Isoform-specific expression of p120 affects cell motility and migration during development and tumor progression. The DIPA coiled-coil protein is a novel binding partner to the conserved isoform 1–specific head domain of p120 family members. Zebrafish data suggest that DIPA is mechanistically linked to p120 isoform–specific function in development.
Abstract
p120-catenin (p120) modulates adherens junction (AJ) dynamics by controlling the stability of classical cadherins. Among all p120 isoforms, p120-3A and p120-1A are the most prevalent. Both stabilize cadherins, but p120-3A is preferred in epithelia, whereas p120-1A takes precedence in neurons, fibroblasts, and macrophages. During epithelial-to-mesenchymal transition, E- to N-cadherin switching coincides with p120-3A to -1A alternative splicing. These isoforms differ by a 101–amino acid “head domain” comprising the p120-1A N-terminus. Although its exact role is unknown, the head domain likely mediates developmental and cancer-associated events linked to p120-1A expression (e.g., motility, invasion, metastasis). Here we identified delta-interacting protein A (DIPA) as the first head domain–specific binding partner and candidate mediator of isoform 1A activity. DIPA colocalizes with AJs in a p120-1A- but not 3A-dependent manner. Moreover, all DIPA family members (Ccdc85a, Ccdc85b/DIPA, and Ccdc85c) interact reciprocally with p120 family members (p120, δ-catenin, p0071, and ARVCF), suggesting significant functional overlap. During zebrafish neural tube development, both knockdown and overexpression of DIPA phenocopy N-cadherin mutations, an effect bearing functional ties to a reported mouse hydrocephalus phenotype associated with Ccdc85c. These studies identify a novel, highly conserved interaction between two protein families that may participate either individually or collectively in N-cadherin–mediated development.
INTRODUCTION
p120-catenin (p120) is an Armadillo (Arm) repeat protein that regulates cell–cell adhesion through direct interaction with the cytoplasmic domain of classical cadherins (Reynolds, 2007). p120 binds the juxtamembrane domain and suppresses cadherin endocytosis by blocking interaction with factors that target the complex for internalization, degradation, or recycling (Yap et al., 1998; Thoreson et al., 2000; Davis et al., 2003; Xiao et al., 2005; Sato et al., 2011; Lohia et al., 2012; Nanes et al., 2012). Thus p120’s main role in the complex is to stabilize cadherin expression at the cell surface (Ireton et al., 2002; Davis et al., 2003; Xiao et al., 2003). β-Catenin binds an adjacent motif on the cadherin tail and, through α-catenin, functionally connects the complex to the actin cytoskeleton (Drees et al., 2005; Desai et al., 2013). p120 may cooperate on this level by recruiting RhoGTPase machinery to control local actin dynamics (Anastasiadis et al., 2000; Noren et al., 2000; Wildenberg et al., 2006; Yanagisawa et al., 2008; Dohn et al., 2009; Molina-Ortiz et al., 2009; Smith et al., 2012; Zebda et al., 2013) and/or by association with microtubules (Yanagisawa et al., 2004; Ichii and Takeichi, 2007; Meng et al., 2008). Although not well understood, p120 has been shown to interact with transcription factors (e.g., Kaiso, Glis2) and presumably has important roles in the nucleus (Daniel and Reynolds, 1999; Prokhortchouk et al., 2001; Kim et al., 2004; Park et al., 2005; Hosking et al., 2007). A particularly interesting facet of p120 function is the controlled expression of distinct p120 isoforms during epithelial-to-mesenchymal transition (EMT), a phenomenon consistent with important roles for these isoforms in development and cancer.
CTNND1 encodes four distinct amino-terminal ATG start sites, giving rise to isoforms 1–4, as well as four alternatively spliced exons (A–D; Aho et al., 1999). Despite a theoretical potential for many alternative splicing combinations, isoforms 1 and 3 (generally in combination with exon A) are by far the most common (i.e., p120 isoforms 1A and 3A; Mo and Reynolds, 1996; Keirsebilck et al., 1998; Aho et al., 1999). Relative to isoform 3, full-length p120-1 features a 101–amino acid N-terminal extension termed the “head domain” containing a highly conserved coiled-coil motif (Figure 1A). The remaining regions of the protein are identical to p120-3. They consist of a central Arm repeat domain, which is required for cadherin binding, and flanking regulatory domains whose roles are not fully understood. Thus all isoforms contain the Arm domain and bind cadherins, but only isoform 1 contains the head domain. p120 is the prototypical member of small family of similarly structured proteins that includes p120, δ-catenin, p0071, and Armadillo repeat gene deleted in velocardiofacial syndrome (ARVCF). The head and Arm domains are highly conserved, whereas other regions are not (Anastasiadis and Reynolds, 2000). Although all members can bind and stabilize classical cadherins (Kaufmann et al., 2000; Hatzfeld et al., 2003; Carnahan et al., 2010; Yang et al., 2010), recent evidence suggests that functional redundancy may be more limited than originally assumed (Kurley et al., 2012).
FIGURE 1:
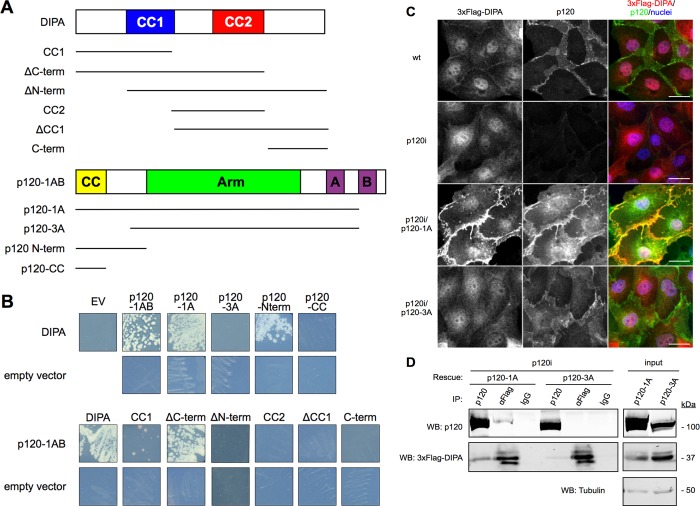
DIPA interacts specifically with p120 isoform 1. (A) Schematic of full-length DIPA, p120-1AB, and respective truncation mutants. Isoforms are named p120-1 through p120-4, depending on N-terminal start site. Designations A–D are included if the exon is present (e.g., p120-1A, p120-1BC). No letter is used if referring to multiple isoforms for which the alternatively spliced exons are either unknown or irrelevant (e.g., p120-1 isoforms). A and B, alternatively spliced exons A and B; Arm, Armadillo repeat domain; CC, p120 coiled-coil; CC1, DIPA coiled-coil 1; CC2, DIPA coiled-coil 2. (B) Growth of yeast indicates a positive interaction between human DIPA and p120. DIPA and p120 constructs were fused to the C-terminal end of the GAL4 activation domain and GAL4 DNA-binding domain, respectively. (C) Immunofluorescence of 3xFlag-DIPA (M2 anti-Flag mAb) and either endogenous p120 or exogenous human p120-1A or p120-3A (F1αSH anti-p120 pAb) in MDCK cells. wt, wild-type; p120i, p120 shRNA knockdown; p120i/p120-1A, p120 shRNA knockdown with human p120-1A rescue; p120i/p120-3A, p120 shRNA knockdown with human p120-3A rescue. Scale bars, 25 μm. (D) Western blot of immunoprecipitated lysates from p120i/p120-1A or p120i/p120-3A MDCK cells with exogenous 3xFlag-DIPA. Antibodies used for immunoprecipitation are indicated at the top of each lane. p120, pp120 mAb; α-Flag, M2 anti-Flag mAb; immunoglobulin G (IgG), KT3 isotype control mAb. Tubulin is used as a loading control for whole-cell lysate fractions (10% input). Images are representative of at least three independent experiments.
p120 typically undergoes isoform switching during EMT. Epithelial cells tend to preferentially express p120-3A at the expense of p120-1A, whereas the opposite is true for mesenchymal cell types and neurons (Mo and Reynolds, 1996; Saito et al., 2012). During EMT, the E- to N-cadherin switch is coordinated with an isoform 3 to isoform 1 switch in p120 alternative splicing. Thus p120-1A expression is linked to N-cadherin expression, suggesting that the head domain confers unique characteristics along the lines of mesenchymal cell behavior. Indeed, relative to isoform 3A, forced expression of p120-1A in epithelial cells significantly enhances cell motility and invasive cell behavior (Aono et al., 1999; Yanagisawa et al., 2008; Slorach et al., 2011). Of interest, ectopic overexpression of p120-3 in the mammary cell line EpH4.9 after Zeppo-1–induced EMT reverses the EMT-associated invasive phenotype, raising the possibility that isoforms 1 and 3 directly mediate these changes in cell behavior (Slorach et al., 2011). Human biopsy data show that p120-1 expression positively correlates with metastasis and independently predicts poor prognosis (Yanagisawa et al., 2008; Miao et al., 2009; Talvinen et al., 2010). These and other lines of evidence suggest a prominent role for the head domain in a range of aggressive cellular activities associated with expression of p120-1A.
