals in the affected group. Each scatter provides quantitative corroboration of the clinical evaluation of the shape change resulting from the inversion
The paucity of cases with duplications meant that we could not even carry out a compromise control-duplication axial comparison to provide more objective evidence of the effect of normative facial inversion on deletion cases. Instead, we applied normative inversion to individuals with Wolf–Hirschhorn, Williams–Beuren, Silver–Russell, Rubinstein–Taybi and Smith–Magenis syndromes to demonstrate the similarity of their inversions to individuals in the published literature with corresponding duplications/mutations at 4p16.3, 7q11.23, 11p15, 16p13.3 and 17p11.2 (supplementary Figs. S2–S6). In each of these supplementary figures, normative inversion is the only process that has been applied and only to the faces of the individuals with Wolf–Hirschhorn, Williams–Beuren, Silver–Russell, Rubinstein–Taybi and Smith–Magenis syndromes. To demonstrate the general effect of normative inversion, we also generated animated morphs between an average syndromic face and its normative inversion for each of Rubinstein-Taybi, Silver-Russell, Smith–Magenis, Williams–Beuren and Wolf–Hirschhorn syndromes (Supplementary videos SV1–SV5).
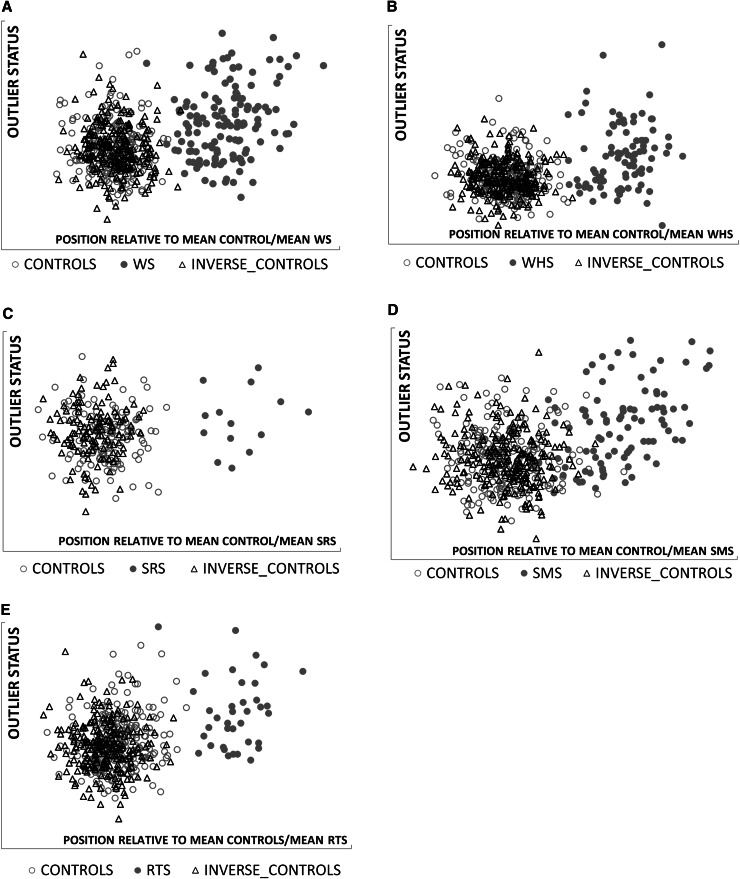
In order to check the effect of normative inversion on unaffected controls, we used multi-folded cross validation, employing closest mean classification to determine discriminating differences between 387 controls and their inverted forms. The average discrimination rate of 20 %, much lower than even chance classification, demonstrates that as a group normative inversions of control faces are indistinguishable from originals i.e., they fall within typical facial growth and development. Finally, to detect any possibility of facial inversion producing features similar to those of Williams–Beuren syndrome, we also tested closest mean classification of normatively inverted controls in a control-Williams–Beuren syndrome combined DSM. The resulting classification clearly demonstrates that inversion does not introduce Williams–Beuren-like facial characteristics (Fig. 6a). This was repeated for Wolf–Hirschhorn, Silver–Russell, Rubinstein–Taybi and Smith–Magenis syndromes, all with similar negative results (Fig. 6b–e).
