Abstract
Protein adsorption onto polymer surfaces is a very complex, ubiquitous, and integrated process, impacting essential areas of food processing and packaging, health devices, diagnostic tools, and medical products. The nature of protein–surface interactions is becoming much more complicated with continuous efforts toward miniaturization, especially for the development of highly compact protein detection and diagnostic devices. A large body of literature reports on protein adsorption from the perspective of ensemble-averaged behavior on macroscopic, chemically homogeneous, polymeric surfaces. However, protein–surface interactions governing the nanoscale size regime may not be effectively inferred from their macroscopic and microscopic characteristics. Recently, research efforts have been made to produce periodically arranged, nanoscopic protein patterns on diblock copolymer surfaces solely through self-assembly. Intriguing protein adsorption phenomena are directly probed on the individual biomolecule level for a fundamental understanding of protein adsorption on nanoscale surfaces exhibiting varying degrees of chemical heterogeneity. Insight gained from protein assembly on diblock copolymers can be effectively used to control the surface density, conformation, orientation, and biofunctionality of prebound proteins in highly miniaturized applications, now approaching the nanoscale. This feature article will highlight recent experimental and theoretical advances made on these fronts while focusing on single-biomolecule-level investigations of protein adsorption behavior combined with surface chemical heterogeneity on the length scale commensurate with a single protein. This article will also address advantages and challenges of the self-assembly-driven patterning technology used to produce protein nanoarrays and its implications for ultrahigh density, functional, and quantifiable protein detection in a highly miniaturized format.
I. Importance of Understanding Protein–Surface Interactions
I.1. Everyday Applications Relying on Protein–Surface Interactions
The nature of protein interactions with polymeric surfaces impacts important application areas such as food processing and packaging, health devices, diagnostic tools, and medical products.1,2 Therefore, insight into the adsorption properties of proteins onto different polymeric surfaces can guide the choice of materials for safer food packaging and human-aid products. In addition, surface-bound proteins are often employed for solid-state immunoassays in diagnostics and detection. For rapid and simultaneous screening, solid-state arrays such as protein chips and microarrays are favored over their traditional counterparts that require a large volume of reagents and detect only one sample at a time.3,4 Microarray surfaces typically need to be modified with certain proteins that will react with specific analytes.5 Therefore, understanding protein interactions with various surfaces is crucial for developing new protein array applications.
I.2. State-of-the-Art Protein Detection: Microarrays
The surfaces with which proteins interact are becoming more complex with the continuous development of new interfaces and low-dimensional materials that enables smaller and smaller application architectures.6 Miniaturized protein detection permits portable, low-cost, low-reagent volume, and high-throughput analyses. Hence, many in vitro measurements nowadays resort to the use of protein microarrays as state-of-the-art detection systems. The far-reaching applications of protein arrays range from proteomics, drug discovery, and diagnostics to treatment development. Protein arrays are typically fabricated with glass and polymeric materials. Periodic, compactly packed spots in the microarrays function as individually addressable detection chambers in which many proteins can be monitored in parallel. Typical spot sizes in commercial protein microarrays range between 300 and 500 μm in diameter, requiring greater than a 0.3–2 nL/4–8 nL reagent volume per spot for contact/noncontact printing delivery, respectively. Figure 1A displays different types of proteins arrays such as antibodies, antigens, aptamers, and peptide arrays along with their applications in protein profiling, protein binding studies, drug discovery, and diagnostics. An example of such protein microarrays is demonstrated in Figure 1B in which a slide printed with three types of proteins is used to screen multiple analytes tagged with fluorescent dyes with distinctive colors.
Figure 1.
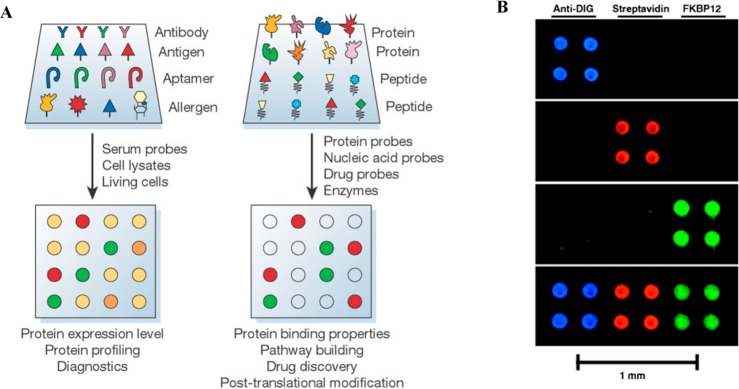
(A) Examples of analytical protein microarrays in which different types of ligands with high affinity and specificity (antibodies, antigens, DNA or RNA aptamers, carbohydrates, and small molecules) are printed on a surface. These protein chips can be subsequently used for monitoring the protein expression level, protein profiling, and clinical diagnostics. (B) Protein microarrays fabricated on glass slides to identify protein–small molecule interactions. Fluorescence emission of blue, red, and green indicates the presence of the specific protein–small molecule interactions through coupled Alexa 488, Cy5, and Cy3 dyes. Images in A and B are reproduced with permission from ref (7) (copyright 2003 Nature Publishing Group) and ref (4) (copyright 2000 American Association for Advancement of Science).
I.3. Challenges in Protein Microarrays
Current applications of microarray technology can greatly benefit from key improvements toward (i) increasing the array spot density for higher throughput and (ii) attaining uniformity in the number and biological activity of bound proteins between all printed spots of an array. Higher spot density in protein microarrays may facilitate applications involving large-scale screening and trace-level detection. In large-scale screening with the number of samples in the tens of thousands range, recent advances in nanoscience can be used to push the spot dimension of protein arrays further down to the nanoscale regime, offering a potential solution to current problems in microarray technology. Such ultrahigh-density protein nanoarrays can permit much lower volume assays (a few to tens of picoliters per spot) than existing microarrays and have the potential to contribute to large-scale protein screening.
Uniformity in the density and biofunctionality of bound proteins among all spots in an array can enable quantitative signal analysis that can be further used for meaningful comparisons between different samples assayed in many spots. Although microarray technologies have drawn significant attention for their potential in quantitative protein analyses, precise control over the two aforementioned critical requirements is still difficult to accomplish during protein printing onto array surfaces. Understanding protein–surface interactions is essential to overcoming these challenges in current microarray technologies. These aspects will be discussed in more detail in the next section.
II. Methods of Assembling Proteins on Surfaces
II.1. Existing Methods for Surface Patterning of Proteins
In microarrays, surface immobilization of capture proteins is a prerequisite for subsequent analyses involving analyte proteins. Extensive research is underway to develop techniques to print (or spot) proteins with a precise spatial periodicity, surface density, and uniformity. Controlling these factors during array production is of importance in achieving accurate and quantitative protein detection.8,9 In recent years, advances in the areas of microfabrication and nanofabrication have influenced various techniques used to deliver proteins to underlying surfaces. Methods developed so far to localize proteins on surfaces include manual and robotic delivery,10 microcontact printing,11,12 capillary force lithography,13 imprint and nanoimprint lithography,14,15 particle lithography,16 microfluidic channel networks,17 focused-ion-beam patterning,18 inkjet deposition,19 and dip-pen and related scanning probe lithography.20,21 Table 1 classifies these techniques on the basis of the pattern size and transfer approach to partitioning proteins on surfaces. Microcontact printing with preconstructed stamps allows the simultaneous spotting of proteins and generates surface-bound proteins typically localized in micrometer scale patches.11,12 When 3D topographical control of patterns is needed, networks of microfluidic channels fabricated by standard photolithography procedures can be alternatively used to achieve protein deposition on the micrometer scale.17 For protein delivery under highly viscous buffer conditions or in complex mixture solutions, the inkjet printing approach has been demonstrated to be effective.19
Table 1. Parallel and Serial Transfer Methods Used to Create Protein Patterns on Surfaces.
| pattern size |
||
|---|---|---|
| transfer type | micrometer or larger | nanometer |
| parallel processing | manual and robotic delivery10 | nanoimprint lithography15 |
| microcontact printing11,12 | particle lithography16 | |
| imprint lithography14 | self-assembly8,9,22−28 | |
| capillary force lithography13 | ||
| microfluidic channel networks17 | ||
| serial processing | inkjet deposition19 | dip-pen lithography20 |
| focused ion beam patterning18 | scanning probe lithography21 | |
In addition to these techniques yielding micrometer-scale protein patterns, recent advances in nanoscience have pushed the spot dimensions of protein arrays further down to the nanoscale regime, offering an even higher spot density and compactness to microarray technology. Both parallel and serial approaches have been developed to produce nanoscale protein arrays. For example, proteins are inked onto an elastomeric stamp containing prefabricated nanosized features and subsequently transferred to array surfaces.12,15 Although the use of electron beam or extreme UV lithographical tools is required to produce stamps with nanofeatures, this contact printing method can allow the parallel transfer of proteins to periodically spaced nanoscale regions on array surfaces. In particle lithography, closely packed particles serve as convenient frames on a substrate whose void spaces can be used for protein capture.16 The delicate interplay between the adhesion forces acting on proteins, particles, and substrates allows for the complete removal of only the particles from the substrates while leaving bound proteins behind. In another example, scanning probe microscopy (SPM)-based lithographic methods enable the surface printing of proteins in nanoscale geometry.20,21 In these serial approaches, proteins are written line by line and spot by spot onto solid surfaces via probe tips with a nanoscale positional control. Although considered to be slower than the parallel approaches of microcontact printing and microchannel networks, these methods can provide much smaller feature sizes than can the parallel techniques without prefabricated templates and have an advantage of real-time error proofing of each nanoscale feature.
II.2. Critical Factors in Surface Protein Patterning
Surface patterning conditions are directly coupled to the accuracy and precision of protein detection because a small shift in the protein signal can serve as an indicator of dramatic changes in the final biological outcome. Variability in the protein density, uniformity, and activity can considerably hamper signal quantification. Protein array technologies promise a quantitative analysis of protein samples in a direct and straightforward manner, which is difficult to accomplish through traditional methods such as immunohistochemistry and radioactive assays. The realization of this crucial factor in array technologies, however, is largely based on controlling the printing conditions of proteins on array surfaces. However, as discussed before, many conventional approaches to fabricating microarrays face great difficulties in controlling the exact number of proteins and maintaining uniformity in protein density on all printed spots. The same challenges are also presented to the aforementioned nanoscale patterning schemes. It is inherently more difficult to develop facile techniques to produce nanoarrays by partitioning proteins with nanoscale periodicity while maintaining a high level of uniformity in the protein surface density. Therefore, the quantification and standardization of protein arrays remain a significant challenge to this date, regardless of the array feature sizes.