Here we identify and characterize a novel p120 binding partner that interacts selectively with p120 isoform 1 via its head domain. The products of a p120 yeast two-hybrid (Y2H) screen were examined for differential binding to p120 isoforms 1 and 3, leading to identification of delta-interacting protein A (DIPA; encoded by CCDC85B), a 202–amino acid protein with two internal coiled-coil domains and no other recognizable domains (Figure 1A). DIPA was named after a putative interaction with the hepatitis delta antigen, the biological significance of which has since been discredited (Brazas and Ganem, 1996; J. Taylor, personal communication). It is suggested to be a nuclear transcriptional repressor with various roles in development and cancer-related pathways (Bezy et al., 2005; Du et al., 2006; Iwai et al., 2007). For example, in the context of p53 activation, DIPA down-regulates canonical Wnt signaling by competing with β-catenin for interaction with TCF4 (Iwai et al., 2007). During adipocyte differentiation, DIPA indirectly regulates PPARγ through inhibition of C/EBP-β and -γ transcriptional activity (Bezy et al., 2005). DIPA has also been shown to interact with p78/MCRS1/MSP58 at centrosomes and to repress SRF and AP-1 signaling (Du et al., 2006).
DIPA belongs to a family comprised of Ccdc85a, Ccdc85b (DIPA), and Ccdc85c. Each contains a pair of conserved coiled-coil motifs that are believed to participate in DNA transcriptional regulation. The C-terminal sequences following the coiled-coil domains are not conserved and do not contain known functional domains. Of interest, a mouse Ccdc85c mutation was identified recently as the causal factor for the hemorrhagic hydrocephalus (hhy) mutant mouse, which exhibits a genetic defect in brain development characterized by subcortical heterotopia and hemorrhagic hydrocephalus (Mori et al., 2012). Although the underlying mechanism(s) are not well understood, the process is triggered by severe truncation of the mouse CCDC85C protein and appears to involve developmental defects in neuronal migration and/or cell–cell adhesion (Ferland et al., 2009). Another mouse model of heterotopia, called hydrocephaly with hop gait (hyh), contains a mutation in the gene encoding soluble N-ethylmaleimide–sensitive factor attachment protein α (α-SNAP), an apical vesicle-trafficking protein that is essential for p120 stability. Remarkably, α-SNAP knockdown in cultured cells results in near-complete loss of p120 (Chae et al., 2004; Naydenov et al., 2012). Thus p120 and DIPA are each associated indirectly with proteins causally implicated in heterotopia and hydrocephalus development.
We now show that all DIPA family members interact reciprocally with the conserved isoform 1–specific head domains of each p120 family member, suggesting a range of at least partly redundant interactions between the various members of each family. In addition to the nuclear localization reported previously, we find that DIPA precisely colocalizes with cadherin complexes at adherens junctions (AJs) in a manner dependent on p120 isoform 1 (or its family-member counterparts). Finally, mRNA- and morpholino-based manipulation of DIPA in zebrafish results in an open neural tube phenotype similar to that for N-cadherin loss of function, suggesting roles for the DIPA and p120 family members in development, motility, and/or adhesion.
RESULTS
DIPA binds to p120-1A but not p120-3A
To identify roles unique to p120-1, we rescreened the products of a Y2H screen against p120-1AB for head domain–specific p120 binding partners. The original prey library comprised cDNA from four breast cancer epithelial cell lines: T47D, MDA-MB468, MCF7, and BT20. Among the 23 putative binding partners tested, DIPA was found to interact with p120-1 but not p120-3 (Figure 1B). Reciprocal interaction sites were then fine mapped by Y2H assay using p120 and DIPA truncation mutants (Figure 1A). The p120 head domain (p120-Nterm) was indeed sufficient for binding DIPA, indicating specificity for p120-1. The amino-terminal coiled-coil segment (p120-CC) by itself, however, was not (Figure 1B). For DIPA, both of the coiled-coil motifs were required for full-strength binding to p120-1AB, but the CC1 DIPA mutant was sufficient for a weaker interaction. Of note, an N-terminal truncation mutant of DIPA (ΔN-term) lacking the first 42 amino acids did not bind to p120-1A (Figure 1B). Empty vector controls for autoactivation were negative. Thus DIPA appears to interact selectively with the p120-1 head domain.
The interaction was further examined in Madin–Darby canine kidney (MDCK) cells, including p120-knockdown cell lines reconstituted with either p120-1 or p120-3. MDCK cells exhibit well-defined cell–cell junctions that are ideal for high-resolution colocalization studies. Figure 1C shows the localization of epitope-tagged DIPA (3xFlag) expressed from a lentiviral vector in wild-type (WT), p120 knockdown (p120i), and p120i cells reconstituted with p120-1A or p120-3A. Although diffusely distributed in WT cells, 3xFlag-DIPA was mostly nuclear, with fine staining at intercellular junctions (Figure 1C, top row). On knockdown of p120, most of the junctional staining of 3xFlag-DIPA disappeared (Figure 1C, second row). In contrast, forced expression of p120-1A induced striking recruitment of 3xFlag-DIPA and precise colocalization of the two proteins at AJs (Figure 1C, third row). Of importance, 3xFlag-DIPA was virtually absent from junctions of p120-3A reconstituted cells (Figure 1C, bottom row).
The p120-1A- and p120-3A-reconstituted cell lines were then used for coimmunoprecipitation studies (Figure 1D). Note that 3xFlag-DIPA coimmunoprecipitated with p120-1A and vice versa but not with p120-3A. Together the data suggest that DIPA is selectively recruited to AJs by p120-1A.
Recruitment of endogenous DIPA to AJs is p120 dependent
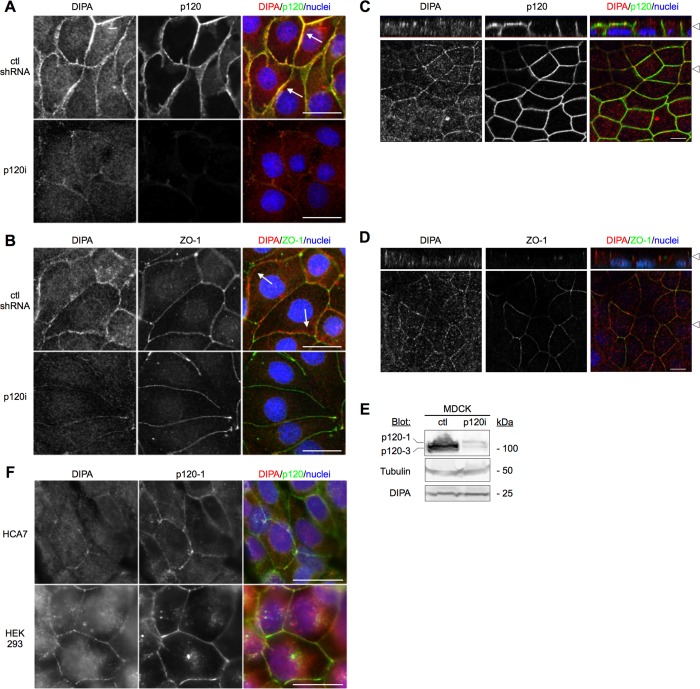
Figure 2 illustrates selective, p120-dependent recruitment of endogenous DIPA to AJs. Although p120 knockdown almost entirely eliminated AJs (Figure 2A; Davis et al., 2003; Dohn et al., 2009), tight junctions were unaffected, as illustrated by zonula occludens-1 (ZO-1) staining (Figure 2B). However, recruitment of endogenous DIPA to junctions was abrogated by loss of p120 but not by control shRNA expression. Confocal imaging shows that endogenous DIPA and p120 colocalize precisely, whereas ZO-1 and DIPA staining are discordant in areas where AJs and tight junctions separate from one another (Figure 2,C and D). Although no longer visible at junctions after p120 knockdown, overall DIPA levels were unchanged, as illustrated by Western blotting (Figure 2E). Endogenous DIPA and p120 colocalization can also be visualized in other epithelial cell lines, as illustrated in HCA7 (human colon carcinoma) and HEK293 (human kidney epithelial) cells (Figure 2F).
FIGURE 2:
Endogenous DIPA colocalizes with p120, and its junctional localization requires p120 expression. (A) Immunofluorescence of endogenous DIPA (3E3 anti-DIPA mAb) and p120 (F1αSH anti-p120 pAb) in MDCK cells with either control shRNA (ctl shRNA) or p120 knockdown (p120i). (B) Immunofluorescence of endogenous DIPA (3E3 anti-DIPA mAb) and ZO-1 (anti-ZO-1 pAb) using the same cells as in A. White arrows in merged images show yellow colocalization of DIPA and p120 in A but distinctly separate localization of DIPA and ZO-1 in B. (C) The same cells shown in A viewed with confocal microscopy to show colocalization of DIPA with p120 in z-stack images (top) and a single x-y plane (bottom). White arrowheads indicate the plane in which the corresponding images were taken. (D) The same cells shown in B imaged with confocal microscopy as in C. (E) Western blot for DIPA (3E3 anti-DIPA mAb) from whole-cell lysates of MDCK with control shRNA (ctl) or p120 knockdown (p120i). Tubulin (DM1α anti–α-tubulin mAb) is shown as a loading control. (F) Immunofluorescence detection of endogenous DIPA (3E3 anti-DIPA mAb) and p120-1 (6H11 isoform-specific anti-p120 mAb) in HCA7 (top) and HEK293 (bottom) human cell lines. Scale bars, 25 μm. Images are representative of at least three independent experiments.
p120-1–dependent recruitment of DIPA to different classical cadherins
MDCK cells express at least four different catenin-binding cadherins: E, N, P, and K (Wu et al., 1993; Stewart, 2000; Chen et al., 2011), including significant levels of E- and N-cadherin, the most prominent cadherins in epithelial and mesenchymal cells, respectively. Because p120-1 is often elevated in N-cadherin–expressing cells (at the expense of p120-3), we considered that DIPA binding to p120-1 might affect, or be affected by, interaction with different classical cadherins. To that end, we knocked down E- and N-cadherin, separately and together, and monitored effects on DIPA staining. The abundance and subcellular distribution of DIPA, however, were not altered by knockdown of E-cadherin, N-cadherin, or both E- and N-cadherin (Supplemental Figure S1). Thus it appears that DIPA localization does not discriminate between the different cadherins, so long as they bind p120-1. Indeed, it has been known for some time that the different p120 isoforms bind equally to all classical cadherins. Note that E-cadherin knockdown is associated with elevated N-cadherin levels and vice versa, a well-documented phenomenon called “p120 sharing.” The pool of p120 left stranded by removal of one cadherin is redistributed to stabilize higher levels of other classical cadherins (e.g., P- and K-cadherin; Reynolds and Carnahan, 2004). On the other hand, p120 knockdown destabilizes all classical cadherins, in which case DIPA is lost from intercellular junctions.