Fig. 6.
Closest mean classification of inverted controls against original controls and affected subgroups. Each scatter shows the closest mean classification of the normative inversion of controls with respect to the means of the original control and syndrome subgroup. In each case, the inverted controls cluster with the original controls and do not show evidence of facial features of the syndrome. a: Williams–Beuren syndrome b: Wolf–Hirschhorn syndrome c: Silver–Russell syndrome d: Smith–Magenis syndrome e: Rubinstein–Taybi syndrome
Conclusion
We have demonstrated the efficacy of normative inversion of facial form in the investigation of gene-dosage effects at 4p16.3, 7q11.23, 11p15, 16p13.3 and 17p11.2. There is a resemblance between normatively inverted faces of individuals with a duplication or (epi-)mutation and the facial characteristics of deletion or mutation cases, and vice versa. On chromosome 4p16.3, the genes TACC3, FGFR3, and LETM1 have been shown to be dosage sensitive (Cyr et al. 2011). Two related genes on 7q11.23, GTF2IRD1 and GTF2I, have been implicated in the cause of craniofacial dysmorphism in Williams–Beuren syndrome and there is evidence of dosage-sensitivity (Tassabehji et al. 2006). BAZ1B has also been implicated in craniofacial development in Williams–Beuren syndrome (Ashe et al. 2008), although its role is still unclear (Yoshimura et al. 2009). Dosage sensitivity of CREBBP was established by the identification of low-level mosaic individuals with a typical Rubinstein–Taybi syndrome phenotype (Gervasini et al. 2007). Mouse studies of RAI1 have also identified opposite behavioural phenotypes with respect to either deletion or duplication (Carmona-Mora and Waltz 2010). Here, we have demonstrated that face shape inversion can be used to investigate how diametric changes in gene dosage influence craniofacial form.
In studying duplication cases, clinicians often remain uncertain about their pathogenicity, and it is often difficult to determine reliably whether or not the facial dysmorphism in a patient is consistent. It might be useful, therefore, to compare the inverted face of an individual with a duplication of uncertain significance to faces of individuals with deletions or mutations in genes of the same region in whom the phenotype has been more clearly defined. This approach would, for example, be useful for screening individuals with uncertain CNVs recorded in on-line databases such as Decipher (Firth et al. 2009). Conversely, the inverted faces of individuals with a deletion or a mutation could be a useful visualisation of possible facial features associated with duplications of associated genomic regions, especially those containing known dosage sensitive genes, which should assist recognition.
The inverted facial form we have prescribed is simply defined but by the same token is a rather gross transformation to apply across the entire face. More localised application, for example to facial profile or perinasal and periorbital regions, will sometimes be more appropriate. Larger numbers of age, sex and ethnicity matched controls will improve the accuracy of matched norms and normative inversions. Animal studies, such as the recent linkage of non-coding regions to facial form (Attanasio et al. 2013), using normative transformation of facial and cranial structures will be an appropriate route for determining where, to what degree, and at what stage, specific genes produce dosage-sensitive effects on facial, cranial and potentially brain development (Crespi 2013). Our results have taken an initial step in demonstrating the use of normative inversion of human faces in the study of gene dosage sensitivity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
One Microsoft WORD file containing six figures (SUPPLEMENTARY_MATERIAL_Hammond_FEB_18th_2014.docx).
One zipped file of separate PDF files describing permission rights for the supplementary figures previously published elsewhere.
Five animated morphs from an average syndromic face to its normative inversion for each of Rubinstein-Taybi, Silver-Russell, Smith-Magenis, Williams-Beuren and Wolf-Hirschhorn syndromes.
Acknowledgments
The authors sincerely thank the parents and children who agreed to be part of the study and who provided signed informed consent; the Smith-Magenis (France, UK and USA), Rubinstein–Taybi (Netherlands, Norway and UK), Williams–Beuren (UK and USA) and Wolf–Hirschhorn (UK and USA) syndrome support groups who allowed images to be captured during their meetings; and UNIQUE (www.rarechromo.org) who also assisted with recruitment.
Funding
This work was partly supported by the US National Institute on Alcoholism and Alcohol Abuse with a grant to PH (2U01AA014809) as part of the CIFASD consortium (www.cifasd.org).
