Abstract
Despite intensive research effort, the rational design of improved nanoparticulate drug carriers remains challenging, in part due to a limited understanding of the determinants of nanoparticle entry and transport in target cells. Recent studies have shown that Niemann-Pick C1 (NPC1), the lysosome membrane protein that mediates trafficking of cholesterol in cells, is involved in the endosomal escape and subsequent infection caused by filoviruses, and that its absence promotes the retention and efficacy of lipid nanoparticles encapsulating siRNA. Here, we report that NPC1 deficiency results in dramatic reduction in internalization and transfection efficiency mediated by degradable cationic gene delivery polymers, poly(β-amino ester)s (PBAEs). PBAEs utilized cholesterol and dynamin-dependent endocytosis pathways, and these were found to be heavily compromised in NPC1-deficient cells. In contrast, the absence of NPC1 had minor effects on DNA uptake mediated by polyethylenimine or Lipofectamine 2000. Strikingly, stable overexpression of human NPC1 in chinese hamster ovary cells was associated with enhanced gene uptake (3-fold) and transfection (10-fold) by PBAEs. These findings reveal a role of NPC1 in the regulation of endocytic mechanisms affecting nanoparticle trafficking. We hypothesize that in-depth understanding sites of entry and endosomal escape may lead to highly efficient nanotechnologies for drug delivery.
Keywords: nanoparticles, endocytosis, NPC1, PBAE, polymer, intracellular trafficking gene delivery
The tremendous medical promise of gene therapy remains unmet due to the lack of safe and effective delivery vehicles.1 Though regarded as efficient gene carriers, viral vectors are associated with safety risks including insertional mutagenesis and adverse immune responses.2 Nonviral gene vectors offer the possibility of improved safety but have generally failed to attain satisfactory delivery efficacy in clinical testing.3,4 One challenge to the rational design of synthetic nanocarriers is the limited understanding of the relationship between nanoparticle structure and function, as well as a lack of insight into the rate-limiting steps and key cellular regulators affecting delivery efficacy.5 Although several groups are working toward characterization of different physicochemical parameters of nanomedicines on cellular uptake, the endosomal machinery involved in the internalization, transport, and escape of nanoparticles to the cytosol remains elusive.6,7
Cellular uptake of nanoparticles is critical for intracellular delivery and has been the subject of various studies.8 Typical endocytic mechanisms include macropinocytosis, clathrin-dependent endocytosis, caveolae-mediated endocytosis, and a number of clathrin- and caveolae-independent pathways such as RhoA-dependent, Arf6-dependent, Cdc42-dependent, and flotillin-dependent endocytosis.9 A handful of nanocarriers appear to rely predominantly on a single pathway. Certain kinds of poly(lactic-co-glycolic acid) (PLGA) nanoparticles, for instance, were reported to be internalized by vascular smooth muscle cells via clathrin-dependent endocytosis,10−12 whereas DOXIL and Abraxane nanoparticles enter tumor cells via caveolae-mediated endocytosis.13,14 Macropinocytosis, meanwhile, appeared to be the major pathway used for entry in HeLa cells by certain siRNA-containing nanoparticles composed of lipid-like materials (LNPs).15 However, for many nanoparticles used for nonviral gene delivery, including poly-l-lysine (PLL) and polyethylenimine (PEI)-based polyplexes, as well as various lipoplexes and liposomes, evidence exists for cellular entry via multiple endocytic mechanisms.16−23
Poly(β-amino ester)s (PBAEs) are biodegradable cationic polymers that mediate gene delivery in a variety of in vitro and in vivo contexts, including suicide gene therapy of several animal models of cancer24−26 as well as genetic modification of stem cells for treatment of ischemia.27 These polymers have demonstrated superior performance and less toxicity in several difficult-to-transfect cell types28−30 compared to commercially available transfection reagents such as Lipofectamine 2000 (LF 2000). The versatility of the polymerization chemistry31 has allowed a broad set of structures to be synthesized and screened in a high-throughput manner,32−36 permitting systematic investigation of key parameters affecting gene delivery potency such as polymer molecular weight distribution,37 hydrophobicity of the side chains,38 and amine end-group structure.39−42 The endocytic mechanisms used by PBAEs for internalization remain unreported.
Here, we show that while PBAE/DNA nanoparticles appear to use multiple pathways to enter cells, efficient cellular uptake and gene transfection depend on the cholesterol transport function of Niemann-Pick C1 (NPC1). We have identified a unique trafficking pathway selective to PBAEs that, upon being misregulated, has a drastic effect on the ability of the polymer to deliver DNA inside cells. This study indicates that NPC1 plays an important role in regulating endocytic mechanisms affecting internalization and efficacy of certain nanoparticles.
Results
Identification of U18666A as an Inhibitor of PBAE-Mediated Gene Transfection
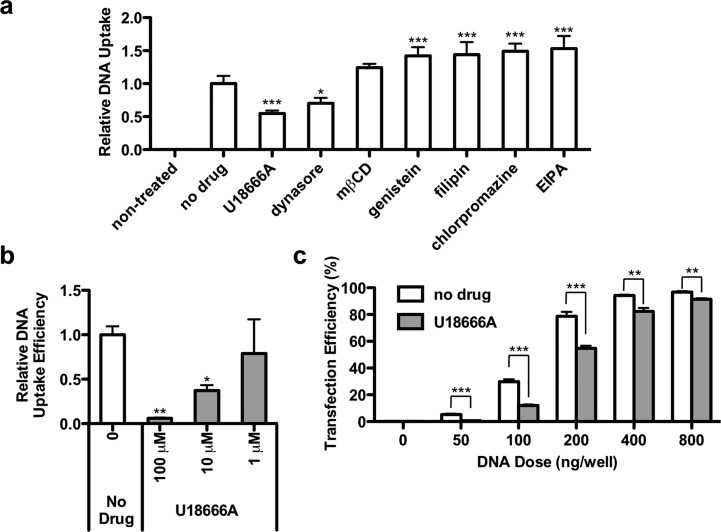
To assess the mechanisms of cellular entry used by poly(β-amino ester)s, we screened a set of inhibitors for their potential to reduce cellular internalization of fluorescently labeled DNA by C32–122, one of the top-performing amine end-modified PBAEs. Immortalized mouse embryonic fibroblast (MEF) cells were pretreated with various inhibitors for 1 h, then transfected with C32–122 polyplexes containing Cy5-labeled DNA in the presence of these inhibitors. Based on optimization experiments, the inhibitors were used at concentrations below the threshold of inducing notable cytotoxicity (data not shown). After 3 h, the cells were washed multiple times, fixed, stained with DAPI, and analyzed by high-throughput confocal microscopy for cell-associated fluorescent signal (Figure 1a). Treatment with inhibitors of clathrin-mediated endocytosis (chlorpromazine), macropinocytosis (EIPA), and caveolae-mediated endocytosis (genistein, filipin, and methyl-β-cyclodextrin) had no significant effect or caused increased uptake, but treatment with dynasore, an inhibitor of dynamin-dependent pathways including clathrin-mediated and caveolae-mediated endocytosis, significantly reduced relative DNA uptake mediated by PBAEs. The significant increases in uptake observed for some inhibitors such as EIPA and chlorpromazine suggest that the cells may compensate for incomplete suppression by promoting or upregulating alternate endocytic pathways.43 However, DNA uptake was inhibited to the greatest extent in the presence of the small molecule U18666A, an inhibitor of cholesterol trafficking.
