Abstract
The shortnose sturgeon, Acipenser brevirostrum, oft considered a phylogenetic relic, is listed as an “endangered species threatened with extinction” in the US and “Vulnerable” on the IUCN Red List. Effective conservation of A. brevirostrum depends on understanding its diversity and evolutionary processes, yet challenges associated with the polyploid nature of its nuclear genome have heretofore limited population genetic analysis to maternally inherited haploid characters. We developed a suite of polysomic microsatellite DNA markers and characterized a sample of 561 shortnose sturgeon collected from major extant populations along the North American Atlantic coast. The 181 alleles observed at 11 loci were scored as binary loci and the data were subjected to multivariate ordination, Bayesian clustering, hierarchical partitioning of variance, and among-population distance metric tests. The methods uncovered moderately high levels of gene diversity suggesting population structuring across and within three metapopulations (Northeast, Mid-Atlantic, and Southeast) that encompass seven demographically discrete and evolutionarily distinct lineages. The predicted groups are consistent with previously described behavioral patterns, especially dispersal and migration, supporting the interpretation that A. brevirostrum exhibit adaptive differences based on watershed. Combined with results of prior genetic (mitochondrial DNA) and behavioral studies, the current work suggests that dispersal is an important factor in maintaining genetic diversity in A. brevirostrum and that the basic unit for conservation management is arguably the local population.
Introduction
Sturgeons (Acipenseridae) are one of two living groups of chondrostean fishes; the other group being the paddlefishes (Polydontidae). The fossil record suggests these were dominant fishes of the Permian period (∼200 Myr [1]) and owing to morphological similarities to their extinct relatives, modern sturgeons often are described as being evolutionarily static [2], [3]. Acipenserids also are notable for their anadromous and amphidromous life histories, unique benthic and life-history specializations, and the propensity for inter-genus and inter-species hybridization, the latter resulting in various levels of polyploidy which is slightly at odds with the fact that estimated mutation rates within the mitochondrial and nuclear genomes of acipenserids are reduced compared to other fishes [4], [5], [6]. The continued existence of these relic fishes is in jeopardy throughout North America, Europe, and Asia where nearly all sturgeon species have experienced overfishing, habitat degradation or loss, and obstruction of spawning areas. Much effort has been directed at understanding ecological factors associated with sturgeon biology [7] and behavior [8], [9] to address the prevailing conservation biology tenet that management planning must be framed in terms of providing conditions that will facilitate potential adaptation. For resource managers to plan for an evolutionary future for such “trust species,” they must have the means to identify evolutionarily distinct and significant lineages (e.g., species, metapopulations, populations, distinct population segments). However, some of the same characters that herald scientific interest (e.g., polyploidization, lengthy lifespans, long times to maturation, and intermittent semi-annual spawning) exacerbate efforts to identify and protect evolutionarily distinct lineages within each species.
Documenting heritable genetic information is a hallmark of contemporary conservation strategies for delineating management units and has previously been applied to sturgeons where recent molecular systematics studies have called into question the taxonomic foundations of the Acipenseriform classification, which has been historically based on morphological characters [10], [11], [12], [13]. Different gene regions, including some physically linked within the mitochondrial (mt) DNA molecule, have yielded differing phylogenetic interpretations of the Acipenseridae (see [13] for review). Efforts to resolve the molecular systematics of the Acipenseridae using nuclear (n) DNA sequences have either focused on a single gene region (18S rRNA, [5]) or on interspecific comparisons of repetitive DNAs observed as a result of genomic DNA digestion (HindIII [14], PstI [6]). The former was complicated by the polyploid genome and provided no additional phylogenetic resolution, whereas the latter studies concur to some degree with molecular phylogeny observed with mtDNA sequences [6]. Unfortunately, not all sturgeon species have detectable levels of some satellite DNAs [14] and furthermore, the level of intraspecific variability has been shown to be greater than the interspecific divergence among species belonging to the same phylogenetic clade [6]. Given such equivocal results, some revision of Acipenseriform classification is needed to guide conservation efforts. Ultimately, this lack of genomic resources for sturgeons hinders mechanistic study (but see [15]).
Gene duplication and subsequent functional divergence is a fundamental process of adaptive evolution [16] and is particularly relevant in the Acipenseridae where the presence of evolutionary polyploidy ranging in a series from 4n-8n-12n times the ancestral haploid number [17] presents significant challenges for investigating the evolutionary processes shaping the nuclear genomes [4], [18], [19]. It is unclear whether these polyploid events resulted from complete genome duplications (autopolyploid), hybridization between species of different ploidy levels combined with genome doubling (allopolyploidy), or a combination of these processes [20]. Following the polyploid events that gave rise to extant sturgeon species, the random and gradual diploidization process [21] is assumed to have resulted in functional diploidy [22]; however, the degree to which their various polyploid nuclear genomes exhibit disomic inheritance is unknown.
The shortnose sturgeon Acipenser brevirostrum is an amphidromous species endemic to the large coastal rivers of eastern North America. This species is distinguished among all the living acipenserids by exhibiting the largest number of chromosomes, 372 [18]. A. brevirostrum was listed as an “endangered species” under the US Endangered Species Preservation Act of 1967 and remains so despite re-assessment in response to a 1994 petition to de-list populations in tributaries to the Gulf of Maine. Of significant note is that of 19 putative population units identified based on the species' perceived strong fidelity to natal rivers [23], [24] some river populations continue to exist, although much reduced, but in other rivers, the species has been extirpated. In most instances, spawning status is either unknown or indicated to be of limited extent [25], [23] further complicating the prediction of biological units that could respond to conservation measures.
To date, all published information on phylogeographic- and population-level structuring in A. brevirostrum has been assessed through nucleotide sequence variation detected in the maternally-inherited mtDNA. This is presumably due to the difficult nature of interpreting allelic data from the functionally polyploid (putatively hexaploid) nuclear genome [26]. The mtDNA research has primarily been focused on a moderately polymorphic 440 base pair segment of the control region (CR) adjacent to the tRNA proline gene. These findings are well documented in the peer-reviewed literature [27], [28], [29], [26], [30], [31] and are consistent both among studies and between researchers. Although results reflect a shallow gene genealogy (gene tree) for the A. brevirostrum mtDNA CR, analyses of haplotype frequencies at the level of putative individual populations showed significant differences among nearly all river/estuarine systems in which reproduction is known to occur. One prior study [31] concluded that although higher level genetic relationships exist (e.g., Northeast vs. Mid-Atlantic; Northeast vs. Southeast; Mid-Atlantic vs. Southeast; and other Mid-Atlantic regional subdivisions), A. brevirostrum appear to function in discrete populations, and that relatively low female-mediated gene flow exists between the majority of populations. This implies that effective dispersal among drainages within regions has been sufficient to prevent deep divergence within this species over evolutionary time scales.
Acipenser brevirostrum has been shown to possess the highest number of chromosomes (N = 362–372) among all the Acipenseriformes karyotyped to date [18]. These authors, however, could not determine the species' exact level of polysomy (hexaploid or dodecaploid). Contemporary cytogenetic techniques (including signals from fluorescent in situ hybridization) suggest A. brevirostrum is a hexaploid species [19]. While immensely complex, nuclear DNA-based approaches to A. brevirostrum conservation could identify significant levels of informative genetic variation because certain duplicated loci and repetitive DNA may lack functional constraints, thus allowing rapid accumulation of differentiation in DNA sequences [32]. Moreover, if the observed patterns of nuclear DNA diversity and variation differed from those empirically determined for the maternally-inherited mtDNA, this would inform biologists of the degrees of site philopatry or sex-biased dispersal for A. brevirostrum. However, no phylogeographic or population informative nuclear markers have been identified for A. brevirostrum [33].
To address this important research need and for the first time, allow an extensive assessment of the phylogeographic structure of A. brevirostrum from a multilocus nuclear DNA perspective, we characterized the inheritance of polysomic microsatellite DNA loci in shortnose sturgeon collected throughout the species' range using loci derived specifically from this species [34]. Because of the complex modes of inheritance underlying the putatively hexaploid genome, we scored each allele (fragment) as a dominant marker with two states, presence or absence, resulting in the production of a binary character matrix. We report on the findings of an extensive statistical comparison of the patterns in allelic variation to identify and assess the reproductive status of populations and to delineate functional units of management to aid in recovery planning.
Methods
Sample collection
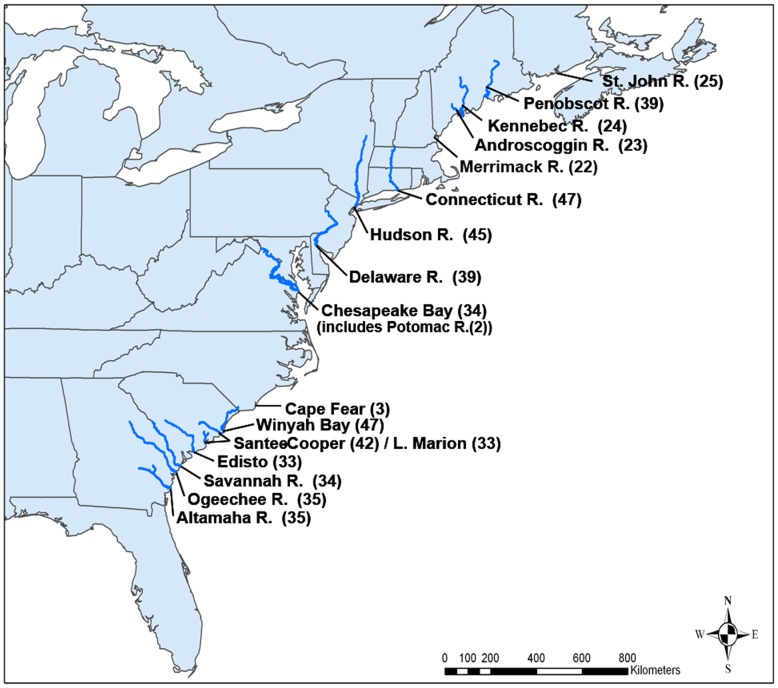
While no tissue sampling was collected as part of this study, Acipenser brevirostrum (n = 561) were sampled from 17 river and estuarine systems representing the species' range (Figure 1) by researchers approved and permitted by the Institutional Animal Care and Use Committee of the U.S. Department of Commerce, National Oceanic and Atmospheric Administration, National Marine Fisheries Service. Sampling followed the guidelines mandated under NOAA Technical Memorandum NMFS-OPR-18 or NOAA Technical Memorandum NMFS-OPR-45. The mandated non-invasive procedures were that tissue (1.0 cm2 fin-clip) taken from soft pelvic fin was stored in 95% absolute ethanol or SDS/urea.
