In Pichia pastoris, the PTS2 receptor, Pex7, is selectively degraded in a regulated manner. The shuttling of Pex7, and consequently its degradation, depends on the receptor recycling pathways used by Pex5 and Pex20 and relies on an interaction between Pex7 and Pex20. The shuttling and stability of Pex7 are divergent from those of Pex5 and Pex20.
Abstract
Peroxisomal matrix protein import uses two peroxisomal targeting signals (PTSs). Most matrix proteins use the PTS1 pathway and its cargo receptor, Pex5. The PTS2 pathway is dependent on another receptor, Pex7, and its coreceptor, Pex20. We found that during the matrix protein import cycle, the stability and dynamics of Pex7 differ from those of Pex5 and Pex20. In Pichia pastoris, unlike Pex5 and Pex20, Pex7 is constitutively degraded in wild-type cells but is stabilized in pex mutants affecting matrix protein import. Degradation of Pex7 is more prevalent in cells grown in methanol, in which the PTS2 pathway is nonessential, in comparison with oleate, suggesting regulation of Pex7 turnover. Pex7 must shuttle into and out of peroxisomes before it is polyubiquitinated and degraded by the proteasome. The shuttling of Pex7, and consequently its degradation, is dependent on the receptor recycling pathways of Pex5 and Pex20 and relies on an interaction between Pex7 and Pex20. We also found that blocking the export of Pex20 from peroxisomes inhibits PTS1-mediated import, suggesting sharing of limited components in the export of PTS receptors/coreceptors. The shuttling and stability of Pex7 are divergent from those of Pex5 and Pex20, exemplifying a novel interdependence of the PTS1 and PTS2 pathways.
INTRODUCTION
Peroxisomes are essential organelles responsible for fatty acid β-oxidation and scavenging of reactive oxygen species (Ma et al., 2011). The importance of peroxisomes has recently been extended, as they also have roles as a signaling platform involved in aging, antivirus defense, and regulation of mTORC1 and autophagy (Dixit et al., 2010; Facciotti et al., 2012; Zhang et al., 2013). Proteins are targeted to peroxisomes through the use of peroxisomal targeting signals (PTSs) of either type 1 or 2. The delivery of PTS1 and PTS2 proteins to the peroxisome lumen depends on the PTS receptors, Pex5 for PTS1 proteins or Pex7 for PTS2 proteins, together with the PTS2 pathway coreceptors, Pex18/21 in Saccharomyces cerevisiae, Pex20 in other fungi, or Pex5L in higher eukaryotes, respectively (Purdue et al., 1998; Titorenko et al., 1998; Otzen et al., 2005; Woodward and Bartel, 2005; Léon et al., 2006).
The molecular mechanisms involved in the dynamic shuttling of Pex5 and Pex18/Pex20 during the matrix protein import cycle have been elucidated (Léon et al., 2006; Platta et al., 2009; Hensel et al., 2011; Liu and Subramani, 2013). After unloading the cargo in the peroxisome lumen, the receptors face two possible fates: either export to the cytosol to be used for another round of import or, when the receptor recycling pathway is blocked, degradation by the ubiquitin-proteasome system (UPS) via the receptor accumulation and degradation in the absence of recycling (RADAR) pathway (Léon et al., 2006). Whereas the recycling of Pex5, Pex18, and Pex20 relies on monoubiquitination on conserved cysteine residues near the N-termini of these proteins, the degradation of these receptors is triggered by polyubiquitination on one or more conserved lysines also located near the N-terminus (Léon et al., 2006; Léon and Subramani, 2007; Platta et al., 2007; Hensel et al., 2011). Monoubiquitination and polyubiquitination of S. cerevisiae Pex5 and Pichia pastoris Pex20 is achieved by the E2 ubiquitin-conjugation enzymes, Pex4 (mono) or Ubc4/5 (poly), in conjunction with the RING subcomplex composed of three E3 ligases, Pex2, Pex10, and Pex12 (Platta et al., 2009; Liu and Subramani, 2013). In S. cerevisiae, each E3 ligase is assigned a distinct function for either monoubiquitination or polyubiquitination of Pex5 and Pex18, whereas in P. pastoris, all three E3 ligases are essential for both monoubiquitination and polyubiquitination of Pex5 and Pex20 (Platta et al., 2009; El Magraoui et al., 2013; Liu and Subramani, 2013). Monoubiquitinated Pex5 can be recognized by AAA ATPases, Pex1 and Pex6, and recycled from the peroxisome membrane to the cytosol, where the ubiquitin moiety is removed by an ubiquitin protease, allowing for the next round of cargo import (Platta et al., 2007; Debelyy et al., 2011; Grou et al., 2012; Miyata et al., 2012). How the polyubiquitinated receptors are extracted from the peroxisome membrane is still elusive, since the RADAR pathway can be activated in the absence of Pex1 or Pex6 (Léon et al., 2006).