Figure 1.
U18666A inhibits C32–122-mediated uptake of DNA in MEFs. (a) MEFs were transfected with C32–122 polyplexes containing Cy3-labeled DNA in the presence of various endocytic pathway inhibitors (5 μM U18666A, 50 μM dynasore, 1 mM methyl-β-cyclodextrin, 10 μM genistein, 5 μM filipin, 5 μM chlorpromazine, 10 μM EIPA). MEFs were pretreated with the inhibitors 1 h prior to transfection. After 3 h, the cells were washed, fixed, treated with a nuclear stain, and analyzed by high-throughput confocal microscopy to quantify relative DNA uptake (mean ± SD, n = 3); * indicates p < 0.05, *** indicates p < 0.001, compared to no drug control. (b) Relative DNA uptake efficiencies in MEFs (mean ± SD, n = 3) as quantified by FACS 3 h following transfection of C32–122/Cy5-labeled DNA in the presence of the indicated concentration of U18666A; * indicates p < 0.05, ** indicates p < 0.01, compared to no drug control. (c) Overall MEF transfection efficiency (mean ± SD, n = 4) as quantified by FACS 24 h following transfection of C32–122/GFP-encoding DNA in the presence of 5 μM U18666A; ** indicates p < 0.01, *** indicates p < 0.001 for the comparisons indicated.
Using fluorescence-activated cell sorting (FACS) analysis, we quantified relative DNA uptake in these cells following PBAE-based transfection in the presence of various doses of U18666A. U18666A treatment was associated with dose-dependent reductions in DNA internalization by C32–122 (Figure 1b). To determine if U18666A inhibits overall gene transfection by PBAEs, we transfected MEFs with various doses of C32–122 polyplexes containing GFP-encoding plasmid DNA in the presence of the inhibitor and assessed GFP expression efficiency by FACS at 24 h. As expected, U18666A substantially inhibited gene transfection by C32–122 (Figure 1c).
Studies with NPC1-Deficient Mouse Embryonic Fibroblasts
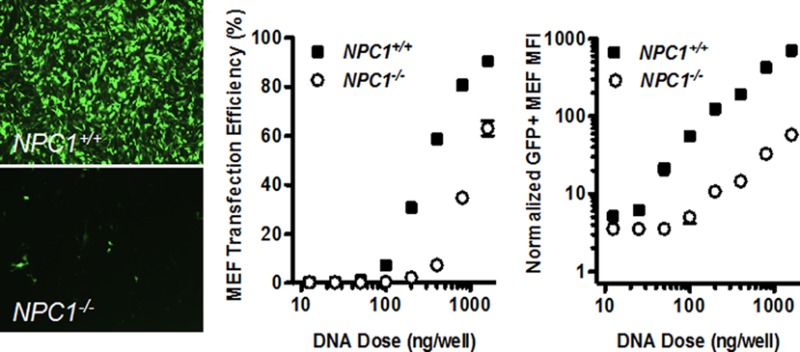
Because U18666A inhibits cholesterol synthesis and trafficking, we hypothesized that Niemann-Pick C1, an endo/lysosomal protein involved in cholesterol trafficking, may play a role in C32–122-mediated DNA transfection. Using MEF cell lines obtained from wild-type or NPC1–/– mice, we compared gene transfection efficiencies 1 day after treatment with C32–122 polyplexes containing GFP-encoding plasmid DNA. Fluorescence microscopy showed that, for equivalent DNA doses, GFP expression was dramatically lower in NPC1–/– MEFs relative to the wild-type cells (Figure 2a). When FACS analysis was used to compare the C32–122/DNA dose–response profiles of the two cell lines, we observed that the ED50 concentration was approximately 3-fold greater for the NPC1–/– cells than that for the wild-type cells (Figure 2b). Even more strikingly, GFP expression levels of positively transfected cells were reduced over 10-fold in NPC1 knockout cells (Figure 2c).
Figure 2.
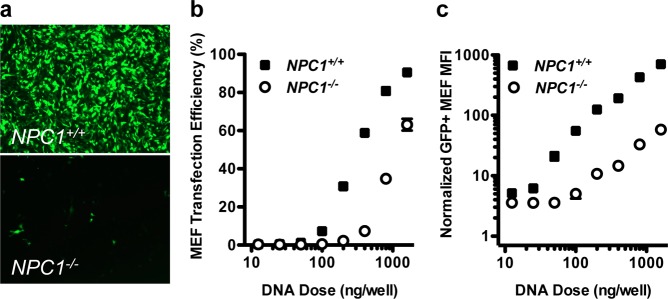
NPC1 knockout inhibits C32–122-mediated DNA transfection of MEFs. NPC1+/+ and NPC1–/– MEFs were incubated for 3 h with various doses of C32–122 polyplexes containing GFP-encoding plasmid DNA, and GFP expression was assessed by fluorescence microscopy and FACS after 24 h. (a) Representative images showing decreased GFP expression in NPC1–/– MEFs relative to wild-type MEFs, and FACS analysis of (b) GFP expression efficiency and (c) geometric mean fluorescent intensity (MFI) of GFP-expressing cells normalized to nontreated cells (mean ± SD, n = 4).
To determine whether NPC1 knockout reduces DNA uptake by C32–122, high-throughput confocal microscopy was used to examine the fluorescent intensity levels of NPC1+/+ and NPC1–/– MEFs treated with polyplexes incorporating Cy3-labeled DNA (Figure 3a). We observed that NPC1 deficiency was associated with significantly (3-fold) decreased uptake across a range of DNA doses tested (Figure 3b).
Figure 3.
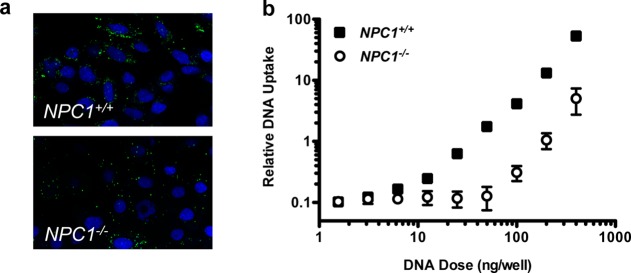
NPC1 knockout inhibits C32–122-mediated internalization of DNA in MEFs. NPC1+/+ and NPC1–/– MEFs were transfected with various doses of C32–122 polyplexes containing Cy3-labeled plasmid DNA (green). After 3 h, the cells were washed, fixed, treated with the nuclear stain Hoescht (blue), and analyzed by high-throughput confocal microscopy. (a) Representative images showing inhibition of uptake in NPC1–/– MEFs, and (b) quantification of DNA uptake (mean ± SD, n = 3).