III. Controlled Nanoscale Protein Adsorption through Self-Assembly
III.1. Bottom-Up Protein Assembly on Surfaces
More recently, a bottom-up method based entirely on self-assembly has been demonstrated to generate surface-patterned proteins using block copolymers.8,9,22−28 In the diblock copolymer-based approach for protein assembly, chemical or physical modifications of surfaces are not necessary. In the past, various processes such as UV lithography, microcontact printing, and focused ion beam milling have been employed in order to create chemically defined surface areas for localized protein adsorption on solid surfaces.11,12,18 Self-assembled monolayers have also been used for the chemical patterning of substrates for subsequent protein adsorption.29 For the physical modification of solid substrates ahead of protein deposition, alterations to delineate the surface areas are performed by laser ablation,30 reactive ion etching,31 and sputtering.32 Unlike the majority of these past efforts to modify substrates, phase-separated diblock copolymers can inherently serve as the basis for individually addressable spots with nanoscopic dimensions. The rapid and controlled organization of proteins on these spots can be subsequently achieved through self-selective interaction processes between a given set of proteins and polymers, which in turn provides the driving force for highly periodic and aligned nanoscale patterns of proteins instantaneously produced over large areas of substrates. This method of protein self-assembly on diblock copolymers does not require top-down fabrication techniques using external fields, prefabricated stamps/masks, or highly specialized lithographic fabrication in a clean room setting. The self-assembly approach eliminates costly and time-consuming steps used to modify physical and chemical properties of polymeric supports as well as to introduce specially defined surface areas prior to protein arrangements.
III.2. Phase-Separated Block Copolymer Nanotemplates
A class of polymeric materials called diblock copolymers is known to provide chemically heterogeneous, self-assembling periodic structures through microphase separation.33−35 Diblock copolymers are formed by covalently joining two chemically immiscible polymeric blocks end-to-end. Because of the immiscibility and differential wetting properties associated with the two components of these materials, microphase separation occurs in diblock copolymers in directions both perpendicular and parallel to the underlying support. The unique microphase separation behavior in ultrathin films of a block copolymer, polystyrene-block-poly(methyl methacrylate) (PS-b-PMMA), has previously been shown to expose both block components to the air/polymer interface under carefully balanced thermodynamic conditions.36 This phenomenon generates spatially periodic, self-assembled, nanoscale polymeric domains consisting of the different chemical constituents of the two polymeric components whose scale and geometry reflect the chemical and physical properties of the polymer.36−39 Their phase diagram dictates the packing nature and orientation of the resulting polymer chains, and their phase separation behavior can be predicted on the basis of mean field theory.33,34 Therefore, the repeat spacing and surface geometry of the diblock copolymer can be readily controlled on the nanoscale, for example, by changing the molecular weight and composition of the two blocks. Figure 2A displays a phase diagram of a linear diblock copolymer (a–b) predicted by theoretical work while varying the volume fraction (f), segment–segment (Flory–Huggins) interaction parameter (χ), and degree of polymerization (N). Possible block copolymer morphologies predicted for the ordered state include spheres, cylinders, gyroids, and lamellars. The packing structures experimentally observed in poly(isoprene-styrene) block copolymers shown in Figure 2B agree well with the theoretical prediction.
Figure 2.
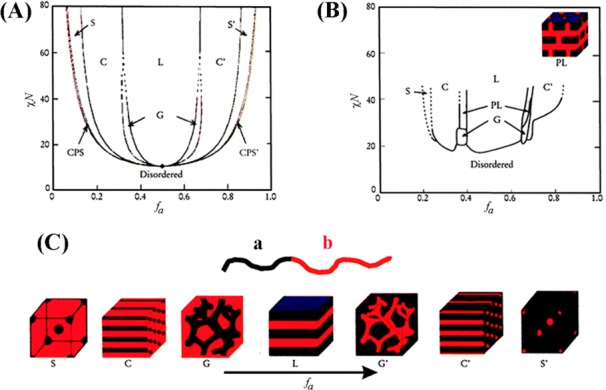
Phase diagram of a linear a–b block copolymer. (A) The diagram is a theoretical phase prediction based on the self-consistent mean-field theory, and (B) the diagram is the experimental phase portrait of poly(isoprene-styrene) block copolymers. χ, N, and fa refer to the segment–segment interaction parameter, the degree of polymerization, and the composition of the a segment, respectively. (C) Morphologies of CPS, S, C, G, L, and metastable PL shown in the phase diagrams correspond to a close-packed sphere, sphere, cylinder, gyroid, lamellar, and perforated layer, respectively. Images in A and C) and (B) are reproduced with permission from (A, C) ref (35) (copyright 2008 AIP Publishing LLC) and (B) ref (40) (copyright 1995 American Chemical Society).
III.3. Self-Assembled Proteins with Nanoscale Periodicity
These chemically alternating and self-assembling polymeric domains can serve as convenient self-constructed templates for the nanoscale arrangement of desired biocomponents. Recent research efforts have successfully demonstrated that subsequent nanoscale surface organizations of proteins onto the phase-separated polymeric nanodomains can be straightforwardly accomplished via self-assembly driven by the strong interaction preferences of proteins with one of the diblocks.8,9,22−27 Because of the nanoscale size of the polymeric templates, the resulting protein arrangements on diblock copolymer templates exhibit unique adsorption characteristics that differ from the macroscopic and microscopic adsorption behavior. The built-in nanoscale chemical heterogeneity of diblock copolymers also separates these platforms from the conventional arrays such as glass slides uniformly treated with polymers and chemically homogeneous polymeric arrays. Polystyrene, polycarbonate, poly(dimethylsiloxane), poly(vinylidene difluoride), and polyolefin are commonly used to construct conventional microarrays and microwell plates. They are also employed as polymeric coating layers on top of glass slides. Chemical heterogeneity in block copolymers plays a significant role in nanoscale protein adsorption, and the presence of interfaces defined by two chemically immiscible polymer blocks is known to influence protein adsorption onto polymer surfaces.22,24 Such findings on the protein adsorption behavior on block copolymer surfaces can provide much needed fundamental insight into protein adsorption on surfaces whose size scale is comparable to that of an individual protein. When used as protein arrays, the new technique can conveniently offer quantification and standardization capabilities. These interesting adsorption phenomena, observed from globular proteins on nanoscale diblock copolymer surfaces, are discussed next.
IV. Unique Protein Adsorption Behavior
IV.1. Characteristic Adsorption Behavior of Proteins on Diblock Copolymers
Nanoscale surface adsorption onto diblock copolymers has been first studied at the individual protein level by examining model protein molecules of immunoglobulin G (IgG) and bovine serum albumin (BSA) assembled on the surface of phase-separated nanodomains of PS-b-PMMA. In these experiments, the size of the underlying templates reaches the size of the individual proteins, and the adsorption properties need to be investigated by a technique capable of yielding a spatial resolution at the single-protein-molecule level. Hence, atomic force microscopy (AFM) is primarily used to examine the nanoscale protein adsorption behavior. Figure 3 displays AFM images showing the highly selective adsorption behavior of IgG proteins onto the PS nanodomains of the diblock copolymer surface.22−24,26
Figure 3.
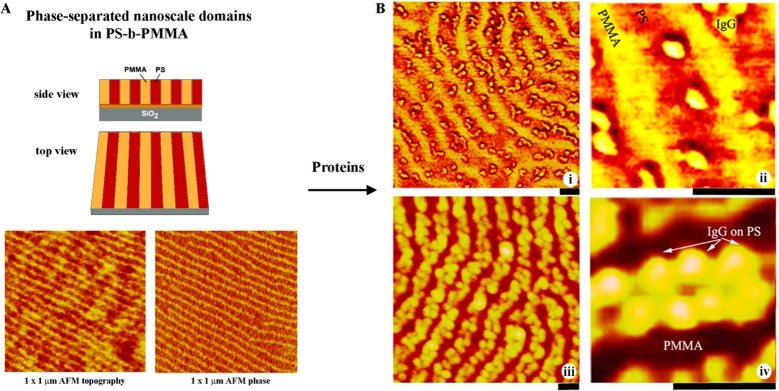
(A) AFM images of phase-separated PS-b-PMMA showing alternating domains of PS and PMMA with a repeat spacing of 45 nm on the surface. (B) AFM results of IgG molecules on PS-b-PMMA at low surface density shown in panels i and ii clearly display their preferred interaction with PS. Individual IgG molecules seen as spherical objects in the images self-assembled onto PS domains close to the interface between PS and PMMA. The diameter of IgG is approximately 15 nm. AFM results in panels iii and iv, taken by using a monolayer-forming condition, display the surface-packing nature of individual IgG molecules on PS. Two IgG molecules occupy the PS domain along its short axis, which is consistent with the size of the protein and the periodic spacing of the polymer domain. The thick black line under each image is a 45 nm scale bar. Adapted with permission from ref (22) (copyright 2005 American Chemical Society).
The proteins entirely avoid adsorption onto the neighboring PMMA domains. When the protein loading condition is adjusted to yield a higher number density of proteins on the surface, the protein molecules self-assemble in a closely surface-packed configuration on the PS nanodomains, leaving the neighboring PMMA domains completely devoid of adsorbed proteins.22,25 All available PS areas are packed with a single layer of adsorbed proteins under this condition. This protein loading state, defined as a monolayer-forming condition, can be experimentally controlled by adjusting the protein concentration and deposition time. Such a strong adsorption preference for PS is observed for many other proteins such as human serum albumin (HSA), fibronectin (Fn), horseradish peroxidase (HRP), mushroom tyrosinase (MT), and protein G (PG). Table 2 lists the proteins confirmed for their selective adsorption onto the PS domains of PS-b-PMMA and their key properties related to surface adsorption. Identification numbers for the RCSB Protein Data Bank, pdb id, are also provided in Table 2, and the crystal structures of the proteins can be accessed using the pdb id.
