MRM2 (RRMJ2, FTSJ2) and MRM3 (RMTL1, RNMTL1) are human methyltransferases involved in the modification of mitochondrial 16S rRNA. Inactivation of MRM2 or MRM3 in human cells by RNAi results in respiratory incompetence owing to diminished mitochondrial translation and the aberrant assembly of the large subunit of the mitochondrial ribosome.
Abstract
Defects of the translation apparatus in human mitochondria are known to cause disease, yet details of how protein synthesis is regulated in this organelle remain to be unveiled. Ribosome production in all organisms studied thus far entails a complex, multistep pathway involving a number of auxiliary factors. This includes several RNA processing and modification steps required for correct rRNA maturation. Little is known about the maturation of human mitochondrial 16S rRNA and its role in biogenesis of the mitoribosome. Here we investigate two methyltransferases, MRM2 (also known as RRMJ2, encoded by FTSJ2) and MRM3 (also known as RMTL1, encoded by RNMTL1), that are responsible for modification of nucleotides of the 16S rRNA A-loop, an essential component of the peptidyl transferase center. Our studies show that inactivation of MRM2 or MRM3 in human cells by RNA interference results in respiratory incompetence as a consequence of diminished mitochondrial translation. Ineffective translation in MRM2- and MRM3-depleted cells results from aberrant assembly of the large subunit of the mitochondrial ribosome (mt-LSU). Our findings show that MRM2 and MRM3 are human mitochondrial methyltransferases involved in the modification of 16S rRNA and are important factors for the biogenesis and function of the large subunit of the mitochondrial ribosome.
INTRODUCTION
Mitochondria supply energy to the cell through the process of oxidative phosphorylation (OXPHOS) and are involved in many other important cellular processes, such as calcium homeostasis, heme biosynthesis, apoptosis, and possibly aging. Mitochondria have retained an independent organellar translation machinery, which is necessary for the synthesis of the 13 polypeptides of the OXPHOS system encoded by mitochondrial DNA (mtDNA). The RNA components of the mitochondrial translation machinery (2 mt-rRNAs and 22 mt-tRNAs) are also encoded by mtDNA, whereas all protein components, including the proteins of the mitochondrial ribosomal subunits, translation factors, aminoacyl tRNA synthetases, RNA-modifying enzymes, and other auxiliary factors, are encoded by nuclear genes and delivered to mitochondria from the cytosol.
Human mitoribosomes consist of a large subunit (mt-LSU) and a small subunit (mt-SSU) of 39 and 28S, respectively, with the entire ribosome having a sedimentation coefficient of 55S (O'Brien, 2003). Mitochondrial factors must exist to facilitate mitoribosome production in order to modulate its function in response to various stimuli, similar to ribosomes in other systems. In mammalian mitochondria only a handful of such proteins have been identified (most of these only very recently), and their exact roles remain elusive (Metodiev et al., 2009, 2014; Dennerlein et al., 2010; Camara et al., 2011; He et al., 2012; Rorbach et al., 2012; Kotani et al., 2013; Wredenberg et al., 2013; Dalla Rosa et al., 2014). Effective ribosome production and function also depends on the maturation of rRNAs through a number of posttranscriptional nucleotide modifications (Venema and Tollervey, 1999). rRNA modifications consist mostly of base methylation, 2′-O-ribose methylation, and pseudouridylation, which can occur at various stages of ribosome production and are conserved between Prokaryota and Eukaryota. Base methylation is performed by specific methyltransferases (MTases; Motorin and Helm, 2011). However, the processes of 2′-O-ribose methylation and pseudouridine conversion, although common to both Prokaryota and Eukaryota, involve different mechanisms in these two groups. In eukaryotes, enzymes that modify cytosolic rRNAs depend on a number of small nucleolar RNAs (snoRNAs) to mark their target nucleotide by forming a transient duplex with the precursor. In contrast, prokaryotic rRNA modifications are performed by several site-specific enzymes that recognize their target without a guide RNA (Motorin and Helm, 2011).
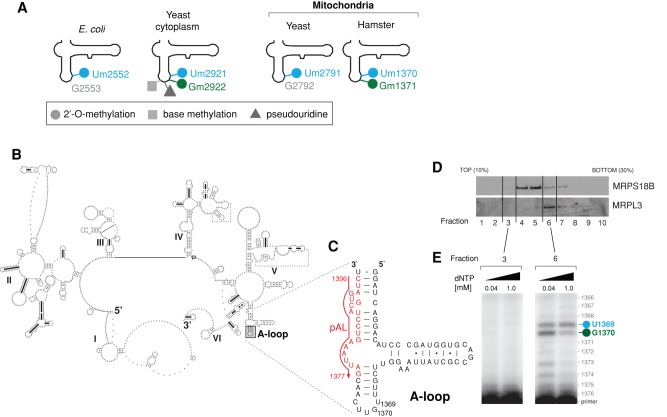
The modifications made to rRNAs have been mapped for mammalian mitoribosomes using hamster cells (Dubin, 1974; Dubin and Taylor, 1978; Dubin et al., 1978; Baer and Dubin, 1980, 1981). This analysis showed that only nine rRNA nucleotides are modified in mitochondrial rRNAs, compared with >200 in eukaryotic cytosolic rRNAs and >30 in bacterial rRNAs (Piekna-Przybylska et al., 2008). Five bases of the mammalian mt-SSU rRNA (12S rRNA) are methylated: m5U429, m4C839, m5C841, m62A936, and m62A937 (using human 12S numbering), whereas the mt-LSU rRNA (16S rRNA) contains three 2′-O-ribose methylated nucleotides— Gm1145, Um1369, and Gm1370—as well as one pseudouridylated base, Psi1397 (using human 16S numbering). The Um1369 and Gm1370 modifications of the mitochondrial 16S rRNA are predicted to lie in the LSU A-loop, which is an essential component of the peptidyl transferase center (PTC) implicated in the interaction of the ribosome with an aminoacyl (A)-site tRNA (Decatur and Fournier, 2002). This structure is well conserved through evolution between bacterial, cytosolic, and organellar ribosomes. Significantly, highly conserved nucleotides in the A-loop often contain 2′-O-methylation of ribose, namely, 2′-O-methyluracil (Escherichia coli, Um2552; yeast cytosol, Um2921) and 2′-O-methylguanosine (yeast cytosol, Gm2922; Figure 1A). Previous studies also revealed that the yeast mitochondrial LSU rRNA A-loop contains 2′-O-methylated ribose (Um2791; Figure 1A) and that this modification is crucial for ribosome function (Pintard et al., 2002a). However, nothing is known about the specific function of these modifications or whether these nucleotides are also modified in human mitochondria (Rorbach and Minczuk, 2012).
FIGURE 1:
Modification of the 16S rRNA A-loop in human mitochondria. (A) Comparison of the modification status of LSU A-loops across the tree of life. The various modifications reported previously in the LSU rRNA A-loop from E. coli (Caldas et al., 2000b), yeast mitochondria (Pintard et al., 2002a), and hamster (Dubin, 1974; Dubin and Taylor, 1978; Baer and Dubin, 1981) and the yeast cytosol are indicated. (B) The predicted secondary structure of human mitochondrial 16S rRNA. Human mitochondrial 16S rRNA was folded using the thermodynamics-based method of Zuker and Turner (Fields and Gutell, 1996) and downloaded from the Comparative RNA Web (CRW) database (Cannone et al., 2002). The position of the A-loop is indicated by a gray box. (C) Strategy of primer extension used to detect modifications of the A-loop in human mitochondrial 16S rRNA. The sequence encompassing the 16S rRNA A-loop is shown (16S nucleotide positions: 1325–1397). The position of the primer (pAL) used for reverse transcription primer extension (RT-PEx) is indicated in red. (D) Fractionation of the mitochondrial ribosome. Mitochondria that were isolated from HeLa cells were lysed, separated through a 10–30% (wt/vol) isokinetic sucrose gradient, and fractionated. Ten fractions were collected and analyzed by Western blotting using antibodies specific for a component of mt-LSU (MRPL3) and a component of mt-SSU (MRPS18B). (E) Detection of the A-loop modifications in human mitochondrial 16S rRNA. RNA isolated from fractions 3 and 6 of the sucrose gradient from D was subjected to RT-PEx using a radioactively labeled primer as shown in C, using two concentrations of dNTPs as indicated. Samples were analyzed on a 15% denaturing polyacrylamide gel and subjected to autoradiography. The specific pausing sites for 16S rRNA nucleotide positions U1369 (blue) and G1370 (green) are indicated.
Studies in yeast and mouse models indicated that, similar to the situation in bacteria, mitochondrial rRNA modifications depend on site-specific enzymes rather than being guided by small RNAs (Sirum-Connolly et al., 1995; Metodiev et al., 2009). TFB1M (and to some extent its paralogue TFB2M) is capable of the dimethylation of two very highly conserved adenines in a stem-loop near the 3′ end of 12S rRNA (m62A936 and m62A937) in mammalian mitochondria (Seidel-Rogol et al., 2003; Metodiev et al., 2009). NSUN4 is an m5C MTase responsible for methylation of m5C841 of 12S rRNA (Metodiev et al., 2014). A recent study by Lee et al. (2013) identified three novel putative mitochondrial methyltransferases, MRM1 (encoded by MRM1), MRM2 (also known as RRMJ2, encoded by FTSJ2) and RMTL1 (encoded by RNMTL1), with suggested roles in modification of the three known modified 2′-O-ribose bases of the 16S rRNA. The same study suggested a role for RMTL1 in methylation of G1370 of human 16S rRNA.