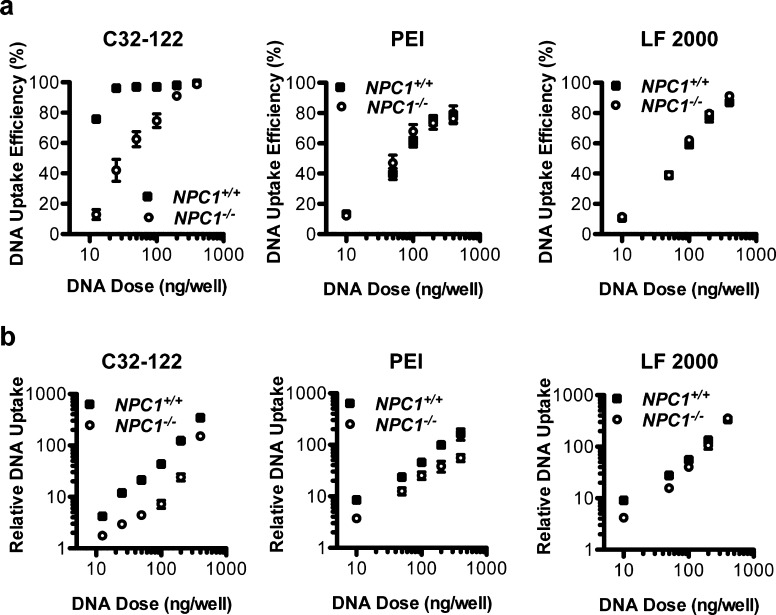
Characterization of Cy5-labeled DNA internalization by FACS analysis confirmed that NPC1 knockout significantly reduced both the efficiency and the fluorescent intensity of MEFs transfected using C32–122 (Figure 4). To determine whether NPC1 deficiency also affected DNA internalization mediated by other transfection reagents, we conducted analogous experiments with PEI and LF 2000. NPC1 knockout had no significant impact on labeled DNA uptake efficiency or the overall uptake level by LF 2000, while NPC1 knockout was associated with a modest reduction in overall uptake, but had little effect on internalization efficiency by PEI (Figure 4).
Figure 4.
Effects of NPC1 knockout on internalization of DNA in MEFs following transfection with C32–122, PEI, and LF 2000.NPC1+/+ and NPC1–/– MEFs were incubated with various doses Cy5-labeled plasmid DNA complexed with C32–122, polyethylenimine, or Lipofectamine 2000. After 3 h, the cells were washed, fixed, and analyzed by FACS to determine (a) DNA uptake efficiency (mean ± SD, n = 4) and (b) geometric mean Cy5 fluorescent intensity normalized to nontreated cells (mean ± SD, n = 4).
Studies with Markers of Endocytosis
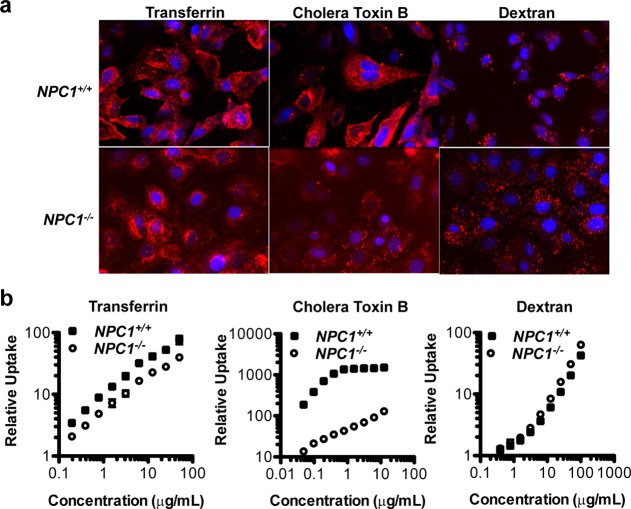
We next assessed the relative uptake of various known markers of endocytic pathways in NPC1+/+ and NPC1–/– MEFs. These markers included fluorescently labeled transferrin, cholera toxin B, and 10 000 MW dextran, which have been shown to undergo internalization via clathrin-dependent endocytosis, caveolin-mediated endocytosis, and macropinocytosis, respectively.8 After a 3 h incubation with the cell lines and multiple washes, we observed dramatically reduced uptake of cholera toxin B in the NPC1-deficient cells (Figure 5a). Furthermore, transferrin uptake appeared to be slightly decreased and dextran uptake slightly increased. Quantification of these experiments by FACS analysis corroborated these trends (Figure 5b). When we compared NPC1-deficient cells to wild-type cells, cholera toxin B internalization was reduced ∼20–30-fold; transferrin uptake was reduced about 2-fold; and dextran uptake was increased by ∼50%.
Figure 5.
NPC1 knockout alters relative endocytic pathway activities in MEFs. (a) MEFs were incubated for 3 h with media containing various AF647-labeled (red) markers: 50 μg/mL transferrin (left), 55 ng/mL cholera toxin B (middle), or 100 μg/mL 10 000 MW dextran (right), traditional markers of clathrin-dependent, endocytosis, caveolae-mediated endocytosis, and macropinocytosis, respectively. After 3 h, the cells were washed, fixed, treated with the nuclear stain Hoescht (blue), and analyzed by high-throughput confocal microscopy. (b) MEFs were incubated for 3 h with media containing various doses of transferrin, cholera toxin B, or dextran, each labeled with AF647. After 3 h, the cells were washed and fixed, and relative uptake was assessed by FACS as the geometric mean AF647 fluorescent intensity normalized to nontreated cells (mean ± SD, n = 4).
To determine whether PBAE polyplexes share common uptake pathways with these markers during endocytosis, we incubated the C32–122 polyplexes containing labeled DNA in the presence of each one of these markers and used confocal microscopy to characterize the extent of colocalization in wild-type MEFs at various time points. Although there was evidence of DNA colocalization with all three markers, the greatest extent of colocalization occurred with cholera toxin B (Figure 6). These results suggest that PBAE polyplexes and cholera toxin B rely on common uptake pathways for internalization.
Figure 6.