Figure 1. Map of a portion of the North American Atlantic Coast depicting the general location and sample size of 17 river and estuary collections of shortnose sturgeon (Acipenser brevirostrum) surveyed at 11 polysomic microsatellite DNA loci.
Sample sizes are in parentheses.
The ambiguous reproductive status of A. brevirostrum in the Potomac and Merrimack Rivers affected categorization of specimens. We chose to treat the Potomac River collection (n = 2) both as of unknown origin and as part of the large Chesapeake Bay-proper collection. The Merrimack River sample consisted of males collected at the same location and time; however, because eggs (n = 4) and embryos (n = 2) were collected in Spring 2009, we considered the Merrimack River collection as a reproducing population. Genomic DNA from ethanol preserved samples was extracted with the Gentra Puregene DNA kit (Qiagen, Valencia, CA) following the manufacturers guidelines for whole non-mammalian blood and resuspended in TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). Previously extracted genomic DNA from other researchers was extracted using the methods described in [28]. DNA concentrations were determined by fluorescence assay [35], and the integrity of the DNA was visually inspected on 1% agarose gels [36]. SDS-urea preserved samples were also processed using these procedures with the exception that cell lysis was not necessary and samples were subjected directly to protein precipitation and alcohol purification. All DNA samples were quantified as described above and diluted to 100 ng/µl for use in PCR amplification.
A suite of 11 microsatellite loci previously identified from A. brevirostrum [34] was surveyed: AbrB438, AbrD10, AbrD114, AbrD135, AbrD141, AbrD193, AbrD236, AbrD332, AbrD345, AbrD379, and AbrD557. Optimized PCR mixes consisted of the following: 100–200 ng of genomic DNA, 1× PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl), 2.0 mM MgCl2, 0.2 mM dNTPs, 0.4 mg/ml Bovine Serum Albumin, 2.0 U Taq DNA polymerase (Promega Corporation, Madison, WI, USA), 0.25 µM forward and 0.5 µM reverse primer, and 0.3 µM fluorescent labeled M13 primer in a total volume of 25 µl. Amplifications were carried out on either PTC-200 or PTC-225 Thermal Cyclers (Bio-Rad Laboratories, Hercules, CA) using the following cycling conditions: initial denaturing at 94°C for 15 min; 29 cycles of 94°C for 1 min; annealing temperature (56–66°C) for 45 sec, 72°C for 45 sec; 5 cycles of 94°C for 1 min; 53°C for 45 sec; 72°C for 45 sec; and final extension at 72°C for 10 min. Fluorescently labeled fragment analysis was performed using an ABI 3130XL Genetic Analyzer and binning of alleles was performed using GENESCAN 2.1 Analysis software and GENOTYPER 3.6 fragment analysis software (Applied Biosystems, Grand Island, NY). Each locus was independently scored, and each amplicon meeting the signal strength conditions specified (at least 10% relative to the strongest allele) and fitting into the appropriate size category (based on repeat motif and an assumed step-wise mutational pattern) was classified as an allele.
Statistical analyses of polyploid microsatellite markers
For polyploid individuals, gene duplication, multiple alleles, and the mode of inheritance can lead to practical and statistical complications in allelic identification and interpreting summary and population-level statistics [37]. Genetic stock identification studies on other Acipenserids using polysomic (tetrasomic) markers [38], [39] estimated gene dosages by relative peak intensity from electropherograms. Because of the large number of alleles (fragments) observed in the putatively hexaploid A. brevirostrum, allele dosage could not be reliably estimated from GENESCAN runs preventing the application of standard population genetic diversity statistics that require genotype or allele frequencies for their calculation. Two prior groups [39], [40] provided a validated approach to this dilemma and as in those studies, we scored each allele (fragment) as its own psuedodominant locus with one of two states, presence or absence, resulting in the production of a binary character (or allele) matrix. When codominant markers are screened in higher order polyploid species, and scored as psuedodominant loci (i.e., as binary character state), it is not possible to estimate either allele frequencies or heterozygosities directly. Allele (loci) frequencies at 11 polysomic microsatellite DNA markers were estimated on the A. brevirostrum allelotype matrix using the method of [41] as implemented by GenAlEx 6.3 [42]. Only alleles with an overall frequency of >1% were used for these and other statistical analyses.
Allelotypes were analyzed for patterns of population ‘genetic’ structure and for regions of genetic discontinuity at both the individual and collection levels. Pair-wise genetic distances of each individual fish to all other individuals (simple match coefficient; [43]) were calculated using the binary distance routine in GenAlEx. When calculated across multiple loci for a given pair of samples, this is equivalent to the tally of state differences between the two DNA allelotype profiles. Principal coordinate analyses (PCO) were used to graphically compare the individual distances without imposing the appearance of a bifurcating evolutionary history (ordinated with PAlaeontological STatistics ver. 1.76, PAST, [44]).
A second individual-based analysis designed to infer population structure among collections and identify genetic discontinuity in the allelotype matrix was performed using the model-based clustering method of the program STRUCTURE 2.3.2 [45]. Due to complex migration patterns assumed to exist among disjunct populations, a sequential method of inferring clusters (k) was used by first identifying the “uppermost” hierarchical level of population structure followed by subsequent analysis of each cluster to identify within-cluster structure [46]. In the initial phase, k = 1 to k = 20 clusters were considered for the 17 collections using a burn-in of 20,000 followed by 50,000 iterations, and 20 independent runs for each k. The optimum number of clusters in the initial phase was identified using Δk as described by Evanno et al. [46]. Subsequent analysis of each cluster tested k = 1 to k = C+3 (the number of collections (C) included in the subset plus three), with a burn-in of 20,000 followed by 50,000 iterations, and 20 runs for each k. In the within-cluster analyses, k also was determined using the Evanno et al. method [46].
Direct comparisons were made of two pair-wise population-scale distance measures, Jaccard's and ΦPT, for assessing the underlying structure contained in the allelotype matrix. Jaccard's similarity coefficient [47] was calculated using PAST as it is one of the most commonly used and recommended [48] ecologically-based measures of similarity between populations. A non-parametric Analysis of Similarity (ANOSIM) [49] was performed on Jaccard's distance metric (1-Jaccard similarity) measured for 17 collections of A. brevirostrum using PAST. The test statistic, R (resulting from the distance values being converted to rank values) and its significance were computed by permutation of group membership, with 10,000 replicates. R values were presumed to be proportional to genetic distance. AMOVA ΦPT calculated in GenAlEx using the binary data was considered as a coefficient of dissimilarity [43], [42]. Pair-wise ΦPT values and significance levels of the variance components (H0 = no genetic difference among populations; ΦPT = 0), based on 10,000 permutations, were measured for all collections. The relationship between the ecological-based (Jaccard's) distance and the population genetic distance (ΦPT) matrices for all 17 collections was statistically assessed with a matrix regression analysis [50] performed by the MXCOMP routine in NTSYS-PC 1.8 [51].
Because a small number of effective migrants can have a profound effect on a small population, as the effects of drift can be large in the absence of balancing gene exchange, estimation of the number of migrants rather than simply the rate [52] is an important conservation consideration. We estimated the effective migrants per generation using ΦPT for the amount of genetic differentiation at these selectively neutral microsatellite loci according to S. Wright's classical relationship [53]. Although this approximation of gene flow has been shown to be quite robust under a range of demographic conditions [54], [55] the empirical validity of substituting the binary character analog of FST, ΦPT, into this equation is, to our knowledge, untested.
The evolutionary history among the 17 collections (geographic populations) of A. brevirostrum was inferred by analysis of the pair-wise population ΦPT matrix using two methods. The non-metric multidimensional scaling (NMDS, [55]) option of PAST was used for visualization of the non-parametric monotonic relationship among the dissimilarity matrix, the Euclidean distance between collections, and the location of each collection in low-dimensional space. In addition, the Neighbor-Joining method [57], a clustering procedure based on the minimum-evolution criterion for phylogeographic trees (the topology resulting in the minimum total branch length at each step of the algorithm) was performed with MEGA4, [58]. The associated pair-wise ΦPT distance matrix was subjected to clustering with 5000 bootstrap replicates using the program PAST.
To compare the genetic variation observed for nDNA (this study) with mtDNA [31] genomes of A. brevirostrum, two comparisons were made to visualize patterns among the 14 collections common to both studies. Ordination of the nDNA ΦPT and mtDNA ΦST matrices was independently performed using the MDS option of PAST. A separate analysis of the relationship between the ΦST and ΦPT pair-wise distance matrices was statistically assessed with the Mantel matrix regression analysis [50] performed by the MXCOMP routine in NTSYS-PC 1.8 [51].
Maximum likelihood assignment tests were used to determine the likelihood of each individual's multilocus allelotype being found in the collection from which it was sampled using the program AFLPOP 1.1 [59]. AFLPOP makes no assumption of Hardy-Weinberg equilibrium and has been shown to be robust when applied to binary-coded polyploid allelotypes [40]. Allelotype patterns observed from other analyses were used to group collections into various management units and to test the appropriateness of these groupings using changes in assignment success. Constituencies of evolutionarily significant groupings of populations (e.g., regions, management units, distinct population segments) were ultimately investigated using hierarchical structuring of genetic variation measured (AMOVA) for numerous combinations of collections [60].
Results
Population structure of shortnose sturgeon
A total of 181 alleles were observed at the 11 loci resulting in a binary character matrix of 181 loci (columns) by 561 individuals (rows) (summarized in Table 1). The numbers of alleles with frequencies ≥1% observed at the 11 loci ranged from 55 (Cape Fear, n = 3) to 152 (Hudson, n = 45). Estimated heterozygosity (assuming Hardy-Weinberg equilibrium) ranged from 10.4% (Cape Fear) to 19.3% (Hudson). A lower estimate of heterozygosity was observed among southeastern populations compared to mid-Atlantic and northeastern populations.
Table 1. Microsatellite allele (a.k.a. pseudodominant locus) counts, percentage of loci (alleles) polymorphic, number of private alleles, and number of common alleles across all loci for shortnose sturgeon (Acipenser brevirostrum) populations across the North American range.
| Population | Saint John | Penobscot | Androscoggin | Kennebec | Merrimack | Connecticut | Hudson | Delaware | Chesapeake Bay | Cape Fear | Winyah Bay | Santee-Cooper | Lake Marion | Edisto | Savannah | Ogeechee | Altamaha |
| Sample size (n) | 25 | 39 | 23 | 24 | 22 | 47 | 45 | 39 | 34 | 3 | 47 | 42 | 33 | 33 | 34 | 35 | 36 |
| 1No. Alleles (loci) | 118 | 131 | 126 | 130 | 105 | 121 | 152 | 134 | 127 | 55 | 119 | 111 | 95 | 112 | 113 | 112 | 107 |
| Polymorphism (%) | 65.2 | 72.4 | 69.1 | 70.7 | 58.0 | 66.9 | 84.0 | 74.0 | 70.2 | 22.7 | 65.2 | 60.8 | 51.9 | 61.9 | 61.9 | 60.8 | 59.1 |
| 2No. Private Alleles | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3No. Common Alleles (< = 25%) | 11 | 16 | 13 | 16 | 6 | 12 | 24 | 12 | 11 | 0 | 7 | 3 | 4 | 4 | 5 | 6 | 2 |
The analyses were conducted on the binary character matrix.