Ethical standard
EDI/EDH JREC 00/E042.
References
- Ashe A, Morgan DK, Whitelaw NC, Bruxner TJ, Vickaryous NK, Cox LL, Butterfield NC, Wicking C, Blewitt ME, Wilkins SJ, et al. A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol. 2008;9:R182. doi: 10.1186/gb-2008-9-12-r182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, et al. Fine tuning of Craniofacial morphology by distant-acting enhancers. Science. 2013;342(6157):1241006-1–1241006-8. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanz V, O’Toole A, Vetter T, Wild HA. On the other side of the mean: the perception of dissimilarity in human faces. Perception. 2000;29:885–891. doi: 10.1068/p2851. [DOI] [PubMed] [Google Scholar]
- Carmona-Mora P, Walz K (2010) Retinoic Acid Induced 1 RAI1: a dosage sensitive gene related to neurobehavioral alterations including autistic behaviour. Curr Gen 11:607–617 [DOI] [PMC free article] [PubMed]
- Crespi B. Diametric gene-dosage effects as windows into neurogenetic architecture. Curr Opin Neurobiol. 2013;23:143–151. doi: 10.1016/j.conb.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Cyr AB, Nimmakayalu M, Longmuir SQ, Patil SR, Keppler-Noreuil KM, Shchelochkov OA. A novel 4p16.3 microduplication distal to WHSC1 and WHSC2 characterized by oligonucleotide array with new phenotypic features. Am J Med Genet A. 2011;155A(9):2224–2228. doi: 10.1002/ajmg.a.34120. [DOI] [PubMed] [Google Scholar]
- Dikow N, Maas B, Gaspar H, Kreiss-Nachtsheim M, Engels H, Kuechler A, Garbes L, Netzer C, Neuhann TM, Koehler U, et al. The phenotypic spectrum of duplication 5q35.2–q35.3 encompassing NSD1: Is it really a reversed Sotos syndrome? Am J Med Genet Part A. 2013;161(9):2158–2166. doi: 10.1002/ajmg.a.36046. [DOI] [PubMed] [Google Scholar]
- Eggermann T. Silver–Russell and Beckwith–Wiedemann syndromes: opposite (epi) mutations in 11p15 result in opposite clinical pictures. Horm Res. 2009;71(2):30–35. doi: 10.1159/000192433. [DOI] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan A, Clayton P, Corpas S, Rajan M, Van Vooren S, Moreau Y, Pettet RM, Carter NP. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco LM, de Ravel T, Graham BH, Frenkel SM, Van Driessche J, Stankiewicz P, Lupski JR, Vermeesch JR, Cheung SW. A syndrome of short stature microcephaly and speech delay is associated with duplications reciprocal to the common Sotos syndrome deletion. Eur J Hum Genet. 2010;18(2):258–261. doi: 10.1038/ejhg.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervasini C, Castronovo P, Bentivegna A, Mottadelli F, Faravelli F, Giovannucci-Uzielli ML, Pessagno A, Lucci-Cordisco E, Pinto AM, Salviati L et al (2007) High frequency of mosaic CREBBP deletions in Rubinstein-Taybi syndrome patients and mapping of somatic and germ-line breakpoints. Genomics 90:567–573 [DOI] [PubMed]
- Hammond P, Hutton TJ, Allanson JE, Campbell LE, Hennekam RCM, Murphy KC Holden S, Patton MA, Shaw A, Temple IK, et al. 3D analysis of facial morphology. Am J Med Gen. 2004;126A(4):339–348. doi: 10.1002/ajmg.a.20665. [DOI] [PubMed] [Google Scholar]
- Hammond P, Hutton TJ, Allanson J, Buxton B, Campbell L, Clayton-Smith J, Donnai D, Karmiloff-Smith A, Metcalfe K, Murphy KC, Patton M, Pober B, et al. Discriminating power of localised 3D facial morphology. Am J Hum Genet. 2005;77:999–1010. doi: 10.1086/498396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond P, Hannes F, Suttie M, Devriendt K, Vermeesch JR, Faravelli F, Forzano F, Parekh S, Williams S, McMullan D, et al. Fine grained facial phenotyping in Wolf–Hirschhorn syndrome. Eur J Hum Genet. 2012;20:33–40. doi: 10.1038/ejhg.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond P, Suttie M. Large-scale objective facial phenotyping of 3D facial morphology. Hum Mut. 2012;33(5):817–825. doi: 10.1002/humu.