As a mobile receptor like Pex5 and Pex18/20, Pex7 also shuttles between the cytosol and peroxisome lumen (Elgersma et al., 1998; Nair et al., 2004). However, the conserved cysteine and lysine residues found at the N-terminus of Pex5 and Pex18/20 that are essential for monoubiquitination or polyubiquitination, respectively, do not exist in Pex7. Until now, it was not known whether the shuttling of Pex7 during the matrix protein import cycle also depends on ubiquitination. Endogenous Pex7 in Arabidopsis is subject to degradation mediated by the small GTPase RabE1c, probably via the UPS, when a nonfunctional GFP-Pex7 accumulates on the peroxisome membrane (Cui et al., 2013).
In P. pastoris, peroxisome proliferation can be induced by growth in either oleate or methanol medium (Gould et al., 1992). The number, morphology, and content of peroxisomes in these two growth conditions are very different. As compared with methanol-grown cells, P. pastoris cells grown in oleate contain more abundant peroxisomes, which are smaller and dispersed randomly in the cytosol without forming big peroxisome clusters (Joshi et al., 2012). Both the PTS1 and PTS2 pathways are required to metabolize oleate. However, few lumenal proteins have a PTS2, and elimination of the PTS2 import pathway, using Δpex7 or Δpex20 cells, does not affect growth in methanol medium (Elgersma et al., 1998; Léon et al., 2006), showing that the PTS2 pathway is dispensable in this medium. How the PTS1 and PTS2 pathways collaborate or differentially regulate each other to reconstitute the peroxisomal matrix in response to different carbon sources remains largely obscure.
In this study, we found that Pex7 undergoes constitutive UPS-mediated degradation when cells are grown in methanol but not in oleate medium. In methanol-grown cells, Pex7 was stabilized and accumulated in the absence of Pex14, Pex8, Pex2, Pex4, or Pex6, whereas the steady-state levels of Pex5 and Pex20 in these mutants was either unchanged (Pex14, Pex8, Pex2) or down-regulated (Pex4 and Pex6), as shown previously (Collins et al., 2000; Léon et al., 2006). These features distinguish the Pex7 degradation pathway from the known RADAR pathway responsible for the degradation of Pex5 and Pex20. Surprisingly, further analyses revealed that the down-regulation of Pex7 depended not only on Pex20, but also on Pex5. Moreover, blocking the export of Pex20 abolished both the import of PTS1 and PTS2 proteins, suggesting shared pathways or components among the export pathways for the three PTS receptors.
RESULTS
Regulated degradation of Pex7
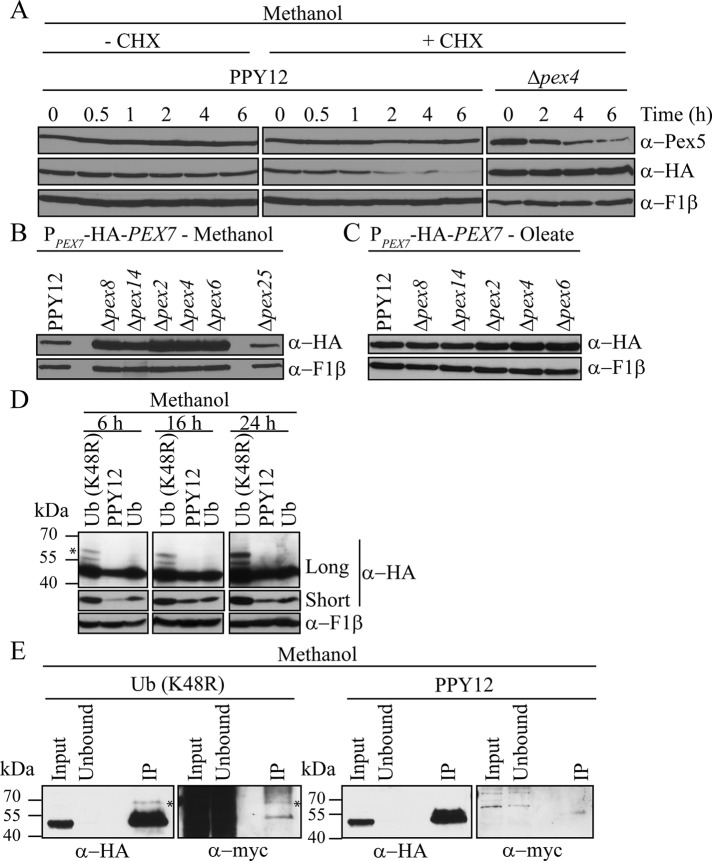
Previous studies showed that in the absence of the receptor recycling machinery (Pex4/Pex22 and Pex1/6/15), Pex5 and Pex20 are degraded by the RADAR pathway when cells are grown in oleate or methanol medium (Collins et al., 2000; Léon et al., 2006). To investigate whether Pex7 is degraded by the same pathway, we analyzed the turnover rate of Pex7 after 3 h of growth in methanol induction, followed by treatment with cycloheximide, a protein synthesis inhibitor. The expression level of Pex7 was monitored by expressing hemagglutinin (HA)-Pex7 from the endogenous PEX7 promoter (PPEX7-HA-Pex7) in