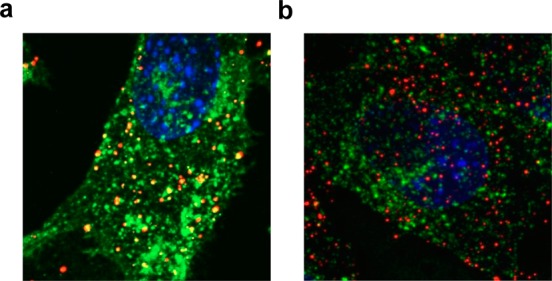
Colocalization of internalized DNA delivered by C32–122 with markers of distinct endocytic pathways in MEFs. NPC1+/+ MEFs were transfected with C32–122 polyplexes containing Cy3-labeled DNA in the presence of AF647-labeled (a) 55 ng/mL cholera toxin B or (b) 50 μg/mL transferrin. After 3 h, the cells were washed, fixed, treated with the nuclear stain Hoescht (blue), and analyzed by confocal microscopy for intracellular colocalization of DNA (green) with each marker (red).
Studies with CHO Cell Lines Varying in NPC1 Expression
To further elucidate the effects of NPC1, we examined C32–122/DNA uptake and transfection in chinese hamster ovary (CHO) epithelial cell lines with varying NPC1 expression (Figure 7). These cell lines included wild-type CHO cells, NPC1-deficient CHO cells (null), CHO cells expressing NPC1 with a mis-sense mutation in the sterol-sensing domain resulting in defective cholesterol trafficking44 (P692S) and wild-type CHO cells stably overexpressing human NPC145 (hNPC1). As was observed with the immortalized MEFs, NPC1 deficiency in CHO cells inhibited both DNA uptake and transfection mediated by C32–122. In the P692S CHO cells, uptake and transfection were inhibited to a nearly identical extent as the null cells, confirming the specific dependence on NPC1’s cholesterol-trafficking function for PBAE-mediated gene delivery. Surprisingly, we observed increased C32–122-mediated DNA uptake and transfection in the CHO cells stably overexpressing human NPC1 (Figure 7). These cells have been reported to have a 1.5-fold increase in total cellular cholesterol and a 2.9-fold increase in cholesterol at the plasma membrane.45 It is possible that increased cholesterol localization at the plasma membrane may enhance the activities of certain endocytic mechanisms used by PBAE/DNA nanoparticles. These observations raise the possibility of modulating key cellular factors to improve nonviral gene delivery.
Figure 7.
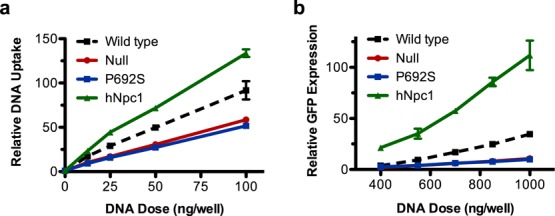
C32–122-mediated DNA uptake and transfection in CHO cell lines varying in NPC1 expression. CHO cells varying in NPC1 expression were incubated for 3 h with the indicated doses of (a) Cy5-labeled plasmid DNA or (b) GFP-encoding plasmid DNA complexed with C32–122 polymer. FACS analysis was used to assess the (a) geometric mean Cy5 fluorescent intensities (MFI) of treated cells relative to nontreated cells immediately after washing or (b) GFP geometric MFI of treated cells relative to nontreated cells after 24 h (mean ± SD, n = 4).
Discussion
Although research within the nonviral gene delivery field continues to focus on materials engineering, the rational design of more advanced nanocarriers will likely benefit from a deeper understanding of the cellular mechanisms involved in nanoparticle internalization and transfection. Toward this end, we examined the uptake of a promising class of degradable cationic polymers, PBAEs, using a variety of tools including pharmacological pathway inhibitors, labeled uptake markers, and gene knockout cell lines. The inhibitor experiments identified dynasore and U18666A as suppressors of polyplex internalization in a mouse embryonic fibroblast cell line (Figure 1). Dynasore inhibits a broad set of dynamin-dependent pathways including clathrin-mediated and caveolae-mediated endocytosis, suggesting that PBAEs may use more than one pathway to enter these cells. U18666A is an amphipathic steroid with multiple actions, inhibiting both the synthesis of cholesterol46−49 and its trafficking from late endosomes and lysosomes to the plasma membrane and the endoplasmic reticulum.50−52 It has also been widely studied as a means of inducing a model of Niemann-Pick type C1 disease53−55 and may directly interact with and inhibit the sterol-sensing site of the NPC1 protein.54,56,57
NPC1 is a 13 transmembrane protein present on the late endosomes responsible for the egress of cholesterol from cells. NPC1 has recently been identified as an essential factor involved in endosomal escape of certain filoviruses,58,59 yet its absence improves the retention and efficacy of lipid/siRNA nanoparticles.6 Examination of NPC1-deficient MEFs indicates that NPC1 plays a significant role in gene transfection and uptake by poly(β-amino ester)s such as C32–122 but only a minor one in DNA internalization by PEI and LF 2000 (Figures 2–4). We demonstrate that NPC1 knockout in MEFs significantly alters normal activities of various endocytic pathways, with dramatically decreased caveolae-mediated endocytosis, slightly reduced clathrin-mediated endocytosis, and slightly upregulated macropinocytosis (Figure 5). This observation is supported by examination of marker colocalization experiments, which indicate that while PBAE/DNA polyplexes likely use multiple mechanisms to enter MEFs, caveolae-mediated endocytosis was the primary route (Figure 6).
Both CHO cells that are devoid of NPC1, or stably express NPC1 with a mutated sterol-sensing domain, led to decreases in uptake and transfection efficiency of PBAEs, further supporting our previous observation that cholesterol transport significantly impacts gene delivery (Figure 7). Although reduced uptake was identified as one factor in the diminished transfection efficiency of NPC1-deficient cells, NPC1 may also contribute to downstream steps such as endosomal escape and/or nuclear transport of the polyplexes since a 3-fold decrease in uptake led to a 10-fold decrease in transfection efficiency in CHO cells (Figure 7).
Because NPC1 plays a role in regulating the trafficking of cholesterol and because cholesterol has been shown to be critically involved in both caveolin- and clathrin-mediated endocytosis,60−64 we had hypothesized that NPC1 knockout would affect these processes. To our knowledge, although NPC1 knockout has been associated with altered intracellular localization of cholera toxin B and lactosylceramide (LacCer),56,65 two markers of caveolin-mediated endocytosis, and with defective recycling of transferrin receptor,66 quantitative analysis of uptake for multiple endocytic markers has not yet been reported. Despite previous reports that fluid-phase uptake of horseradish peroxidase is impaired in NPC1 knockout cells67,68 or cause no change in macropinocytosis-based internalization, our results indicate that macropinocytic internalization of dextran is slightly increased in NPC1-deficient cells. Additionally, these results provide further perspective on earlier work by our group suggesting that lipid/siRNA nanoparticles are retained in cells longer in NPC1-deficient cells due to defects in recycling, despite being internalized by macropinocytosis. In contrast to both PBAE/DNA nanoparticles (which likely enter via caveolae) and lipid/siRNA nanoparticles (which enter via macropinocytosis),6 cholesterol in general enters through the clathrin-dependent pathway and exits the late endosomes via NPC1-mediated trafficking.69
Conclusions
The investigation of nonviral DNA transfection of NPC1-deficient cells reported here demonstrates the importance of cholesterol trafficking for certain delivery formulations such as poly(β-amino ester)s. In this study, one of the most striking observations was that stable overexpression of NPC1 significantly improved DNA internalization and transfection by PBAEs (Figure 7). These findings raise the tantalizing prospect of enhancing nonviral gene delivery through active modulation of cellular factors known to play key roles in internalization, intracellular trafficking, or endosomal progression such as lysobisphosphatidic acid, Rab5, and perhaps NPC1. Manipulation of NPC1 or other such targets would of course need to be performed in a manner that is specific, targeted, and temporary and, due to the potential risks, would likely only be considered for patients suffering from genetic disorders for which there is no current viable therapy.