In this work we corroborate and extend the data on the roles of MRM2 (RRMJ2) and RMTL1, which we renamed MRM3, in mitochondria. We additionally show further independent evolutionary and experimental evidence on the involvement of MRM2 and MRM3 in the modification of nucleotides of the 16S rRNA A-loop, specifically Um1369 and Gm1370, respectively. Both proteins are found to be associated with mitoribosomes, and their down-regulation leads to defects in mitochondrial protein synthesis and respiratory deficiency, likely due to the disruption of mt-LSU assembly. Our findings provide additional support for the notion that MRM2 and MRM3 are important factors for the proper functioning of the mitochondrial translational machinery.
RESULTS
Modifications of the human mitochondrial LSU rRNA A-loop
We began by experimentally determining the modification status of the A-loop nucleotides in human mitochondrial 16S rRNA. To this end, we performed reverse transcription primer extension (RT-PEx) analysis as previously described (Pintard et al., 2002a). This assay is based on specific pausing of radioactive primer extension by a reverse transcriptase when a 2′-O-ribose modification is encountered. On the basis of the predicted secondary structure of the human mitochondrial 16S rRNA, we localized the A-loop to between nucleotides 1364 and 1378 (mtDNA positions 3034 and 3048, respectively; Figure 1B). We then extended a primer that binds to the 16S rRNA region downstream from the A-loop (1396–1377; Figure 1C). To enrich the 16S rRNA template, we isolated mitochondria from HeLa cells and then performed sucrose gradient sedimentation followed by isolation of mitochondrial RNA from fractions containing mtLSU (Figure 1D, fraction 6). RT-PEx identified two discrete pause sites at nucleotides U1369 and G1370 (Figure 1E), consistent with these nucleotides being modified in a similar manner to the corresponding modifications identified previously in hamster LSU rRNA (Um1370 and Gm1371). This result indicates that the human mitochondrial rRNA A-loop also contains well-conserved modifications of U1369 and G1370.
Identification and cellular localization of putative human 2′-O-MTases of the mitochondrial LSU 16S rRNA
Having confirmed the presence of U1369 and G1370 modifications in the LSU A-loop nucleotides in human mitochondria, we set out to determine the enzymatic machinery responsible for introducing these modifications. Thus far, snoRNAs have not been detected in mitochondria and, similar to the case for bacteria, yeast mitochondrial rRNA modifications depend on site-specific enzymes that are not guided by small RNAs (Sirum-Connolly et al., 1995; Pintard et al., 2002a). Therefore we assumed that human mitochondrial U1369 and G1370 will be modified by site-specific enzymes. All known 2′-O-MTases identified thus far belong to either the FtsJ-like (class I) or SpoU-type (class IV) protein families (Schubert et al., 2003).
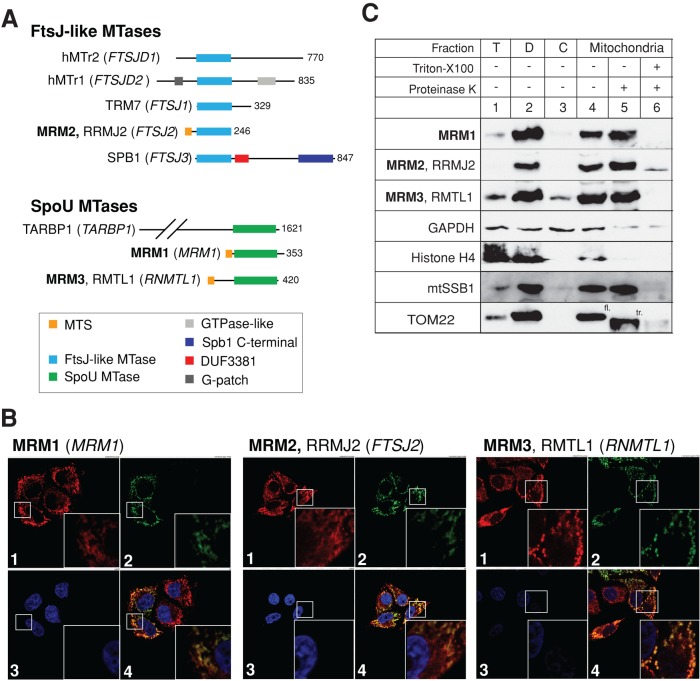
We first searched the human genome for homologues of FtsJ-like MTases using E. coli FtsJ/RrmJ uridine 2′-O-MTase (which modifies U2552 in LSU rRNA) and Mrm2p (which modifies U2791 in the yeast mitochondrial LSU 21S rRNA) as queries in PSI-BLAST searches. Both of these ribose-methylated nucleotides correspond to U1369 in the human mitochondrial 16S rRNA (Figure 1A). This search identified five human proteins (coding genes given in parentheses): hMTr1 (FTSJD2), hMTr2 (FTSJD1), TRM7 (FTSJ1), MRM2/RRMJ2 (FTSJ2), and SPB1 (FTSJ3; Figure 2A). The role of hMTr1 (FTSJD2) and hMTr2 (FTSJD1) in human cells was established previously, and both of these proteins function outside of mitochondria (Belanger et al., 2010; Werner et al., 2011). Of the remaining three proteins, MRM2/RRMJ2 is the closest homologue of the yeast mitochondrial Mrm2p and E. coli FtsJ/RrmJ. Previous ancestry analysis suggested that MRM2/RRMJ2 was brought into the eukaryotic cell via the mitochondrial endosymbiotic merger, followed by gene duplication events that resulted in the occurrence of TRM7 and SPB1 (Pintard et al., 2002b; further details in the Supplemental Text). The availability of three-dimensional structures of E. coli FtsJ/RrmJ uridine 2′-O-MTase (Bugl et al., 2000) and human MRM2 (unpublished structure, Protein Data Bank ID: 2NYU) permitted a comparison of the catalytic sites of these two enzymes, revealing a high degree of similarity (Supplemental Figure S1). The MRM2 protein sequence also has a high probability of possessing a mitochondrial targeting sequence (MTS) as analyzed by several computer programs (MitoProtII, 0.63; TargetP, 0.60; Predotar, 0.64; MultiLoc, 0.88). The human MRM2 protein is therefore a putative uridine 2′-O-MTase and a plausible candidate for modification of U1369 in the human mitochondrial 16S rRNA.
FIGURE 2:
Identification and mitochondrial localization of human FtsJ-like and SpoU-type MTases. (A) Human FtsJ-like and SpoU-type MTases. Several FtsJ-like and SpoU-type MTases identified in the human genome are shown schematically. Three MTases, MRM1, MRM2, (FTSJ2) and MRM3 (RMTL1), have a predicted mitochondrial targeting sequence (MTS). Other domains present in the MTases analyzed are also indicated. (B) The intracellular localization of MRM1, MRM2, and MRM3 MTases by immunofluorescence. The cDNA encoding the STREP2- and FLAG-tagged variants of each MTase was transiently transfected into HeLa cells. Mitochondria were stained with MitoTracker Red CMXRos (red, 1). MRM1, MRM2, or MRM3 were detected using an anti-FLAG antibody and visualized using secondary antibodies conjugated with FITC (green, 2). Cell nuclei were stained with DAPI (blue, 3). Colocalization of the mitochondria-specific red signal and MTase-specific green signal appears yellow on digitally merged images (4). (C) Localization of MRM1, MRM2, and MRM3 MTases in subcellular fractions. HeLa cells were fractionated into debris (D, lane 2), cytosol (C, lane 3), and mitochondria (lanes 4–6). The mitochondrial fraction was treated with 25 μg/ml proteinase K in the absence (lane 5) or presence of 1% Triton X-100 (lane 6). T, total cell lysate. The fractions were analyzed by Western blotting using antibodies to each MTase. The distribution of MTases was compared with that of the following marker proteins: mtSSB1 (mitochondrial matrix), TOM22 (mitochondrial outer membrane), GAPDH (cytosol), and histone H4 (nucleus). fl, TOM22 of full length; tr, truncated TOM22.
To identify other candidate enzymes for the modification of nucleotides in the human mitochondrial 16S rRNA A-loop, we also searched the human genome for homologues of MTases from the SpoU superfamily (Anantharaman et al., 2002). Several enzymes from this group have been identified as 2′-O-MTases, including the bacterial guanosine 2′-O-MTase RlmB responsible for the formation of Gm at nucleotide 2251 of the E. coli LSU rRNA (Lovgren and Wikstrom, 2001) and Pet56p (Mrm1p, YOR201C), which introduces a Gm modification in the yeast mitochondrial LSU rRNA at position 2270 (Sirum-Connolly and Mason, 1993). Using E. coli RlmB and yeast Pet56p as queries in PSI-BLAST homology searches, we identified three human proteins (coding genes given in parentheses): TARBP1/TRP-185 (TARBP1), MRM1 (MRM1), and RMTL1 (RNMTL1; Figure 2A). The TARBP1 protein was previously characterized as a nuclear-localized factor that regulates RNA polymerase II binding to HIV-1 RNA regulatory element (TAR; Wu-Baer et al., 1995). The functions of MRM1 and RMTL1 in human cells have not been extensively studied. Given the strong homology to well-characterized SpoU-type guanosine 2′-O-MTases (Supplemental Figure S2) and the observation that both protein sequences exhibit a high probability of mitochondrial import (MRM1 and MRM3: MitoProtII, 0.97 and 0.96; TargetP, 0.92 and 0.89; Predotar, 0.84 and 0.63; MultiLoc, 0.19 and 0.96), MRM1 and RMTL1 appear to be good candidates for guanosine 2′-O-methyl modification of mitochondrial 16S rRNA. For consistency, we renamed the RMTL1 protein MRM3 (mitochondrial rRNA MTase 3).
To study experimentally the cellular localization of MRM1, MRM2, and MRM3, we transiently expressed the corresponding cDNAs, containing a FLAG tag, in HeLa cells. Immunofluorescence analysis revealed that the recombinant proteins colocalized with mitochondria (Figure 2B). Cell fractionation experiments confirmed the mitochondrial localization of the three proteins, as the endogenous proteins cofractionated with the well-characterized mitochondrial matrix protein mtSSB1 and were resistant to proteinase K treatment (Figure 2C). In similar conditions the outer membrane protein TOM22 was truncated by proteinase K. These results demonstrate that MRM1, MRM2, and MRM3 are localized within the mitochondria of human cells.