Number of different fragments.
Number of bands unique to a single population.
Number of common alleles with frequency ≤25%.
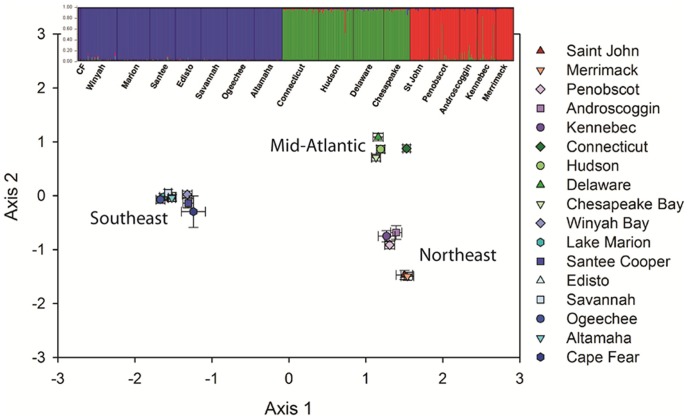
At the individual level, the PCO scatter plot (Figure 2) indicated presence of three regional groups among the 17 surveyed river/estuarine systems. These regional groupings were: 1) Northeast - including five rivers from the Gulf of Maine (GOM), namely Saint John River, Canada; Penobscot, Kennebec, Androscoggin and Merrimack rivers; 2) Mid-Atlantic - including the Connecticut, Hudson, and Delaware rivers, and the Chesapeake Bay proper; and 3) Southeast – including the Cape Fear River, Winyah Bay, the Santee-Cooper, Edisto, Savannah, Ogeechee, Altamaha rivers, and Lake Marion. The number of inferred clusters (k) determined by STRUCTURE for the initial (uppermost hierarchical level) analysis was three, corresponding to the Northeast, Mid-Atlantic, and Southeast regional groupings identified by PCO (Figure 2). The PCO analysis clearly illustrated that the Southeastern cluster of individuals was the most divergent group of fish, separated from the other two regional groupings by a stronger zone of genetic discontinuity than the degree of separation between populations in the Northeast and Mid-Atlantic regions.
Figure 2. Combined graphical representation of principal coordinates (scatter plot) and STRUCTURE (histogram) analyses of 561 shortnose sturgeon (Acipenser brevirostrum) from 17 locations along the North American coast, surveyed at 11 polysomic microsatellite DNA loci.
For the STRUCTURE histograms, each individual is represented by a single vertical bar, broken into k colored segments, the length of which is proportional to the membership fraction in each of the k clusters. Black lines partition the river samples.
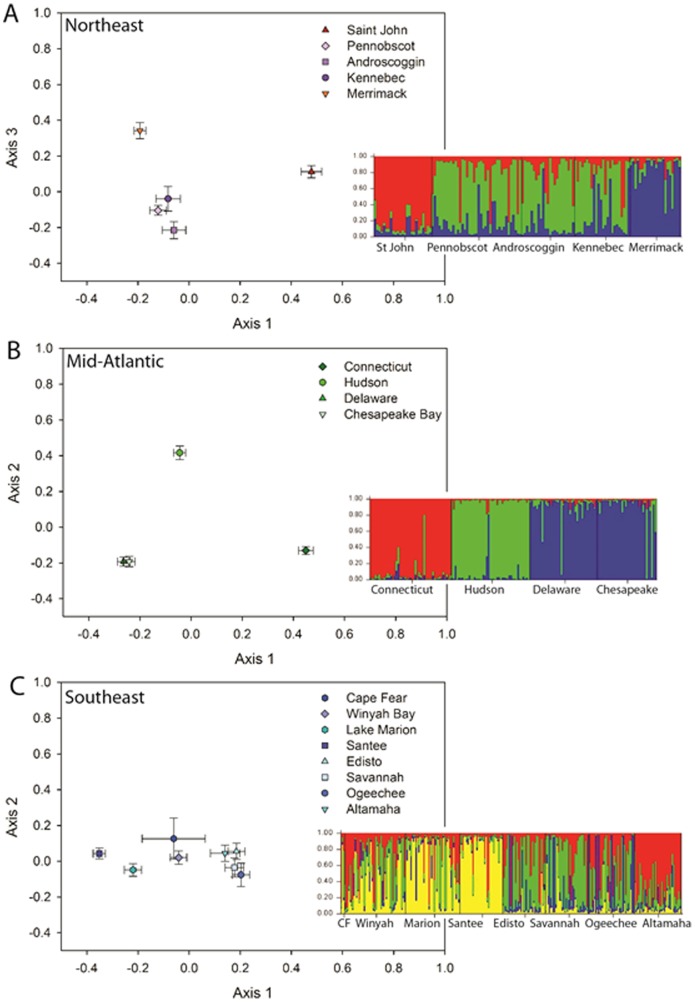
Due to the apparent complex migration patterns (zones of genetic discontinuity) existing among the three regions, a sequential method of inferring the number of clusters was employed to identify within-cluster structure (Figure 3; Evanno et al. 2005). Subsequent PCO analysis of the Northeast regional grouping of collections indicated a high degree of relatedness among the A. brevirostrum sampled from the Penobscot, Androscoggin, and Kennebec Rivers (Figure 3A), whereas fish sampled from the Saint John and Merrimack Rivers appeared well-differentiated from each other and from the other GOM rivers. The sequential STRUCTURE analysis of the Northeast cluster suggested panmixia among the A. brevirostrum sampled from the Penobscot, Androscoggin, and Kennebec Rivers and a moderate degree of differentiation between this group of collections and Saint John River to the north and Merrimack River to the south (Figure 3A). The level of differentiation of the Saint John and Merrimack Rivers from the other GOM collections did not appear as great as that seen among the mid-Atlantic collections or between the southeastern rivers and other collections (Figure 2).
Figure 3. Combined graphical representation of sequential principal coordinates (scatter plots) and STRUCTURE (histograms) analyses of 561 shortnose sturgeon (Acipenser brevirostrum) surveyed at 11 polysomic microsatellite DNA loci.
For the STRUCTURE histograms, each individual is represented by a single vertical bar, broken into k colored segments, the length of which is proportional to the membership fraction in each of the k clusters. Black lines partition the river samples. A) Northeast region collections; B) Mid-Atlantic region collections; C) Southeast region collections.
PCO analysis of the Mid-Atlantic regional grouping indicated that A. brevirostrum sampled from Delaware River and Chesapeake Bay were genetically indistinguishable (mean and SE values were tightly clustered) and differentiated from Connecticut and Hudson River samples (Figure 3B). Likewise, the sequential STRUCTURE analysis also subdivided the mid-Atlantic cluster into three subclusters (Figure 3B). The two fish recently collected in Potomac River (during efforts to determine the existence of a reproducing population in this system) were genetically indistinguishable from other fish from Chesapeake Bay. This subsequent examination also revealed minimal overlap among the Connecticut River, Hudson River, and Delaware River/Chesapeake Bay collections; a pattern not readily discernible in the full analysis (Figure 2). Both PCO and sequential STRUCTURE analysis of A. brevirostrum from the Southeastern cluster of rivers suggested a low level of genetic differentiation among this group of populations, minimal structuring, and a moderate level of gene flow among collections throughout that region (Figure 3C).
At the population level, pair-wise ΦPT values (Table 2, above diagonal) revealed that most (118/136; 87%) pair-wise comparisons among collections were statistically greater than zero (P<0.0004) indicating the presence of multiple populations displaying statistically significant differences in allele frequencies throughout the range of A. brevirostrum. Given the range and magnitude (0.040–0.249) of the ΦPT values in question, the non-significant findings associated with this collection can likely be attributed to the inadequate sample size. The six remaining non-significant ΦPT values were observed among geographically proximal collections within the Northeast and Southeast groupings (clusters). Pair-wise ΦPT values were greatest among collections compared among the three major clusters and lowest among collections within these groupings. Low ΦPT estimates were observed among collections in the Northeast (average 0.06) and Southeast (0.047) clusters. Moderately high ΦPT estimates were observed among the four collections in the mid-Atlantic cluster (averaging 0.077) although the value between the Delaware River and Chesapeake Bay collections was 0.018. Considering the hierarchical AMOVA results (Table 3) in light of the individual-based PCO and STRCTURE analyses, seven distinct populations or groups of populations of A. brevirostrum were consistently supported: 1) St. John River in Canada, 2) three Maine rivers, 3) Merrimack River, 4) Connecticut River, 5) Hudson River, 6) Delaware River/Chesapeake Bay; and 7) southeast (all collections between the Cape Fear and Altamaha Rivers).