Table 2. Proteins Exhibiting Exclusive Adsorption onto PS Domains of PS-b-PMMA and Their Properties.
| proteins | molecular weight (kDa) | isoelectric point (pI) | pdb id |
|---|---|---|---|
| PG | 33 | 4.85 | 1PGAa |
| HRP | 44 | 7.2 | 2ATJb |
| HSA | 47 | 5.3–6.0 | 4G04 |
| BSA | 67 | 4.7 | 4F5S |
| MT | 120 | 4.7–5.3 | 3NM8c |
| IgG | 150 | 6.1–8.5 | 1IGT |
| Fn | 440 | 5.5–6.0 | 1FNFd |
B1 IgG binding domain of PG.
Recombinant HRP in complex with benzhydroxamic acid.
Tyrosinase from Bacillus megaterium.
Fn fragment encompassing the 7th through 10th type III repeats.
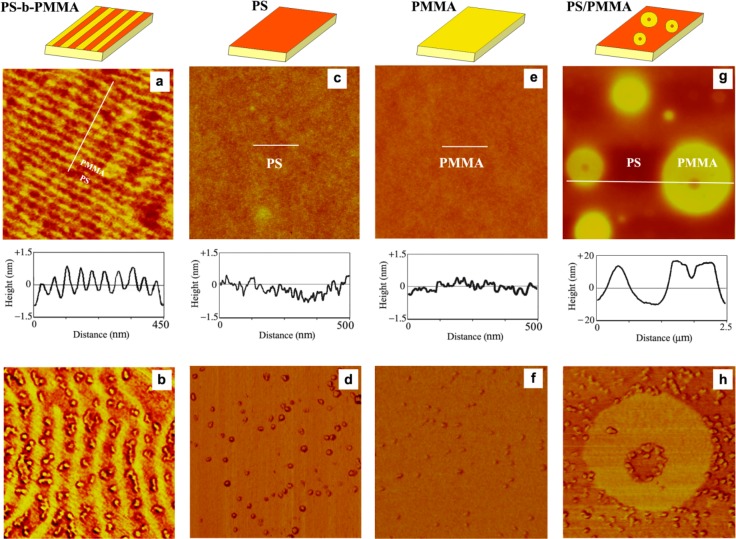
Further examination of the protein assembly shown in Figure 3 reveals that a large percentage of proteins prefer adsorption to the PS regions close to the chemical interfaces defined by the two neighboring PS and PMMA nanodomains. This tendency of proteins to favor PS regions close to PS/PMMA interfaces is consistently observed even when the loading parameters are tuned using a low enough concentration to provide conditions for preferential protein adsorption in the middle of the PS nanodomains, as far away from the two neighboring interfaces as possible.22,25 When comparing the number of adsorbed protein molecules on the PS-b-PMMA surface to those on homopolymer PS or PMMA surfaces, the number of proteins on the PS-b-PMMA surface is found to be much larger than those on the homopolymer surfaces.24 The number of adsorbed proteins per given surface area is referred to as the protein surface density (PSD) herein. The PSD is determined to be from highest to lowest in the following order: PS-b-PMMA block copolymer > PS/PMMA blend > homopolymer PS > homopolymer PMMA. This observation is noteworthy considering the fact that approximately half of the available surface on PS-b-PMMA contains the nonpreferred PMMA domains. These interesting findings are displayed in Figure 4 with the representative AFM images of homopolymer, blend, and block copolymer surfaces as well as those treated with IgG.
Figure 4.
AFM images showing various polymeric surfaces before (top) and after (bottom) the deposition of protein molecules. PS-b-PMMA diblock copolymer, PS homopolymer, PMMA homopolymer, and PS/PMMA blend ultrathin films are used as templates, and the topographic roughness of each surface is presented in the corresponding height profile taken along the inserted white line. AFM images correspond to (a) PS-b-PMMA (750 × 750 nm2), (b) 4 μg/mL IgG on PS-b-PMMA (380 × 380 nm2), (c) PS (2 × 2 μm2), (d) 4 μg/mL IgG on PS (500 × 500 nm2), (e) PMMA (2 × 2 μm2), (f) 4 μg/mL IgG on PMMA (500 × 500 nm2), (g) PS/PMMA blend (2.5 × 2.5 μm2), and (h) 10 μg/mL IgG on PS/PMMA blend (750 × 750 nm2). The high adsorption preference of proteins to chemical interfaces is clearly seen in the AFM panel (h). Reproduced with permission from ref (24) (copyright 2008 American Chemical Society).
Similar to the protein adsorption behavior seen in the diblock copolymer, a large number of proteins on PS/PMMA blends gather tightly on the PS region next to the PS/PMMA interface (Figure 4h). The surface density of PS/PMMA interfaces (herein referred to as ISD for interfacial surface density) follows the order diblock copolymer > blend > homopolymer templates. The separation distance between two neighboring PS/PMMA interfaces is in the reverse order of ISD: copolymer (tens of nanometers) < blend (several to tens of micrometers) < homopolymer (∞). The adsorption measurements on various polymeric templates of varying size scale with the same chemical makeup indicate that a high PSD is expected on high ISD surfaces. The significance of chemical interfaces on protein adsorption is substantiated by measuring the normalized PSD on blends as a function of the distance to the closest PS/PMMA interface. For both cases of IgG and Fn adsorption, the PSD decreases exponentially with the distance from the PS/PMMA interface as shown in Figure 5A (i.e., adsorption sites on PS away from the interfaces are favored much less by proteins). In addition, when the number of adsorbed proteins is analyzed on blends as a function of the separation distance between the two nearest PS/PMMA interfaces, the PSD is inversely proportional to the separation distance between two neighboring interfaces as displayed in Figure 5B.
Figure 5.
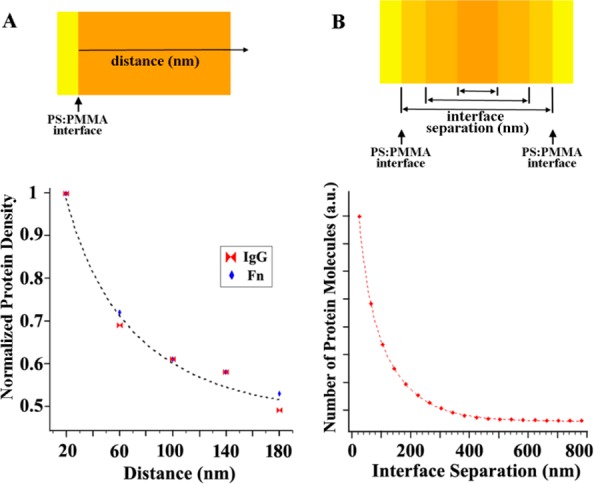
(A) Graph illustrating normalized PSD vs the spatial location of proteins away from the interface of PS and PMMA. The PSD decreases exponentially with the distance of proteins from the interface. (B) Correlation of protein density to the interfacial separation distance between two nearest PS/PMMA interfaces. The number of adsorbed proteins is inversely proportional to the separation distance between two neighboring PS/PMMA interfaces. Reproduced with permission from ref (24) (copyright 2008 American Chemical Society).
IV.2. Protein Assembly on the Nanoscale with Two-Dimensional Spatial Control
The micellar assembly of amphiphilic block copolymers above a critical polymer concentration is a well-known behavior whose exact structures and configurations are determined by the composition of the diblock polymer, the length of each polymer segment, the polarity of the solvent, and the relative solubility of each polymer block in the solvent. For these studies, polymeric systems such as polystyrene-b-poly(acrylic acid), poly(ethylene-propylene)-b-poly(ethylene oxide), polystyrene-b-poly(2-vinylpyridine) (PS-b-P2VP), and polystyrene-b-poly(4-vinylpyridine) (PS-b-P4VP) are extensively researched to understand their fascinating micellar properties and dependence on diblock copolymer characteristics.41−43 In addition to the spontaneously formed original micelles, additional structures are also reported by altering the micelles via simple exposure to a solvent vapor that exhibits biased interactions with the two polymeric blocks. Examples of such structures can be found in PS-b-P2VP and PS-b-P4VP micelles annealed in ethanol, tetrahyrdofuran, and chloroform vapors, leading to a rich spectrum of morphologies with varying repeat spacings tunable in two dimensions.25,27,44−46 In an example of PS-b-P4VP annealing in ethanol, new structures of open and reverted micelles emerge from the original template. The former structure results from the rearrangement of PS chains to expose the underlying PVP fully, whereas the latter form is produced by the reverting PS chains and their partial covering of the underlying PVP chains.25 Figure 6 displays a series of eight different PS-b-P4VP templates identified during various stages of chloroform annealing, whose structures span from the original micellar to the cylindrical phase.
Figure 6.
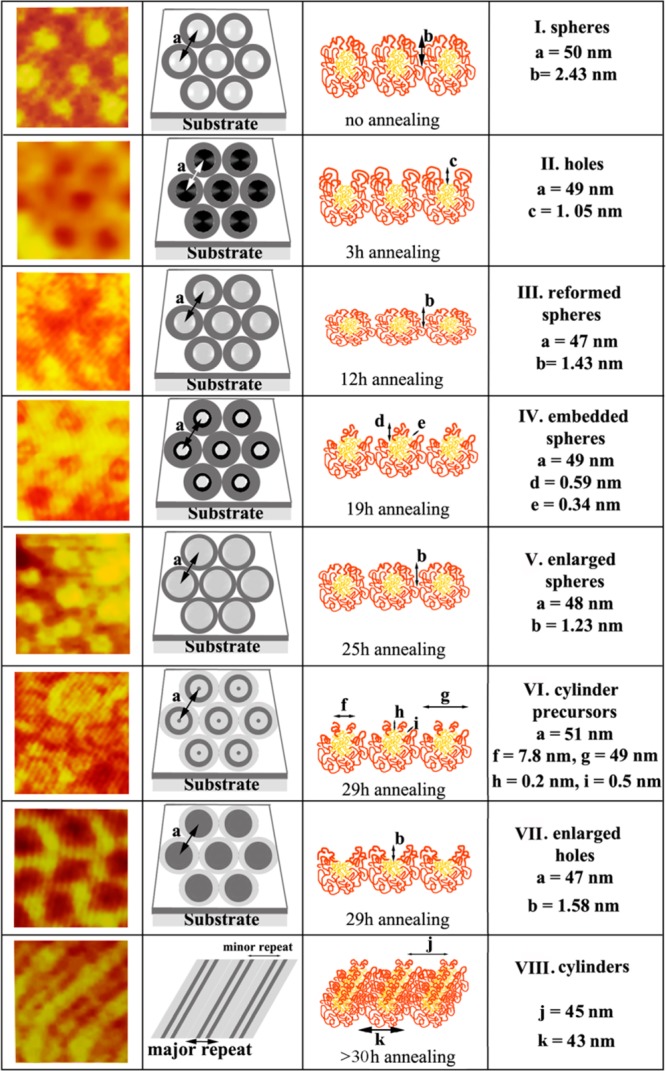
AFM images (125 × 125 nm2) showing hexagonally packed nanostructures identified during chloroform annealing of PS-b-P4VP nanodomains: (I) original spheres, (II) holes, (III) reformed spheres, (IV) embedded spheres, (V) enlarged spheres, (VI) cylinder precursors, (VII) enlarged holes, and (VIII) cylinders. Adapted with permission from ref (44) (copyright 2012 American Chemical Society).