DIPA knockdown does not noticeably alter AJ structure or function
Although p120-1 and p120-3 have been extensively associated with mesenchymal and epithelial cell activities, respectively, the basis for these differences at the molecular level is largely unknown. The selective binding of DIPA to p120-1, on the other hand, is likely to have functional consequences. However, stable knockdown or overexpression of DIPA did not noticeably alter AJ structure or cell morphology (Supplemental Figure S2). DIPA overexpression and knockdown efficiency are shown in Supplemental Figure S2A along with p120 isoforms 1 and 3, β-catenin, E-cadherin, and N-cadherin. Moreover, perturbing DIPA levels had no effect on transepithelial electrical resistance (TER), a sensitive measure of junctional integrity (Supplemental Figure S2C).
DIPA family members bind to p120 family members
Like p120, family members δ-catenin, p0071, and ARVCF are endowed with highly conserved N-terminal head domains (Carnahan et al., 2010). To determine whether DIPA interacts with other members of the p120 family, we performed direct Y2H assays with full-length DIPA. Figure 3A shows that all p120 family members interact with DIPA by this method. To test for interaction in MDCK cells, C-terminal GFP fusion constructs of each family member were expressed in a stable p120-knockdown MDCK cell line. p120-1A-GFP, δ-catenin-GFP, p0071-GFP, and ARVCF-GFP all colocalized with and recruited endogenous DIPA to the cell membrane (Figure 3B). Neither p120-3A-GFP nor GFP alone could engage DIPA (Figure 3B). Thus the interaction with DIPA is conserved across all immediate p120 family members.
FIGURE 3:
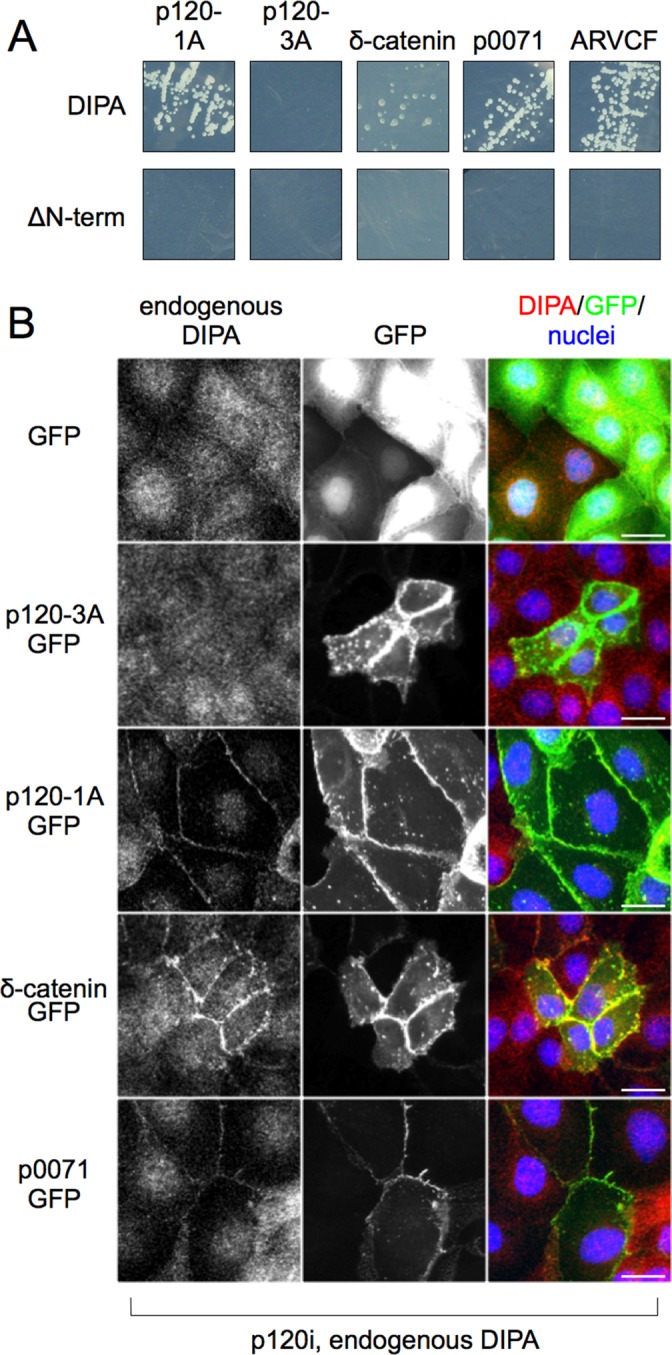
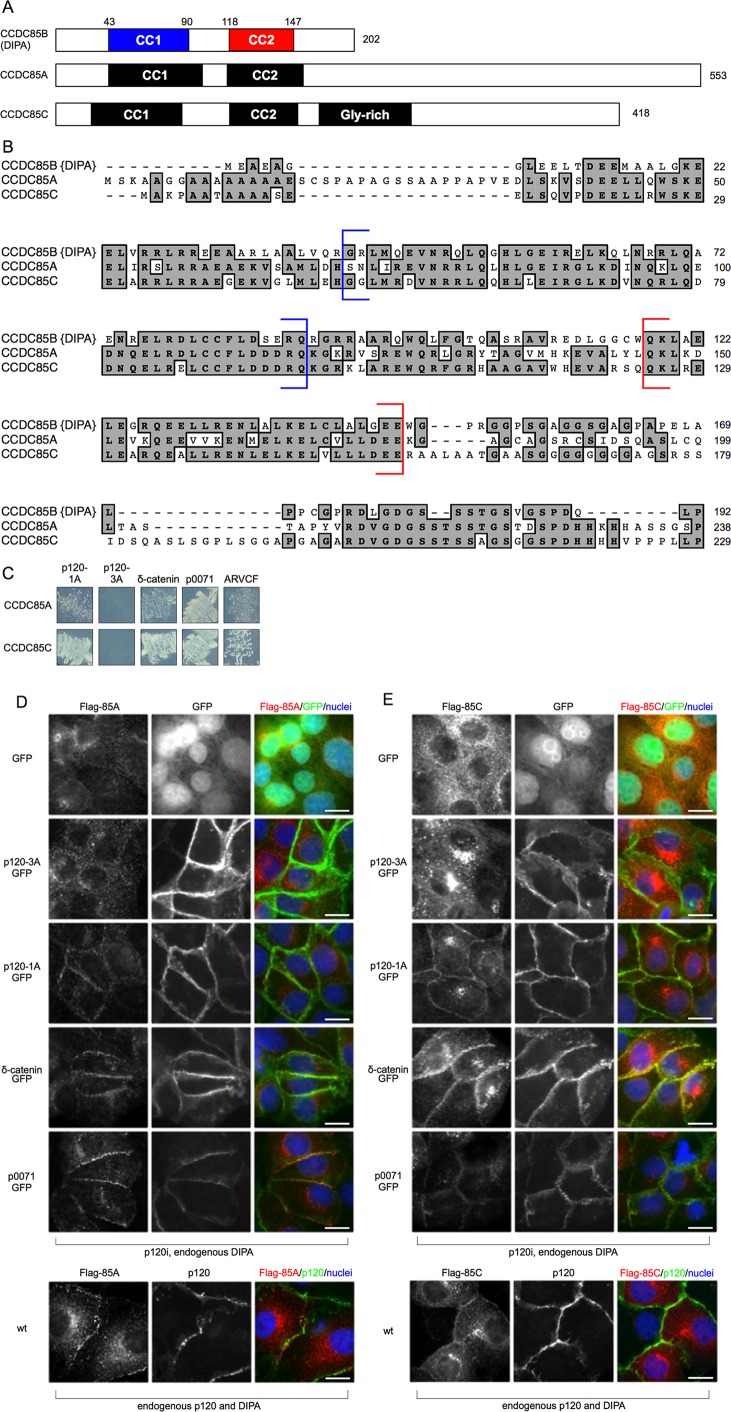
DIPA interacts with p120 family members. (A) Direct Y2H assays with full-length DIPA and Flag-ΔN-term-DIPA vs. p120-1A, p120-3A, δ-catenin, p0071, and ARVCF. Flag-ΔN-term-DIPA does not contain the first 42 amino acids of full-length DIPA. Both DIPA constructs were fused to the C-terminus of the GAL4 activation domain, and all p120 and family member constructs were fused to the C-terminus of the GAL4 DNA-binding domain. Y2H experiments using δ-catenin required the addition of 12.5 mM 3-amino-1,2,4-triazole (3-AT) to the medium to control for autoactivation. (B) Immunofluorescence detection of p120-knockdown MDCK cells with endogenous DIPA (3E3 anti-DIPA mAb) and one of five exogenous GFP fusion proteins: GFP alone, p120-3A, p120-1A, δ-catenin, or p0071. Scale bars, 25 μm. Images are representative of at least three independent experiments.