Table 2. Pair-wise ΦPT among putative shortnose sturgeon populations (above diagonal) and estimates of the effective number of migrants per generation, Nem (below diagonal), for 17 collections of shortnose sturgeon (Acipenser brevirostrum) surveyed at 11 polysomic microsatellite loci.
| Collection | Saint John | Penobscot | Androscoggin | Kennebec | Merrimack | Connecticut | Hudson | Delaware | Chesapeake Bay | Winyah Bay | Santee-Cooper | Lake Marion | Edisto | Savannah | Ogeechee | Altamaha |
| Saint John | - | 0.068 | 0.077 | 0.068 | 0.100 | 0.191 | 0.162 | 0.175 | 0.155 | 0.253 | 0.289 | 0.269 | 0.269 | 0.278 | 0.280 | 0.277 |
| Penobscot | 3.43 | - | 0.015 | 0.003 | 0.065 | 0.116 | 0.094 | 0.107 | 0.095 | 0.189 | 0.219 | 0.207 | 0.189 | 0.201 | 0.205 | 0.203 |
| Androscoggin | 3.00 | 16.42 | - | 0.013 | 0.087 | 0.113 | 0.091 | 0.099 | 0.093 | 0.200 | 0.248 | 0.226 | 0.209 | 0.210 | 0.227 | 0.226 |
| Kennebec | 3.43 | 83.08 | 18.98 | - | 0.058 | 0.114 | 0.073 | 0.096 | 0.088 | 0.186 | 0.222 | 0.205 | 0.188 | 0.201 | 0.207 | 0.204 |
| Merrimack | 2.25 | 3.60 | 2.62 | 4.06 | - | 0.201 | 0.153 | 0.184 | 0.167 | 0.268 | 0.307 | 0.297 | 0.279 | 0.295 | 0.293 | 0.296 |
| Connecticut | 1.06 | 1.91 | 1.96 | 1.94 | 0.99 | - | 0.086 | 0.100 | 0.118 | 0.239 | 0.272 | 0.263 | 0.256 | 0.261 | 0.273 | 0.273 |
| Hudson | 1.29 | 2.41 | 2.50 | 3.17 | 1.38 | 2.66 | - | 0.067 | 0.075 | 0.179 | 0.217 | 0.208 | 0.188 | 0.196 | 0.210 | 0.201 |
| Delaware | 1.18 | 2.09 | 2.28 | 2.35 | 1.11 | 2.25 | 3.48 | - | 0.018 | 0.188 | 0.228 | 0.217 | 0.200 | 0.206 | 0.216 | 0.212 |
| Chesapeake Bay | 1.36 | 2.38 | 2.44 | 2.59 | 1.25 | 1.87 | 3.08 | 13.64 | - | 0.183 | 0.234 | 0.210 | 0.200 | 0.213 | 0.216 | 0.206 |
| Winyah Bay | 0.74 | 1.07 | 1.00 | 1.09 | 0.68 | 0.80 | 1.15 | 1.08 | 1.12 | - | 0.049 | 0.034 | 0.037 | 0.046 | 0.031 | 0.032 |
| Santee-Cooper | 0.62 | 0.89 | 0.76 | 0.88 | 0.56 | 0.67 | 0.90 | 0.85 | 0.82 | 4.85 | - | 0.044 | 0.043 | 0.043 | 0.046 | 0.069 |
| Lake Marion | 0.68 | 0.96 | 0.86 | 0.97 | 0.59 | 0.70 | 0.95 | 0.90 | 0.94 | 7.10 | 5.43 | - | 0.089 | 0.095 | 0.091 | 0.085 |
| Edisto | 0.68 | 1.07 | 0.95 | 1.08 | 0.65 | 0.73 | 1.08 | 1.00 | 1.00 | 6.51 | 5.56 | 2.56 | - | 0.005 | 0.008 | 0.020 |
| Savannah | 0.65 | 0.99 | 0.94 | 0.99 | 0.60 | 0.71 | 1.03 | 0.96 | 0.92 | 5.18 | 5.56 | 2.38 | 49.75 | - | 0.00 7 | 0.052 |
| Ogeechee | 0.64 | 0.97 | 0.85 | 0.96 | 0.60 | 0.67 | 0.94 | 0.91 | 0.91 | 7.81 | 5.18 | 2.50 | 31.00 | 35.46 | - | 0.020 |
| Altamaha | 0.65 | 0.98 | 0.86 | 0.98 | 0.59 | 0.67 | 0.99 | 0.93 | 0.96 | 7.56 | 3.37 | 2.69 | 12.25 | 4.56 | 12.25 | - |
Non-significant pair-wise ΦPT probability values (H0 = No genetic difference among populations; ΦPT = 0) based on 10,000 permutations values are in bold italics. Cape Fear River sample is not included due to inadequate sample size.
Table 3. Hierarchical AMOVA results for biogeographically relevant groups among 17 collections of shortnose sturgeon (Acipenser brevirostrum) surveyed at 11 polysomic microsatellite DNA markers.
| 3-regions | 5-regions | 7-regions | ||||
| Φ-statistic | % variance | Φ-statistic | % variance | Φ-statistic | % variance | |
| Among regions (ΦRT) | 0.158 | 16 | 0.164 | 16 | 0.170 | 17 |
| Among populations within regions (ΦPR) | 0.057 | 5 | 0.042 | 4 | 0.031 | 3 |
| Within populations (ΦPT) | 0.206 | 79 | 0.199 | 80 | 0.196 | 80 |
Non-parametric analysis of similarity (ANOSIM) of Jaccard's (ecological) distance resulted in a similar pattern of genetic differentiation in the pairwise R values (Table S1; below diagonal) and probability values (above diagonal) as observed with ΦPT values. This correspondence was further reflected in the Mantel matrix regression analysis of the relationship between ΦPT and Jaccard's distance measures, which indicated a strong, nearly absolute, correlation between the two distance metrics (r = 0.98, P<0.0001; Figure S1). As a result of the strength of this relationship, all subsequent distance-based analyses performed utilized the ΦPT statistic because of its relationship to the standard diversity index used with codominant markers (FST).
Multivariate (NMDS) analysis of the underlying structure contained in the population-level pair-wise ΦPT matrix (data not shown) revealed a congruent pattern among the 17 collections with that observed with the individual-based PCO and in the STRUCTURE analysis (Figure 2) resulting in three major groupings of populations (Northeast, Mid-Atlantic, and Southeast) with varying degrees of clustering. The collections representing the southeastern populations and those from the GOM appeared tightly clustered (genetically similar). The Mid-Atlantic collections appeared to form three discrete groupings with the Delaware River and Chesapeake Bay collections the most closely related and more so than either was to the Hudson or Connecticut River collections. The Merrimack and Saint John River collections appeared moderately differentiated from the Maine collections.
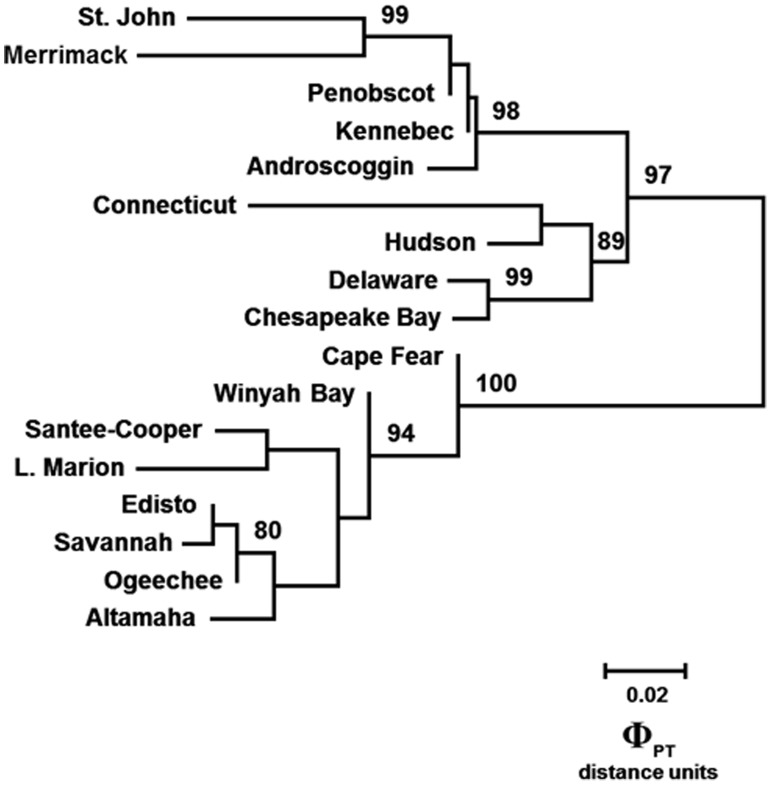
The underlying genetic structure of the ΦPT matrix also was depicted with an unrooted neighbor-joining (N-J) tree (Figure 4), which illustrated high levels of differentiation among A. brevirostrum collections that mirrored those identified by the sequential PCO and STRUCTURE analyses. The deep level of differentiation (genetic discontinuity) among the three major groupings of collections was strongly supported in the backbone of the phenogram and the evolutionary distinctiveness of the populations or groups of populations was confirmed by high bootstrap support, in particular, the absolute (100%) bootstrap support distinguishing the southeast clade from all other collections, and the high bootstrap support for clades containing the Northeast (98%) and Mid-Atlantic (89%) collections. The high degree of genetic similarity observed with other analyses among the Penobscot, Androscoggin and Kennebec Rivers collections and between the Delaware River and Chesapeake Bay collections (99% bootstrap support) was confirmed with high bootstrap support for these pairings.
Figure 4. The evolutionary history among 17 collections of shortnose sturgeon (Acipenser brevirostrum) surveyed at 11 polysomic microsatellite DNA loci inferred from the pair-wise ΦPT distance matrix using the Neighbor-Joining method [56].
Phylogenetic analyses were conducted in MEGA4 [57].
The effective number of A. brevirostrum migrants per generation, Nem (based on ΦPT), estimated between all pairs of collections (Table 2; below diagonal) was consistent with the patterns of differentiation observed with the individual- and population-based analyses (e.g., PCO, STRUCTURE, NMDS, and N-J). Clear zones of genetic discontinuity were evident as Nem among the three major regions was generally low, ranging from an average of 1.89 migrants between the Northeast and Mid-Atlantic collections to 0.89 between the Northeast and Southeast. The average Nem between the Mid-Atlantic and Southeast populations was 0.95. Estimates of Nem among collections within the three major regions were considerably higher ranging from 2.25–83.08 among the Northeast collections and 1.87–13.64 among the Mid-Atlantic collections. The range of Nem values among collections within the Southeast was 2.38–>100.
Quantitative estimates of hierarchical gene diversity among all collections also identified statistically significant genetic structuring; 16% (P<0.001) of the genetic variation occurred among collections (river/estuary populations) and 84% (P<0.001) was attributed to differentiation within collections. Of 11 hierarchical AMOVA analyses (Table S2), four models provided optimal delineation of genetic differentiation although the increase in variance among groupings over other models was minimal (1%). These four models all resulted in 17% (P<0.001) of the genetic variation occurring among groupings, 3% (P<0.001) occurring among populations within groupings, and 80% of the genetic variation was due to variation within collections. All four models, based on the subclusters identified by the PCO and STRUCTURE analyses, were variations either including or omitting the Saint John and Merrimack Rivers as distinct populations. All attempts to manipulate the southeast grouping of populations resulted in a decrease in variation among grouping components and an increase in the amount of within population variation.
The average correct assignment to collection of origin was 58.6% and ranged from 0% (Cape Fear River) to 97.8% (Connecticut River) (Table S3). With the exception of the Cape Fear River collection (n = 3), assignment to each collection was statistically greater than would be expected by chance (P<0.05). When the 17 rivers were pooled by the geographic regions identified in Figure 3, correct assignment to major grouping averaged 99.8% (Table 4). When pooled into two Northeast population groupings (i.e., Saint John separate from all other GOM collections), assignment to the GOM grouping was 100% (data not shown). A five-group model, (where the Northeast region included all GOM collections except the Saint John River) resulted in correct assignment to regional grouping in 99.1% of comparisons (Table S4).