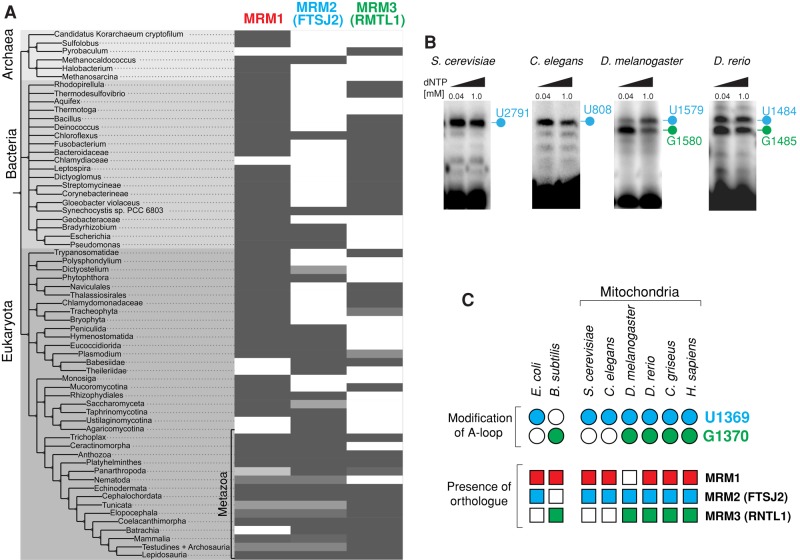
Orthologues of the MRM proteins and modification of mitochondrial LSU rRNA across evolution
Next we set out to assign which of the identified MRM candidates are responsible for specific modifications of the LSU A-loop in human mitochondria. To this end, we investigated the correlation between the evolutionary occurrence of MRM orthologues and the presence of modified nucleotides corresponding to U1369 and U1370 in the LSU rRNA A-loop in prokaryotic organisms and mitochondria from other species. First, we examined the distribution of MRM1, MRM2, and MRM3 via reciprocal best BLAST hits across the 147 fully sequenced genomes (Reference Proteomes, 2013_04 release; Gabaldon et al., 2009; Figure 3A, Supplemental Figure S3, and Supplemental Text). Homologues of the three MRM proteins were found to be broadly distributed across Archaea, Bacteria, and Eukaryota (Supplemental Text). Second, we queried published data for reports on 2′-O-ribose methylation in the LSU A-loops of prokaryotic organisms and mitochondria, in which posttranscriptional modifications of rRNA are introduced by site-specific enzymes. This latter step of analysis, however, was hindered by the fact that very few eukaryotic species have been analyzed for the presence of modifications in their mitochondrial LSU rRNAs. Therefore we experimentally analyzed the mitochondrial LSU rRNA A-loop modifications in several eukaryotes (Caenorhabditis elegans, Drosophila melanogaster, Danio rerio) using RT-PEx of mitochondrial or total RNA preparations from these organisms. We first confirmed the previously published result that Saccharomyces cerevisiae contains only one modification of the LSU A-loop, at position U2791, corresponding to human U1369 (Figure 3B). A single modification at the corresponding position (U808) was also present in the A-loop of C. elegans LSU mt-rRNA. In D. melanogaster and D. rerio, in addition to this conserved U nucleotide (U1579 and U1484, respectively), the G nucleotide that corresponds to human mitochondrial 16S G1370 was also modified (G1580 and G1485, respectively; Figure 3B). Finally, comparing the evolutionary distribution of the MRM proteins with the presence of particular modifications in the A-loop revealed the following relationships: 1) The nucleotide corresponding to human mitochondrial LSU rRNA U1369 is modified in species in which MRM2 is present (Figure 3C, blue), supporting our sequence/structure analysis-based prediction that this protein is responsible for modification of this conserved uridine in the human 16S A-loop (see earlier discussion); 2) modification of the position corresponding to human mitochondrial LSU rRNA G1370 correlates with the evolutionary presence of MRM3 (Figure 3C, green), suggesting that this protein is responsible for modification of this conserved guanosine in the human 16S rRNA A-loop; and 3) the evolutionary distribution of MRM1 does not coincide well with modification of either U1369 or G1370 (Figure 3C, red).
FIGURE 3:
Evolutionary distribution of MTase orthologues and specific modification of the A-loop of LSU rRNA. (A) Evolutionary analysis of human MRM1, MRM2, and MRM3 across higher taxa. The tree shows higher taxa for human MRM1, MRM2, and MRM3 based on National Center for Biotechnology Information taxonomy and reciprocal BLAST hits computed for 147 fully sequenced genomes (Reference Proteomes, 2013_04 release). The percentage of hits within a taxon is shown by a grayscale, with the darker tones representing the higher percentages. Light gray may indicate the presence of evolutionary divergence within the taxon, incomplete proteome data, or limitations of the reciprocal BLAST approach. (B) Detection of A-loop modifications in mitochondrial LSU rRNAs of several eukaryotic species. RNA isolated from S. cerevisiae, C. elegans, D. melanogaster, and D. rerio was subjected to RT-PEx using a radioactively labeled primer binding upstream from the A-loop. Samples were analyzed on a 15% denaturing polyacrylamide gel and subjected to autoradiography. The specific pausing sites for the LSU rRNA A-loop are indicated. Nucleotide positions that correspond to human 16S rRNA U1369 are indicated in blue, whereas those corresponding to human 16S rRNA U1370 are in green. (C) Correlation between the presence of MTase orthologues and specific modifications of the A-loop. The presence of MRM1 (red), MRM2 (blue), or MRM3 (green) in analyzed species is indicated by filled squares. Open squares indicate that an orthologue has not been identified. The modification status of LSU nucleotides corresponding to the human 16S residues U1369 (blue) and G1370 (green) is represented by circles. Open circles indicate a lack of modification; filled circles indicate that the nucleotide is modified. The data for Bacillus subtilis are those of Hansen et al. (2002), and other references can be found in the text.
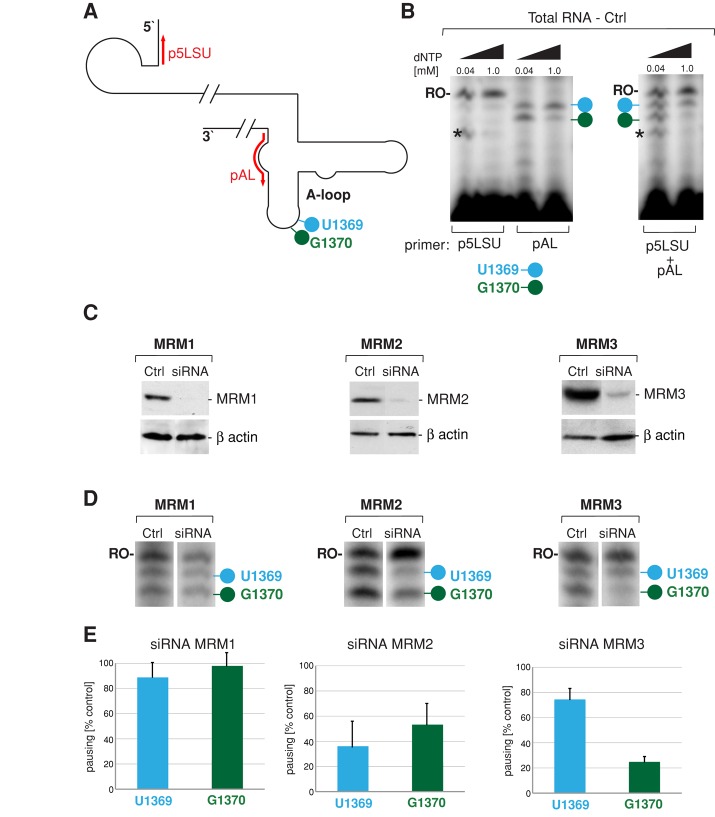
RNA interference silencing of MRM2 or MRM3 affects modification of the 16S rRNA A-loop in human mitochondria
To confirm our computational predictions experimentally, we studied the modifications of the human 16S rRNA A-loop upon small interfering RNA (siRNA) silencing of the three MRM candidates using RT-PEx. Two primers were used for each PEx reaction: the A-loop–specific primer (Figure 4A, pAL) was designed to detect reverse transcriptase pausing at U1369 and G1370, whereas a primer that anneals at the 5′ end of 16S rRNA and produces a runoff transcript (Figure 4A, p5LSU) was used to provide standardization. Initial tests demonstrated that it is feasible to use total RNA preparations from HeLa cells and a combination of the two primers in RT-PEx reactions (Figure 4B). We silenced the three MRM proteins in HeLa cells using three different siRNA oligonucleotides in each case (Supplemental Figure S4) and selected the most efficient oligonucleotides for further analysis (Figure 4C). After 9 d of depletion of each MRM transcript in HeLa cells (achieved by three subsequent transfections of siRNA in a 3-d interval), we applied RT-PEx to analyze the A-loop modifications of 16S mt-rRNA. The degree of pausing at U1369 and G1370 (Figure 4D, blue and green) was compared with the amount of runoff transcription at the 5′ end of 16S mt-rRNA (Figure 4D, “RO-”). Quantitative analysis of three independent siRNA/RT-PEx experiments (Figure 4E) revealed that 1) siRNA silencing of MRM1 does not affect any of the 16S rRNA A-loop modifications, 2) down-regulation of MRM2 results in a significant loss of the U1369 modification, although the G1370 is also affected, and 3) knockdown of MRM3 predominantly affects modification of G1370, with only a minor effect on U1369. Taken together, these data indicate that MRM2 and MRM3, but not MRM1, play a role in posttranscriptional modification of the A-loop residues of 16S mt-rRNA. In particular, RT-PEx experiments clearly confirm our previous in silico prediction that MRM3 is responsible for modification of the G1370 residue of the 16S mt-rRNA A-loop. Furthermore, MRM2 has been suggested to be a U1369 2′-O-ribose methylase by sequence/three-dimensional (3D) structure homology–based computational comparisons (Supplemental Figure S1), as well as the evolutionary cooccurrence of the A-loop uridine modification with MRM2 homologues (Figure 3C). The strong effect of MRM2 silencing upon the U1369 modification is therefore compatible with this prediction.