DIPA family members Ccdc85a and Ccdc85c share high homology in their N-terminal sequences, each of which contains two coiled-coil motifs (Figure 4, A and B). Thus we performed Y2H assays with full-length Ccdc85a or Ccdc85c versus p120-1A, p120-3A, δ-catenin, p0071, or ARVCF. Both DIPA family members exhibited specific interactions with p120-1A and not p120-3A (Figure 4C). They also interacted with all three p120 family members (Figure 4C). To test for interaction in cells, we stably introduced Flag-tagged Ccdc85a or Ccdc85c into p120-knockdown MDCK cells. p120-1A, p120-3A, δ-catenin, or p0071 tagged with GFP were then introduced and analyzed as before for colocalization (Figure 4, D and E). Exogenous Ccdc85a and Ccdc85c were expressed at similar levels and were effectively recruited to junctions by p120-1A-GPF, δ-catenin-GFP, and p0071-GFP. GFP alone or GFP-p120-3A, on the other hand, had no effect. In wild-type cells, both overexpressed Ccdc85a and Ccdc85c localized to AJs (Figure 4, D and E, bottom). ARVCF-GFP vectors were generated several times and directly validated by sequencing but could not be expressed. Considering the positive Y2H data and the similarity between the ARVCF N-terminal region and the other p120 family members, however, it is likely that DIPA, Ccdc85a, and Ccdc85c interact with ARVCF as well.
FIGURE 4:
(A–C) DIPA family members bind to p120-1A and its family members. (A) Schematic of DIPA, Ccdc85a, and Ccdc85c proteins with structured domains and relative sizes. CC1, coiled-coil 1. CC2, coiled-coil 2; Gly-rich, glycine-rich domain. Numbers indicate amino acids. (B) ClustalW alignment of approximately the first 250 amino acids of the human DIPA family members shows at least 54% similarity and 73% identity in the coiled-coil regions. Shaded boxed areas indicate identical amino acids for at least two of the proteins. Blue and red brackets enclose the DIPA CC1 and CC2, respectively. National Center for Biotechnology Information accession numbers: Ccdc85b (DIPA), AAB05928.1; Ccdc85a, AAI31558.1; Ccdc85c, NP_001138467.1. (C) Direct yeast two-hybrid assays with full-length Ccdc85a and Ccdc85c vs. p120-1A, p120-3A, δ-catenin, p0071, and ARVCF. Both Ccdc85a and Ccdc85c were fused to the C-terminus of the GAL4 activation domain, and all p120 and family member constructs were fused to the C-terminus of the GAL4 DNA-binding domain. Y2H experiments using δ-catenin required the addition of 12.5 mM 3-AT to the medium to control for autoactivation. (D, E) Immunofluorescence detection of p120-knockdown MDCK cells with Flag-tagged Ccdc85a (D) or Flag-tagged Ccdc85c (E; M2 anti-Flag mAb) and one of five exogenous GFP fusion proteins: GFP alone, p120-3A, p120-1A, δ-catenin, or p0071. Bottom, Flag-tagged Ccdc85a (D) and Flag-tagged Ccdc85c (E; M2 anti-Flag mAb) immunofluorescence with endogenous p120 (F1αSH anti-p120 pAb) in MDCK cells. White arrows indicate junctional colocalization. Scale bars, 25 μm. Images are representative of at least three independent experiments.
Overexpression or morpholino knockdown of DIPA in zebrafish causes a neural tube closure defect and neuroepithelial disorganization
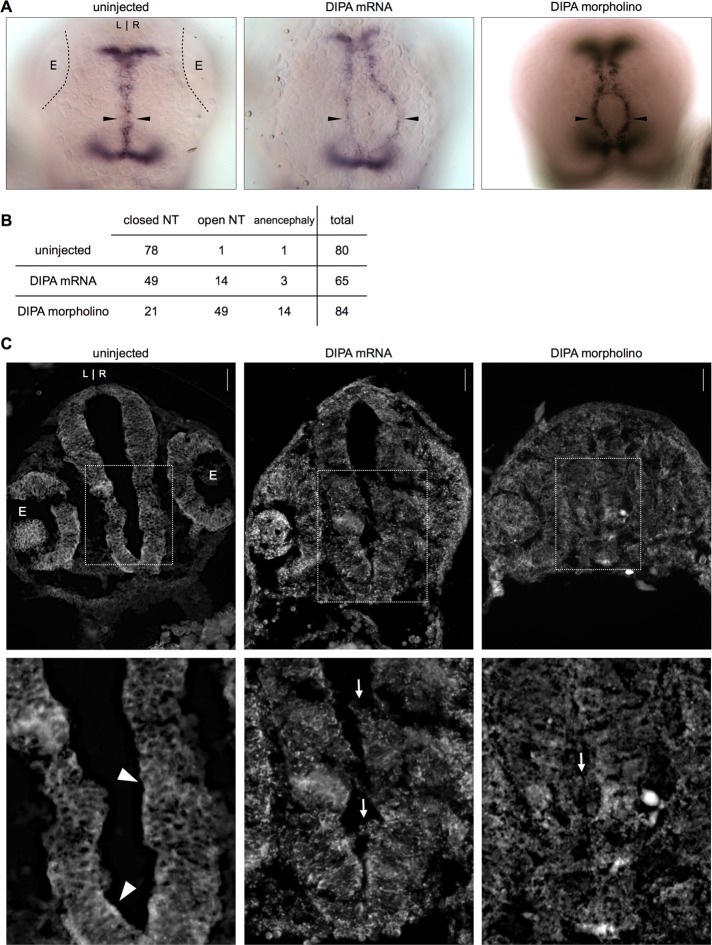
To examine DIPA function in vivo, we manipulated its expression genetically in a zebrafish model. Published observations reveal that DIPA family member Ccdc85c and the p120-trafficking protein α-SNAP both regulate neuronal development (Chae et al., 2004; Naydenov et al., 2012), and so we considered a model where mutant N-cadherin phenotypes are well established. The trafficking and stabilization of N-cadherin have been shown to be critical for early neuron integrity and migration, and their failure can lead to hydrocephalus and heterotopia (Shikanai et al., 2011; Sival et al., 2011; Kawauchi, 2012). Closing of the zebrafish dorsal neural tube similarly requires N-cadherin for cell–cell adhesion and migration and provides a relatively simple in vivo basis for comparison of the effects of DIPA manipulation to those associated with N-cadherin (Lele et al., 2002; Hong and Brewster, 2006; Chalasani and Brewster, 2011). Zebrafish embryos were injected at the one- to two-cell stage with either human DIPA mRNA (0.5 ng) or DIPA morpholino (4 ng). Uninjected sibling embryos were used as control. We allowed the embryos to develop for 48 h postfertilization (hpf) and fixed them at the long-pec stage for in situ hybridization with a zebrafish Wnt1 probe. Figure 5A shows the Wnt1 mRNA staining, which outlines the fused dorsal neural tube in the diencephalon, midbrain, and hindbrain. Nearly one-fourth (n = 14 of 65, 21.5%) of the embryos injected with DIPA mRNA exhibited an open neural tube phenotype, with Wnt1 expression remaining as separate bilateral stripes. More than half of the embryos injected with DIPA morpholino (n = 49 of 84, 58.3%) exhibited a similar phenotype (Figure 5, A and B). Although we cannot exclude off-target effects, the similarity of the neural tube defect upon DIPA morpholino and mRNA expression is quite striking and suggests that the same target (DIPA) is affected. The control and each treatment produced a small number of anencephalic embryos in which the Wnt1 staining does not delineate the midbrain. The DIPA mRNA- and morpholino-injected zebrafish phenotype is similar to that seen in the N-cadherin/parachute mutant zebrafish (Lele et al., 2002).
FIGURE 5:
DIPA overexpression or morpholino knockdown phenocopies an N-cadherin mutant neural tube defect in zebrafish embryos. (A) The dorsal view of AB zebrafish heads at 48 hpf shows neural tube staining with Wnt1 in situ hybridization of uninjected embryos and embryos injected with either DIPA mRNA or DIPA morpholino at the one- to two-cell stage. Embryos are oriented with anterior toward the top. Black arrowheads show closed or open neural tubes. E, eyes; L, left; R, right. Images are representative of at least three independent experiments. (B) Table showing exact numbers of embryos counted for each variable. Chi-squared tests yield p < 0.01 when comparing either mRNA or morpholino to control. (C) N-cadherin–stained cross sections of the AB zebrafish neural tube at 24–30 hpf after no injection, DIPA mRNA injection, or DIPA morpholino injection. White boxes in top row represent the borders of the zoomed images in the bottom row. E, eyes; L, left; R, right. Arrowheads, organized neuroepithelium. Arrows, disorganized cell aggregates. Scale bars, 50 μm.