The significance of these chemical annealing methods in the surface organization of proteins is the ability to provide additional nanostructures beyond those equilibrium structures that the mean field phase diagrams predict. These intricate polymeric nanotemplates can provide much needed versatility and flexibility to meet the increasing demand for self-assembled, nanoscale guides and platforms for organizing proteins into 2D arrays. The feasibility of using amphiphilic block copolymers to create self-assembled proteins into hexagonally arranged spots with 2D spatial controls has been first demonstrated by protein self-organization on several PS-b-P4VP templates.25,27 Figure 7 displays such protein patterns on PS-b-P4VP templates, spontaneously formed upon protein deposition into the 2D periodic structures attained from annealing the original templates under ethanol vapor.
Figure 7.
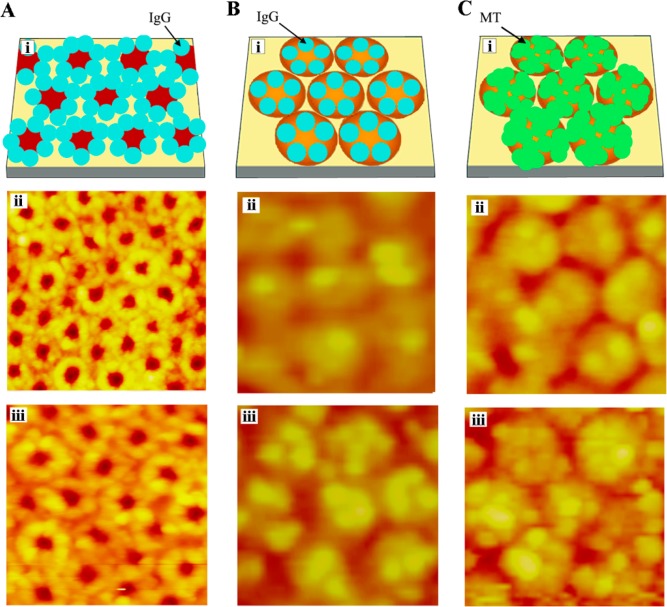
Protein assembly behavior on hexagonally packed PS-b-P4VP micellar nanodomains with a periodicity of 42 nm. (A) 20 μg/mL IgG molecules on open PS-b-P4VP templates. The AFM scan size corresponds to (ii) 300 × 300 nm2 and (iii) 180 × 180 nm2, respectively. (B) 180 × 180 nm2 AFM panels of (ii) 4 μg/mL and (iii) 10 μg/mL IgG molecules on reverted PS-b-P4VP. (C) 180 × 180 nm2 AFM panels of (ii) 4 μg/mL and (iii) 20 μg/mL MT molecules assembled on reverted PS-b-P4VP. Reproduced with permission from ref (25) (copyright 2007 American Chemical Society).
IV.3. Understanding the Unique Protein Adsorption Behavior on the Nanoscale
Protein interactions are known to occur typically through van der Waals (dispersion) interactions, electrostatic forces, hydrogen bonding, and hydrophobic/hydrophilic interactions.1,47 Under the experimental conditions used for protein assembly on diblock copolymers,22−27 the protein molecular weight does not have a large effect on the selective and complete protein segregation to PS. Varying the surface charges of proteins by adjusting the pH also does not yield a difference in the PS-favoring assembly behavior of proteins. Therefore, van der Waals and electrostatic interactions do not seem to dominate the nanoscale protein segregation observed in the block copolymer systems. During protein adsorption, hydrogen bonds can be established for hydroxyl–carbonyl, amide–carbonyl radicals, hydroxyl–hydroxyl, and amide–hydroxyl bonds. Although an important criterion in general protein interaction cases, hydrogen bonding is not as influential in the diblock copolymer systems where the same polymeric surfaces are tested repeatedly for protein adsorption with no specific modification of surface chemical groups.
Diblock copolymers used in the aforementioned studies have polymeric components with different hydrophobicities. The hydrophobic/hydrophilic interactions between the polymer and protein surfaces may be responsible for the interesting segregation phenomena of proteins on selective polymer domains. For example, the water contact angles (θwater) of bulk PMMA, P4VP, and PS are approximately 75, 62, and 90°, respectively.48,49 However, hydrophobic interactions based on the bulk values alone may not fully explain protein adsorption in the size range where even water molecule behavior is different from bulk behavior. Other factors relevant to the system are the roughness and evaporation on the nanoscale. The Cassie50 and Wenzel51 models describe significant changes in the contact angle caused by the physical roughness of surfaces whose phenomena are well studied with respect to bulk and micrometer-scale roughness.52 Capillary effects on evaporation may also be considered under ambient imaging conditions because the effect is manifested differently on hydrophobic versus hydrophilic surfaces. This aspect has also been extensively studied on macroscale and microscale surfaces.53 However, very little is known about the possible effect of nanometer scale surface roughness or nanoscale chemical heterogeneity on the contact angle, surface dewetting, and capillary evaporation.54
IV.4. In-Depth Look at Nanoscale Protein–Surface Interactions: Hydrophobic Interactions
Protein adsorption onto PS-b-PMMA belongs in the nanoscale category because the undulating height difference between the two domains of PS and PMMA is 10 Å.36 Although no experimental contact angle values of water on alternating nanoscale surfaces of PS and PMMA exist, the amplitude (10 Å) and period of the diblock polymer nanodomains (45 nm) in Figure 4A can be compared to the prediction made in a molecular dynamics (MD) simulation study.55 On the basis of this comparison, the above-mentioned bulk θwater values can be reasonably applied to the nanoscale system. When considering θwater values as given above, it can be determined that the proteins in Table 2 prefer the more hydrophobic polymeric nanodomains of the diblocks.
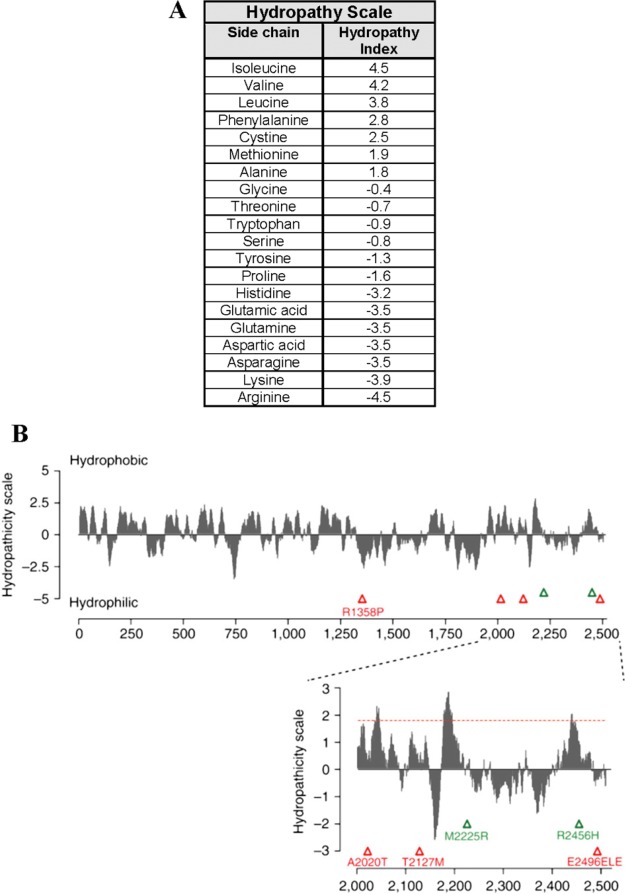
The hydropathy index (HI) on the Kyte–Doolittle56 and Hopp–Woods57 scales calculates average hydrophobicity/hydrophilicity values in a moving window of predetermined size along the amino acid sequence of a protein. A hydropathy plot, charting the average HI value as a function of the amino acid sequence, displays protein regions with high hydrophobicity/hydrophilicity (positive HI regions on the Kyte–Doolittle scale and negative regions on the Hopp–Woods scale and vice versa). Therefore, hydropathy plots can be conveniently used to compare the overall hydrophobicity of proteins and to find a region of amino acid sections showing high hydrophobicity/hydrophilicity.56−58 Figure 8A displays Kyte–Doolittle hydropathy scale values for various amino acid residues. Figure 8B introduces a hydrophobicity analysis based on this scale. The analysis is carried out in the ProtScale program (Expasy) with a moving window of 19 residues for a human ion channel protein called PIEZO1. Although effective in delineating the overall hydrophobic character of a protein based on the sequence of amino acids, the current HI models may not be effectively used to explain the protein adsorption behavior monitored on the diblock copolymer surfaces. During surface adsorption, the hydrophobic contributions of amino acids consisting of the exterior parts of proteins (i.e., protein surfaces) will be significantly greater than those constituting the interior. Therefore, further work is needed to describe protein adsorption better on nanoscale surfaces by including folded protein structures as a part of the HI consideration.
Figure 8.
(A) Hydropathy scale for amino acid residues used for Kyte–Doolittle HI calculations. (B) Example of a Kyte–Doolittle hydrophobicity analysis carried out in the ProtScale program (Expasy). The hydropathy plot for human PIEZO1 protein is produced with a moving window of 19 amino acid residues in which regions above/below the hydropathy scale of 0 indicate hydrophobic/hydrophilic domains of PIEZO1. Images in A and B are reproduced with permission from ref (56) (copyright 1982 Elsevier) and ref (58) (copyright 2013 Nature Publishing Group), respectively.