FIGURE 4:
siRNA depletion of MRM2 and MRM3 perturbs LSU rRNA A-loop modifications. (A) Strategy for primer extension to measure levels of the A-loop modifications in human mitochondrial 16S rRNA. The location of primers used for reverse transcription primer extension (RT-PEx) are indicated in red. The pAL primer detects the modifications of U1369 (blue) and G1370 (green), whereas the p5LSU primer is used to standardize the loading of 16S rRNA. (B) Detection of the A-loop modifications of human mitochondrial 16S rRNA in total RNA preparations. Radioactively labeled primers as per A were extended separately (left) or simultaneously (right) in a RT-PEx reaction using total RNA isolated from HeLa cells using two concentrations of dNTPs as indicated. Reaction products were separated on a 15% (wt/vol) denaturing polyacrylamide gel and subjected to autoradiography. The specific pausing sites upon extension of the pAL primer at nucleotide positions U1369 and G1370 are indicated by blue and green lollipops, respectively. RO-, the runoff product resulting from extending the p5LSU primer. The asterisk denotes an unspecified band resulting from RT-PEx of the p5LSU primer using the lower dNTP concentration. (C) RNA interference knockdown of MTases. Steady-state protein levels of the endogenous MRM1, MRM2, and MRM3 proteins analyzed by Western blotting in control HeLa cells transfected with an unrelated siRNA (Ctrl) or cells transfected with selected siRNAs specific for a particular MTase for 9 d. β-Actin was used as loading control. (D) 16S rRNA A-loop modifications upon siRNA knockdown of MRM1, MRM2, or MRM3 Total RNA isolated from control HeLa cells or cells transfected with siRNA to MTases was analyzed using RT-PEx as depicted in A and B. Specific pausing at modified nucleotides U1369 and G1370 is indicated by blue or green lollipop, respectively. The runoff product used for normalization is indicated as RO-. (E) Relative ratios between A-loop–specific and runoff 16S rRNA RT-PEx products in cells treated with siRNA to MTases. RT-PEx reactions as per D were quantified using ImageQuant software, and the ratios U1369/RO and G1370/RO were plotted relative to controls for three independent siRNA transfection experiments. n = 3; error bars, 1 SD.
Depletion of MRM2 and MRM3 perturbs mitochondrial function
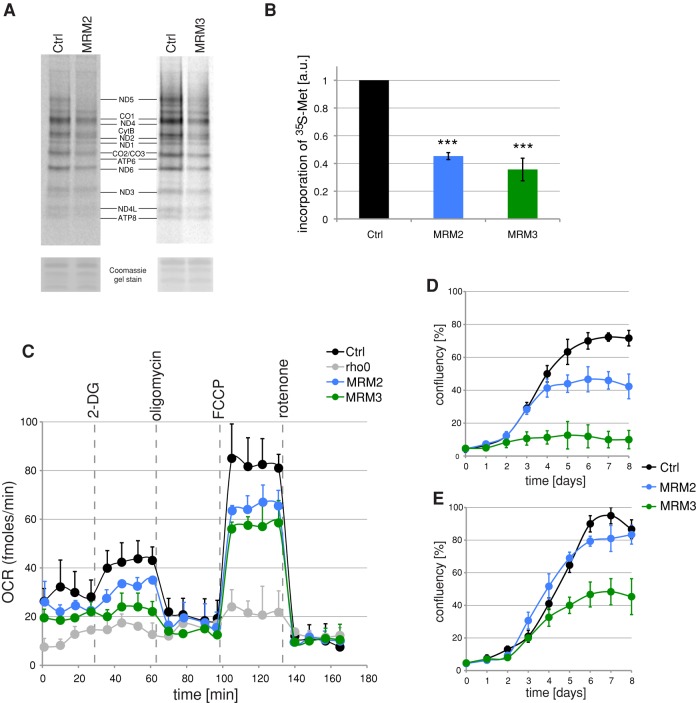
We next set out to determine the functional consequences of a reduction of methylation of the LSU A-loop after depletion of MRM2 and MRM3. De novo labeling of mitochondrial protein synthesis products, followed by gel electrophoresis and autoradiography, showed a significant inhibition of translation in mitochondria of cells subjected to MRM2 or MRM3 RNA interference (RNAi; Figure 5A). The overall rate of mitochondrial protein synthesis in the MRM2 or MRM3 siRNA transfectants was ∼50 and 35%, respectively, compared with that of cells transfected with an unrelated control siRNA (Figure 5B). In both cases, the observed decrease in mitochondrial translation was accompanied by a reduction of the cellular oxygen consumption rate (OCR), consistent with perturbed function of the OXPHOS complexes (Figure 5C). When cells were grown in media containing galactose as the sole carbon source, which forces dependence on oxidative phosphorylation for ATP production, the MRM2- or MRM3-depleted cells showed significantly reduced growth rate, with a very pronounced effect in the case of MRM3 (Figure 5D). Furthermore, MRM3-depleted cells showed reduced growth rate when cultured in standard medium containing glucose (Figure 5E), effects observed previously under conditions of perturbed mitochondrial translation (Rorbach et al., 2008).
FIGURE 5:
Mitochondrial translation and OXPHOS function after siRNA depletion of MRM2 or MRM3. (A) Mitochondrial translation in cells treated with siRNA to MRM2 or MRM3. Products of mitochondrial translation were labeled with [35S]methionine in control cells transfected with an unrelated siRNA (Ctrl) or in cells transfected for 9 d with siRNAs specific for MRM2 or MRM3. Mitochondrial proteins were separated by 4–12% gradient SDS–PAGE and visualized by autoradiography. To validate equal protein loading, the gel was stained with Coomassie dye. (B) Radiolabeled products of mitochondrial translation as per A were quantified using ImageQuant software after exposure to a PhosphorImager screen. ***p < 0.001; two-tailed Student's t test; n = 3; error bars, 1 SD. (C) OCRs in MRM2- or MRM3-depleted cells. OCR measured in an extracellular flux Seahorse instrument in control cells transfected with an unrelated siRNA or in cells treated with siRNAs to MRM2 or MRM3 for 9 d. The wells were sequentially injected with 20 mM 2-DG to inhibit glycolysis, 100 nM oligomycin to inhibit ATP-synthase, 1 μM FCCP to uncouple the respiratory chain, and 200 nM rotenone to inhibit complex I. n = 4; error bars, 1 SD. (D, E) Cell growth upon inactivation of MRM2 or MRM3. Growth curves obtained by Incucyte kinetic imaging system of parental HEK293 cells (Ctrl) or cells transfected with siRNA to MRM2 or MRM3 for 8 d in galactose (D) or glucose (E) medium.
MRM2 and MRM3 interact with the mitochondrial ribosome
Having shown that MRM2 and MRM3 are responsible for modification of nucleotides of the human 16S rRNA A-loop and that depletion of the proteins impairs mitochondrial translation, we were interested to determine whether these proteins associate with the mitochondrial ribosome. To investigate this possibility, we first performed pull-down experiments using FLAG-tagged versions of MRM2 or MRM3, which were inducibly expressed in HEK293T cells. MRM2-FLAG was found to coimmunoprecipitate substantial quantities of components of both the mt-LSU (MRPL3 and MRPL12) and the mt-SSU (MRPS18B and DAP3/MRPS29). On the other hand, MRM3-FLAG was found to efficiently coimmunoprecipitate components of mt-LSU but only very marginal amounts of components of mt-SSU (Figure 6, A and B). The lack of other highly abundant mitochondrial proteins in the elution fractions, such as subunit NDUFB8 of complex I, suggests specific interaction of MRM2 and MRM3 with the mitochondrial ribosome (Figure 6, A and B).
FIGURE 6:
Interaction of MRM2 and MRM3 with the mitochondrial ribosome. (A, B) IP of components of the mitochondrial ribosome using tagged version of MRM3 and MRM3. Mitochondria from HEK293T cells expressing MRM2.FLAG (A) or MRM3.FLAG (B) were lysed, and IP was performed using anti-FLAG antibodies. The lysates (Input) and eluates were analyzed by Western blotting with antibodies as indicated. NDUFB8, antibody against a subunit of mitochondrial complex I. (C, D) IP of endogenous MRM2 and MRM3 using tagged components of the mitoribosome. Mitochondria from HEK293T cells expressing a component of mt-LSU (ICT1.FLAG; C) or a component of mt-SSU (MRPS27.FLAG; D) were lysed, and IP was performed using anti-FLAG antibodies. The lysates (Input) and eluates were analyzed by Western blotting using antibodies as indicated. NDUFB8, antibody against a subunit of mitochondrial complex I. (E, F) Cosedimentation of MRM2 and MRM3 with mitochondrial ribosome in sucrose gradients. Total lysates of HEK293T cells expressing MRM2.FLAG (E) or untransfected HEK293T cells (F) were separated through a 10–30% (wt/vol) isokinetic sucrose gradient and fractionated. The fractions were analyzed by Western blotting using antibodies specific for the FLAG tag (MRM2), endogenous MRM3, components of mt-LSU (MRPL12 or MRPL3), and a component of mt-SSU (MRPS18B). (G) Summary of MRM2 and MRM3 interactions with the mitoribosome. Interpretation of the IP and sucrose gradient experiments.