To compare the observed neural tube phenotype to changes in N-cadherin at the cellular level, we repeated the same injection procedure but fixed embryos at 24–30 hpf. At this developmental stage, the individual cells are larger and junctions are more easily identified (Lo Sardo et al., 2012). Embryos were frozen, sectioned, and stained for N-cadherin (Figure 5C). We did not detect any overall difference in N-cadherin abundance, but its cellular distribution and the tissue architecture were markedly altered in embryos injected with either mRNA or morpholino. In 23 of 24 uninjected embryos, N-cadherin staining demonstrated clear cell junctions, and the ventricle neuroepithelium formed a sharp, distinct barrier (Figure 5C, bottom, arrowheads). In contrast, the cell junctions of injected embryos were not as well defined, and the neural tube structure was compromised at the level of the diencephalon, such that cells were even aggregating within the ventricle (Figure 5C, bottom, arrows). We observed this cell sloughing at the diencephalon or midbrain in seven of 12 mRNA-injected and 12 of 12 morpholino-injected embryos. This same phenotype was observed previously in N-cadherin/parachute mutants (Lele et al., 2002). Other zebrafish with genetic alterations affecting N-cadherin processing and stability also have abnormal neuroepithelial cell sloughing (Doll et al., 2011; Lo Sardo et al., 2012).
DISCUSSION
We demonstrate here that DIPA family members interact selectively with the conserved head domain of all four immediate p120 family members. Moreover, each of the full-length p120 homologues can selectively recruit DIPA, and DIPA homologues, to AJ-associated cadherin complexes. In addition, manipulating DIPA expression during zebrafish development results in a neural tube defect similar to that caused by N-cadherin mutation in zebrafish. Together these data reveal a novel, highly conserved interaction between two protein families and suggest potential relationships in N-cadherin-associated disease.
Our data show for the first time that DIPA binds and colocalizes precisely with p120 isoform 1 at AJs. Although similarly localized in other cell lines, the findings are especially evident in MDCK cells owing to sharply defined AJs that can be readily distinguished from other junction types (e.g., tight junctions, desmosomes). Of interest, the low level of p120-1 in these cells may be a limiting factor in terms of DIPA recruitment to AJs, as forced expression of DIPA does not increase the AJ signal but instead leads to additional accumulation in the nucleus (compare Figures 1C and 2A). In contrast, cooverexpression of DIPA and p120-1 (but not p120-3) reveals a striking increase in isoform 1–dependent recruitment to AJs (Figure 1C; compare bottom two rows). Presumably, overexpressed isoform 1 competes with endogenous isoform 3 for limited catenin-binding sites, and the increased DIPA recruitment to AJs is permitted in part by the increased AJ-bound p120 isoform 1.
p120 association, however, does not affect overall DIPA protein levels and/or stability. Although DIPA immunofluorescence staining decreases markedly in the absence of p120, Western blotting shows that DIPA protein levels are unchanged (Figure 2E). The reduced staining therefore reflects diffuse cytoplasmic redistribution of DIPA following removal of p120. As expected, p120 knockdown has no effect on tight junctions (e.g., ZO-1 staining; Figure 2B) but induces loss of AJs due to simultaneous destabilization of the entire MDCK complement of classical cadherins (i.e., E-, N-, K-, and P-cadherin; Wu et al., 1993; Stewart, 2000; Chen et al., 2011). Endogenous p120 family members are apparently insufficient to compensate for loss of p120. Thus mislocalization of DIPA under these circumstances stems from the near-complete loss of AJs (as opposed to the inability to bind endogenous p120 family members). Forced expression of p120 family members (in p120-depleted cells) restores cadherin stability and simultaneously rescues recruitment of DIPA to AJs. Consistent with this result, DIPA membrane localization is not altered by E- and/or N-cadherin knockdown because of compensation by other classical cadherins, which maintain both the integrity of AJs and the recruitment of DIPA through direct binding to p120 isoform 1 (Reynolds and Carnahan, 2004). Of importance, all DIPA family members are selectively recruited to endogenous p120-1, and the Y2H data argue that this observation can be effectively extended to the other p120 homologues. Thus all DIPA and p120 family members interact reciprocally, suggesting a flexible interface for a conserved and presumably important function.
Although the function of the interaction itself is unknown, mouse Ccdc85c is disrupted in the hhy mouse and encodes a protein that normally localizes to apical junctions of radial glia, neuroepithelial precursors lining the ventricles during cortical brain development (Merkle and Alvarez-Buylla, 2006; Mori et al., 2012). Our data predict that “apical” wild-type mouse CCDC85C protein is, in fact, recruited to AJs via interaction with p120 isoform 1, or, alternatively, the equivalent isoform of a p120 family member. The mutant form of the Ccdc85c gene, on the other hand, manifests as a homozygous truncation lacking the domain required for interaction with p120. The mutation causes a malformation of the developing cortex, resulting in subcortical heterotopia and hydrocephaly, indicating a role for Ccdc85c, and possibly the Ccdc85c/p120 interaction, in congenital alterations associated with epilepsy and mental retardation in humans. Of interest, p120 itself is implicated in another mouse model of congenital hydrocephalus. A point mutation in the Napa gene (encoding α-SNAP) is the causal defect in the hyh mouse (Hong et al., 2004). The mutation affects α-SNAP mRNA stability, resulting in low levels of expression (Chae et al., 2004). α-SNAP is normally involved in polarized vesicle trafficking, and α-SNAP depletion dramatically reduces cellular levels of p120, thereby compromising the stability of E-cadherin (Naydenov et al., 2012). Thus it is possible that the hhy and hyh phenotypes are mechanistically related at the level of developmental pathways involving the interaction between DIPA and p120 family members. Of note, p120 isoform 1 is enriched in highly motile neuroblasts that arise within the proliferative ventricular zone (Chauvet et al., 2003) and depend on rapid recycling of N-cadherin as they move distally through the cortex, a process guided by scaffolds of radial glial cells (Kawauchi, 2012). Developmental defects in neuroblast migration and/or radial glial cell integrity are known to result in heterotopia such as seen in hhy and hyh mutant mice (Chae et al., 2004; Shikanai et al., 2011; Mori et al., 2012).
Given the involvement of N-cadherin in these processes, we examined the effects of perturbing DIPA expression in zebrafish, a model in which N-cadherin functional defects are particularly well characterized. Zebrafish Ccdc85b (DIPA) is expressed in the midbrain and hindbrain during neurulation in early and mid somitogenesis (10–16 hpf; Thisse and Thisse, 2008). At this same stage in development, the N-cadherin/parachute (pac) mutant zebrafish exhibits defects in neural tube closure (Lele et al., 2002) due to a loss-of-function mutation in N-cadherin (Lele et al., 2002). We show that both overexpression and knockdown of DIPA in zebrafish embryos result in essentially the same neural tube defect (Figure 5), which in turn bears a striking similarity to the pac mutant, in that the dorsal neural tube is split bilaterally, with a dramatic gap toward the posterior midbrain. Notably, the phenotype is distinct from that of the e-cadherin/half-baked zebrafish mutant (Kane et al., 2005). The neuroepithelial disorganization shown in Figure 5C is observed in fish with either N-cadherin mutations or alterations in N-cadherin–binding proteins (Lele et al., 2002; Doll et al., 2011; Lo Sardo et al., 2012). These fish also progress at 48 hpf to form neural tube defects similar to our phenotype (Figure 5A), and the ectopic neuroepithelial cells have been compared with paraventricular heterotopia formation in the hyh and hhy mutants (Hong et al., 2004; Doll et al., 2011; Mori et al., 2012). It is possible that DIPA is specifically required for N-cadherin function at this stage, consistent with the selective interaction of DIPA with p120 isoform 1 and the generally observed coordination between isoform 1 and N-cadherin expression.
As to the exact role of DIPA in this process, we speculate that it may contribute to the dynamic nature of N-cadherin–mediated activity. For example, neural tube cells acquire an elongated phenotype during neurulation. Although catenin complexes are uniformly distributed around the cell periphery, adhesive activity is restricted to the poles (Hong and Brewster, 2006). Neural tube cells maintain these domains of adhesion and nonadhesion to enable rapid directional migration. Perhaps overexpression or depletion of DIPA disrupts this balance, resulting in migration defects and delayed neural tube closure. The migration characteristics of neural tube cells may be highly dependent on tightly controlled activity of both p120 isoform 1 and DIPA, whereas dedicated epithelial cells (e.g., MDCK cells) are programmed for functions like ion transport, which instead depend on the more stable adhesive properties of E-cadherin, preferential expression of p120 isoform 3, and constitutive maintenance of barrier function. Thus, although MDCK cells were ideal for characterizing the physical interactions between p120 isoform 1 and DIPA family members, the absence of a morphological response to DIPA knockdown is perhaps to be expected. DIPA is also implicated preliminarily in adipogenesis, Wnt signaling, and centrosome activity via various factors, including C/EBP-β, TCF4, and p78/MCRS1/MSP58 (Bezy et al., 2005; Du et al., 2006; Iwai et al., 2007). Given these observations, it is possible that the effect of DIPA on adhesion-related events may be mechanistically unrelated to p120, or alternaltively, that DIPA function is not limited to adhesion but instead comprises a spectrum of activities that are not well understood.
In summary, a strong preference for expression of p120 isoform 1 has long been associated with dynamic morphological activities such as those observed in neurons, macrophages, and fibroblasts. The importance of this isoform is suggested by the striking coordination of p120 alternative splicing during EMT to ensure that the E- to N-cadherin switch associated with EMT is accompanied by expression of p120-1. The isoform 1–specific head domain has indeed been linked to a wide variety of events associated with cell migration and invasion, including metastasis (Slorach et al., 2011). Here we identify DIPA as the first isoform 1–specific p120 binding partner and show that all members of the DIPA family (Ccdc85a, Ccdc85b/DIPA, and Ccdc85c) interact reciprocally with all members of the p120 family (p120-1, δ-catenin, p0071, and ARVCF) via conserved coiled-coil and head domains, respectively. Consistent with these data, DIPA overexpression or depletion in zebrafish phenocopies a neural tube closure defect associated with N-cadherin loss of function. On the basis of our data and the neuronal migration–associated disorders linked previously to Ccdc85c and α-SNAP mutations, we speculate that DIPA family members may mediate a broad range of p120-1 and/or N-cadherin–associated developmental phenotypes.