Materials and Methods
Materials
1,4-Butanediol diacrylate and 5-amino-1-pentanol were purchased from Alfa Aesar (Ward Hill, MA, USA). Dodecylamine was purchased from Sigma-Aldrich (St. Louis, MO, USA). (PEO)4-bisamine (“122”) was acquired from Molecular Biosciences (Boulder, CO, USA). All chemical reagents were used without further purification. Plasmids encoding green fluorescent protein (gWiz-GFP) and firefly luciferase (gWiz-Luc) were purchased from Aldevron (Fargo, ND, USA). jetPEI (Polyplus Transfection, Illkirch, France) and Lipofectamine 2000 were purchased from VWR (Radnor, PA, USA) and Invitrogen (Carlsbad, CA, USA), respectively. Transferrin, cholera toxin B, and 10 000 MW dextran, each labeled with AlexaFluor 647, were purchased from Invitrogen. Cytochalasin D, dynasore hydrate, chlorpromazine hydrochloride, filipin III, genistein, methyl-β-cyclodextrin, 5-(N-ethyl-N-isopropyl)amiloride, and U18666A were obtained from Sigma-Aldrich. Immortalized mouse embryonic fibroblast cell lines were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen).
Polymer Synthesis
To synthesize C32–122, acrylate-terminated C32 poly(β-amino ester) was first prepared in a 5 g batch by reacting 1,4-butanediol diacrylate (“C”) and 5-amino-1-pentanol (“32”) (1.2:1.0 diacrylate/amine molar ratio) without solvent at 90 °C for 24 h with stirring. After cooling to room temperature and dissolving the polymer in 10 mL of anhydrous THF, it was added to a vial containing 10 mmol of (PEO)4-bisamine (10 mmol in 40 mL of anhydrous THF). Following overnight stirring at room temperature, the amine end-modified polymer was purified by precipitation in anhydrous diethyl ether (1:3 v/v THF/ether) and dried under vacuum for 24 h. C32–122 was then dissolved at 100 mg/mL in dimethyl sulfoxide (DMSO) and stored at −20 °C with desiccant until use.
DNA Transfection Experiments
One day before transfection, cells (100 μL) were seeded into each well of a 96-well polystyrene tissue culture plate (HeLa: 12 500 per well; MEFs: 9000 per well). For studies using the pharmacological inhibitors, conditioned medium was removed 1 h prior to transfection and replaced with fresh, prewarmed medium containing the indicated concentration of pharmacological inhibitor. For GFP transfection experiments, gWiz-GFP (5 mg mL–1) was diluted to 160 μg mL–1 in 25 mM sodium acetate (NaOAc) buffer at pH 5.2; for DNA uptake experiments, gWiz-Luc was labeled with Cy3 or Cy5 using the LabelIt kit (Mirus, Madison, WI, USA) following the manufacturer’s instructions and was diluted in NaOAc buffer as above. PBAEs (100 mg mL–1) were thawed immediately prior to transfection and diluted in NaOAc buffer to a concentration of 3.2 mg mL–1 (20:1 w/w polymer/DNA). To form DNA–polymer nanoparticles, polymer solution (200 μL) was added to the diluted DNA (200 μL), mixed by repeated pipetting, and allowed to incubate for 10 min at room temperature. Depending on the dose, polymer–DNA complexes were diluted in NaOAc as needed, and then were gently mixed in a deep 96-well plate with prewarmed fresh medium (360 μL). For inhibition experiments, this medium was prepared with the indicated concentration of pharmacological inhibitor, whereas for colocalization experiments, labeled transferrin, cholera toxin B, or dextran were present. Conditioned medium was removed using a 12-channel aspirating wand and replaced with the complexes diluted in medium (100 μL). jetPEI and Lipofectamine 2000 were used according to the manufacturers’ protocols.
For GFP transfection experiments, following a 3 h incubation at 37 °C, complexes were removed with the aid of a multichannel aspiration wand and replaced with fresh medium (100 μL), and the cells were analyzed for GFP expression by fluorescence-activated cell sorting after 24 h at 37 °C. For DNA uptake experiments, cells were washed three times at the indicated time point and prepared for analysis by either confocal microscopy (high-throughput or high-resolution microscopy) as previously described or using FACS.6
FACS Analysis
After aspirating conditioned medium and washing cells three times with PBS, cells were detached using 25 μL per well of 0.25% trypsin–EDTA (Invitrogen). Following a 5 min incubation at 37 °C, fresh medium (50 μL) was added to the cells, which were mixed thoroughly and then transferred to a 96-well round-bottom plate. Cells were then pelleted, resuspended in fixation buffer (4% v/v formaldehyde in PBS), incubated for 10 min at room temperature, pelleted again, and finally resuspended in ice-cold FACS running buffer (2% v/v FBS in PBS) containing 1:200 v/v propidium iodide (Invitrogen). The cells were kept at 4 °C until FACS analysis using a BD LSR II (Becton Dickinson, San Jose, CA, USA). Except for experiments with Cy3-labeled DNA, propidium iodide (PI) staining was used to exclude dead cells from the analysis. PI staining was also used to determine the viabilities of treated cells relative to nontreated control cells, where the relative viability was calculated as the ratio of live (unstained) treated cells per well to the mean number of live nontreated cells per well. For GFP expression analysis, 2D gating was used to separate increased autofluorescence signals from increased GFP signals to more accurately count positively expressing cells. Gating and analysis were performed using FlowJo v8.8 software (TreeStar, Ashland, OR, USA). Geometric mean fluorescent intensities of transfected cells were normalized to those of the corresponding nontransfected control cells.
Statistics
Data are expressed as mean ± SD for groups of at least three replicates. Data in Figure 1a,b were analyzed for statistical significance by one-way ANOVA with Bonferroni multiple comparison correction. Differences between drug-treated and control groups in Figure 1c were analyzed for statistical significance by unpaired, two-tailed Student’s t test with 95% confidence. Statistical tests were implemented in GraphPad Prism 5 (*p < 0.05, **p < 0.01, and ***p < 0.001).