We next performed reciprocal pull-down experiments in which either FLAG-tagged ICT1 (a component of mt-LSU) or FLAG-tagged MRPS27 (a component of mt-SSU) were used as the bait. In this experiment, endogenous MRM2 was coimmunoprecipitated by proteins from both the large and small ribosomal subunits. In contrast, MRM3 was pulled down only by ICT1, a large-subunit protein (Figure 6, C and D). The specificity of detected interactions was confirmed by probing the elution fractions for NDUFB8 as before. It was reported previously that ICT1.FLAG and MRPS27.FLAG not only are able to pull-down components of mt-SSU or mt-LSU, respectively, but that each of these proteins is also capable of pulling down the entire 55S monosome (Richter et al., 2010). Indeed, proteins of mt-LSU were present in the elution fraction of the MRPS27.FLAG immunoprecipitation, consistent with copurification of the intact mitochondrial monosome. Because MRM2 was present in both the ICT1.FLAG and the MRPS27.FLAG elution fractions, it is possible that the protein interacts with either of the subunits or with the mitochondrial monosome. The lack of MRM3 in the MRPS27.FLAG elution indicates that the protein does not bind to the fully assembled 55S mitoribosome.
These observations were further investigated by analysis of mitoribosomes using sucrose gradient centrifugation. We carried out a gradient analysis of HEK293T cells using antibodies against MRPL12 and MRPS18B to visualize the large and small mitoribosomal subunits, respectively. We could not readily detect endogenous MRM2 comigrating with markers of the mitochondrial ribosomal subunits, likely due to the low abundance of the mitoribosome-associated protein (Supplemental Figure S5). Therefore we expressed MRM2 tagged with a FLAG tag in HEK293T cells, allowing more sensitive detection by anti-FLAG monoclonal antibody. Consistent with our initial coimmunoprecipitation (co-IP) experiments (Figure 6A, C and D), a small proportion of MRM2.FLAG comigrated with mt-LSU (Figure 6E, fractions 10 and 11) and larger protein assemblies (Figure 6E, fractions 14 and 15). In a similar sucrose gradient analysis, endogenous MRM3 was found to be partially associated with mt-LSU (Figure 6F, fractions 9–11), fully corroborating the results of the co-IP analysis (Figure 6, B– D).
In conclusion, our co-IP experiments suggest that a portion of the mitochondrial pool of MRM2 associates with mt-LSU. The MRM2 co-IP pattern is also consistent with association of this protein with the 55S monosome. Sucrose gradient sedimentation analysis of a tagged protein confirms association of MRM2 with mt-LSU. The presence of MRM2 in the higher sucrose gradient fractions further suggests association with larger mitochondrial ribosome assemblies, likely the 55S monosome (often not easily detectable by the sucrose gradient in proliferating cells; Dennerlein et al., 2010; Richter et al., 2010, 2013). The results of co-IP experiments and sucrose gradient sedimentation indicate that a fraction of MRM3 interacts with mt-LSU but not with the fully assembled monosome (Figure 6G).
Loss of MRM2 and MRM3 alters the sedimentation profile of mt-LSU
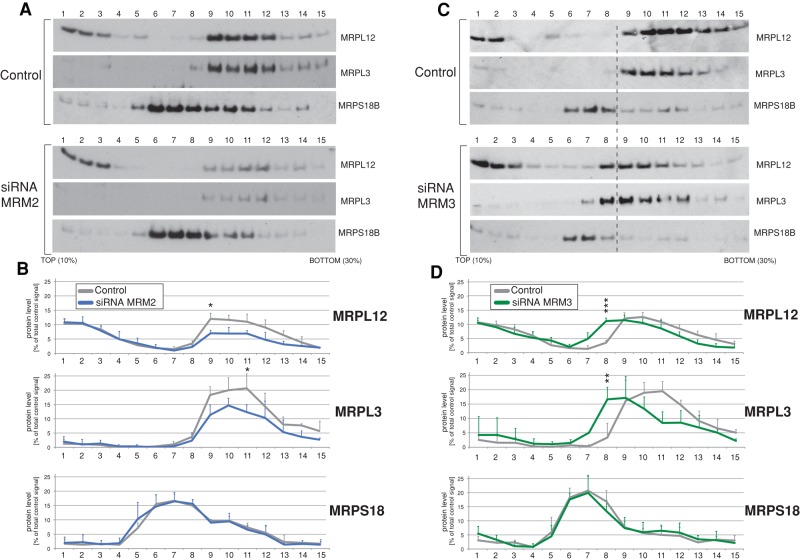
To further characterize the molecular basis for the observed translation defect upon MRM2 or MRM3 knockdown, especially in the context of their interaction with the mitoribosome, we examined the integrity of mitochondrial ribosomes by ultracentrifugation of cell extracts through sucrose gradients. Silencing of MRM2 expression for 9 d revealed 20–30% reduction in levels of assembled 39S mitoribosomal subunits (Figure 7, A and B), manifested as reduction in signal from the MRPL12 protein incorporated into mt-LSU (Figure 7, A and B, fractions 9–12) relative to the pool of free MRPL12 (Figure 7, A and B, fractions 1–3). The profile of the mt-SSU marker, MRPS18B, was not markedly changed after MRM2 depletion. Next we examined the levels of mt-rRNAs in individual sucrose gradient fractions of cell lysates from controls and cells treated with RNAi to MRM2 (Supplemental Figure S6). We detected significant reduction of steady-state levels of 16S rRNA in siRNA-treated cells in fractions corresponding to mtLSU, consistent with defect in assembly or reduced stability of this mitoribosomal subunit. An analogous experiment was performed using cells treated with siRNA against MRM3. In this experiment, a significant proportion of the mt-LSU markers (MRPL3 and MRPL12) relocated to less dense fractions compared with control cells (Figure 7, C and D, fractions 7 and 8), implying that the assembly and/or stability of mt-LSU is impaired. The profile of the mt-SSU marker MRPS18B was again unchanged, suggesting that assembly of mt-SSU is unaffected by depletion of MRM3. There were no apparent changes in the stability of mt-rRNA in MRM3 siRNA-treated cells (Supplemental Figure S6). Taken together, these observations suggest that both MRM2 and MRM3 are important for mt-LSU integrity, mitochondrial protein synthesis, and consequently OXPHOS function.
FIGURE 7:
Mitochondrial ribosome integrity upon siRNA depletion of MRM2 or MRM3. (A) The mitochondrial ribosome profile in MRM2-depleted cells. Total cell lysates of control HeLa cells transfected with an unrelated siRNA or HeLa cells transfected with siRNA to MRM2 were separated on a 10–30% (wt/vol) isokinetic sucrose gradient. Fractions obtained for control and RNAi-treated cells were analyzed by Western blotting using antibodies to mt-LSU (MRPL3, MRPL12) and mt-SSU (MRPS18B). (B) Quantification of the gradient distribution of mitoribosomal components upon MRM2 silencing. Western blot signals for MRPL3, MRPL12, and MRPS18B for control and MRM2-depleted cells were quantified using ImageQuant software. The y-axis represents the fraction of the total signal relative to control cells. n = 3. All significant value changes are indicated; *p < 0.05, **p < 0.01, ***p < 0.001; two-tailed unpaired Student's t test. Error bars, 1 SD. (C) The mitochondrial ribosome profile in MRM3-depleted cells. Total cell lysates of control HeLa cells or HeLa cells transfected with siRNA to MRM3 were analyzed as per A. (D) Quantification of the gradient distribution of mitoribosomal components upon MRM2 silencing. Western blot signal was quantified and analyzed as per B.
DISCUSSION
MRM2 and MRM3 modify the LSU rRNA A-loop in human mitochondria
In this study we set out to determine whether and how the LSU rRNA A-loop is posttranscriptionally modified in human mitochondria. We confirmed that the 16S rRNA nucleotides U1369 and G1370, located at the top of the hairpin structure of the A-loop, are modified in human cells (Figure 1), consistent with previous descriptions of mt-rRNA modifications in other mammalian species (Dubin, 1974; Dubin and Taylor, 1978; Baer and Dubin, 1981). We identified three members of an MTase family, MRM1, MRM2, and MRM3 (renamed from RMTL1, encoded by RNMTL1), and found that all three proteins localize to mitochondria in human cells (Figure 2). Human MRM2 has been identified as a putative uridine 2′-O-MTase responsible for modification of U1369 and MRM3 as a putative guanine 2′-O-MTase that modifies G1370, based, first, on sequence/structure homology analysis (Supplemental Figures S1 and S2) and the evolutionary cooccurrence of orthologues with particular A-loop modifications in bacteria and mitochondria (Figure 3), and, second, on use of siRNA knockdown experiments (Figure 4).
The mitochondrial localization of MRM1, MRM2, and MRM3 in HeLa cells observed in our studies is in full agreement with the report by Lee et al. (2013) and also with systematic studies of the whole human proteome (Human Protein Atlas; Uhlen et al., 2010) and the mitochondrial proteome (Mitocarta; Pagliarini et al., 2008).