MATERIALS AND METHODS
Yeast two-hybrid
The human CTNND1 gene cDNA encoding p120-1AB (accession number AF062328) was cloned into the lexA vector pB27 and screened against a human breast cancer epithelial cell line prey cDNA library (T47D, MDA-MB468, MCF7, and BT20 cells). Y2H screens were performed by Hybrigenics SA (Paris, France) as previously described (Formstecher et al., 2005).
For our own direct Y2H binding assays, we used a protocol from Kendirgi et al. (2005), with several exceptions. Briefly, DIPA, p120, and their family members were used to screen for protein interactions with pGAD424 (activation domain) and pGBT9 (DNA binding domain) vectors in PJ69-4A yeast strain (James et al., 1996). Transformations were conducted using the lithium acetate method (Ito et al., 1983), and cells were plated on yeast nitrogen base media without amino acids (Difco, Detroit, MI) supplemented with 2% glucose and 75 mg/l methionine, uracil, adenine, and histidine (Sigma-Aldrich, St. Louis, MO). Colonies able to survive at 30°C for at least 48 h were plated on the same media but lacking adenine and histidine. Plates were imaged after an additional 5–9 d at 30°C. For experiments using pGBT9-δ-catenin, we added 12.5 mM 3-AT to prevent autoactivation (3-amino-1,2,4-triazole; A8056, Sigma-Aldrich).
Tissue culture and TER
All cell lines were grown in DMEM supplemented with 10% fetal bovine serum (Hyclone, Thermo Scientific, Waltham, MA), penicillin (100 U/ml), streptomycin (100 mg/ml; Life Technologies, Gaithersburg, MD), and ciprofloxicin (10 μg/ml; Sigma-Aldrich). The cell lines used for immunofluorescence or immunoblotting were MDCK kidney epithelial cells (canine), HCA7 colon carcinoma cells (human), or HEK293 embryonal kidney cells (human) as indicated. TER was measured as described in Kim et al. (2012). Briefly, TER was measured using a Millicell Electrical Resistance System (Millipore, Billerica, MA) with MDCK cells plated on Transwell filters (Corning, Corning, NY). Cells were cultured on 12-mm filter inserts for at least 7 d, with one Transwell chamber left blank to determine the intrinsic resistance of the membrane. To obtain final TER values, the blank value was subtracted from the measured value, and the results were recorded as Ω cm2.
Cloning, transfection, and infection
Full-length human DIPA (encoded by the CCDC85B gene) and human CCDC85A were purchased from the Dana-Farber/Harvard Cancer Center DNA Resource Core (Cambridge, MA) in the form of pENTR223-CCDC85B-fusion (clone HsCD00288507) and pENTR-223.1-CCDC85A-fusion (clone HsCD00431731). Human CCDC85C was synthesized by GeneArt (Life Technologies). To create Flag-tagged and Y2H expression genes, these DIPA family genes were recombined into the LZRS-3xFlag-Gateway-IRES-Neo and pGAD-Gateway vectors using LR recombinase from Life Technologies. Viral production and cell transduction protocols are described elsewhere (Ireton et al., 2002; Davis et al., 2003). To knock down canine DIPA, short hairpin RNA (shRNA) constructs were ordered from IDT (Coralville, IA) as oligomers, annealed, and cloned into pLentiLox3.7-Puro. The specific targets are 5′-GGGAGAACCTGGCGCTTAA-3′, 5′-GACTGAGGCTCATCTTCCT-3′, and 5′-GCCTGGCTCTGGGTGAGGA-3′. The p120 knockdown and add-back constructs are described by Dohn et al. (2009). To create C-terminally tagged GFP-fusion proteins, p120-1A, p120-3, δ-catenin, and p0071 were PCR amplified, cloned into pENTR2B, and recombined into pLentiLox3.7-Gateway-GFP with LR recombinase from Life Technologies.
Immunofluorescence microscopy and antibodies
For immunofluorescence labeling, cells were plated and cultured for 2 d on glass coverslips before fixing with 3% paraformaldehyde and permeabilization with 0.2% Triton X-100 in phosphate-buffered saline (PBS; pH 7.4). Primary antibody and hybridoma supernatant incubations were performed at room temperature for 30 min in 3% milk/PBS at 0.5–1.0 mg/ml. After three washes with PBS, the coverslips were incubated with Alexa Fluor goat anti-mouse 594 and/or Alexa Fluor goat anti-rabbit 488 (Life Technologies) secondary antibodies in 3% milk/PBS at 2.5 μg/ml for 30 min at room temperature. The coverslips were finally washed three times with PBS, mounted on glass slides with Prolong Gold (Life Technologies), and visualized using a Zeiss Axioplan 2 microscope (Zeiss, Thornwood, NY).
Anti-Flag monoclonal antibody (mAb; M2 cat. #F1804) was purchased from Sigma-Aldrich. Anti-tubulin (DM1α) and KT3 mAbs (Tomonari, 1988) were obtained from the Vanderbilt Antibody and Protein Resource (Nashville, TN). Anti-p120 mAb pp120 was purchased from BD Transduction (San Jose, CA), and anti–ZO-1 polyclonal Ab (pAb) was from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies for DIPA (3E3 mAb and pAb) are described in Markham et al. (2012), and their use is indicated in the figure legends. The specific use of p120 antibodies F1αSH (Wu et al., 1998), pp120 (BD Biosciences, San Jose, CA), and 6H11 (Wu et al., 1998) is also indicated in figure legends.
Immunoblotting and immunoprecipitation
Procedures for immunoblot analysis and immunoprecipitation have been described previously in detail (Daniel and Reynolds, 1999). Briefly, cells were lysed in a buffer containing 0.5% Nonidet P-40, 10 mM Tris (pH 7.4), 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 1 mM sodium vanadate, 0.1 trypsin inhibitor units of aprotinin, and 5 mg/ml leupeptin. Whole-cell lysates were separated by SDS–PAGE and transferred to nitrocellulose membranes. Blots were briefly blocked at 4°C with 5% nonfat dried milk in Tris-buffered saline (TBS), pH 7.4, and incubated overnight at 4°C with primary antibody (0.2–2.0 mg/ml) in 3% milk/TBS. The membranes were then washed five times with TBS before incubation with the secondary anti-mouse 680 and/or anti-rabbit 800 antibody (LiCor, Lincoln, NE) in Odyssey blocking buffer/TBS for 30 min at room temperature. Blots were finally washed three times with TBS plus 0.1% Tween-20 and three times with TBS and then processed with the Odyssey immunodetection system (LiCor) according to the manufacturer's protocol. For immunoprecipitation, primary antibody (4 μg/ml) was incubated with clear whole-cell lysates for at least 4 h at 4°C. Protein G Sepharose (GE Healthcare, Piscataway, NJ) was then added and incubated at 4°C for an additional hour. The Sepharose-bound immunocomplexes were then washed five times in NP-40 lysis buffer with inhibitors (see previous description) and solubilized in Laemmli sample buffer (Daniel and Reynolds, 1999).
Zebrafish injections and Wnt1 in situ hybridization
Zebrafish maintenance and injection were described in Doll et al. (2011). Briefly, zebrafish embryos were raised at 28.5°C on a 14/10 light/dark cycle. Embryos were staged according to hours postfertilization as previously described (Kimmel et al., 1995). The wild-type strain AB was used in all experiments (Walker, 1999). An antisense morpholino oligonucleotide (MO) complementary to the translation initiation site was used to deplete ccdc85b function (DIPA2 5′-AGTGCTGGGTAGTAAGTGATTACAT). DIPA mRNA was created by in vitro transcription using the mMessage mMachine SP6 kit and manufacturer's protocol (Life Technologies). For morpholino or RNA injection, the stock solution (10 mg/ml for MO, 1 mg/ml for RNA) was diluted in distilled water, and embryos (one- to two-cell stage) were pressure injected with ∼1 nl of MO/RNA and phenol red per embryo. A total of 4 ng/embryo of the MO or 0.5 ng/embryo of RNA concentration was used.
Whole-mount in situ hybridization was performed as described previously (Gamse et al., 2003; Thisse and Thisse, 2008) using reagents from Roche Applied Bioscience (Indianapolis, IN). The wnt1 antisense RNA probe was synthesized from Hind III–linearized template using T3 RNA polymerase in the presence of digoxigenin-UTP. Embryos were incubated at 70°C with the antisense probe in hybridization solution containing 50% formamide. Hybridized probes were detected using alkaline phosphatase–conjugated antibodies and visualized by 4-nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate staining (Thisse and Thisse, 2008).
Supplementary Material
Acknowledgments
We thank Nichole Lobdell, Erin Booton, and Qiang Guan for expert technical assistance and the Vanderbilt Antibody and Protein Resource for assistance in generating DIPA antibodies. Public Health Service Award T32 GM07347 from the National Institute of General Medical Studies for the Vanderbilt Medical-Scientist Training Program supported training for N.O.M. This work was also supported by Vanderbilt GI SPORE Grant P50-CA95103 to R.J.C., Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institutes of Health Award R01HD054534 to J.T.G., and National Center for Advancing Translational Sciences/National Institutes of Health Award UL1 TR000445 to J.T.G. Further support came from Vanderbilt Cancer Center Support Grant P30-CA11947 and National Institutes of Health Grants R01Ca55724 and R01Ca111947 to A.B.R. Vanderbilt shared resources contributing to the work include the Functional Genomics Shared Resource and the Vanderbilt Antibody and Protein Resource.