Acknowledgments
The authors would like to thank Dr. Peter Lobel and Liang Huang from Rutgers University and Dr. Daniel Ory from Washington University for NPC1-deficient cells as well as Alnylam Pharmaceuticals for the use of their high-throughput confocal microscope. We would also like to thank Dr. Sovan Sarkar for healthy discussion. We would like to thank the Koch Institute Swanson Biotechnology Flow Cytometery Core facility and the confocal core facility at Whitehead institute for their support. This work was supported by National Cancer Institute Grant U54CA151884, 5R01HL107550-03 grant and Program of Excellence in Nanotechnology (PEN) Award, Contract #HHSN268201000045C from the National Heart, Lung, and Blood Institute, National Institutes of Health. This work was also supported by the Skolkovo Foundation Grant as part of the Center for RNA therapeutics and biology.
Author Contributions
¶ A.A.E. and G.S. contributed equally.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Kay M. A. State-of-the-Art Gene-Based Therapies: The Road Ahead. Nat. Rev. Genet. 2011, 12, 316–328. [DOI] [PubMed] [Google Scholar]
- Giacca M.; Zacchigna S. Virus-Mediated Gene Delivery for Human Gene Therapy. J. Controlled Release 2012, 161, 377–388. [DOI] [PubMed] [Google Scholar]
- Mintzer M. A.; Simanek E. E. Nonviral Vectors For Gene Delivery. Chem. Rev. 2009, 109, 259–302. [DOI] [PubMed] [Google Scholar]
- Edelstein M. Gene Therapy Clinical Trials Worldwide. http://Www.Wiley.Com/Legacy/Wileychi/Genmed/Clinical/ (Accessed March 2011).
- Guo X.; Huang L. Recent Advances in Nonviral Vectors for Gene Delivery. Acc. Chem. Res. 2012, 45, 971–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay G.; Querbes W.; Alabi C.; Eltoukhy A.; Sarkar S.; Zurenko C.; Karagiannis E.; Love K.; Chen D.; Zoncu R.; et al. Efficiency of SiRNA Delivery by Lipid Nanoparticles Is Limited by Endocytic Recycling. Nat. Biotechnol. 2013, 31, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangarkar C.; Dinh A.-T.; Mitragotri S. Endocytic Pathway Rapidly Delivers Internalized Molecules to Lysosomes: An Analysis of Vesicle Trafficking, Clustering and Mass Transfer. J. Controlled Release 2012, 162, 76–83. [DOI] [PubMed] [Google Scholar]
- Sahay G.; Alakhova D. Y.; Kabanov A. V. Endocytosis of Nanomedicines. J. Controlled Release 2010, 145, 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty G. J.; Mcmahon H. T. Mechanisms of Endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. [DOI] [PubMed] [Google Scholar]
- Vasir J. K.; Labhasetwar V. Quantification of the Force of Nanoparticle–Cell Membrane Interactions and Its Influence on Intracellular Trafficking of Nanoparticles. Biomaterials 2008, 29, 4244–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyam J.; Labhasetwar V. Dynamics of Endocytosis and Exocytosis of Poly(d,l-Lactide-co-Glycolide) Nanoparticles in Vascular Smooth Muscle Cells. Pharm. Res. 2003, 20, 212–220. [DOI] [PubMed] [Google Scholar]
- Qaddoumi M. G.; Ueda H.; Yang J.; Davda J.; Labhasetwar V.; Lee V. H. The Characteristics and Mechanisms of Uptake of PLGA Nanoparticles in Rabbit Conjunctival Epithelial Cell Layers. Pharm. Res. 2004, 21, 641–648. [DOI] [PubMed] [Google Scholar]
- Sahay G.; Kim J. O.; Kabanov A. V.; Bronich T. K. The Exploitation of Differential Endocytic Pathways in Normal and Tumor Cells in the Selective Targeting of Nanoparticulate Chemotherapeutic Agents. Biomaterials 2010, 31, 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele E.; Spinelli G. P.; Tomao F.; Tomao S. Albumin-Bound Formulation of Paclitaxel (Abraxane Abi-007) in the Treatment of Breast Cancer. Int. J. Nanomed. 2009, 4, 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love K. T.; Mahon K. P.; Levins C. G.; Whitehead K. A.; Querbes W.; Dorkin J. R.; Qin J.; Cantley W.; Qin L. L.; Racie T.; et al. Lipid-like Materials for Low-Dose, In Vivo Gene Silencing. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman J.; Bragonzi A.; Conese M. Role of Clathrin- and Caveolae-Mediated Endocytosis in Gene Transfer Mediated by Lipo- and Polyplexes. Mol. Ther. 2005, 12, 468–474. [DOI] [PubMed] [Google Scholar]
- Luhmann T.; Rimann M.; Bittermann A. G.; Hall H. Cellular Uptake and Intracellular Pathways of PLL-G-PEG-DNA Nanoparticles. Bioconjugate Chem. 2008, 19, 1907–1916. [DOI] [PubMed] [Google Scholar]
- Van Der Aa M. A.; Huth U. S.; Hafele S. Y.; Schubert R.; Oosting R. S.; Mastrobattista E.; Hennink W. E.; Peschka-Suss R.; Koning G. A.; et al. Cellular Uptake of Cationic Polymer–DNA Complexes via Caveolae Plays a Pivotal Role in Gene Transfection in Cos-7 Cells. Pharm. Res. 2007, 24, 1590–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Gersdorff K.; Sanders N. N.; Vandenbroucke R.; De Smedt S. C.; Wagner E.; Ogris M. The Internalization Route Resulting in Successful Gene Expression Depends on Both Cell Line and Polyethylenimine Polyplex Type. Mol. Ther. 2006, 14, 745–753. [DOI] [PubMed] [Google Scholar]
- Wong A. W.; Scales S. J.; Reilly D. E. DNA Internalized via Caveolae Requires Microtubule-Dependent, Rab7-Independent Transport to the Late Endocytic Pathway for Delivery to the Nucleus. J. Biol. Chem. 2007, 282, 22953–22963. [DOI] [PubMed] [Google Scholar]
- Grosse S.; Aron Y.; Thévenot G.; François D.; Monsigny M.; Fajac I. Potocytosis and Cellular Exit of Complexes as Cellular Pathways for Gene Delivery by Polycations. J. Gene Med. 2005, 7, 1275–1286. [DOI] [PubMed] [Google Scholar]
- Rejman J.; Conese M.; Hoekstra D. Gene Transfer by Means of Lipo- and Polyplexes: Role of Clathrin and Caveolae-Mediated Endocytosis. J. Liposome Res. 2006, 16, 237–247. [DOI] [PubMed] [Google Scholar]