Table 4. Assignment to three groupings of origin model consisting of 17 collections of shortnose sturgeon (Acipenser brevirostrum) surveyed at 11 polysomic microsatellite DNA markers.
| allocated to | Northeast | Mid-Atlantic | Southeast |
| Northeast | 132 | 0 | 0 |
| Mid-Atlantic | 1 | 165 | 0 |
| Southeast | 0 | 0 | 254 |
| Assignment % | 99.1 | 100 | 100 |
Mis-assigned individuals are distributed vertically.
Comparison of the patterns nDNA and mtDNA
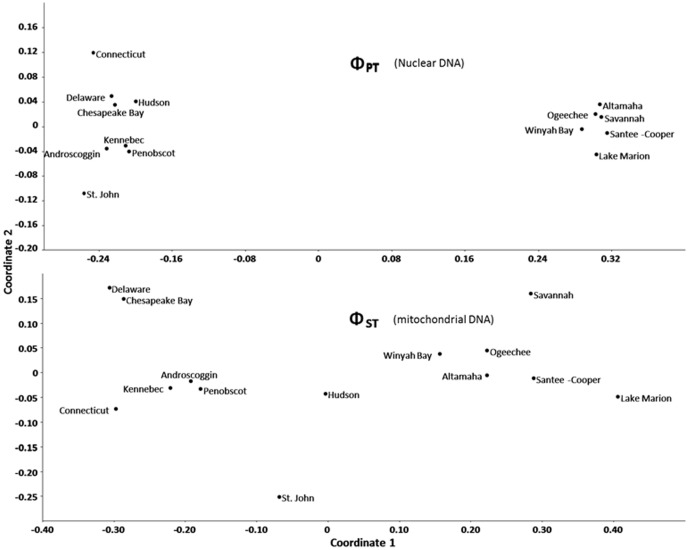
Mantel analysis comparing the pair-wise ΦPT (this study) and ΦST (from Table 5 of Wirgin et al. [31]) distance matrices for 14 shared collections of Atlantic coast collections of A. brevirostrum, identified a strong statistical relationship (correlation coefficient r = 0.84, P<0.0001; Figure S2) between the variation detected in the two genomes. Furthermore, nDNA and mtDNA data yielded concurrent depictions of the presence of three major groupings representing the northeastern, mid-Atlantic, and southeastern populations (Figure 5). Moreover, similar levels and patterns of genome differentiation were observed among the mid-Atlantic groups: Connecticut River, Hudson River, and Delaware River/Chesapeake Bay. The respective scatter plots also suggest the presence of at least three regional metapopulations: Northeast (i.e., Penobscot, Kennebec, and Androscoggin Rivers), Mid-Atlantic (Delaware River and Chesapeake Bay), and Southeast (Altamaha, Winyah Bay, Santee-Cooper, Edisto, Savannah, Ogeechee, and Cape Fear Rivers, and Lake Marion).
Figure 5. Results of independent multidimensional scaling analyses of pair-wise a) ΦPT (nuclear DNA; Table 4, this study) and b) ΦST (mitochondrial DNA; Table 5 of Wirgin et al. [30]) matrices for 14 Acipenser brevirostrum collections that are in common between the two studies.
Discussion
Phylogeography
Allelotypes
This study represents the first report of nuclear DNA variation in the higher-order polyploid Acipenser brevirostrum. The markers uncovered sufficient diversity that each of the 561 individuals surveyed possessed a unique multilocus allelotype. Alleles detected at these loci, scored as binary allelotypes, allowed robust qualitative and quantitative assessment of population structuring, similar to that conducted on disomically-inherited codominant markers. This study applied multivariate ordination and Bayesian clustering, assessed statistical significance (i.e., value>zero) of pair-wise distance metrics, performed matrix regression analyses, estimated the evolutionary history among populations via phylogenetic algorithm (phenogram), conducted hierarchical partitioning of variation (AMOVA), and assessed the success of likelihood assignment testing. Notably, the results of all statistical approaches used to assess phylogeographic structure resulted in consistent findings.
Identifying units of management
Owing to their morphological similarity to lower Jurassic (∼200 MYBP) Acipenseriformes [61], Acipenser species such as A. brevirostrum are considered to be evolutionarily static and are often referred to as phylogenetic relics. Intraspecific examination of the nuclear genome has revealed the presence of considerable allelic diversity and differentiation that appears to be reflective of the actions of various evolutionary processes. Phylogeographically, these findings suggest the presence of similar levels of genetic diversity and variation among the collections punctuated with a series of genetic discontinuities of varying “depth” across the species' range that could indicate demographic independence, regional adaptation, and reflect vicariant geographic events. Populations sampled within these regional groupings exhibited shallow but statistically significant differentiation, which were congruent with theoretical estimates of gene flow. Moreover, patterns of population relatedness were consistent with the observations of [23] that populations at both ends of the species' range are more dispersive than those in the middle. A possible explanation for elevated geneflow within the northern and southern collections could be the greater geographic proximity of rivers in these areas relative to those in the mid-Atlantic region leading to higher levels of straying.
Upon inspection of the patterns of allelotypic variation at the individual and population scales in the nDNA two major (“deep”) zones of genetic discontinuity are inferred from all analyses conducted 1) separating the Northeastern and Mid-Atlantic collections and 2) delineating the Mid-Atlantic and Southeastern populations. Moreover, narrower (“shallow”) zones of genetic discontinuity were evident among Saint John, Merrimack and the three Maine rivers in the Northeast region, and between the Connecticut and Hudson River collections and between Hudson River and an apparent Delaware River/Chesapeake Bay metapopulation (unconfirmed) within the mid-Atlantic region. This implies there are seven demographically and evolutionary distinct lineages across the range and that within the United States portion of the A. brevirostrum range, six lineages are relevant in conservation considerations. In addition to support for recognition of these zones of discontinuity in clustering, the phylogeographic analysis, AMOVAs, and assignment testing, all suggested there are low levels of theoretical gene exchange between collections on either side of these genetic discontinuities.
The presence of demographically distinct and evolutionary significant lineages delineated by zones of genetic discontinuity is consistent with the findings of researchers assessing behavioral patterns in A. brevirostrum. Parker [9] and Parker and Kynard (unpublished data) found that under common garden conditions, A. brevirostrum were locally adapted to particular rivers. These researchers demonstrated differences in the innate dispersal patterns in early life stages of A. brevirostrum from Connecticut River versus sturgeon of Savannah River origin, suggesting that A. brevirostrum are likely behaviorally adapted to unique features of their watershed. Parker and Kynard [62] also found that A. brevirostrum from different rivers can have different migration strategies. Similar adaptive differences have been inferred for other sturgeon species including Acipenser fulvescens (Wolf and Menominee rivers [Kynard and Parker unpublished data]), A. transmontanus (Sacramento and Kootenai rivers [63]), and A. oxyrinchus oxyrinchus/A. o. desotoi (Hudson and Suwannee rivers [24]).
Inter-genomic comparison
Comparison of nuclear genetic diversity observed in this study with that identified previously in mtDNA sequence variation ([31] and references therein) illustrates that the patterns of variation in the two A. brevirostrum genomes were qualitatively consistent. Similarly, the observation from the NMDS scatter plots in the two studies (Figure 5) that pair-wise ΦST values on average were approximately twice the pair-wise ΦPT values for the same suite of collections is consistent with hyper-polymorphism associated with microsatellite evolution ([64], [65]) and the expectation of allele size homoplasy associated with polyploidy and an artifact of scoring alleles as binary types. Ultimately, this is the quantitative ‘penalty’ realized because allelotypic diversity is likely to be an underestimation of the actual differentiation that exists among populations; particularly for those that have experienced extended reproductive isolation. Although quantitative variation and molecular variation are correlated, adaptive population structuring often far exceeds neutral population structuring even for populations diverging over contemporary time ([66], [67]). Therefore, the estimates of allelic differentiation detected at neutral loci in this study are likely an underestimation of the actual divergence. Lastly, a component of the greater mtDNA haplotype differentiation relative to nuclear DNA differentiation could reflect gender-mediated gene flow between adjacent populations and resulting in reduced philopatry (i.e., sex-biased dispersal) of males throughout the range.
The phylogeographic and genomically-congruent patterns predicting genetic structuring of A. brevirostrum have profound management implications, foremost being the strong indication of regional structure. Specific cases within each region provide evidence that the fundamental units of management generally are the populations located in rivers and estuaries. However, because interpretations of the delineation of those distinct population segments differ depending upon the genome under investigation (7–9 based on mtDNA versus 5 if based on nDNA), we are faced with the need to specify evolutionarily relevant thresholds for deciding statistical significance of haplotype or allelotype frequencies. Ultimately, the approach that may offer maximum flexibility in reaching recovery goals may be to allocate (or nest) biological units that are most likely to respond to management within regions. The following two sections examine specific cases and the suggestion that a metapopulation approach may serve best to illustrate the structure of gene flow in this species.
Cases in point
Saint John's River
Differentiation among the Northeast populations is on the whole less than observed for the Mid-Atlantic populations but greater than among the Southeast populations. Within the Northeast group, the Saint John River, Canada collection constitutes a population that is appreciably differentiated from the GOM collections at a number of levels. Foremost, the degree of gene differentiation between Saint John River and the GOM is “shallow”. This “shallowness” of may be attributed to the fact that these populations are relatively young (due to recent deglaciation of the region) as compared to more southerly distributed populations based on the observed levels of divergence. In addition, designation as a distinct management unit may be warranted because 1) the Saint John River A. brevirostrum population's age-to-reproduction is different than other GOM sturgeon, 2) it is the northernmost reproducing population, and 3) it experiences differences in control of exploitation, management of habitat, and conservation status.
Gulf of Maine
The GOM collections analyzed in this study included a recently obtained sampling from the Merrimack River. These A. brevirostrum exhibited patterns of nDNA variation that suggest the collection is genetically differentiated from the other GOM collections. Such an interpretation should be made with caution because the sample consisted of 22 males collected at the same location and time, and because the level of differentiation is not as great as that observed among other collections proposed as distinct evolutionary lineages. Choosing to consider this collection as a distinct evolutionary lineage would serve the precautionary principle and conserve biodiversity.