The assignment of MRM3 to the modification of a conserved guanosine, G1370, in the human 16S rRNA A-loop is strongly supported by sequence and structure homology studies, as well as by the evolutionary correlation between the presence of the orthologue with the guanine modification in the A-loop and, most important, by siRNA down-regulation experiments. The same conclusion regarding the role of MRM3 was reached by Lee et al. (2013). Our computational sequence/structure analysis and evolutionary distribution studies, combined with experimental data on the LSU A-loop modification across various eukaryotic species, is consistent with MRM2 being responsible for modification of U1369 in the human 16S rRNA A-loop. However, experiments in which MRM2 was depleted using siRNA found that, in addition to the primary effect on the U1369 modification, G1370 methylation is also affected by depletion of MRM2, albeit to a lesser degree. One explanation for this could be interdependence between these two modification events. In this situation, the U1369 residue would be modified earlier during mitoribosome biogenesis, with the later modification of G1370 being dependent on this event (see later discussion). To some extent, this would be similar to the sequence of events during modification of the A-loop during the biogenesis of the cytosolic ribosome in yeast. The universally conserved U2921 modification (corresponding to U1369), located within the A-loop of the yeast 28S LSU rRNA, is formed at an earlier stage of the biogenesis pathway, whereas the G2922 modification (the equivalent of G1370) is introduced at a later step, during the maturation of LSU (Lapeyre and Purushothaman, 2004). An alternative explanation may be provided by the technical features of the PT-PEx assay used in these experiments. For example, 1) the lack of U1369 modification could cause changes in the conformation of the RNA template, altering the degree of stalling by the reverse transcriptase at the preceding G1370 site, or 2) the lack of modification at G1370 could lead to more of the reverse transcriptase reaching U1369, influencing the results at this site. Future experiments should involve an alternative method for detection of the posttranscriptional modifications in mt-rRNA, such as mass spectrometry (Suzuki and Suzuki, 2014).
The role of MRM2 and MRM3 proteins in mitoribosome biogenesis and function
Chemical modifications of rRNAs are mostly clustered in conserved and functionally critical regions of the ribosome. The 2′-O-methyluracil and 2′-O-methylguanosine modifications at positions 1369 and 1370, respectively, are predicted to lie in the A-loop of the LSU rRNA in human mitochondria. The A-loop is an essential component of the peptidyl transferase center, since it is the docking site for the incoming aminoacyl (A)-site tRNA, which binds through its 3′-end CCA tail. In other systems, enzymes responsible for modifications to the A-loop have been shown to play a major role not only in ribosome function, but also in its biogenesis.
Our data indicate that 2′-O-ribose methylation of a conserved uridine residue, U1369, by the human MRM2 protein is critical for proper mitochondrial translation and consequently for mitochondrial respiratory function (Figure 5). The role of MRM2 in these processes was not studied before. We further show that human MRM2 associates both with mt-LSU and the mitochondrial monosome. Two different methods have been used to this end: 1) reciprocal coimmunoprecipitation of tagged and endogenous MRM2 proteins with components of the mitochondrial ribosome and 2) cofractionation of the tagged MRM2 with mitoribosomal subunits upon sucrose gradient sedimentation (Figure 6). An association of MRM2 with the mitochondrial ribosome was also suggested by Lee et al. (2013), but their methodology did not allow for identification of the exact nature of this interaction. Finally, we provide additional data indicating that depletion of MRM2 by RNAi affects the stability of mt-LSU (Figure 7). The base equivalent of U1369 of human mitochondrial 16S rRNA was shown to be 2′-O-ribose methylated in the majority of organisms investigated (Sirum-Connolly et al., 1995), and model organisms have been used to assess the functional role of this modification. In E. coli, strains that are deficient in RrmJ, which is responsible for methylation of the corresponding position (Um2552) in the large 23S rRNA, exhibit growth defects and reduced translation rates. RrmJ is active against ribosomes and free LSU but not free rRNA. This Um2552 methylation is important for stability of the LSU, consistent with a role for this MTase in later stages of bacterial ribosome maturation (Caldas et al., 2000a, b). Similarly, deletion of the yeast MRM2 gene (responsible for methylation of the corresponding position, Um2791, in mitochondrial LSU rRNA) causes a thermosensitive respiratory growth phenotype (Pintard et al., 2002a). Mrm2p cosediments with the large rRNA on sucrose gradients and methylates U2791 only when rRNA is assembled into the LSU, again suggesting that this modification is required for the later steps of the ribosome maturation process (Pintard et al., 2002a). Therefore our results indicate that not only is human MRM2 highly similar to the bacterial RrmJ and yeast Mrm2p on the sequence level, but it also retained an analogous function during ribosome biogenesis. However, further research is required to study the exact functional role of the U1369 modification. It has been shown that, in bacteria, 2′-O-ribose methylation of the A-loop uridine not only is required for proper ribosome biogenesis, but also plays a role in regulating translational accuracy. The lack of this modification to the bacterial 23S rRNA A-loop causes a decrease in programmed +1 and −1 translational frameshifting and a reduction in readthrough of UAA and UGA stop codons (Widerak et al., 2005). It would be interesting to see whether the Um1369 modification in human 16S rRNA fulfils a similar function, especially in the context of the discovery that programmed −1 frameshifting by the mammalian mitoribosome is necessary for translation termination of two mitochondrial open reading frames (Temperley et al., 2010).
Our results also imply that MRM2 interacts with the intact mitochondrial monosome (Figure 6). Neither the bacterial RrmJ nor the yeast Mrm2p exhibits this property. It is possible that human MRM2 has acquired an extra function compared with its orthologues that could, for example, contribute to the very final stages of joining mt-SSU and mt-LSU into a functional mitochondrial monosome. Such an extra function would not be unprecedented, as dual functions of rRNA MTases have been reported. For instance, the bacterial MTase KsgA, which is responsible for dimethylation of adenines at the 3′ end of the SSU rRNA, has also been proposed to play a role in the final stages of monosome assembly. Some earlier data showed that extracts from KsgA-deficient E. coli cells have reduced affinity between the 30 and 50S ribosomal subunits (Poldermans et al., 1980). It has further been proposed that KsgA sterically blocks incompletely assembled small ribosomal subunits from entering the translational cycle prematurely (Xu et al., 2008; Demirci et al., 2010).
Our work also concentrated on 2′-O-ribose methylation of a conserved A-loop guanosine residue, G1370, which is performed by the human MRM3 protein and is indispensable for correct mitochondrial translation and OXPHOS function (Figure 5). Coimmunoprecipitation and sucrose gradient sedimentation analyses indicate that human MRM3 associates with mt-LSU but not with the monosome (Figure 6). These data are in full agreement with a recent study by Lee et al. (2013). However, our study also provides a partial explanation for this phenotype. Down-regulation of MRM3 expression using siRNA results in the accumulation of species that are consistent with mt-LSU pre-ribosomal particles, likely to be late-stage assembly intermediates (Figure 7). The base equivalent of G1370 in the human mt-LSU rRNA is not modified in the E. coli 23S rRNA (G2553), although evidence exists that this residue pairs with C75 of the aminoacyl tRNA in the bacterial ribosomal A-site (Kim and Green, 1999). Yeast mitochondrial 21S rRNA also does not carry a modification at this site (G2792). However, an analogous 2′-O-methylguanosine modification is present in the yeast cytosolic LSU 25S rRNA (Gm2922). Unlike other 25S 2′-O-ribose methylation events, which are guided by numerous small nucleolar RNAs, the formation of Gm2922 is catalyzed by an essential FtsJ-like MTase called Spb1p. Modification of G2922 is a late event occurring on the 27S ribosome intermediate and is essential for proper ribosome production (Harnpicharnchai et al., 2001; Lapeyre and Purushothaman, 2004). Parallels can therefore be drawn between the methylation of guanosine in the A-loop in eukaryotic cytosolic ribosomes (by Spb1p) and in the mammalian mitochondrial ribosome (by MRM3), in that both modifications are performed by site-specific enzymes, with the modification being necessary for the regulation of ribosome biogenesis and translation. The possibility of this modification occurring at a late stage in assembly is supported by structural data indicating that both residues U1369 and G1370 are potentially accessible to the enzymes even on the fully assembled mt-LSU (Greber et al., 2014; Supplemental Figure S7).
In conclusion, MRM2 and MRM3 are novel human genes that are indispensable for mitochondrial translation via their contributions to the modification of 16S rRNA and the biogenesis of mt-LSU. The function of MRM3 in mt-LSU is also supported by the recent study of Lee et al. (2013). Further studies are required to distinguish whether it is the presence of the MRM2 and MRM3 proteins in mitochondria, their catalytic activity necessary for modification of 16S mt-rRNA, or possibly both that are crucial for mitochondrial ribosome biogenesis. In addition, further analysis of these proteins and the identification of other factors involved in mitochondrial rRNA modifications would provide greater insights into the largely unknown area of mitochondrial ribosome biogenesis pathways.
MATERIALS AND METHODS
Plasmids
To construct plasmids encoding FLAG.STREP2-tagged MRM1, MRM2, and MRM3 proteins, cDNAs encoding each protein (MRM1: cDNA clone MGC:90266, IMAGE:6164397; MRM2: cDNA clone MGC:138620, IMAGE:40031660; MRM3: cDNA clone MGC:60073, IMAGE:5248052) were modified by PCR to introduce unique restriction sites, and the resulting fragments were cloned into pcDNA5-FST2 as described previously (Minczuk et al., 2011).
Maintenance and transfection of mammalian cell lines
Human HeLa cells (routinely used by us for immunofluorescence localization of mitochondrial proteins) were cultured in DMEM containing 2 mM l-glutamine (Invitrogen, Carlsbad, CA) and supplemented with 10% fetal calf serum (FCS; PAA Laboratories, Pasching, Austria). For immunofluorescence experiments, HeLa cells were transfected with pcDNA5-FST2 harboring MRM1, MRM2, or MRM3 using Lipofectamine 2000 (Invitrogen) according to the manufacturer′ s instructions.
The Flp-In T-Rex human embryonic kidney 293T (HEK293T) cell line (Invitrogen), which allows for the generation of stable doxycycline-inducible expression of transgenes by FLP recombinase-mediated integration, was used to express the MRM1-, MRM2-, or MRM3-FLAG.STREP2 proteins. HEK293T cells were grown in DMEM containing 2 mM l-glutamine (Invitrogen) and 10% tetracycline-free FCS (Autogen Bioclear, Calne, UK) and supplemented with 100 μg/ml Zeocin (Invitrogen) and 15 μg/ml blasticidin (Invivogen, San Diego, CA). Cells were transfected using Lipofectamine 2000 according to the manufacturer's instructions. Twenty-four hours after transfection, the selective antibiotics hygromycin (100 μg/ml, Invivogen) and blasticidin (15 μg/ml) were added, and this selective medium was replaced every 3–4 d.