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, Kutalik Z, Martinet D, Shen Y, Valsesia A, Beckmann ND, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478(7367):97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski LR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genetics. 2005;1(6):627–633. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenfant P, Xuding Liu, Hudson Melissa L, Ying Qiao, Hrynchak M, Riendeau N, Hildebrand Jeanette M, Cohen Ira L, Chudley Albert E, Cynthia Forster-Gibson, Mickelson Elizabeth CR, Evica Rajcan-Separovic, Suzanne Lewis ME, Holden JA. Association of GTF2i in the Williams syndrome critical region with autism spectrum disorders. J Aut Dev Disord. 2012;42:1459–1469. doi: 10.1007/s10803-011-1389-4. [DOI] [PubMed] [Google Scholar]
- Merla G, Brunetti-Pierri N, Micale L, Fusco C. Copy number variants at Williams syndrome 7q11.23 region. Hum Genet. 2010;128:3–26. doi: 10.1007/s00439-010-0827-2. [DOI] [PubMed] [Google Scholar]
- Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, et al. Molecular mechanism for duplication 17p11.2—the homologous recombination reciprocal of the Smith–Magenis microdeletion. Nat Genet. 2000;24(1):84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- Potocki L, Bi W, Treadwell-Deering D, Carvalho CM, Eifert A, Friedman EM, Glaze D, Krull K, Lee JA, Lewis RA, Mendoza-Londono R, et al. Characterization of Potocki-Lupski Syndrome (dup17p11.2p112) and delineation of a dosage-sensitive critical interval that can convey and autism phenotype. Am J Hum Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Kim KH, Angle B, Troxell R, Gorski JL, Westemeyer M, Frydman M, Senturias Y, Earl D, Torchia B, et al. Further evidence of contrasting phenotypes caused by reciprocal deletions and duplications: duplication of nsd1 causes growth retardation and microcephaly. Mol Syndromol. 2013;3(6):247–254. doi: 10.1159/000345578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert C (2009) The genomic basis of the Williams syndrome. Cell Mol Life Sci 66:1178–1197 [DOI] [PMC free article] [PubMed]
- Tassabehji M, Hammond P, Karmiloff-Smith A, Thompson P, Durkin ME, Thorgeirsson S, Metcalfe K, Rucka A, Hutton T, Hogan T, et al. GTF2IRD1 in craniofacial development of humans and mice. Science. 2006;310:1184–1187. doi: 10.1126/science.1116142. [DOI] [PubMed] [Google Scholar]
- Valsesia A, Macé A, Jacquemont S, Beckmann JS, Kutalik Z. The growing importance of CNVs: new insights for detection and clinical interpretation. Front Genet. 2013;4(92):1–19. doi: 10.3389/fgene.2013.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Gen. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Kitagawa H, Fujiki R, Tanabe M, Takezawa S, Takada I, Yamaoka I, Yonezawa M, Kondo T, Furutani Y, et al. Distinct function of 2 chromatin remodeling complexes that share a common subunit, Williams syndrome transcription factor (WSTF) Proc Natl Acad Sci USA. 2009;106:9280–9285. doi: 10.1073/pnas.0901184106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang H, Lu X, Beasley J, Mulvihill JJ, Liu R, Li S, Lee JY. Reversed clinical phenotype due to a microduplication of Sotos syndrome region detected by array CGH: microcephaly developmental delay and delayed bone age. Am J Med Genet A. 2011;155A(6):1374–1378. doi: 10.1002/ajmg.a.33769. [DOI] [PubMed] [Google Scholar]
- Žilina O, Reimand T, Tammur P, Tillmann V, Kurg A, Õunap K. Patient with dup(5)(q35.2-q35.3) reciprocal to the common Sotos syndrome deletion and review of the literature. Eur J Med Genet. 2013;56(4):202–206. doi: 10.1016/j.ejmg.2013.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.