Protein adsorption favoring hydrophobic surfaces is well known and extensively reported for macroscale and microscale surfaces where the statistical adsorption behavior of proteins is collectively monitored.1,59−61 A kinetic evaluation of protein adsorption to chemically homogeneous polymeric surfaces also exist in the literature using techniques such as surface plasmon resonance spectroscopy (SPR),62,63 total internal reflectance fluorescence (TIRF),59,60 and quartz crystal microbalance with dissipation monitoring (QCM-d).64 However, the above-discussed results of protein adsorption onto nanoscale block copolymer surfaces cannot be explained simply by hydrophobicity competition between the polymeric blocks. From the AFM investigations carried out on the individual molecule level, protein adsorption is determined to be not only favored on the interfacial areas of the preferred polymeric domain but also greatly facilitated by the presence of polymeric interfaces.24 These results suggest that the co-occurrence of hydrophobicity and hydrophilicity in the size regime of an individual protein is one of the determining factors in nanoscale protein self-assembly on diblock copolymers.
IV.5. Extended Discussion of Nanoscale Protein–Surface Interactions: Chemical Heterogeneity on the Nanoscale
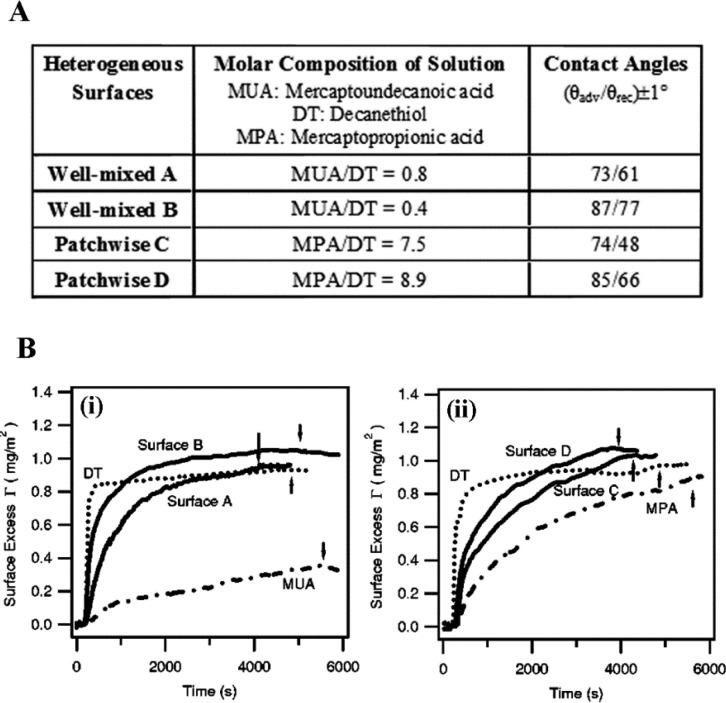
The effects of nanoscale size and chemical heterogeneity on protein adsorption have also been studied on substrates using organic molecules as surface modifiers.61,65 When SPR is used to monitor time-dependent, ensemble-averaged adsorption behavior of BSA proteins, the amounts and rates of BSA adsorption differ on the chemically homogeneous and heterogeneous surfaces as shown in Figure 9A. When compared to those found on the homogeneous mercaptopropionic acid (MPA) and decanethiol (DT) surfaces, the BSA surface coverage (Γmax) is observed to be higher on the patchwise heterogeneous surfaces containing mixtures of MPA and DT. The heterogeneous surface effect on Γmax is seen more dramatically on the patchwise MPA/DT substrate than on the well-mixed mercaptoundecanoic acid (MUA)/DT surfaces, (Figure 9B). This outcome corroborates the aforementioned observations made from individual protein studies, underscoring the importance of the length scale in chemical heterogeneity being commensurate with the dimension of the proteins (i.e., nanoscale chemical heterogeneity).
Figure 9.
(A) Chemical compositions and contact angles used as heterogeneous substrates for BSA adsorption. (B) SPR sensorgram showing the BSA surface coverage over time on (i) well-mixed and (ii) patchwise heterogeneous surfaces where the arrows indicate rinsing with pure buffer. For comparison, BSA adsorption on homogeneous, single-component surfaces of DT, MUA, and MPA is displayed with dashed lines. Adapted with permission from ref (65) (copyright 2004 AIP Publishing LLC).
The inherently amphiphilic nature of proteins may explain the unique observations of favored protein adsorption onto the chemically heterogeneous surfaces in both the diblock copolymer and the organic compound-modified systems. The surface of a protein is extremely complex, containing varying degrees of hydrophobic/hydrophilic residues, chemical moieties, and charges. The precise interpretation and prediction of protein adsorption, therefore, can be even more difficult when chemically heterogeneous, nanoscale surfaces are employed for protein assembly. Although very few studies are found in the literature on the topic of nanoscale protein–surface interactions combined with chemical heterogeneity, an MD study has recently revealed the key role of amphiphilic amino acids in facilitating the adsorption of cytochrome C (Cyt C) onto mixed-composition surfaces composed of heterogeneous segments.66 A related, atomistic MD study shows that different groups of surface amino acid residues are responsible for the surface adsorption of lysozyme (Lyz) to various 1-octanethiol (OT)-terminated and 6-mercapto-1-hexanol (MH)-terminated surfaces.67 The simulation results in Figure 10 clearly demonstrate that the most energetically favorable conformations of adsorbed proteins vary greatly between the homogeneous OT and MH surfaces as well as between the heterogeneous substrates modeling mixed surfaces of OT/MH in varying ratios.
Figure 10.
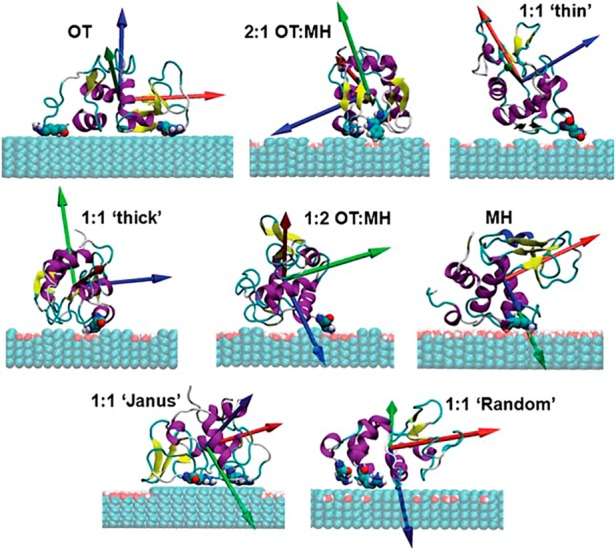
The most energetically favorable Lyz binding orientations on assorted OT- and MH-terminated surfaces. The blue and red spheres on the substrate correspond to OT- and MH-containing areas, respectively. The three principal axes (PA) of Lyz are indicated as red (PA1), blue (PA2), and green (PA3) vectors in the order of longest to shortest axis of the protein. Reproduced with permission from ref (67) (copyright 2012 Royal Society of Chemistry).
V. Toward Protein Nanoarray Applications
V.1. Surface-Bound versus Free Proteins and Their Bioactivities
Protein nanoarrays may provide a very high PSD compared to that of their conventional counterparts, but their usefulness in arrayed detection will largely depend on the degree to which the functionality of the surface-bound proteins is maintained. Biological functionalities of proteins adsorbed on surfaces will differ from their native activities in buffer. Unlike free proteins, surface-bound proteins cannot readily change their conformations to expose ligand binding sites or reactive pockets toward other biomolecules. Hence, the steric hindrance of functional protein sites due to the presence of an underlying substrate is often attributed to the reduced activities observed in many randomly adsorbed protein systems.23,24,26 In contrast, when proteins are linked to a surface via orientation-specific coupling methods using chemical and biological moieties, protein activity has been reported to increase in some cases.68,69 This increase in the activities of orientation-controlled, surface-bound proteins results from the effective downstream bioreactions guided in space toward active protein sites along the well-defined molecular axis. Protein reactions in solution, however, rely on Brownian motion for the stochastic chances of collisions. Ideally, functional protein arrays should have precise control over the orientation of surface-bound protein molecules, ensuring full biofunctionality via directionally guided interactions between prebound proteins on the array surfaces and analyte proteins in samples.
Although bioactivity differences between surface-bound proteins and free proteins in solution can be qualitatively probed using plate readers and microarrays, the quantification of their surface-dependent functionalities (e.g., protein activity comparisons between various surface-bound and free states) requires information on the exact number of proteins involved in the measurements. The quantitative evaluation and comparison of protein activities are often hampered by this difficulty in precisely determining the PSD in different environments. The development of block-copolymer-based protein nanoarrays has permitted a quantitative activity comparison of an enzymatic protein, HRP, in the surface-bound versus solution states.26 Similar quantitative analysis can be carried out to determine HRP activities on different types of polymeric surfaces.24,25 In these experiments, the exact numbers of surface-bound proteins are first determined by AFM via topological inspections of individual proteins on each surface of interest. This PSD information is then coupled with spectroscopic outcomes for quantitative activity evaluation between the same number of protein molecules on various surfaces and in solution.
Figure 11 displays UV–vis absorbance spectra resulting from HRP in surface-bound and solution environments. The enzymatic activity is monitored spectroscopically on the basis of the well-known absorbance changes of a chromogenic indicator, 3,3′,5,5′-tetramethylbenzidine (TMB). Active HRP catalyzes the oxidation of TMB in the presence of H2O2 via one-electron and two-electron oxidation processes.70 The one-electron transfer process produces a free-radical cation whereas the two-electron process forms a complex of diimine and diamine. The presence of these oxidized TMB products is responsible for the distinctive color changes of the assay solution and the characteristic absorbance peaks in their UV–vis spectra. Figure 11A displays HRP activity differences between the PS-b-PMMA surface-bound state and the free-in-solution state. When the same number of HRP molecules is compared between the two cases, approximately 85% of their free-state activity is retained after surface adsorption to PS-b-PMMA.26 Similarly, when HRP activity is compared between the PS-b-P4VP bound state versus free-in-solution state, 78% of the free-state activity is maintained after adsorption to the micellar copolymer surface.25 These outcomes suggest that high percentages of enzymatic activity are conserved in both cases of random protein adsorption onto nanoscale surfaces. These spectroscopic results also confirm the AFM observations discussed earlier regarding higher PSD found on diblocks than on homopolymers. Data in Figure 11B clearly indicate that chemically heterogeneous PS-b-PMMA surfaces are more effective at achieving a higher loading density of protein molecules than chemically homogeneous PS templates.24 Little is known about changes in protein activity after nanoscale surface adsorption, especially in a quantitative manner. Therefore, the above-mentioned research efforts provide valuable insights into the effect of different polymeric surfaces on protein functionalities when compared to their free-state activities.