Potomac River
Recent captures of adult A. brevirostrum in the Potomac ([68], [69]) and Merrimack Rivers (this study) have raised questions concerning the status of this endangered species and available habitat in these river systems. Fishery managers require empirically supported prioritization schemes to protect existing diversity. It is important, therefore to determine if these observations are indicative of a discrete natal population or simply the result of migratory foraging behavior of fish from nearby populations. Analysis of mtDNA sequences from four Potomac River A. brevirostrum identified haplotypes also found in fish from the Chesapeake Bay and Delaware River [28]. This suggested the fish (or their mothers) were genetically related to other collections made in the Chesapeake Bay proper and Delaware River. Grunwald et al. 2002 suggested the fish might have been migrants from the Chesapeake and Delaware Canal [28]. Individual-based analyses of the two Potomac River A. brevirostrum sampled for the present study strongly suggested the fish are genetically part of a Delaware River/Chesapeake Bay metapopulation (Figure 3B). It will require significant additional resources to determine whether a reproducing population of A. brevirostrum exists within the Chesapeake Bay proper or whether the collections made there simply represent fish on foraging forays from the Delaware River population. Without recolonization and reestablishment of a reproducing population somewhere among the Chesapeake Bay rivers, a large gulf will continue to exist among northern and southern A. brevirostrum populations.
Savannah River supplemental stocking program
Early resource management efforts for A. brevirostrum included the release of hatchery-reared juveniles to supplement Savannah River populations [70]. Although conducted as an experimental program from 1984–1992, approximately 97,000 juveniles were introduced. This management effort has potentially altered the population genetic structure of the targeted river system as well as the southeast metapopulation. Smith et al. [70] estimated that 39% of the Savannah River's ‘breeding’ adults were of hatchery origin. It has been suggested that because A. brevirostrum were released as larger juveniles during this supplemental stocking program to allow marking, the fish were not imprinted to the natal stream and may have effectively strayed to adjacent river systems, i.e., traveled to non-natal rivers for reproduction, thereby having a homogenizing effect on impacted river populations [29]. However, it is unknown if effective introgression of hatchery-origin fish into the wild populations of any southeast river has occurred. No strong signal supporting or refuting a discernible impact on the southeast metapopulation by the Savannah River supplemental stocking is apparent from this study. Due to the geographic proximity of the Edisto and Ogeechee rivers to the Savannah River, the degree of evolutionary similarity among these rivers (e.g., in assignment testing) could be the result of natural straying or a homogenizing effect of the supplemental stocking program.
Metapopulations
The current paradigm is that A. brevirostrum stray less often than the congeneric A. oxyrinchus [71]. However, at least two additional groupings involving adjacent river and estuarine systems have been identified in this study that exhibit strong signatures of functioning metapopulations (i.e., Maine rivers and Delaware River/Chesapeake Bay). Moreover, biologists tagging and tracking fish in Connecticut River recaptured two A. brevirostrum that originally were tagged and released in Hudson River [72]. Although this latter finding does not confirm effective movement (i.e., gene flow) between these river systems, evidence suggests some degree of straying and recolonization from adjacent southeastern rivers is possible. Indeed, if the strong signal of metapopulation structure observed among the southeastern rivers in this study resulted from the straying of stocked Savannah River A. brevirostrum, the effective population sizes of the affected rivers are indeed minuscule and the supplementation program must have been one of the most successful such efforts on record.
In addition to the demographically discrete and evolutionarily significant lineages identified for A. brevirostrum within the U.S., three metapopulations and other individual river populations delineated within each discrete lineage may be considered distinct management/recovery units for future recovery planning purposes. The three possible metapopulations are the: 1) Maine rivers (i.e., Penobscot, Kennebec, and Androscoggin rivers), 2) Delaware River and Chesapeake Bay proper, and 3) the Southeast assemblage (Cape Fear River, Winyah Bay, Santee-Cooper, Edisto, Savannah, Ogeechee, and Altamaha rivers, and Lake Marion). Population biology theory predicts that lower dispersal and associated gene flow leads to decreased genetic diversity in small isolated populations, which generates adverse consequences for fitness, and subsequently for demographic stability. Given recent tagging data suggesting A. brevirostrum migrate to adjacent rivers to a greater extent than previously believed [70], [71], [73], [74], [75], concomitant with the identification of at least three metapopulations within the species' range, this interpretation appears to bode well for the demographic fitness of some southern A. brevirostrum populations.
Conservation Implications
This study presents evidence that sufficient levels of genetic diversity are present in the A. brevirostrum nuclear genome to discriminate evolutionarily significant lineages of management relevance. The variation detected was highly phylogeographically congruent with the inferences based on the mtDNA control region ([31] and references therein). Moreover, these nDNA analyses detected statistically significant differences in allelotype frequencies among most collections. Four regional zones of genetic discontinuity were inferred from the patterns of genetic variation across the range of A. brevirostrum (two shallow zones in the Northeast and two deep zones dividing three regions) that likely delineate seven demographically discrete and evolutionarily significant lineages, each with differing adaptive potential for this species. These zones of inferred genetic discontinuity represented deeper levels of differentiation and a higher degree of reproductive isolation than that typically attributed to population-level differentiation; groups of populations of evolutionary significance which may warrant distinct conservation considerations. Perhaps more notable than the delineation of evolutionarily significant lineages, was the identification of at least one putative metapopulation within each of the three major regional groupings of populations, a finding that is encouraging as migration may help stave localized extinctions. It should be noted that while an increased level of gene flow is present within these putative metapopulations, a demographic connection has yet to be established and documentation of such a link should be considered a high priority research need. Moreover, many of the populations within an evolutionarily significant lineage were genetically differentiated to some degree and each geographic population is subjected to differing threats [75]. Based on patterns observed from these multilocus allelotypes, the basic unit for management and conservation (for recovery planning) of A. brevirostrum is arguably the individual (local) population.
Assuming conservation of local populations within a metapopulation is the acceptable focus, restoration of effective connectivity among currently fragmented A. brevirostrum populations should be a prominent recovery goal; the allelotypic patterns observed in this study provide guidance by facilitating understanding of where and how such efforts could be attempted. For example, the best available information suggests that individual rivers and estuaries along the North Carolina coast do not currently support reproducing populations of A. brevirostrum. If the distance to North Carolina (or elsewhere) rivers that could support a reproducing population exceeds the vagility of sturgeon inhabiting the southeast or Delaware River/Chesapeake Bay metapopulations, targeted translocations or restorative supplementation may represent plausible restoration strategies. In contrast, rivers geographically south of the Altamaha River historically occupied by shortnose sturgeon (i.e., Satilla, St. Marys, and St. John Rivers), no longer support reproducing A. brevirostrum populations. The shoreline distances of these rivers to the Altamaha River are similar to that observed among the major rivers comprising the Southeast metapopulation. Given the recent findings of moderate physical migration [74] and our implications for effective migration (gene flow) among A. brevirostrum inhabiting many of the rivers in the Southeast metapopulation, it seems logical that if one or more of these southernmost rivers provided suitable spawning and rearing habitat, A. brevirostrum would have effectively colonized this region. Therefore, habitat characterization and/or restoration in these southernmost rivers could facilitate range expansion.
Gene diversity estimates for A. brevirostrum have been shown to be moderately high in both nuclear (this study) and mitochondrial ([29], [30], [31]) genomes. Although rates of genetic diversity loss in polyploids versus diploids (functional sensu) has not been characterized for sturgeon, the nDNA and mtDNA studies performed to date suggest that dispersal is a very important factor maintaining genetic diversity in shortnose sturgeon. The gene diversity estimates may be indicative of larger effective population sizes than previously assumed. However, even at a very local spatial scale in a metapopulation consisting of moderate-density populations interconnected by considerable dispersal rates, genetic diversity can erode and directly affect the fitness of individuals. Gene flow estimates do not capture the intra-specific variation in individual behavior related to vagility, which is strongly affected by habitat fragmentation and population/metapopulation history. From a biodiversity conservation perspective, future success in A. brevirostrum management could benefit from both in-depth demographic and genetic analyses. It should be considered a high-priority research need to better delineate population structure within the evolutionarily significant lineage framework suggested by these data.
Supporting Information
Scatter plot illustrating the significant correlation ( r = 0.98; P <0.0001; Mantel analysis) between Jaccard and ΦPT pair-wise distances for 17 collections of shortnose sturgeon ( Acipenser brevirostrum ) surveyed at 11 polysomic microsatellite DNA loci.
(TIF)
Scatter plot depicting the Mantel matrix regression analysis comparing the mtDNA ΦST matrix for 14 Atlantic coast collections of shortnose sturgeon ( Acipenser brevirostrum ) (Wirgin et al. [30] ) and the nuclear DNA ΦPT pair-wise distance matrix (this study) for the same collections surveyed at 11 polysomic microsatellite DNA loci. The correlation coefficient (r) for this analysis was 0.84 (P<0.0001).
(TIF)
Pair-wise R values (below diagonal) and Bonferroni-corrected probability values (above diagonal) from the non-parametric Analysis of Similarity (ANOSIM) (Clark 1993) on Jaccard's distance metric (1-Jaccard similarity) measured among 17 collections of shortnose sturgeon ( Acipenser brevirostrum ) surveyed at 11 polysomic microsatellite loci.
(DOC)
Hierarchical structuring of genetic variation was measured for numerous combinations of shortnose sturgeon ( Acipenser brevirostrum ) collections using analysis of molecular variance (AMOVA). Significance levels of the variance components were based on 1000 permutations. Abbreviations are as follows: NE = Northeast regional grouping includes Saint John River (SJ), Canada, Penobscot, Kennebec, Androscoggin and Merrimack rivers; Mid-Atlantic regional grouping includes the Connecticut (CT), Hudson (H), and Delaware (DE) rivers, and the Chesapeake Bay proper (CB); and the 3) SE = Southeast regional grouping includes the Cape Fear River (CF), Winyah Bay (WB), Santee-Cooper (S-C), Edisto (E), Savannah (S), Ogeechee (O), and Altamaha (ALT) rivers, and Lake Marion (LM).
(DOC)
Assignment to collection of origin for 17 shortnose sturgeon ( Acipenser brevirostrum ) collections surveyed at 11 polysomic microsatellite DNA markers. Mis-assigned individuals are distributed horizontally.
(DOC)
Assignment to proposed grouping (five groupings model) in shortnose sturgeon ( Acipenser brevirostrum ) surveyed at 11 polysomic microsatellite DNA markers. The overall correct assignment rate to proposed grouping was 99.1% (522/527). Mis-assigned individuals are distributed vertically. Northeast regional grouping includes Saint John River (SJ), Canada, Penobscot, Kennebec, Androscoggin and Merrimack rivers; and the Southeast regional grouping includes the Cape Fear River (CF), Winyah Bay (WB), Santee-Cooper (S-C), Edisto (E), Savannah (S), Ogeechee (O), and Altamaha (ALT) rivers, and Lake Marion (LM).