Cell fractionation
Cell fractionation was performed as described by Minczuk et al. (2011), omitting the sucrose gradient step.
FLAG-tag affinity purification
Mitochondria were isolated from HEK293T cells expressing MRM2.FLAG, MRM3.FLAG, ICT1.FLAG, or MRPS27.FLAG essentially as described in Richter et al. (2010). Pelleted mitochondria were resuspended in lysis buffer (Sigma-Aldrich, St. Louis, MO), and IP was performed with a FLAG M2 affinity gel (Sigma-Aldrich) following manufacturer's recommendations. Elution was conducted with FLAG peptide.
Immunodetection of proteins
The localization of proteins by immunofluorescence in fixed HeLa cells was performed as described (Minczuk et al., 2010). The following antibodies were used: mouse anti-Flag immunoglobulin G (IgG; 1:200; Sigma-Aldrich) and AlexaFluor488–conjugated anti-mouse IgG (1:200; Life Technologies, Paisley, UK). Immunofluorescence images were captured using a Zeiss LSM 510 META confocal microscope.
For immunoblot analysis, equal amounts of protein corresponding to total cell lysates or gradient-separated protein fractions were subjected to SDS–PAGE, semidry transferred to nitrocellulose membranes, blocked in 5% (wt/vol) nonfat milk (Marvel) in phosphate-buffered saline (PBS) for 1 h, and incubated with specific primary antibodies in 5% (wt/vol) nonfat milk in PBS for 1 h or overnight. The blots were further incubated with horseradish peroxidase (HRP)–conjugated secondary antibodies in 5% nonfat milk in PBS for h and visualized using ECL+ (Amersham, Little Chalfont, UK).
The primary antibodies used were rabbit anti-MRM1 (1:1000; Sigma-Aldrich), mouse anti-FTSJ2 1:100 (MyBioSource, San Diego, CA), rabbit anti-RNTL1 (1:1000; Sigma-Aldrich), mouse anti-FLAG IgG (1:5000; Sigma-Aldrich), mouse anti-DAP3/MRPS29 (1:1000; Abcam, Cambridge, UK), goat anti-MRPL3 (1:1000; Abcam), mouse anti-MRPL12 (1:1000; Abcam), rabbit anti-MRPS18B (1:1000; ProteinTech, Manchester, UK), mouse anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH; (1:10,000; Abcam), rabbit anti–histone H4 (1:1000; Abcam), rabbit anti–SSB1 IgG (1:4000; kindly donated by D. Kang, Kyushu University), and mouse anti-TOM22 (1:1000, Abcam).
Secondary antibodies were anti-rabbit IgG-HRP (1:2000; Promega, Fitchburg, WI), anti-mouse IgG-HRP (1:2000; Promega), and anti-goat IgG-HRP (1:1000; Sigma-Aldrich).
siRNA gene silencing
Stealth siRNAs were obtained from Invitrogen and delivered to cells using Lipofectamine RNAiMAX. Stealth RNAi duplexes were generated using the following sequences:
MRM1 (siRNA1): CCAUUUGCAGCCAGAGGAAGGGUUU
MRM1 (siRNA2): GAUAAGGUCAUCACCAGCCGGAGAA
MRM1 (siRNA3): GCUAUGGAGGUGAUGGACGUGUUCU
MRM2/FTSJ2 (siRNA1): GACCCGACAUCUCAGGGACCCAUUU
MRM2/FTSJ2 (siRNA2): GGGCUGGAAGUCAAAGCCGUCGGUU
MRM2/FTSJ2 (siRNA3): GAGCAGAUGUGAUUCUGAGCGACAU
MRM3/RMTL1 (siRNA1): GACAAUAGUAAAGUCCAGGCCAUUU
MRM3/RMTL1 (siRNA2): GCAGGCUCAUUUCAGACGCUCUCAA
MRM3/RMTL1 (siRNA3): CCUUUAUGCCCAGGCUGAGAUGUCU
RNA isolation, Northern blotting, and reverse transcription primer extension
Total RNA from HeLa cells was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. For Northern blots, RNA was resolved on 1% (wt/vol) agarose gels containing 0.7 M formaldehyde in 1× 3-(N-morpholino)propanesulfonic acid buffer, transferred to a nylon membrane in 2× saline–sodium citrate, and hybridized with radioactively labeled PCR fragments corresponding to appropriate regions of mtDNA or nuclear 18S rDNA. For reverse transcription primer extension (RT-PEx), the specific 5′ 32P-labeled primer (0.1 pmol) was incubated with 0.5–1 μg of the total RNAs in a 10-μl solution containing 10 mM Tris-HCl (pH 8.0) and1 mM EDTA at 80°C for 2 min and cooled slowly in a PCR block. Subsequently, reverse transcription reactions were performed using an Omniscript RT Kit (Qiagen, Hilden, Germany), with the following components added to the annealed RNA primer: 10× reaction buffer, dNTP mix to achieve a final concentration of 0.04 or 1 mM, 4 U of Omniscript reverse transcriptase, and 10 U of RNasin and RNase-free water to a final volume of 20 μl. The reaction mixture was incubated at 37°C for 1 h, followed by addition of nucleic acid loading dye (2× solution of 95% formamide, 18 mM EDTA, and 0.025% SDS, xylene cyanol, and bromophenol blue). The reaction mixture was separated by 12.5% (vol/vol) PAGE containing 7 M urea. Dried gels were exposed to a storage phosphor screen (GE Healthcare, Little Chalfont, UK) and scanned using a Typhoon phosphor imaging system. The signals were quantified using ImageQuant software, and for each set of siRNA experiments the ratio between A-loop–specific and 5′-end 16S rRNA runoff RT-PEx products (U1369/RO or U1370/RO; Figure 4) was set as 100% for the control transfection (cells transfected with siRNA to green fluorescent protein). The same ratios for the MTase siRNA–transfected cells were then calculated as a percentage of the controls.
The following primers were used in RT-PEx:
Homo sapiens: pAL: GATCACGTAGGACTTTAATC (Figures 1 and 4)
p5LSU: CTGGTAGTAAGGTGGAGTG (Figure 4)
S. cerevisiae: CCCAACTCACGTAACATTTTAATTG (Figure 3)
C. elegans: CTAATATCACGTCAGATTAATTAATAG (Figure 3)
D. melanogaster: AACTCAGATCATGTAAGAATTTAAAA (Figure 3)
D. rerio: ATCACGTAGGACTATTAATC (Figure 3)
Measurement of mitochondrial respiration
HeLa cells transfected with siRNA against MRM proteins were seeded at 3 × 104 cells/well in 200 μl of growth medium in XF 24-well cell culture microplates (Seahorse Bioscience, North Billerica, MA) and incubated at 37°C in 5% CO2 for 36–40 h. One hour before the assay, growth medium was removed and replaced with assay medium (low-buffered DMEM, 10 mM l-glutamine, 1 mM sodium pyruvate, 2 mM glucose) with one wash of assay medium and left to stabilize in a 37°C non-CO2 incubator. Analysis was performed in quadruplicate using a XF24 Extracellular Flux Analyzer (Seahorse Bioscience). The wells were sequentially injected with 20 mM 2-deoxyglucose (2-DG) to inhibit glycolysis, 100 nM oligomycin to inhibit ATP-synthase, 500-1000 nM carbonylcyanide-4-trifluorometho-xyphenylhydrazone (FCCP) to uncouple the respiratory chain, and 200 nM rotenone to inhibit complex I. The OCR was measured for each well every 5 min before and after each injection. Test compounds 2-DG, oligomycin, FCCP, and rotenone were all obtained from Sigma-Aldrich.
[35S]methionine metabolic labeling of mitochondrial proteins
HeLa cells transfected with siRNA against MRM proteins were grown as described and, then the standard medium was replaced with methionine/cysteine free DMEM (Sigma-Aldrich) supplemented with 2 mM l-glutamine, 48 μg/ml cysteine, and 50 μg/ml uridine. The cells were incubated for 2 × 10 min in this medium and then transferred to methionine/cysteine-free DMEM containing 10% (vol/vol) dialyzed FCS and emetine dihydrochloride (100 μg/ml) to inhibit cytosolic translation. Cells were incubated for 10 min before addition of 120 μCi/ml of [35S]methionine. Labeling was performed for 15 min, and then the cells were washed twice with standard growth medium. Protein samples (30 μg) were separated on 4–12% SDS–PAGE gels, and products were visualized and quantified using a PhosphorImager system with ImageQuant software (Molecular Dynamics, GE Healthcare).
Analysis of the mitochondrial ribosome profile on density gradients
Total cell lysates (0.7 mg) were loaded on a linear sucrose gradient (2 ml 10–30% [wt/vol]) in 50 mM Tris–HCl (pH 7.2), 10 mM Mg(OAc)2, 80 mM NH4Cl, 0.1 M KCl, and 1 mM phenylmethylsulfonyl fluoride and centrifuged for 2 h, 15 min at 100,000 × gmax at 4°C (39,000 rpm; Beckman Coulter TLS-55 rotor). Twenty fractions (of 100 μl each) were collected, and 10-μl aliquots were analyzed by Western blotting.
Supplementary Material
Acknowledgments
This work was supported by the Medical Research Council, United Kingdom (J.R., P.B., P.A.G, T.J.J.N, S.F.P., D.P., and M.M.), and the Bavarian Ministry of Sciences, Research and the Arts in the framework of the Bavarian Molecular Biosystems Research Network (A.H. and F.P). We are grateful to Robert Lightowlers and Zosia Chrzanowska-Lightowlers for providing the ICT1.FLAG and MRPS27.FLAG HEK293T cell lines. We thank Alexey Amunts for his help in preparation of Supplemental Figure S7 and our colleagues from the Mitochondrial Biology Unit Mitochondrial Genetics group for help with the manuscript.