- Khalil I. A.; Kogure K.; Futaki S.; Harashima H. High Density of Octaarginine Stimulates Macropinocytosis Leading to Efficient Intracellular Trafficking for Gene Expression. J. Biol. Chem. 2006, 281, 3544–3551. [DOI] [PubMed] [Google Scholar]
- Anderson D. G.; Peng W.; Akinc A.; Hossain N.; Kohn A.; Padera R.; Langer R.; Sawicki J. A. A Polymer Library Approach to Suicide Gene Therapy for Cancer. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16028–16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W.; Anderson D. G.; Bao Y.; Padera R. F. Jr.; Langer R.; Sawicki J. A. Nanoparticulate Delivery of Suicide DNA to Murine Prostate and Prostate Tumors. Prostate 2007, 67, 855–862. [DOI] [PubMed] [Google Scholar]
- Huang Y. H.; Zugates G. T.; Peng W.; Holtz D.; Dunton C.; Green J. J.; Hossain N.; Chernick M. R.; Padera R. F. Jr.; Langer R.; et al. Nanoparticle-Delivered Suicide Gene Therapy Effectively Reduces Ovarian Tumor Burden in Mice. Cancer Res. 2009, 69, 6184–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F.; Cho S. W.; Son S. M.; Bogatyrev S. R.; Singh D.; Green J.; Mei Y.; Park S.; Bhang S. H.; Kim B. S.; et al. Genetic Engineering of Human Stem Cells for Enhanced Angiogenesis Using Biodegradable Polymeric Nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 3317–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J.; Shi J.; Chiu E.; Leshchiner E.; Langer R.; Anderson D. Biodegradable Polymeric Vectors for Gene Delivery to Human Endothelial Cells. Bioconjugate Chem. 2006, 17, 1162–1169. [DOI] [PubMed] [Google Scholar]
- Green J.; Zhou B.; Mitalipova M.; Beard C.; Langer R.; Jaenisch R.; Anderson D. Nanoparticles for Gene Transfer to Human Embryonic Stem Cell Colonies. Nano Lett. 2008, 8, 3126–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F.; Green J. J.; Dinio T.; Keung L.; Cho S. W.; Park H.; Langer R.; Anderson D. G. Gene Delivery to Human Adult and Embryonic Cell-Derived Stem Cells Using Biodegradable Nanoparticulate Polymeric Vectors. Gene Ther. 2009, 16, 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn D. M.; Langer R. Degradable Poly(β-amino esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA. J. Am. Chem. Soc. 2000, 122, 10761–10768. [Google Scholar]
- Lynn D.; Anderson D.; Putnam D.; Langer R. Accelerated Discovery of Synthetic Transfection Vectors: Parallel Synthesis and Screening of a Degradable Polymer Library. J. Am. Chem. Soc. 2001, 123, 8155–8156. [DOI] [PubMed] [Google Scholar]
- Akinc A.; Lynn D.; Anderson D.; Langer R. Parallel Synthesis and Biophysical Characterization of a Degradable Polymer Library for Gene Delivery. J. Am. Chem. Soc. 2003, 125, 5316–5323. [DOI] [PubMed] [Google Scholar]
- Anderson D. G.; Lynn D. M.; Langer R. Semi-automated Synthesis and Screening of a Large Library of Degradable Cationic Polymers for Gene Delivery. Angew. Chem., Int. Ed. 2003, 42, 3153–3158. [DOI] [PubMed] [Google Scholar]
- Akinc A.; Anderson D. G.; Lynn D. M.; Langer R. Synthesis of Poly(β-amino ester)s Optimized for Highly Effective Gene Delivery. Bioconjugate Chem. 2003, 14, 979–988. [DOI] [PubMed] [Google Scholar]
- Anderson D.; Akinc A.; Hossain N.; Langer R. Structure/Property Studies of Polymeric Gene Delivery Using a Library of Poly(β-amino esters). Mol. Ther. 2005, 11, 426–434. [DOI] [PubMed] [Google Scholar]
- Eltoukhy A. A.; Siegwart D. J.; Alabi C. A.; Rajan J. S.; Langer R.; Anderson D. G. Effect of Molecular Weight of Amine End-Modified Poly(β-amino ester)s on Gene Delivery Efficiency and Toxicity. Biomaterials 2012, 33, 3594–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltoukhy A. A.; Chen D.; Alabi C. A.; Langer R.; Anderson D. G. Degradable Terpolymers with Alkyl Side Chains Demonstrate Enhanced Gene Delivery Potency and Nanoparticle Stability. Adv. Mater. 2013, 25, 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. J.; Zugates G. T.; Tedford N. C.; Huang Y.; Griffith L. G.; Lauffenburger D. A.; Sawicki J. A.; Langer R.; Anderson D. G. Combinatorial Modification of Degradable Polymers Enables Transfection of Human Cells Comparable to Adenovirus. Adv. Mater. 2007, 19, 2836–2842. [Google Scholar]
- Zugates G. T.; Peng W.; Zumbuehl A.; Jhunjhunwala S.; Huang Y. H.; Langer R.; Sawicki J. A.; Anderson D. G. Rapid Optimization of Gene Delivery by Parallel End-Modification of Poly(β-amino ester)s. Mol. Ther. 2007, 15, 1306–1312. [DOI] [PubMed] [Google Scholar]
- Zugates G.; Tedford N.; Zumbuehl A.; Jhunjhunwala S.; Kang C.; Griffith L.; Lauffenburger D.; Langer R.; Anderson D. Gene Delivery Properties of End-Modified Poly(β-amino ester)s. Bioconjugate Chem. 2007, 18, 1887–1896. [DOI] [PubMed] [Google Scholar]
- Sunshine J.; Green J. J.; Mahon K. P.; Yang F.; Eltoukhy A. A.; Nguyen D. N.; Langer R.; Anderson D. G. Small-Molecule End-Groups of Linear Polymer Determine Cell-Type Gene-Delivery Efficacy. Adv. Mater. 2009, 21, 4947–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A. I.Pharmacological Inhibition of Endocytic Pathways: Is It Specific Enough to Be Useful? In Exocytosis And Endocytosis; Springer: Berlin, 2008; pp 15–33. [DOI] [PubMed] [Google Scholar]
- Millard E. E.; Gale S. E.; Dudley N.; Zhang J.; Schaffer J. E.; Ory D. S. The Sterol-Sensing Domain of the Niemann-Pick C1 (NPC1) Protein Regulates Trafficking of Low Density Lipoprotein Cholesterol. J. Biol. Chem. 2005, 280, 28581–28590. [DOI] [PubMed] [Google Scholar]
- Millard E. E.; Srivastava K.; Traub L. M.; Schaffer J. E.; Ory D. S. Niemann-Pick Type C1 (NPC1) Overexpression Alters Cellular Cholesterol Homeostasis. J. Biol. Chem. 2000, 275, 38445–38451. [DOI] [PubMed] [Google Scholar]