(DOC)
Acknowledgments
The authors wish to acknowledge the efforts of Diane Pavek (NPS-NCR) for being the driving force behind this research. We also acknowledge the NOAA-Fisheries funded Shortnose Sturgeon Status Review Team for their contributions to interpretations presented in this report, namely: Jeanette Bowers-Altman, New Jersey Division of Fish and Wildlife; Mark Collins, South Carolina Dept. of Natural Resources; Joel Fleming, Georgia Department of Natural Resources; Kathryn Hattala, New York Department of Environmental Conservation; Wilson Laney, USFWS – South Atlantic Fisheries Coordination Office, Raleigh, NC; Malcolm Mohead, NOAA Fisheries, Office of Protected Resources; Tom Squiers, Maine Department of Marine Resources; Dana Hartley, NOAA – Fisheries, NERO, Gloucester, MA; and Stephania Bolden, NOAA – Fisheries, SERO, St. Petersburg, FL. All tissue sampling followed the guidelines mandated by the U.S. Department of Commerce, National Oceanic and Atmospheric Administration, National Marine Fisheries Service under NOAA Technical Memorandum NMFS-OPR-18 or NOAA Technical Memorandum NMFS-OPR-45. Ike Wirgin, New York University College of Medicine, provided Connecticut, Androscoggin, Delaware, Hudson, and Kennebec rivers tissue and DNA samples. Douglas Peterson and Robert De Vries, University of Georgia, provided the Altamaha River tissue samples. NOS-NOAA archives provided Cooper, Edisto, Savannah, Santee-Cooper, Ogeechee, and Waccamaw rivers tissue samples. Morris Lee and Steve Fernandez, University of Maine contributed Penobscot River tissue samples. The U.S. Fish and Wildlife Service's Bear's Bluff National Fish Hatchery provided tissue samples of the parents and 30 progeny from each of seven captive-bred A. brevirostrum families, samples that were instrumental in determining the inheritance of the polysomic markers. The authors are grateful to Katharine Coykendall, USGS-Leetown Science Center for an IPDS review and Miranda McManus and Allan Strand, College of Charleston, for constructive suggestions on statistical analyses. The senior author is deeply indebted to reviewers, Mark Bain (deceased), Matthew Litvak, David Secor, and John Waldman for their helpful comments on an early version of this report in the form of the genetics section of the NOAA-Fisheries Shortnose Sturgeon Status Review Report. Mike Eackles (USGS-Leetown Science Center) kindly provided laboratory assistance for this project. Use of trade, product, or firm names does not imply endorsement by the U.S. Government.
Funding Statement
The National Capital Region (NCR) of the U.S. National Park Service (NPS), and the U.S. Geological Survey's (USGS) Leetown Science Center provided principal funding for this project through the National Resources Preservation Program. The Northeast Regional Office, Protected Resources Division, NOAA Fisheries, Department of Commerce also provided partial funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schultz RJ (1980) Role of polypoidy in the evolution of fishes. In: Lewis WH, editor. Polyploidy: Biological Relevance. New York, Plenum Press. pp. 313–340. [Google Scholar]
- 2.Gardiner BG (1984) Sturgeons as Living Fossils. In: Eldredge N, Stanley SM, editors. Living Fossils. New York: Springer-Verlag. pp. 148–152. [Google Scholar]
- 3. Bemis WE, Kynard B (1997) Sturgeon rivers: an introduction to acipenseriform biogeography and life history. Environ Biol Fishes 48: 167–183. [Google Scholar]
- 4. Birstein VJ, Hanner R, DeSalle R (1997) Phylogeny of the Acipenseriformes: cytogenetic and molecular approaches. Environ Biol Fishes 48: 127–155. [Google Scholar]
- 5. Krieger J, Fuerst PA (2002) Evidence for a slowed rate of molecular evolution in the order Acipenseriformes. Mol Biol Evol 19: 891–897. [DOI] [PubMed] [Google Scholar]
- 6. Robles F, de la Herrán R, Ludwig A, Ruiz R, Ruiz R, et al. (2004) Evolution of ancient satellite DNAs in sturgeon genomes. Gene 338: 133–142. [DOI] [PubMed] [Google Scholar]
- 7. Ludwig A (2006) A sturgeon view on conservation genetics. Eur J Wildl Res 52: 3–8. [Google Scholar]
- 8. Kynard B, Parker E (2005) Ontogenetic behavior and dispersal of Sacramento River white sturgeon, Acipenser transmontatus, with a note on body color. Environ Biol Fishes 71: 145–146. [Google Scholar]
- 9.Parker EL (2007) Ontogeny and life history of shortnose sturgeon (Acipenser brevirostrum Lesueur 1818): Effects of latitudinal variation and water temperature. PhD dissertation. Univ. of Massachusetts Amherst.
- 10. Birstein VJ, DeSalle R (1998) Molecular phylogeny of Acipenserinae. Mol Phylogenet Evol 9: 141–155. [DOI] [PubMed] [Google Scholar]
- 11. Zhang S, Zhang Y, Zheng X, Chen Y, Deng H, et al. (2000) Molecular phylogenetic systematics of twelve species of Acipenseriformes based on mtDNA ND4L-ND4 gene sequence analysis. Sci China C Life Sci 43: 129–137. [DOI] [PubMed] [Google Scholar]
- 12. Birstein VJ, Doukakis P, DeSalle R (2002) Molecular phylogeny of Acipenseridae: nonmonophyly of Scaphirhynchidae. Copeia 2: 287–301. [Google Scholar]
- 13. Krieger J, Hett AK, Fuerst PA, Artyukhin E, Ludwig A (2008) The molecular phylogeny of the order Acipenseriformes revisited. J Appl Ichthyol 24: 36–45. [Google Scholar]
- 14. Lanfredi M, Congiu L, Garrido-Ramos MA, de la Herrán R, Leis M, et al. (2001) Chromosomal location and evolution of satellite DNA family in seven sturgeon species. Chromosome Res 9: 47–52. [DOI] [PubMed] [Google Scholar]
- 15. Lazzari B, Mariani V, Malinverni R, Caprera A, Giuffra E (2008) A comparative gene index for the white sturgeon Acipenser transmontanus . Mar Genomics 1: 15–21. [DOI] [PubMed] [Google Scholar]
- 16. Ohno S, Wolf U, Atkin NB (1968) Evolution from fish to mammals by gene duplication. Hereditas 59: 169–187. [DOI] [PubMed] [Google Scholar]
- 17. Dingerkus G, Howell WM (1976) Karyotypic analysis and evidence of tetraploidy in the North American paddlefish, Polyodon spathula . Science 194 (4267) 842–844. [DOI] [PubMed] [Google Scholar]
- 18. Kim DS, Nam YK, Noh JK, Park CH, Chapman FA (2005) Karyotype of North American shortnose sturgeon Acipenser brevirostrum with the highest chromosome number in the Acipenseriformes. Ichthyol Res 52: 94–97. [Google Scholar]
- 19. Fontana F, Congiu L, Mudrak VA, Quattro JM, Smith TIJ, et al. (2008) Evidence of hexaploid karyotype in shortnose sturgeon. Genome 51: 113–119. [DOI] [PubMed] [Google Scholar]
- 20. Ollermann LK, Skelton PH (1990) Hexaploidy in yellowfish species (Barbus, Pisces, Cyprinidae) from Southern Africa. J Fish Biol 37: 105–115. [Google Scholar]
- 21. Furlong RF, Holland PWH (2002) Were vertebrates octoploid? Philos Trans R Soc Lond B Biol Sci 357: 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blacklidge KH, Bidwell CA (1993) Three ploidy levels indicated by genome quantification in acipenserformes of North America. J Hered 84: 427–430. [Google Scholar]
- 23. Kynard B (1997) Life history, latitudinal patterns, and status of the shortnose sturgeon, Acipenser brevirostrum . Environ Biol Fishes 48: 319–334. [Google Scholar]
- 24. Kynard B, Parker E (2004) Ontogenetic behavior and migration of Gulf of Mexico sturgeon, Acipenser oxyrinchus desotoi, with notes on body color and development. Environ Biol Fishes 70: 43–55. [Google Scholar]
- 25. Kieffer MC, Kynard B (1996) Spawning of the shortnose sturgeon in the Merrimack River, Massachusetts. Trans Am Fish Soc 125: 179–186. [Google Scholar]
- 26. Waldman JR, Grunwald C, Stabile J, Wirgin I (2002) Impacts of life history and biogeography on the genetic stock structure of Atlantic sturgeon Acipenser oxyrinchus oxyrinchus, Gulf sturgeon A. oxyrinchus desotoi, and shortnose sturgeon A. brevirostrum . J Appl Ichthyol 18: 509–518. [Google Scholar]
- 27. Walsh MG, Bain MB, Squiers T Jr, Waldman JR, Wirgin I (2001) Morphological and genetic variation among shortnose sturgeon Acipenser brevirostrum from adjacent and distant rivers. Estuaries 24: 41–48. [Google Scholar]
- 28. Grunwald C, Stabile J, Waldman JR, Gross R, Wirgin I (2002) Population genetics of shortnose sturgeon Acipenser brevirostrum based on mitochondrial DNA control region sequences. Mol Ecol 11: 1885–1898. [DOI] [PubMed] [Google Scholar]
- 29. Quattro JM, Greig TW, Coykendall DK, Bowen BW, Baldwin JD (2002) Genetic issues in aquatic species management: the shortnose sturgeon (Acipenser brevirostrum) in the southeastern United States. Conserv Genet 3: 155–166. [Google Scholar]
- 30. Wirgin I, Grunwald C, Carlson E, Stabile J, Peterson DL, Waldman J (2005) Range-wide population structure of shortnose sturgeon Acipenser brevirostrum based on sequence analysis of the mitochondrial DNA control region. Estuaries 28: 406–421. [Google Scholar]
- 31. Wirgin I, Grunwald C, Stabile J, Waldman JR (2010) Delineation of discrete population segments of shortnose sturgeon Acipenser brevirostrum based on mitochondrial DNA control region sequence analysis. Conserv Genet 11: 689–708. [DOI] [PubMed] [Google Scholar]
- 32. Wirgin II, Stabile JE, Waldman JR (1997) Molecular analysis in the conservation of sturgeons and paddlefish. Environ Biol Fishes 48: 385–398. [Google Scholar]
- 33. Hett AK, Ludwig A (2005) SRY-related (Sox) genes in the genome of European Atlantic sturgeon (Acipenser sturio). Genome 48: 181–186. [DOI] [PubMed] [Google Scholar]
- 34. Henderson AP, King TL (2012) Development of polysomic microsatellite markers for characterization of population structuring and phylogeography in the shortnose sturgeon (Acipenser brevirostrum). Conserv Genet Resour 4: 853–859. [Google Scholar]