wild-type (PPY12) and ∆pex4 mutant strains. HA-Pex7 could fully complement the growth defects of Δpex7 in oleate medium (Supplemental Figure S1). Surprisingly, after 2 h of treatment with cycloheximide, Pex7 was degraded to an almost undetectable level in wild-type cells, whereas Pex5 was stable (Figure 1A). Furthermore, Pex7 levels remained constant when concomitantly treated with the proteasome inhibitor MG-132, indicating that degradation of Pex7 requires the proteasome (Supplemental Figure S2). In contrast, Pex7 levels were stable for up to 6 h in Δpex4 cells treated with cycloheximide and in untreated wild-type cells, whereas Pex5 was unstable in Δpex4 cells, as expected (Collins et al., 2000; Figure 1A). These results suggest that the fate and stability of Pex7 differ from those of Pex5.
FIGURE 1:
Regulated degradation of Pex7. (A) Pex7 turnover was analyzed in wild-type and Δpex4 cells and induced in methanol medium for 3 h by treatment with 5 mg/ml cycloheximide (CHX) for the indicated intervals of time. F1β serves as a control for equal loading. (B, C) Steady-state levels of HA-Pex7 were analyzed in wild-type (PPY12) and pex mutant strains grown in methanol (B) or oleate medium (C) by immunoblot with α-HA. (D) Cells overexpressing ubiquitin (Ub) or mutated ubiquitin (Ub (K48R)) were induced in methanol medium. Equal-OD samples were collected at 6, 16, and 24 h after transfer to methanol. Long and short exposures are shown. Ubiquitinated Pex7 indicated by asterisk. (E) Coimmunoprecipitation of wild-type and overexpressed His-myc-Ub (K48R) cells grown overnight in methanol medium. Lysed cells were subject to immunoprecipitation of HA-Pex7 with α-HA beads. Samples were analyzed by immunoblot to detect the pull-down of Pex7 (α-HA) or myc-Ub (K48R; α-myc). Ubiquitinated species indicated by asterisk.
To further characterize the stability of Pex7, we monitored the expression level of Pex7 in wild-type (PPY12) and various pex mutant strains affecting peroxisome biogenesis. To our surprise, we found that unlike Pex5 and Pex20, Pex7 was not degraded when expressed overnight in methanol medium in Δpex4 and Δpex6 cells, in which receptor recycling is blocked (Figure 1B). Instead, Pex7 accumulated in these two mutants, compared with the relatively low amounts seen in wild-type cells. The Pex7 level was also stabilized in the absence of Pex14, Pex8 and Pex2, components of the docking and RING subcomplexes, respectively. However, Pex7 did not accumulate in the absence of Pex25, a peroxin that is not involved in matrix protein import. These results suggest that Pex7 is subject to degradation as a consequence of its direct engagement in peroxisomal matrix protein import and that blocking any step of the import cycle results in its stabilization.
However, when expressed in oleate medium, Pex7 levels were consistently and equally high in both wild-type and mutant strains (Figure 1C). Although there was some slight accumulation of Pex7 as compared with wild-type cells in Δpex2, Δpex4, and Δpex6 cells, this was much less than that seen in methanol-induced cells. Thus the expression of Pex7 seems to be differentially regulated based on the physiological necessity of the PTS2 import pathway.