- Phillips W. A.; Avigan J. Inhibition of Cholesterol Biosynthesis in the Rat by 3 beta-(2-Diethylaminoethoxy) Androst5-en-17-one Hydrochloride. Proc. Soc. Exp. Biol. Med. 1963, 112, 233–236. [DOI] [PubMed] [Google Scholar]
- Cenedella R. J. Concentration-Dependent Effects of Ay-9944 and U18666a on Sterol Synthesis in Brain. Variable Sensitivities of Metabolic Steps. Biochem. Pharmacol. 1980, 29, 2751–2754. [DOI] [PubMed] [Google Scholar]
- Sexton R. C.; Panini S. R.; Azran F.; Rudney H. Effects of 3 beta-[2-(Diethylamino)ethoxy]androst-5-en-17-one on the Synthesis of Cholesterol and Ubiquinone in Rat Intestinal Epithelial Cell Cultures. Biochemistry 1983, 22, 5687–5692. [DOI] [PubMed] [Google Scholar]
- Panini S. R.; Sexton R. C.; Rudney H. Regulation of 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase by Oxysterol By-products of Cholesterol Biosynthesis. Possible Mediators of Low Density Lipoprotein Action. J. Biol. Chem. 1984, 259, 7767–7771. [PubMed] [Google Scholar]
- Liscum L.; Faust J. R. The Intracellular Transport of Low Density Lipoprotein-Derived Cholesterol Is Inhibited in Chinese Hamster Ovary Cells Cultured with 3-beta-[2-(Diethylamino)ethoxy]androst-5-en-17-one. J. Biol. Chem. 1989, 264, 11796–11806. [PubMed] [Google Scholar]
- Liscum L. Pharmacological Inhibition of the Intracellular Transport of Low-Density Lipoprotein-Derived Cholesterol in Chinese Hamster Ovary Cells. Biochim. Biophys. Acta 1990, 1045, 40–48. [DOI] [PubMed] [Google Scholar]
- Liscum L.; Collins G. J. Characterization of Chinese Hamster Ovary Cells That Are Resistant to 3-beta-[2-(Diethylamino)ethoxy]androst-5-en-17-one Inhibition of Low Density Lipoprotein-Derived Cholesterol Metabolism. J. Biol. Chem. 1991, 266, 16599–165606. [PubMed] [Google Scholar]
- Liscum L.; Klansek J. J. Niemann-Pick Disease Type C. Curr. Opin. Lipidol. 1998, 9, 131–135. [DOI] [PubMed] [Google Scholar]
- Lange Y.; Ye J.; Rigney M.; Steck T. Cholesterol Movement in Niemann-Pick Type C Cells and in Cells Treated with Amphiphiles. J. Biol. Chem. 2000, 275, 17468–17475. [DOI] [PubMed] [Google Scholar]
- Ko D. C.; Gordon M. D.; Jin J. Y.; Scott M. P. Dynamic Movements of Organelles Containing Niemann-Pick C1 Protein: NPC1 Involvement in Late Endocytic Events. Mol. Biol. Cell 2001, 12, 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y.; Ninomiya H.; Ohsaki Y.; Higaki K.; Davies J. P.; Ioannou Y. A.; Ohno K. Accumulation of Cholera Toxin and Gm1 Ganglioside in the Early Endosome of Niemann-Pick C1-Deficient Cells. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 12391–12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.; Lu P.; Chu J. W.; Sharom F. J. Characterization of Fluorescent Sterol Binding to Purified Human NPC1. J. Biol. Chem. 2009, 284, 1840–1852. [DOI] [PubMed] [Google Scholar]
- Côté M.; Misasi J.; Ren T.; Bruchez A.; Lee K.; Filone C. M.; Hensley L.; Li Q.; Ory D.; Chandran K.; et al. Small Molecule Inhibitors Reveal Niemann-Pick C1 Is Essential for Ebola Virus Infection. Nature 2011, 477, 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E.; Raaben M.; Wong A. C.; Herbert A. S.; Obernosterer G.; Mulherkar N.; Kuehne A. I.; Kranzusch P. J.; Griffin A. M.; Ruthel G.; et al. Ebola Virus Entry Requires the Cholesterol Transporter Niemann-Pick C1. Nature 2011, 477, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy D. I.; Li W. P.; Luby-Phelps K.; Anderson R. G. Caveolin Targeting to Late Endosome/Lysosomal Membranes Is Induced by Perturbations of Lysosomal pH and Cholesterol Content. Mol. Biol. Cell 2012, 23, 864–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil A.; Gaidarov I.; Kobylarz K.; Lampson M. A.; Keen J. H.; Mcgraw T. E. Acute Cholesterol Depletion Inhibits Clathrin-Coated Pit Budding. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 6775–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal S. K.; Skretting G.; Garred O.; Vilhardt F.; Van Deurs B.; Sandvig K. Extraction of Cholesterol with Methyl-beta-Cyclodextrin Perturbs Formation of Clathrin-Coated Endocytic Vesicles. Mol. Biol. Cell 1999, 10, 961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z. J.; Singh R. D.; Sharma D. K.; Holicky E. L.; Hanada K.; Marks D. L.; Pagano R. E. Distinct Mechanisms of Clathrin-Independent Endocytosis Have Unique Sphingolipid Requirements. Mol. Biol. Cell 2006, 17, 3197–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper N. M. Detergent-Insoluble Glycosphingolipid/Cholesterol-Rich Membrane Domains, Lipid Rafts and Caveolae (Review). Mol. Membr. Biol. 1999, 16, 145–156. [DOI] [PubMed] [Google Scholar]
- Sun X.; Marks D. L.; Park W. D.; Wheatley C. L.; Puri V.; O’Brien J. F.; Kraft D. L.; Lundquist P. A.; Patterson M. C.; Pagano R. E.; et al. Niemann-Pick C Variant Detection by Altered Sphingolipid Trafficking and Correlation with Mutations within a Specific Domain of NPC1. Am. J. Hum. Genet. 2001, 68, 1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipalia N. H.; Hao M.; Mukherjee S.; Maxfield F. R. Sterol, Protein and Lipid Trafficking in Chinese Hamster Ovary Cells with Niemann-Pick Type C1 Defect. Traffic 2007, 8, 130–141. [DOI] [PubMed] [Google Scholar]
- Mayran N.; Parton R. G.; Gruenberg J. Annexin II Regulates Multivesicular Endosome Biogenesis in the Degradation Pathway of Animal Cells. EMBO J. 2003, 22, 3242–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Vruchte D.; Lloyd-Evans E.; Veldman R. J.; Neville D. C.; Dwek R. A.; Platt F. M.; Van Blitterswijk W. J.; Sillence D. J. Accumulation of Glycosphingolipids in Niemann-Pick C Disease Disrupts Endosomal Transport. J. Biol. Chem. 2004, 279, 26167–26175. [DOI] [PubMed] [Google Scholar]
- Ikonen E. Cellular Cholesterol Trafficking and Compartmentalization. Nat. Rev. Mol. Cell Biol. 2008, 9, 125–138. [DOI] [PubMed] [Google Scholar]