Abbreviations used:
- co-IP
coimmunoprecipitation
- FCS
fetal calf serum
- LSU
large ribosomal subunit
- MRM
mitochondrial rRNA methyltransferase
- mt
mitochondrial
- MTase
methyltransferase
- MTS
mitochondrial targeting sequence
- OXPHOS
oxidative phosphorylation
- pAL
A-loop–specific primer
- PTC
peptidyl transferase center
- RO
run-off product
- RT-PEx
reverse transcription primer extension
- snoRNA
small nucleolar RNA
- SSU
small ribosomal subunit
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-01-0014) on July 9, 2014.
REFERENCES
- Anantharaman V, Koonin EV, Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J Mol Microbiol Biotechnol. 2002;4:71–75. [PubMed] [Google Scholar]
- Baer R, Dubin DT. The 3′-terminal sequence of the small subunit ribosomal RNA from hamster mitochondria. Nucleic Acids Res. 1980;8:4927–4941. doi: 10.1093/nar/8.21.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer RJ, Dubin DT. Methylated regions of hamster mitochondrial ribosomal RNA: structural and functional correlates. Nucleic Acids Res. 1981;9:323–337. doi: 10.1093/nar/9.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger F, Stepinski J, Darzynkiewicz E, Pelletier J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J Biol Chem. 2010;285:33037–33044. doi: 10.1074/jbc.M110.155283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U. RNA methylation under heat shock control. Mol Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- Caldas T, Binet E, Bouloc P, Costa A, Desgres J, Richarme G. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J Biol Chem. 2000a;275:16414–16419. doi: 10.1074/jbc.M001854200. [DOI] [PubMed] [Google Scholar]
- Caldas T, Binet E, Bouloc P, Richarme G. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem Biophys Res Commun. 2000b;271:714–718. doi: 10.1006/bbrc.2000.2702. [DOI] [PubMed] [Google Scholar]
- Camara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D′ Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The Comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Rosa I, Durigon R, Pearce SF, Rorbach J, Hirst EM, Vidoni S, Reyes A, Brea-Calvo G, Minczuk M, Woellhaf MW, et al. MPV17L2 is required for ribosome assembly in mitochondria. Nucleic Acids Res. 2014;42:8500–8515. doi: 10.1093/nar/gku513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- Demirci H, Murphy Ft, Belardinelli R, Kelley AC, Ramakrishnan V, Gregory ST, Dahlberg AE, Jogl G. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA. 2010;16:2319–2324. doi: 10.1261/rna.2357210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennerlein S, Rozanska A, Wydro M, Chrzanowska-Lightowlers ZM, Lightowlers RN. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem J. 2010;430:551–558. doi: 10.1042/BJ20100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin DT. Methylated nucleotide content of mitochondrial ribosomal RNA from hamster cells. J Mol Biol. 1974;84:257–273. doi: 10.1016/0022-2836(74)90584-1. [DOI] [PubMed] [Google Scholar]
- Dubin DT, Taylor RH. Modification of mitochondrial ribosomal RNA from hamster cells: the presence of GmG and late-methylated UmGmU in the large subunit (17S) RNA. J Mol Biol. 1978;121:523–540. doi: 10.1016/0022-2836(78)90398-4. [DOI] [PubMed] [Google Scholar]
- Dubin DT, Taylor RH, Davenport LW. Methylation status of 13S ribosomal RNA from hamster mitochondria: the presence of a novel riboside, N4-methylcytidine. Nucleic Acids Res. 1978;5:4385–4397. doi: 10.1093/nar/5.11.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields DS, Gutell RR. An analysis of large rRNA sequences folded by a thermodynamic method. Fold Des. 1996;1:419–430. doi: 10.1016/S1359-0278(96)00058-2. [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Dessimoz C, Huxley-Jones J, Vilella AJ, Sonnhammer EL, Lewis S. Joining forces in the quest for orthologs. Genome Biol. 2009;10:403. doi: 10.1186/gb-2009-10-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leitner A, Bieri P, Voigts-Hoffmann F, Erzberger JP, Leibundgut M, Aebersold R, Ban N. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;505:515–519. doi: 10.1038/nature12890. [DOI] [PubMed] [Google Scholar]
- Hansen MA, Kirpekar F, Ritterbusch W, Vester B. Posttranscriptional modifications in the A-loop of 23S rRNAs from selected Archaea and Eubacteria. RNA. 2002;8:202–213. doi: 10.1017/s1355838202013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- He J, Cooper HM, Reyes A, Di Re M, Kazak L, Wood SR, Mao CC, Fearnley IM, Walker JE, Holt IJ. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012;40:6097–6108. doi: 10.1093/nar/gks257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DF, Green R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- Kotani T, Akabane S, Takeyasu K, Ueda T, Takeuchi N. Human G-proteins, ObgH1 and Mtg1, associate with the large mitochondrial ribosome subunit and are involved in translation and assembly of respiratory complexes. Nucleic Acids Res. 2013;41:3713–3722. doi: 10.1093/nar/gkt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B, Purushothaman SK. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol Cell. 2004;16:663–669. doi: 10.1016/j.molcel.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Lee KW, Okot-Kotber C, LaComb JF, Bogenhagen DF. Mitochondrial ribosomal RNA (rRNA) methyltransferase family members are positioned to modify nascent rRNA in foci near the mitochondrial DNA nucleoid. J Biol Chem. 2013;288:31386–31399. doi: 10.1074/jbc.M113.515692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovgren JM, Wikstrom PM. The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J Bacteriol. 2001;183:6957–6960. doi: 10.1128/JB.183.23.6957-6960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Metodiev MD, Spahr H, Loguercio Polosa P, Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG, Ruzzenente B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014;10:e1004110. doi: 10.1371/journal.pgen.1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M, He J, Duch AM, Ettema TJ, Chlebowski A, Dzionek K, Nijtmans LG, Huynen MA, Holt IJ. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39:4284–4299. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M, Kolasinska-Zwierz P, Murphy MP, Papworth MA. Construction and testing of engineered zinc-finger proteins for sequence-specific modification of mtDNA. Nat Protoc. 2010;5:342–356. doi: 10.1038/nprot.2009.245. [DOI] [PubMed] [Google Scholar]
- Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2:611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- O'Brien TW. Properties of human mitochondrial ribosomes. IUBMB Life. 2003;55:505–513. doi: 10.1080/15216540310001626610. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Bujnicki JM, Lapeyre B, Bonnerot C. MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J. 2002a;21:1139–1147. doi: 10.1093/emboj/21.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Lecointe F, Bujnicki JM, Bonnerot C, Grosjean H, Lapeyre B. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 2002b;21:1811–1820. doi: 10.1093/emboj/21.7.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldermans B, Bakker H, Van Knippenberg PH. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′ end of 16S ribosomal RNA of Escherichia coli. IV. The effect of the methylgroups on ribosomal subunit interaction. Nucleic Acids Res. 1980;8:143–151. doi: 10.1093/nar/8.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U, Lahtinen T, Marttinen P, Myohanen M, Greco D, Cannino G, Jacobs HT, Lietzen N, Nyman TA, Battersby BJ. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr Biol. 2013;23:535–541. doi: 10.1016/j.cub.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J. 2010;29:1116–1125. doi: 10.1038/emboj.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach J, Gammage PA, Minczuk M. C7orf30 is necessary for biogenesis of the large subunit of the mitochondrial ribosome. Nucleic Acids Res. 2012;40:4097–4109. doi: 10.1093/nar/gkr1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach J, Minczuk M. The post-transcriptional life of mammalian mitochondrial RNA. Biochem J. 2012;444:357–373. doi: 10.1042/BJ20112208. [DOI] [PubMed] [Google Scholar]
- Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M, Kuhl I, Gaisne M, Bonnefoy N, Smeitink JA, et al. The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res. 2008;36:5787–5799. doi: 10.1093/nar/gkn576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- Sirum-Connolly K, Mason TL. Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science. 1993;262:1886–1889. doi: 10.1126/science.8266080. [DOI] [PubMed] [Google Scholar]
- Sirum-Connolly K, Peltier JM, Crain PF, McCloskey JA, Mason TL. Implications of a functional large ribosomal RNA with only three modified nucleotides. Biochimie. 1995;77:30–39. doi: 10.1016/0300-9084(96)88101-6. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–7357. doi: 10.1093/nar/gku390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperley R, Richter R, Dennerlein S, Lightowlers RN, Chrzanowska-Lightowlers ZM. Hungry codons promote frameshifting in human mitochondrial ribosomes. Science. 2010;327:301. doi: 10.1126/science.1180674. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Werner M, Purta E, Kaminska KH, Cymerman IA, Campbell DA, Mittra B, Zamudio JR, Sturm NR, Jaworski J, Bujnicki JM. 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res. 2011;39:4756–4768. doi: 10.1093/nar/gkr038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widerak M, Kern R, Malki A, Richarme G. U2552 methylation at the ribosomal A-site is a negative modulator of translational accuracy. Gene. 2005;347:109–114. doi: 10.1016/j.gene.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Wredenberg A, Lagouge M, Bratic A, Metodiev MD, Spahr H, Mourier A, Freyer C, Ruzzenente B, Tain L, Gronke S, et al. MTERF3 regulates mitochondrial ribosome biogenesis in invertebrates and mammals. PLoS Genet. 2013;9:e1003178. doi: 10.1371/journal.pgen.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Baer F, Lane WS, Gaynor RB. The cellular factor TRP-185 regulates RNA polymerase II binding to HIV-1 TAR RNA. EMBO J. 1995;14:5995–6009. doi: 10.1002/j.1460-2075.1995.tb00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, O′ Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat Struct Mol Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.