Figure 11.
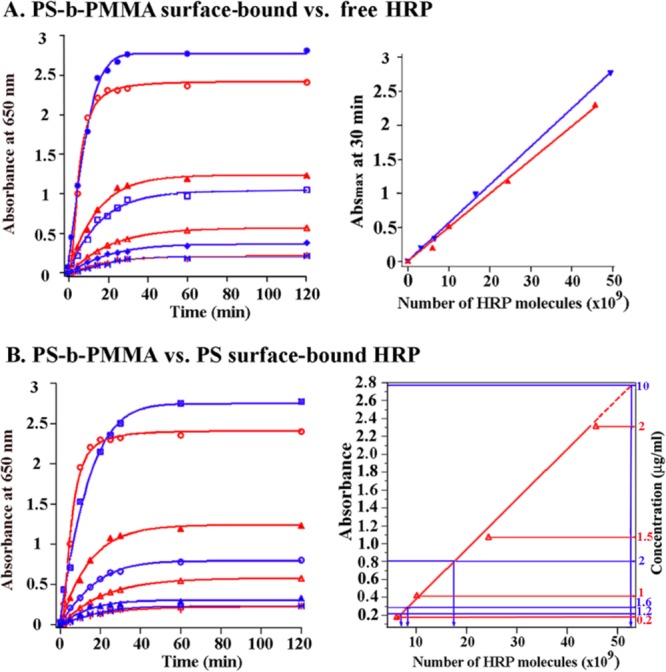
(A) Differences in HRP activity between their free state and PS-b-PMMA bound state are evaluated on the basis of their UV–vis absorbance at λ = 650 nm. Blue data points represent the activities of HRP molecules freely floating in solution, and red data points represent those of HRP molecules immobilized on PS-b-PMMA surfaces. From top to bottom, plots shown in the left panel correspond to 0.15 (f: free-state), 2 (b: bound-state), 1.5 (b), 0.05 (f), 1 (b), 0.02 (f), 0.1 (b), and 0.01 (f) μg/mL of either the free- or bound-state HRP concentration. These concentration conditions correspond to the total number of HRP molecules of 49.5 × 109, 45.7 × 109, 24.4 × 109, 16.5 × 109, 10.1 × 109, 6.6 × 109, 6.1 × 109, and 3.3 × 109 from top to bottom plots. When the enzymatic activities of the same number of HRP molecules in the free versus bound state are compared in the right panel, PS-b-PMMA-bound HRP retained approximately 85% of its free-state activity. (B) PSD comparison of HRP molecules carried out between PS-b-PMMA (red) vs PS (blue) surfaces. UV–vis absorbance values of PS-b-PMMA-bound and PS-bound HRP recorded at λ = 650 nm are measured with respect to time. When absorbance maxima are compared against the number of HRP molecules on the two types of surfaces, the adsorbed amount of HRP on the chemically heterogeneous diblock surface is much greater than that on the chemically homogeneous PS surface at the same HRP deposition concentration. Reproduced with permission from refs (24) and (26) (copyrights 2007 and 2008 American Chemical Society, respectively).
V.2. Advantages of Block-Copolymer-Assisted Assembly of Protein Nanoarrays
Diblock copolymer-assisted protein patterning is based entirely on self-assembly of polymers and proteins. Therefore, nanoscale protein features can be readily attained without the use of photolithography, ebeam lithography, particle lithography, or scanning probe lithography. No prefabricated masks or stamps are necessary, which in turn eliminates the use of specialized equipment in a clean room. In addition, the surface partitioning of proteins into periodic nanoscale patterns is rapid and spontaneous and can be easily produced over a large substrate area. When compared to conventional microarrays, the block-copolymer-based protein nanoarrays provide inherently high spot density with densely packed protein molecules, offering at least 3 orders of magnitude higher spots in a given size array than those provided by commercial microarrays.
A wide range of diblock and triblock copolymers with extensively studied chemical and physical properties for phase separation are available.33−35 Protein patterning based on the nanoscale self-assembly of chemically heterogeneous polymeric templates, therefore, is not limited to the few systems of diblock copolymers demonstrated so far. The large availability and chemical/physical tunability of many polymeric surfaces can be used to facilitate protein assembly. In addition, various equilibrium and kinetically trapped nanodomains available through the block-copolymer-based approach provide a large amount of versatility in the size and shape of nanoscale templates and the subsequently assembled protein patterns.27,44
Diblock-copolymer-based methods can be advantageous for quantitative protein detection because the technique has the unique ability to match the sizes of proteins and polymeric templates.38,39 The nanoscale size and shape of the individually addressable spots can be tuned precisely during the phase separation of diblock copolymers. The subsequent self-assembly of proteins can be directed to yield precisely controlled PSD by matching the size of the nanoscale patterns to that of proteins in 2D close-packed configurations. This capability provides the basis for quantitative protein analysis that cannot be accomplished straightforwardly through conventional means in current protein microarray applications.
The diblock copolymer approach can also be beneficial to improving the sensitivity (or limit) of protein detection. This challenge becomes more important as protein detection devices and platforms reach the nanoscale size regime where high background noise from nonspecifically bound proteins outside the printed areas in the array can contribute to a poor signal-to-noise ratio. Surface passivation (a process to avoid nonspecific protein adsorption on unwanted surface areas) of protein arrays is an important step in discerning analyte signals clearly while ruling out the background noise of detection. The use of block copolymers in protein self-assembly presents an important benefit in this area by driving the complete segregation of proteins only onto preferred polymeric nanodomains. The phenomenon of exclusive protein adsorption can facilitate the effortless self-passivation of nanoarrays because the surface areas occupied by the nonpreferred block function as a built-in passivation layer to deter nonspecific adsorption.
Protein nanoarrays formed on diblock copolymers maintain high functionality, selectivity, and stability over a long time (>3 months when kept at 4 °C).23,26 Figure 12 displays the activity and stability of enzyme molecules on PS-b-PMMA under different assay conditions. In Figure 12A,B, self-assembled enzyme nanoarrays of HRP and MT on PS-b-PMMA are evaluated for their activity and stability in their unmodified state as well as after heat and acid denaturing processes.23 Chromogenic substrates of TMB and pyrocatechol are used to identify functional HRP and MT enzymes on the surfaces, respectively. The ability to maintain the selectivity of proteins is also investigated on the diblock copolymer surfaces using a fluorescence technique (Figure 12C).23 Fluorescence emission panels in Figure 12C demonstrate that the stability and selectivity of protein molecules self-assembled on PS-b-PMMA are effectively retained for downstream protein–protein recognitions and reactions.
Figure 12.
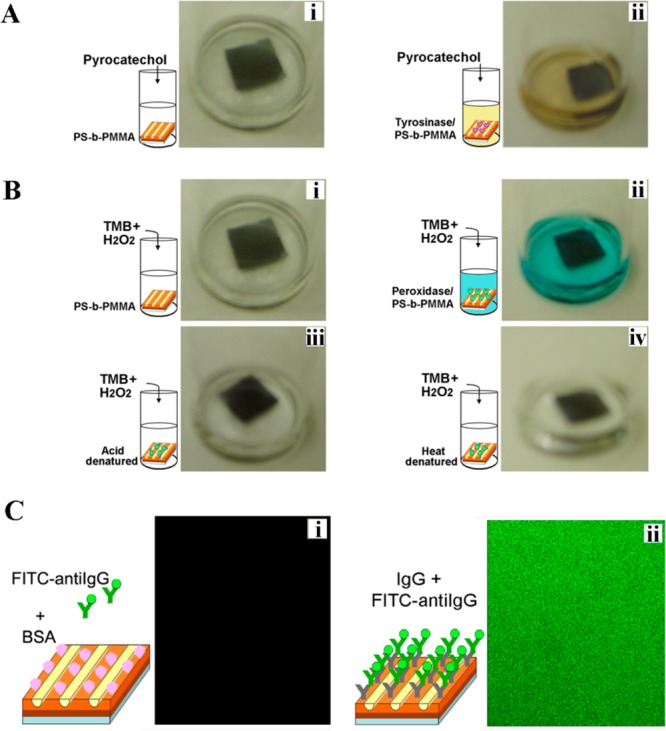
(A) Digital images taken after adding 1 mL of pyrocatechol to vials containing (i) as-annealed PS-b-PMMA where no color change was observed as a result of the absence of the enzyme molecules on the control diblock surface and (ii) PS-b-PMMA with self-assembled MT molecules showing active MT on the surface through the change in the assay color. (B) Digital images taken after adding 1 mL of TMB solution to vials containing (i) an as-annealed PS-b-PMMA substrate with no HRP, (ii) a PS-b-PMMA substrate with self-assembled HRP molecules in which the assay color changed to blue, (iii) a PS-b-PMMA substrate with acid (100 μL of 0.1 N HCl for 1 h)-denatured HRP, and (iv) a PS-b-PMMA substrate with heat (75 °C for 12 h)-denatured HRP. (C) Confocal fluorescence data of protein–protein interaction collected at 400× magnification. The fluorescence panel in i, obtained after the deposition of a 20 μg/mL fluorescein isothiocyanate (FITC)-antiIgG droplet onto BSA-incubated PS-b-PMMA, led to no observable emission. In contrast, clear fluorescence was obtained in panel ii between the self-assembled 20 μg/mL IgG on PS-b-PMMA and the 20 μg/mL FITC-antiIgG analyte introduced subsequently onto the plate. Adapted with permission from refs (23) and (26) (copyright 2007 American Chemical Society).
V.3. Current Challenges and Limits of Protein Nanoarrays
The current findings on nanoscale protein assembly reveal that the nature of protein adsorption differs on the nanometer scale and protein interactions with nanoscale surfaces deviate significantly from the behavior on a larger-scale material, although the chemical makeup of the surface is the same. Despite these insights, very little is yet known for protein-domain specific adsorption behavior on nanoscale surfaces. Few guiding principles exist presently for the design of nanoscale protein interactions with chemically heterogeneous surfaces on the same length scale. Consequently, a fundamental understanding of protein–polymer interaction at the molecular level is still highly warranted to fill the gap in our scientific understanding of nanoscale protein adsorption involving chemically complex surfaces. True nanoscale insights into protein adsorption on polymeric surfaces can shed light on overcoming the current hurdles associated with easy, rapid, low-cost, high-throughput protein assembly and detection.