Abbreviations used:
- AJ
adherens junction
- Arm
Armadillo
- DIPA
delta-interacting protein A
- EMT
epithelial-to-mesenchymal transition
- hhy
hemorrhagic hydrocephalus
- hyh
hydrocephaly with hop gait
- MDCK
Madin–Darby canine kidney
- p120
p120-catenin
- α-SNAP
soluble N-ethylmaleimide–sensitive factor attachment protein α
- Y2H
yeast two-hybrid
- ZO-1
zonula occludens-1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0492 on July 9, 2014.
REFERENCES
- Aho S, Rothenberger K, Uitto J. Human p120ctn catenin: tissue-specific expression of isoforms and molecular interactions with BP180/type XVII collagen. J Cell Biochem. 1999;73:390–399. [PubMed] [Google Scholar]
- Anastasiadis PZ, Moon SY, Thoreson MA, Mariner DJ, Crawford HC, Zheng Y, Reynolds AB. Inhibition of RhoA by p120 catenin. Nat Cell Biol. 2000;2:637–644. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ, Reynolds AB. The p120 catenin family: complex roles in adhesion, signaling and cancer. J Cell Sci. 2000;113:1319–1334. doi: 10.1242/jcs.113.8.1319. [DOI] [PubMed] [Google Scholar]
- Aono S, Nakagawa S, Reynolds AB, Takeichi M. p120(ctn) acts as an inhibitory regulator of cadherin function in colon carcinoma cells. J Cell Biol. 1999;145:551–562. doi: 10.1083/jcb.145.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezy O, Elabd C, Cochet O, Petersen RK, Kristiansen K, Dani C, Ailhaud G, Amri E-Z. Delta-interacting protein A, a new inhibitory partner of CCAAT/enhancer-binding protein beta, implicated in adipocyte differentiation. J Biol Chem. 2005;280:11432–11438. doi: 10.1074/jbc.M411741200. [DOI] [PubMed] [Google Scholar]
- Brazas R, Ganem D. A cellular homolog of hepatitis delta antigen: implications for viral replication and evolution. Science. 1996;274:90–94. doi: 10.1126/science.274.5284.90. [DOI] [PubMed] [Google Scholar]
- Carnahan RH, Rokas A, Gaucher EA, Reynolds AB. The molecular evolution of the p120-catenin subfamily and its functional associations. PLoS One. 2010;5:e15747. doi: 10.1371/journal.pone.0015747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae TH, Kim S, Marz KE, Hanson PI, Walsh CA. The hyh mutation uncovers roles for αSnap in apical protein localization and control of neural cell fate. Nat Genet. 2004;36:264–270. doi: 10.1038/ng1302. [DOI] [PubMed] [Google Scholar]
- Chalasani K, Brewster RM. N-cadherin-mediated cell adhesion restricts cell proliferation in the dorsal neural tube. Mol Biol Cell. 2011;22:1505–1515. doi: 10.1091/mbc.E10-08-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvet N, Prieto M, Fabre C, Noren NK, Privat A. Distribution of p120 catenin during rat brain development: potential role in regulation of cadherin-mediated adhesion and actin cytoskeleton organization. Mol Cell Neurosci. 2003;22:467–486. doi: 10.1016/s1044-7431(03)00030-7. [DOI] [PubMed] [Google Scholar]
- Chen Y, Kung H-N, Chen C-H, Huang S-H, Chen K-H, Wang S-M. Acidic extracellular pH induces p120-catenin-mediated disruption of adherens junctions via the Src kinase-PKCδ pathway. FEBS Lett 585, 705–710. 2011 doi: 10.1016/j.febslet.2011.01.022. [DOI] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R, Sarpal R, Ishiyama N, Pellikka M, Ikura M, Tepass U. Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat Cell Biol. 2013;15:261–273. doi: 10.1038/ncb2685. [DOI] [PubMed] [Google Scholar]
- Dohn MR, Brown MV, Reynolds AB. An essential role for p120-catenin in Src- and Rac1-mediated anchorage-independent cell growth. J Cell Biol. 2009;184:437–450. doi: 10.1083/jcb.200807096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll CA, Burkart JT, Hope KD, Halpern ME, Gamse JT. Subnuclear development of the zebrafish habenular nuclei requires ER translocon function. Dev Biol. 2011;360:44–57. doi: 10.1016/j.ydbio.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. α-catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Wang Q, Hirohashi Y, Greene MI. DIPA, which can localize to the centrosome, associates with p78/MCRS1/MSP58 and acts as a repressor of gene transcription. Exp Mol Pathol. 2006;81:184–190. doi: 10.1016/j.yexmp.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Ferland RJ, Batiz LF, Neal J, Lian G, Bundock E, Lu J, Hsiao Y, Diamond R, Mei D, Banham AH, et al. Disruption of neural progenitors along the ventricular and subventricular zones in periventricular heterotopia. Hum Mol Genet. 2009;18:497–516. doi: 10.1093/hmg/ddn377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamse JT, Thisse C, Thisse B, Halpern ME. The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development. 2003;130:1059–1068. doi: 10.1242/dev.00270. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M, Green KJ, Sauter H. Targeting of p0071 to desmosomes and adherens junctions is mediated by different protein domains. J Cell Sci. 2003;116:1219–1233. doi: 10.1242/jcs.00275. [DOI] [PubMed] [Google Scholar]
- Hong E, Brewster R. N-cadherin is required for the polarized cell behaviors that drive neurulation in the zebrafish. Development. 2006;133:3895–3905. doi: 10.1242/dev.02560. [DOI] [PubMed] [Google Scholar]
- Hong H-K, Chakravarti A, Takahashi JS. The gene for soluble N-ethylmaleimide sensitive factor attachment protein α is mutated in hydrocephaly with hop gait (hyh) mice. Proc Natl Acad Sci USA. 2004;101:1748–1753. doi: 10.1073/pnas.0308268100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosking CR, Ulloa F, Hogan C, Ferber EC, Figueroa A, Gevaert K, Birchmeier W, Briscoe J, Fujita Y. The transcriptional repressor Glis2 is a novel binding partner for p120 catenin. Mol Biol Cell. 2007;18:1918–1927. doi: 10.1091/mbc.E06-10-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichii T, Takeichi M. p120-catenin regulates microtubule dynamics and cell migration in a cadherin-independent manner. Genes Cells. 2007;12:827–839. doi: 10.1111/j.1365-2443.2007.01095.x. [DOI] [PubMed] [Google Scholar]
- Ireton RC, et al. A novel role for p120 catenin in E-cadherin function. J Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai A, Hijikata M, Hishiki T, Isono O, Chiba T, Shimotohno K. Coiled-coil domain containing 85B suppresses the β-catenin activity in a p53-dependent manner. Oncogene. 2007;27:1520–1526. doi: 10.1038/sj.onc.1210801. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, McFarland KN, Warga RM. Mutations in half baked/E-cadherin block cell behaviors that are necessary for teleost epiboly. Development. 2005;132:1105–1116. doi: 10.1242/dev.01668. [DOI] [PubMed] [Google Scholar]
- Kaufmann U, Zuppinger C, Waibler Z, Rudiger M, Urbich C, Martin B, Jockusch BM, Eppenberger H, Starzinski-Powitz A. The armadillo repeat region targets ARVCF to cadherin-based cellular junctions. J Cell Sci. 2000;113:4121–4135. doi: 10.1242/jcs.113.22.4121. [DOI] [PubMed] [Google Scholar]
- Kawauchi T. Cell adhesion and its endocytic regulation in cell migration during neural development and cancer metastasis. Int J Mol Sci. 2012;13:4564–4590. doi: 10.3390/ijms13044564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keirsebilck A, Bonné S, Staes K, van Hengel J, Nollet F, Reynolds A, van Roy F. Molecular cloning of the human p120 ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics. 1998;50:129–146. doi: 10.1006/geno.1998.5325. [DOI] [PubMed] [Google Scholar]
- Kendirgi F, Rexer DJ, Alcázar-Román AR, Onishko HM, Wente SR. Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: a conserved mechanism in the export of Hsp70 mRNA. Mol Biol Cell. 2005;16:4304–4315. doi: 10.1091/mbc.E04-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Park J-I, Spring CM, Sater AK, Ji H, Otchere AA, Daniel JM, McCrea PD. Non-canonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nat Cell Biol. 2004;6:1212–1220. doi: 10.1038/ncb1191. [DOI] [PubMed] [Google Scholar]
- Kim TI, Poulin EJ, Blask E, Bukhalid R, Whitehead RH, Franklin JL, Coffey RJ. Myofibroblast keratinocyte growth factor reduces tight junctional integrity and increases claudin-2 levels in polarized Caco-2 cells. Growth Factors. 2012;30:320–332. doi: 10.3109/08977194.2012.717076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kurley SJ, Bierie B, Carnahan RH, Lobdell NA, Davis MA, Hofmann I, Moses HL, Muller WJ, Reynolds AB. p120-catenin is essential for terminal end bud function and mammary morphogenesis. Development. 2012;139:1754–1764. doi: 10.1242/dev.072769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele Z, Folchert A, Concha M, Rauch GJ, Geisler R, Rosa F, Wilson SW, Hammerschmidt M, Bally-Cuif L. Parachute/N-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development. 2002;129:3281–3294. doi: 10.1242/dev.129.14.3281. [DOI] [PubMed] [Google Scholar]