- 35. Labarca C, Paigen K (1980) A simple, rapid, and sensitive DNA assay procedure. Anal Biochem 102: 344–352. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: a laboratory manual, second edition. Plainview: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 37. Obbard DJ, Harris SA, Pannell JR (2006) Simple allelic-phenotype diversity and differentiation statistics for allopolyploids. Heredity 97: 296–303. [DOI] [PubMed] [Google Scholar]
- 38. Welsh A, Blumberg M, May B (2003) Identification of microsatellite loci in lake sturgeon, Acipenser fulvescens, and their variability in green sturgeon, A. medirostris . Mol Ecol Notes 3: 47–55. [Google Scholar]
- 39. Rodzen JA, May B (2002) Inheritance of microsatellite loci in the white sturgeon (Acipenser transmonatus). Genome 45: 1064–1076. [DOI] [PubMed] [Google Scholar]
- 40. Israel JA, Bando KJ, Anderson EC, May B (2009) Polyploid microsatellite data reveal stock complexity among estuarine North American green sturgeon (Acipenser medirostris). Can J Fish Aquat Sci 66: 1491–1504. [Google Scholar]
- 41. Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3: 91–99. [DOI] [PubMed] [Google Scholar]
- 42. Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalograss Buchloe dactyloides (Nutt) Engelm. Theor Appl Genet 86: 927–934. [DOI] [PubMed] [Google Scholar]
- 44. Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica 4: 9. [Google Scholar]
- 45. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Evanno G, Regnault S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 47. Jaccard P (1901) Étude comparative de la distribution florale dans une portion des Alpes et des Jura. Bull Soc Vaudoise Sci Nat 37: 547–579. [Google Scholar]
- 48. Kosman E, Leonard KJ (2005) Similarity coefficients for molecular markers in studies of genetic relationships between individuals for haploid, diploid, and polyploid species. Mol Ecol 14: 415–424. [DOI] [PubMed] [Google Scholar]
- 49. Clarke KR (1993) Non-parametric multivariate analysis of changes in community structure. Aust J of Ecol 18: 117–143. [Google Scholar]
- 50. Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220. [PubMed] [Google Scholar]
- 51.Rohlf FJ (1993) NTSYS-pc: numerical taxonomy and multivariate analysis system. Version 1.8. New York: Exeter Publications. [Google Scholar]
- 52. Ryman N, Utter R, Laikre L (1995) Protection of intraspecific biodiversity of exploited fishes. Rev Fish Biol Fish 5: 417–446. [Google Scholar]
- 53.Wright S (1969) Evolution and the Genetics of Populations, Vol. 2. The Theory of Gene Frequencies. Chicago: University of Chicago Press. [Google Scholar]
- 54.Nei M (1987) Molecular Evolutionary Genetics. New York: Columbia University Press. [Google Scholar]
- 55.Chakraborty R, Leimar O (1987) Genetic variation within a subdivided population. In Ryman N, Utter F, editors. Population Genetics and Fishery Management. Seattle: University of Washington Press. pp 89–120 [Google Scholar]
- 56.Kruskal JB, Wish M (1978) Multidimensional Scaling, Sage University Paper series on Quantitative Application in the Social Sciences, 07-011. Beverly Hills and London: Sage Publications. [Google Scholar]
- 57. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 58. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 59. Duchesne P, Bernatchez L (2002) AFLPOP: A computer program for simulated and real population allocation based on AFLP data. Mol Ecol Notes 3: 380–383. [Google Scholar]
- 60. Antunes A, Troyer JL, Roelke ME, Pecon-Slattery J, Packer C, et al. (2008) The evolutionary dynamics of the lion Panthera leo revealed by host and viral population genomics. PLoS Genet 4 (11) e1000251 doi:10.1371/journal.pgen.1000251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bemis WE, Findeis EK, Grande L (1997) An overview of Acipenseriformes. Environ Biol Fishes 48: 25–71. [Google Scholar]
- 62.Parker E, Kynard B (2005) Latitudinal differences in ontogenetic behavior between two populations of shortnose sturgeon (Acipenser brevirostrum): A laboratory study. Turners Falls, Massachusetts, USA: United States Geological Survey. [Google Scholar]
- 63. Kynard B, Parker E, Kynard BE (2010) Ontogenetic behavior of Kootenai River white sturgeon, Acipenser transmontanus, with a note on body color: a laboratory study. Environ Biol Fishes 88: 65–77. [Google Scholar]
- 64. Hedrick PW (1999) Perspective: Highly variable loci and their interpretation in evolution and conservation. Evolution 53: 313–318. [DOI] [PubMed] [Google Scholar]
- 65. Balloux F, Brüunner H, Lugon-Moulin N, Hausser J, Goudet J (2000) Microsatellites can be misleading: an empirical and simulation study. Genetics 54: 1414–1422. [DOI] [PubMed] [Google Scholar]
- 66. Koskinen MT, Sundell P, Piironen J, Primmer CR (2002) Genetic assessment of spatiotemporal evolutionary relationships and stocking effects in grayling (Thymallus thymallus, Salmonidae). Ecol Lett 5: 193–205. [Google Scholar]
- 67. Stockwell CA, Hendry AP, Kinnison MT (2003) Contemporary evolution meets conservation biology. Trends Ecol Evol 18: 94–101. [Google Scholar]
- 68. Welsh SA, Mangold MF, Skjeveland JE, Spells AE (2002) Distribution and Movement of Shortnose Sturgeon (Acipenser brevirostrum) in the Chesapeake Bay. Estuaries 25 (1) 101–104. [Google Scholar]
- 69. Kynard B, Breece M, Atcheson M, Kieffer M, Mangold M (2009) Life history and status of shortnose sturgeon (Acipenser brevirostrum) in the Potomac River. J Appl Ichthyol 25 (Suppl. 2) 34–38. [Google Scholar]
- 70. Smith TIJ, McCord JW, Collins MR, Post WC (2002) Occurrence of stocked shortnose sturgeon Acipenser brevirostrum in non-target rivers. J Appl Ichthyol 18: 470–474. [Google Scholar]
- 71.Fernandes SJ, Kinnison MT, Zydlewski GB (2008) Investigation into the distribution and abundance of Atlantic sturgeon and other diadromous species in the Penobscot River, Maine: with special notes on the distribution and abundance of federally endangered shortnose sturgeon (Acipenser brevirostrum). Gloucester, Massachusetts, USA: National Oceanic and Atmospheric Administration. [Google Scholar]
- 72.Savoy T (2004) Population estimate and utilization of the lower Connecticut River by shortnose sturgeon. In Jacobson PM, Dixon DA, Leggett WC, Marcy BC Jr., Massengill RR, editors. The Connecticut River Ecological Study (1965–1973) revisited: ecology of the lower Connecticut River 1973–2003. Am Fish Soc Mon 9. pp. 345–352. [Google Scholar]
- 73. Dadswell MJ (1979) Biology and population characteristics of the shortnose sturgeon, Acipenser brevirostrum LeSueur (1818) (Osteichthyes: Acipenseridae), in the Saint John River estuary, New Brunswick, Canada. Can J Zool 57: 2186–2210. [Google Scholar]
- 74.Fernandes SJ (2008) Population demography, distribution, and movement patterns of Atlantic and shortnose sturgeons in the Penobscot River estuary, Maine. Master's thesis. Univ. of Maine. 88 pp.
- 75.Shortnose Sturgeon Status Review Team. 2010. A Biological Assessment of shortnose sturgeon (Acipenser brevirostrum). Report to National Marine Fisheries Service, Northeast Regional Office. November 1, 2010. 417 pp.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scatter plot illustrating the significant correlation ( r = 0.98; P <0.0001; Mantel analysis) between Jaccard and ΦPT pair-wise distances for 17 collections of shortnose sturgeon ( Acipenser brevirostrum ) surveyed at 11 polysomic microsatellite DNA loci.
(TIF)
Scatter plot depicting the Mantel matrix regression analysis comparing the mtDNA ΦST matrix for 14 Atlantic coast collections of shortnose sturgeon ( Acipenser brevirostrum ) (Wirgin et al. [30] ) and the nuclear DNA ΦPT pair-wise distance matrix (this study) for the same collections surveyed at 11 polysomic microsatellite DNA loci. The correlation coefficient (r) for this analysis was 0.84 (P<0.0001).
(TIF)
Pair-wise R values (below diagonal) and Bonferroni-corrected probability values (above diagonal) from the non-parametric Analysis of Similarity (ANOSIM) (Clark 1993) on Jaccard's distance metric (1-Jaccard similarity) measured among 17 collections of shortnose sturgeon ( Acipenser brevirostrum ) surveyed at 11 polysomic microsatellite loci.
(DOC)
Hierarchical structuring of genetic variation was measured for numerous combinations of shortnose sturgeon ( Acipenser brevirostrum ) collections using analysis of molecular variance (AMOVA). Significance levels of the variance components were based on 1000 permutations. Abbreviations are as follows: NE = Northeast regional grouping includes Saint John River (SJ), Canada, Penobscot, Kennebec, Androscoggin and Merrimack rivers; Mid-Atlantic regional grouping includes the Connecticut (CT), Hudson (H), and Delaware (DE) rivers, and the Chesapeake Bay proper (CB); and the 3) SE = Southeast regional grouping includes the Cape Fear River (CF), Winyah Bay (WB), Santee-Cooper (S-C), Edisto (E), Savannah (S), Ogeechee (O), and Altamaha (ALT) rivers, and Lake Marion (LM).
(DOC)
Assignment to collection of origin for 17 shortnose sturgeon ( Acipenser brevirostrum ) collections surveyed at 11 polysomic microsatellite DNA markers. Mis-assigned individuals are distributed horizontally.
(DOC)
Assignment to proposed grouping (five groupings model) in shortnose sturgeon ( Acipenser brevirostrum ) surveyed at 11 polysomic microsatellite DNA markers. The overall correct assignment rate to proposed grouping was 99.1% (522/527). Mis-assigned individuals are distributed vertically. Northeast regional grouping includes Saint John River (SJ), Canada, Penobscot, Kennebec, Androscoggin and Merrimack rivers; and the Southeast regional grouping includes the Cape Fear River (CF), Winyah Bay (WB), Santee-Cooper (S-C), Edisto (E), Savannah (S), Ogeechee (O), and Altamaha (ALT) rivers, and Lake Marion (LM).
(DOC)