Conventional optical detection methods dominantly used for protein detection are restricted by the inherent resolution limit known as the optical diffraction limit. The size of the smallest resolvable feature (d) is defined by d= 0.61 λ/NA, where λ is the wavelength of light and NA is the numerical aperture of a lens. The block-copolymer-based protein nanoarrays have the full potential to serve as truly nanoscale optical detection platforms in which signals are independently resolved from each addressable spot with a nanoscale diameter. However, optical detection techniques that can overcome the diffraction limit are required for this to be realized widely in bioapplications. Techniques to overcome the optical diffraction limit exist for research purposes whose examples include near-field signal collection in near-field scanning optical microscopy (NSOM) and tip-enhanced high spatial resolution in tip-enhanced Raman spectroscopy (TERS). However, both NSOM and TERS are used mainly for research purposes in a low-throughput setting and are not yet viable for large-scale applications in basic biology and medical detection. Therefore, it is anticipated that the immediate impact of the block-copolymer-based protein nanoarrays lies in their ability to aid in the signal quantification of existing fluorescence detection coupled in a conventional microarray setting. For example, the approach may be effective at providing known numbers of self-assembled proteins on polymeric nanodomains, after which optical signal such as fluorescence or absorbance can be quantitatively measured from each micrometer-scale spot containing a collection of the nanodomains.
As discussed earlier, a large fraction of surface-bound protein molecules maintain their functionality even after random adsorption, and the number of proteins exhibiting biofunctionality is much greater than what is expected from the random protein orientations assumed on the diblock copolymer surfaces. This observation suggests that protein adsorption may be directional in the diblock copolymer cases, dominated by those amino acid groups on the surface of the protein predominantly driving adsorption to the preferred polymeric domain. The high activity can be hypothesized to originate from this specific protein–surface interaction and the subsequent spatial alignment of the protein on diblock copolymer surfaces through its most energetically favorable surface configuration. This assumption implies that different polymeric surfaces may be used to control the orientation of surface-bound proteins in the protein nanoarrays without the use of chemical or biological functional groups. No direct experimental evidence yet exists for this assumption, although the MD simulation cases of Lyz and Cyt C adsorption onto different nanoscale surfaces support the likelihood of directional protein binding.66,67 Further investigation is necessary to understand protein adsorption behavior on nanoscale surfaces fully, to control the surface conformation of bound proteins, and to evaluate accurately and quantitatively protein functionality that is critical in solid-state arrays.
VI. Concluding Remarks and Outlook
Protein adsorption on nanoscopic, chemically heterogeneous surfaces signifies a scientifically rich, basic research area with important technological ramifications in biology and medicine. The intriguing new phenomena of nanoscale protein assembly discovered recently reveal that the nature of protein interactions is significantly different from its behavior on a microscale or macroscale polymeric material. Although protein adsorption has been extensively studied for many decades, little is yet known about protein-domain-specific adsorption behavior on nanoscale surfaces. Neither direct nor indirect data of high-resolution images or spectroscopic/diffraction evidence exist on the nanoscale pertaining to protein-domain-specific information on surface adsorption. Further opportunities still exist to deepen our understanding of protein–polymer interactions, particularly at the subprotein level. In the future, nanoscale surface adsorption behavior of a protein may be more accurately predicted by considering the chemical and structural complexity of varying surface regions within a protein. New experimental and theoretical research efforts may identify kinetic parameters for protein adsorption/desorption onto chemically heterogeneous, nanoscale surfaces that are distinctive from those on macroscale or microscale surfaces. Such efforts may bring about new discoveries on equilibrium and kinetic characteristics of nanoscale protein–surface interactions and may provide a much needed mechanistic understanding of the key experimental observations still left to be explained.
Acknowledgments
The work highlighted in this feature article was funded by the sources described in the acknowledgement sections of the referenced articles. I acknowledge the National Institutes of Health and a National Research Service Award (1R01DK088016) from the National Institute of Diabetes and Digestive and Kidney Diseases for financial support during the writing of this article.
Biography

Jong-in Hahm received her B.S. from Seoul National University, South Korea and a Ph.D. in chemistry from the University of Chicago and completed her postdoctoral training at Harvard University. She then started her first faculty appointment in the Department of Chemical Engineering at Pennsylvania State University. She is currently an associate professor in the Department of Chemistry at Georgetown University. She conducts fundamental and applied research pertaining to 1D, polymeric, and semiconducting nanomaterials interfacing with biomolecules. She was awarded the 2013 WCC Rising Star Award, 2008 Progress/Dreyfus Lectureship Award, and 2007 WISE Lectureship Award from the American Chemical Society.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Latour R. A.Biomaterials: Protein–Surface Interactions. In Encyclopedia of Biomaterials and Biomedical Engineering, 2nd ed.; Informa Healthcare USA: New York, 2008; Vol. 1, pp 270–284. [Google Scholar]
- Ratner B. D.; Bryant S. J. Biomaterials: where we have been and where we are going. Annu. Rev. Biomed. Eng. 2004, 6, 41–75. [DOI] [PubMed] [Google Scholar]
- Talapatra A.; Rouse R.; Hardiman G. Protein microarrays: challenges and promises. Pharmacogenomics 2002, 3, 1–10. [DOI] [PubMed] [Google Scholar]
- MacBeath G.; Schreiber S. L. Printing proteins as microarrays for high-throughput function determination. Science 2000, 289, 1760–1763. [DOI] [PubMed] [Google Scholar]
- Zhu H.; Snyder M. Protein chip technology. Curr. Opin. Chem. Biol. 2003, 7, 55–63. [DOI] [PubMed] [Google Scholar]
- Sauer S.; Lange B. M. H.; Gobom J.; Nyarsik L.; Seitz H.; Lehrach H. Miniaturization in functional genomics and proteomics. Nat. Rev. Genet. 2005, 6, 465–476. [DOI] [PubMed] [Google Scholar]
- Phizicky E.; Bastiaens P. I. H.; Zhu H.; Snyder M.; Fields S. Protein analysis on a proteomic scale. Nature 2003, 422, 208–215. [DOI] [PubMed] [Google Scholar]
- Hahm J. I. Polymeric surface-mediated, high-density nano-assembly of functional protein arrays. J. Biomed. Nanotechnol. 2011, 7, 731–742. [DOI] [PubMed] [Google Scholar]
- Hahm J. I. Functional polymers in protein detection platforms: Optical, electrochemical, electrical, mass-sensitive, and magnetic biosensors. Sensors 2011, 11, 3327–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten B.; Wanker E. E.; Hoheisel J. D.; Angenendt P. Multiplexed approaches in protein microarray technology. Expert Rev. Proteomics 2005, 2, 499–510. [DOI] [PubMed] [Google Scholar]
- Mooney J. F.; Hunt A. J.; McIntosh J. R.; Liberko C. A.; Walba D. M.; Rogers C. T. Patterning of functional antibodies and other proteins by photolithography of silane monolayers. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 12287–12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrksich M.; Whitesides G. M. Patterning self-assembled monolayers using microcontact printing - a new technology for biosensors. Trends Biotechnol. 1995, 13, 228–235. [Google Scholar]
- Shim H.-W.; Lee J.-H.; Hwang T.-S.; Rhee Y. W.; Bae Y. M.; Choi J. S.; Han J.; Lee C.-S. Patterning of proteins and cells on functionalized surfaces prepared by polyelectrolyte multilayers and micromolding in capillaries. Biosens. Bioelectron. 2007, 22, 3188–3195. [DOI] [PubMed] [Google Scholar]
- Clemmens J.; Hess H.; Lipscomb R.; Hanein Y.; Bohringer K. F.; Matzke C. M.; Bachand G. D.; Bunker B. C.; Vogel V. Mechanisms of microtubule guiding on microfabricated kinesin-coated surfaces: chemical and topographic surface patterns. Langmuir 2003, 19, 10967–10974. [Google Scholar]
- Falconnet D.; Pasqui D.; Park S.; Eckert R.; Schift H.; Gobrecht J.; Barbucci R.; Textor M. A novel approach to produce protein nanopatterns by combining nanoimprint lithography and molecular self-assembly. Nano Lett. 2004, 4, 1909–1914. [Google Scholar]
- Garno J. C.; Amro N. A.; Wadu-Mesthrige K.; Liu G.-Y. Production of periodic arrays of protein nanostructures using particle lithography. Langmuir 2002, 18, 8186–8192. [Google Scholar]
- Delamarche E.; Bernard A.; Schmid H.; Michel B.; Biebuyck H. Patterned delivery of immunoglobulins to surfaces using microfluidic networks. Science 1997, 276, 779–781. [DOI] [PubMed] [Google Scholar]
- Jiang J.; Li X.; Mak W. C.; Trau D. Integrated direct DNA/protein patterning and microfabrication by focused ion beam milling. Adv. Mater. 2008, 20, 1636–1643. [Google Scholar]
- Nishioka G. M.; Markey A. A.; Holloway C. K. Protein damage in drop-on-demand printers. J. Am. Chem. Soc. 2004, 126, 16320–16321. [DOI] [PubMed] [Google Scholar]
- Lee K. B.; Park S. J.; Mirkin C. A.; Smith J. C.; Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science 2002, 295, 1702–1705. [DOI] [PubMed] [Google Scholar]