- Lohia M, Qin Y, Macara IG. The Scribble polarity protein stabilizes E-cadherin/p120-catenin binding and blocks retrieval of E-cadherin to the Golgi. PLoS One. 2012;7:e51130. doi: 10.1371/journal.pone.0051130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Sardo V, Zuccato D, Gaudenzi G, Vitali B, Ramos C, Tartari M, Myre MA, Walker JA, Pistocchi A, Conti L, et al. An evolutionary recent neuroepithelial cell adhesion function of huntingtin implicates ADAM10-N-cadherin. Nat Neurosci. 2012;15:713–721. doi: 10.1038/nn.3080. [DOI] [PubMed] [Google Scholar]
- Markham NO, Cooper T, Goff M, Gribben EM, Carnahan RH, Reynolds AB. Monoclonal antibodies to DIPA: a novel binding partner of p120-catenin isoform 1. Hybridoma (Larchmt) 2012;31:246–254. doi: 10.1089/hyb.2012.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135:948–959. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Alvarez-Buylla A. Neural stem cells in mammalian development. Curr Opin Cell Biol. 2006;18:704–709. doi: 10.1016/j.ceb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Miao Y, Liu N, Zhang Y, Liu Y, Yu J-H, Dai S-D, Xu H-T, Wang E-H. p120ctn isoform 1 expression significantly correlates with abnormal expression of E-cadherin and poor survival of lung cancer patients. Med Oncol. 2009;27:880–886. doi: 10.1007/s12032-009-9300-2. [DOI] [PubMed] [Google Scholar]
- Mo YY, Reynolds AB. Identification of murine p120 isoforms and heterogeneous expression of p120cas isoforms in human tumor cell lines. Cancer Res. 1996;56:2633–2640. [PubMed] [Google Scholar]
- Molina-Ortiz I, Bartolomé RA, Hernández-Varas P, Colo GP, Teixidó J. Overexpression of E-cadherin on melanoma cells inhibits chemokine-promoted invasion involving p190RhoGAP/p120ctn-dependent inactivation of RhoA. J Biol Chem. 2009;284:15147–15157. doi: 10.1074/jbc.M807834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N, Kuwamura M, Tanaka N, Hirano R, Nabe M, Ibuki M, Yamate J. Ccdc85c encoding a protein at apical junctions of radial glia is disrupted in hemorrhagic hydrocephalus (hhy) mice. Am J Pathol. 2012;180:314–327. doi: 10.1016/j.ajpath.2011.09.014. [DOI] [PubMed] [Google Scholar]
- Nanes BA, Chiasson-MacKenzie C, Lowery AM, Ishiyama N, Faundez V, Ikura M, Vincent PA, Kowalczyk AP. p120-catenin binding masks an endocytic signal conserved in classical cadherins. J Cell Biol. 2012;199:365–380. doi: 10.1083/jcb.201205029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenov NG, Brown B, Harris G, Dohn MR, Morales VM, Baranwal S, Reynolds AB, Ivanov AI. A membrane fusion protein αSNAP is a novel regulator of epithelial apical junctions. PLoS One. 2012;7:e34320. doi: 10.1371/journal.pone.0034320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Liu BP, Burridge K, Kreft B. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J Cell Biol. 2000;150:567–580. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-I, Kim SW, Lyons JP, Ji H, Nguyen TT, Cho K, Barton MC, Deroo T, Vleminckx K, McCrea PD. Kaiso/p120-catenin and TCF/β-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell. 2005;8:843–854. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Prokhortchouk A, Hendrich B, Jørgensen H, Ruzov A, Wilm M, Georgiev G, Bird A, Prokhortchouk E. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB. p120-catenin: past and present. Biochim Biophys Acta. 2007;1773:2–7. doi: 10.1016/j.bbamcr.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB, Carnahan RH. Regulation of cadherin stability and turnover by p120ctn: implications in disease and cancer. Semin Cell Dev Biol. 2004;15:657–663. doi: 10.1016/j.semcdb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Saito M, Tucker DK, Kohlhorst D, Niessen CM, Kowalczyk AP. Classical and desmosomal cadherins at a glance. J Cell Sci. 2012;125:2547–2552. doi: 10.1242/jcs.066654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Watanabe T, Wang S, Kakeno M, Matsuzawa K, Matsui T, Yokoi K, Murase K, Sugiyama I, Ozawa M, et al. Numb controls E-cadherin endocytosis through p120 catenin with aPKC. Mol Biol Cell. 2011;22:3103–3119. doi: 10.1091/mbc.E11-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai M, Nakajima K, Kawauchi T. N-cadherin regulates radial glial fiber-dependent migration of cortical locomoting neurons. Commun Integr Biol. 2011;4:326–330. doi: 10.4161/cib.4.3.14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sival DA, Guerra M, Dunnen, den WFA, Bátiz LF, Alvial G, Castañeyra-Perdomo A, Rodríguez EM. Neuroependymal denudation is in progress in full-term human foetal spina bifida aperta. Brain Pathol. 2011;21:163–179. doi: 10.1111/j.1750-3639.2010.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slorach EM, Chou J, Werb Z. Zeppo1 is a novel metastasis promoter that represses E-cadherin expression and regulates p120-catenin isoform expression and localization. Genes Dev. 2011;25:471–484. doi: 10.1101/gad.1998111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AL, Dohn MR, Brown MV, Reynolds AB. Association of Rho-associated protein kinase 1 with E-cadherin complexes is mediated by p120-catenin. Mol Biol Cell. 2012;23:99–110. doi: 10.1091/mbc.E11-06-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DB. Differential regulation of endogenous cadherin expression in Madin-Darby canine kidney cells by cell-cell adhesion and activation of beta-catenin signaling. J Biol Chem. 2000;275:20707–20716. doi: 10.1074/jbc.M000467200. [DOI] [PubMed] [Google Scholar]
- Talvinen K, Tuikkala J, Nykänen M, Nieminen A, Anttinen J, Nevalainen OS, Hurme S, Kuopio T, Kronqvist P. Altered expression of p120 catenin predicts poor outcome in invasive breast cancer. J Cancer Res Clin Oncol. 2010;136:1377–1387. doi: 10.1007/s00432-010-0789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Thoreson MA, Anastasiadis PZ, Daniel JM, Ireton RC, Wheelock MJ, Johnson KR, Hummingbird DK, Reynolds AB. Selective uncoupling of p120ctn from E-cadherin disrupts strong adhesion. J Cell Biol. 2000;148:189–202. doi: 10.1083/jcb.148.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonari K. A rat antibody against a structure functionally related to the mouse T-cell receptor/T3 complex. Immunogenetics. 1988;28:455–458. doi: 10.1007/BF00355379. [DOI] [PubMed] [Google Scholar]
- Walker C. Haploid screens and gamma-ray mutagenesis. Methods Cell Biol. 1999;60:43–70. doi: 10.1016/s0091-679x(08)61893-2. [DOI] [PubMed] [Google Scholar]
- Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, Lobdell NA, Settleman J, Reynolds AB. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 2006;127:1027–1039. doi: 10.1016/j.cell.2006.09.046. [DOI] [PubMed] [Google Scholar]
- Wu J, Mariner DJ, Thoreson MA, Reynolds AB. Production and characterization of monoclonal antibodies to the catenin p120ctn. Hybridoma. 1998;17:175–183. doi: 10.1089/hyb.1998.17.175. [DOI] [PubMed] [Google Scholar]
- Wu JC, Gregory CW, DePhilip RM. P-cadherin and E-cadherin are co-expressed in MDCK cells. Biochem Biophys Res Commun. 1993;195:1329–1335. doi: 10.1006/bbrc.1993.2189. [DOI] [PubMed] [Google Scholar]
- Xiao K, Allison DF, Buckley KM, Kottke MD, Vincent PA, Faundez V, Kowalczyk AP. Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J Cell Biol. 2003;163:535–545. doi: 10.1083/jcb.200306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Garner J, Buckley KM, Vincent PA, Chiasson CM, Dejana E, Faundez V, Kowalczyk AP. p120-Catenin regulates clathrin-dependent endocytosis of VE-cadherin. Mol Biol Cell. 2005;16:5141–5151. doi: 10.1091/mbc.E05-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, Copland JA, Anastasiadis PZ. A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J Biol Chem. 2008;283:18344–18354. doi: 10.1074/jbc.M801192200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M, Kaverina IN, Wang A, Fujita Y, Reynolds AB, Anastasiadis PZ. A novel interaction between kinesin and p120 modulates p120 localization and function. J Biol Chem. 2004;279:9512–9521. doi: 10.1074/jbc.M310895200. [DOI] [PubMed] [Google Scholar]
- Yang I, Chang O, Lu Q, Kim K. δ-Catenin affects the localization and stability of p120-catenin by competitively interacting with E-cadherin. Mol Cell. 2010;29:233–237. doi: 10.1007/s10059-010-0030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS, Niessen CM, Gumbiner BM. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebda N, Tian Y, Tian X, Gawlak G, Higginbotham K, Reynolds AB, Birukova AA, Birukov KG. Interaction of p190RhoGAP with C-terminal domain of p120-catenin modulates endothelial cytoskeleton and permeability. J Biol Chem. 2013;288:18290–18299. doi: 10.1074/jbc.M112.432757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.