- Kenseth J. R.; Harnisch J. A.; Jones V. W.; Porter M. D. Investigation of approaches for the fabrication of protein patterns by scanning probe lithography. Langmuir 2001, 17, 4105–4112. [Google Scholar]
- Kumar N.; Hahm J. Nanoscale protein patterning using self-assembled diblock copolymers. Langmuir 2005, 21, 6652–6655. [DOI] [PubMed] [Google Scholar]
- Kumar N.; Parajuli O.; Dorfman A.; Kipp D.; Hahm J. Activity study of self-assembled proteins on nanoscale diblock copolymer templates. Langmuir 2007, 23, 7416–7422. [DOI] [PubMed] [Google Scholar]
- Kumar N.; Parajuli O.; Gupta A.; Hahm J. Elucidation of protein adsorption behavior on polymeric surfaces: towards high density, high payload, protein templates. Langmuir 2008, 24, 2688–2694. [DOI] [PubMed] [Google Scholar]
- Kumar N.; Parajuli O.; Hahm J. Two-dimensionally self-arranged protein nanoarrays on diblock copolymer templates. J. Phys. Chem. B 2007, 111, 4581–4587. [DOI] [PubMed] [Google Scholar]
- Parajuli O.; Gupta A.; Kumar N.; Hahm J. Evaluation of enzymatic activity on nanoscale PS-b-PMMA diblock copolymer domains. J. Phys. Chem. B 2007, 111, 14022–14027. [DOI] [PubMed] [Google Scholar]
- Song S.; Milchak M.; Zhou H. B.; Lee T.; Hanscom M.; Hahm J. I. Nanoscale protein arrays of rich morphologies via self-assembly on chemically treated diblock copolymer surfaces. Nanotechnology 2013, 24, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K. H. A.; Bang J.; Kim D. H.; Knoll W. Self-assembly of protein nanoarrays on block copolymer templates. Adv. Funct. Mater. 2008, 18, 3148–3157. [Google Scholar]
- Wadu-Mesthrige K.; Xu S.; Amro N. A.; Liu G. Y. Fabrication and imaging of nanometer-sized protein patterns. Langmuir 1999, 15, 8580–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E. P.; Wright J. P.; Pham D.; Filipponi L.; Viezzoli A.; Nicolau D. V. Polymer microstructures fabricated via laser ablation used for multianalyte protein microassay. Langmuir 2002, 18, 9539–9546. [Google Scholar]
- Rucker V. C.; Havenstrite K. L.; Simmons B. A.; Sickafoose S. M.; Herr A. E.; Shediac R. Functional antibody immobilization on 3-dimensional polymeric surfaces generated by reactive ion etching. Langmuir 2005, 21, 7621–7625. [DOI] [PubMed] [Google Scholar]
- Aggarwal N.; Lawson K.; Kershaw M.; Horvath R.; Ramsden J. Protein adsorption on heterogeneous surfaces. Appl. Phys. Lett. 2009, 94, 083110. [Google Scholar]
- Bates F. S.; Fredrickson G. H. Block copolymer thermodynamics - theory and experiment. Annu. Rev. Phys. Chem. 1990, 41, 525–557. [DOI] [PubMed] [Google Scholar]
- Fredrickson G. H.; Bates F. S. Dynamics of block copolymers: theory and experiment. Annu. Rev. Mater. Sci. 1996, 26, 501–550. [Google Scholar]
- Bates F. S.; Fredrickson G. H. Block copolymers—designer soft materials. Phys. Today 1999, 52, 32–38. [Google Scholar]
- Morkved T. L.; Lopes W. A.; Hahm J.; Sibener S. J.; Jaeger H. M. Silicon nitride membrane substrates for the investigations of local structures in polymer thin films. Polymer 1998, 39, 3871. [Google Scholar]
- Hahm J.; Sibener S. J. Time-resolved atomic force microscopy imaging studies of asymmetric PS-b-PMMA ultrathin films: Dislocation and disclination transformations, defect mobility, and evolution of nanoscale morphology. J. Chem. Phys. 2001, 114, 4730–4740. [Google Scholar]
- Hahm J.; Sibener S. J. Cylinder alignment in annular structures of microphase-separated polystyrene-b-poly(methyl methacrylate). Langmuir 2000, 16, 4766–4769. [Google Scholar]
- Hahm J.; Lopes W. A.; Jaeger H. M.; Sibener S. J. Defect evolution in ultrathin films of polystyrene-block-polymethylmethacrylate diblock copolymers observed by atomic force microscopy. J. Chem. Phys. 1998, 109, 10111–10114. [Google Scholar]
- Khandpur A. K.; Foerster S.; Bates F. S.; Hamley I. W.; Ryan A. J.; Bras W.; Almdal K.; Mortensen K. Polyisoprene-polystyrene diblock copolymer phase diagram near the order-disorder transition. Macromolecules 1995, 28, 8796–8806. [Google Scholar]
- Sidorov S. N.; Bronstein L. M.; Kabachii Y. A.; Valetsky P. M.; Soo P. L.; Maysinger D.; Eisenberg A. Influence of metalation on the morphologies of poly(ethylene oxide)-block-poly(4-vinylpyridine) block copolymer micelles. Langmuir 2004, 20, 3543–3550. [DOI] [PubMed] [Google Scholar]
- Choucair A.; Eisenberg A. Control of amphiphilic block copolymer morphologies using solution conditions. Eur. Phys. J. E 2003, 10, 37–44. [DOI] [PubMed] [Google Scholar]
- Choucair A.; Lavigueur C.; Eisenberg A. Polystyrene-b-poly(acrylic acid) vesicle size control using solution properties and hydrophilic block length. Langmuir 2004, 20, 3894–3900. [DOI] [PubMed] [Google Scholar]
- Song S.; Milchak M.; Zhou H. B.; Lee T.; Hanscom M.; Hahm J. I. Elucidation of novel nanostructures by time-lapse monitoring of polystyrene-block-polyvinylpyridine under chemical treatment. Langmuir 2012, 28, 8384–8391. [DOI] [PubMed] [Google Scholar]
- Kim T. H.; Huh J.; Hwang J.; Kim H.; Kim S. H.; Sohn B.; Park C. Ordered arrays of PS-b-P4VP micelles by fusion and fission process upon solvent annealing. Macromolecules 2009, 42, 6688–6697. [Google Scholar]
- Peng J.; Kim D. H.; Knoll W.; Xuan Y.; Li B.; Han Y. Morphologies in solvent-annealed thin films of symmetric diblock copolymer. J. Chem. Phys. 2006, 125, 064702. [DOI] [PubMed] [Google Scholar]
- Jones S.; Thornton J. M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansky P.; Russell T. P.; Hawker C. J.; Pitsikalis M.; Mays J. Ordered diblock copolymer films on random copolymer brushes. Macromolecules 1997, 30, 6810–6813. [Google Scholar]
- Laforgue A.; Bazuin C. G.; Prud’homme R. E. A study of the supramolecular approach in controlling diblock copolymer nanopatterning and nanoporosity on surfaces. Macromolecules 2006, 39, 6473–6482. [Google Scholar]
- Cassie A. B. D.; Baxter S. Wettability of porous surfaces. Trans. Faraday Soc. 1944, 40, 546–551. [Google Scholar]
- Wenzel R. N. Resistance of solid surfaces to wetting by water. Ind. Eng. Chem. 1936, 28, 988–994. [Google Scholar]
- Genzer J.; Efimenko K. Creating long-lived superhydrophobic polymer surfaces through mechanically assembled monolayers. Science 2000, 290, 2130–2133. [DOI] [PubMed] [Google Scholar]
- Lei Y.; Leng Y. Hydrophobic drying and hysteresis at different length scales by molecular dynamics simulations. Langmuir 2012, 28, 3152–3158. [DOI] [PubMed] [Google Scholar]
- Giovambattista N.; Lopez C. F.; Rossky P. J.; Debenedetti P. G. Hydrophobicity of protein surfaces: separating geometry from chemistry. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 2274–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal J.; Hummer G. Interfacial thermodynamics of confined water near molecularly rough surfaces. Faraday Discuss. 2010, 146, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J.; Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Hoop T. P.; Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. U.S.A. 1981, 78, 3824–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuisson J.; Murthy S. E.; Bandell M.; Coste B.; Louis-dit-Picard H.; Mathur J.; Fénéant-Thibault M.; Tertian G.; Jaureguiberry J.-P. d.; Syfuss P.-Y.; Cahalan S.; Garçon L.; Toutain F.; Rohrlich P. S.; Delaunay J.; Picard V.; Jeunemaitre X.; Patapoutian A. Dehydrated hereditary stomatocytosis linked to gain-of-function mutations in mechanically activated PIEZO1 ion channels. Nat. Commun. 2013, 4, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz C. F.; Santore M. M. Effect of surface hydrophobicity on adsorption and relaxation kinetics of albumin and fibrinogen: Single-species and competitive behavior. Langmuir 2001, 17, 3006–3016. [Google Scholar]
- Wertz C. F.; Santore M. M. Fibrinogen adsorption on hydrophilic and hydrophobic surfaces: geometrical and energetic aspects of interfacial relaxations. Langmuir 2002, 18, 706–715. [Google Scholar]
- Lau K. H. A.; Bang J.; Hawker C. J.; Kim D. H.; Knoll W. Modulation of protein-surface interactions on nanopatterned polymer films. Biomacromolecules 2009, 10, 1061–1066. [DOI] [PubMed] [Google Scholar]
- Wei Y.; Latour R. A. Determination of the adsorption free energy for peptide-surface interactions by SPR spectroscopy. Langmuir 2008, 24, 6721–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y.; Latour R. A. Correlation between desorption force measured by atomic force microscopy and adsorption free energy measured by surface plasmon resonance spectroscopy for peptide-surface interactions. Langmuir 2010, 26, 18852–18861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M. C. Quartz crystal microbalance with dissipation monitoring: enabling real-time characterization of biological materials and their interactions. J. Biomol. Technol. 2008, 19, 151–158. [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-W.; Gupta V. K. A SPR and AFM study of the effect of surface heterogeneity on adsorption of proteins. J. Chem. Phys. 2004, 121, 2264–2271. [DOI] [PubMed] [Google Scholar]
- Hung A.; Mwenifumbo S.; Mager M.; Kuna J. J.; Stellacci F.; Yarovsky I.; Stevens M. M. Ordering surfaces on the nanoscale: Implications for protein adsorption. J. Am. Chem. Soc. 2011, 133, 1438–1450. [DOI] [PubMed] [Google Scholar]
- Hung A.; Mager M.; Hembury M.; Francesco Stellacci d.; Stevensc M. M.; Yarovsky I. Amphiphilic amino acids: a key to adsorbing proteins to nanopatterned surfaces. Chem. Sci. 2013, 4, 928–937. [Google Scholar]
- Wan J.; Thomas M. S.; Guthrie S.; Vullev V. I. Surface-bound proteins with preserved functionality. Anal. Biomed. Eng. 2009, 37, 1190–1205. [DOI] [PubMed] [Google Scholar]
- Cha T. W.; Guo A.; Zhu X.-Y. Enzymatic activity on a chip: The critical role of protein orientation. Proteomics 2005, 5, 416–419. [DOI] [PubMed] [Google Scholar]
- Azevedo A. M.; Martins V. C.; Prazeres D. M.; Vojinovic V.; Cabral J. M.; Fonseca L. P. Horseradish peroxidase: a valuable tool in biotechnology. Biotechnol. Annu. Rev. 2003, 9, 199–247. [DOI] [PubMed] [Google Scholar]