Pex7 is ubiquitinated
Although other PTS receptors and coreceptors (P. pastoris Pex5 and Pex20 and S. cerevisiae Pex18) have been shown to be polyubiquitinated during degradation by the RADAR pathway (Purdue and Lazarow, 2001; Léon et al., 2006), no ubiquitination of Pex7 has yet been detected. To investigate whether Pex7 degradation is achieved through ubiquitination, we overexpressed ubiquitin (Ub) mutated at position 48 from lysine to arginine, Ub (K48R), in wild-type cells. This mutation prevents the formation of K48-linked branches of polyubiquitin, a common signal for degradation by the UPS, and thus hinders proteasomal degradation. Cells overexpressing histidine (His)-myc-Ub (K48R) showed stabilization of Pex7 compared with wild-type cells at three separate time points (6, 16, and 24 h) after shift to methanol medium (Figure 1D). Stabilization of Pex7 was also seen in cells overexpressing wild-type His-myc-Ub, especially after 6 h on methanol, but not to the same extent as seen with Ub (K48R). Moreover, a higher–molecular mass band, ∼10 kDa larger than Pex7 (marked with an asterisk), was visible upon overexpression of Ub (K48R). This 10-kDa shift is due to addition of a single His-myc-ubiquitin moiety, which has a molecular mass of ∼10 kDa. However, this species is most likely indicative of polyubiquitinated Pex7 since it is present in cells expressing Ub (K48R) but not Ub. Consistent with this, in methanol, Pex7 is degraded in a proteasome-dependent manner (Supplemental Figure S2), which often relies on K48-linked polyubiquitination. Although this higher–molecular mass band was also present when cells were grown in oleate medium, no added stabilization of Pex7 was seen in cells overexpressing Ub or Ub (K48R; Supplemental Figure S3), consistent with the lack of degradation seen in oleate-induced cells. In addition, we note that because Pex7 was monoubiquitinated in both the presence and absence of expression of Ub (K48R; Supplemental Figure S3), this suggests that the stability of Pex7 in oleate may be due to the lack of polyubiquitination.
To confirm that the higher–molecular mass band represents a ubiquitinated species of Pex7, we immunoprecipitated Pex7 by pull-down with HA antibody. His-myc-Ub (K48R)–overexpressing cells displayed a higher–molecular mass band of Pex7, at ∼60 kDa, which was detected with both anti-HA (detecting Pex7) and anti-myc (detecting Ub) antibodies (Figure 1E, left, asterisk). This higher–molecular mass band was not detected in wild-type cells (Figure 1E, right) and represents a ubiquitinated species of Pex7 that can only be detected when the species is stabilized by blocking proteasomal degradation.
Pex7 degradation is regulated based on physiological need
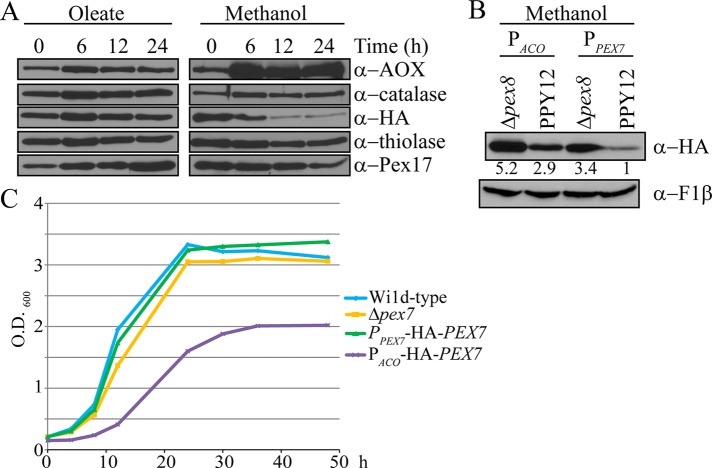
Because Pex7 degradation appeared to be methanol specific, we asked whether a possible remodeling of peroxisomal proteins occurs upon shifting media. Wild-type cells were first induced in oleate medium overnight and then shifted to fresh oleate or methanol medium for growth for up to 24 h (Figure 2A). In cells shifted to methanol medium, proteins required for methanol utilization, such as alcohol oxidase (AOX), were induced, as seen by the greater increase in protein levels compared with the cells shifted to oleate. On the other hand, Pex7 was degraded significantly within 6 h of growth in methanol in wild-type cells but was stable when grown on oleate (Figure 2A). These results suggest that the degradation of Pex7 can be regulated according to physiological need.
FIGURE 2:
Pex7 degradation is regulated based on physiological need. (A) Wild-type cells were induced in oleate medium for 16 h and then transferred to either fresh oleate or methanol medium. Equal-volume samples were taken at the indicated times after transfer and analyzed by immunoblot with the indicated antibodies. (B) Pex7 levels were compared in PPY12 and Δpex8 cells when it was either overexpressed or expressed normally from the PACO or PPEX7 promoter, respectively. Quantification indicates fold change in HA-Pex7 relative to PPY12 + PPEX7-HA-Pex7, normalized to F1β levels. (C) Growth analysis of wild-type cells expressing Pex7 from either the PACO or PPEX7 promoter in methanol medium.
To determine whether there is a physiological need for this degradation, we overexpressed Pex7 from the Acyl-CoA oxidase (ACO) promoter. Overexpressed Pex7, in methanol-grown cells, displayed higher levels of Pex7 in both Δpex7 and Δpex8 background cells as compared with Pex7 expressed from its endogenous promoter, suggesting that Pex7 degradation may be impaired due to such high levels of Pex7 (Figure 2B). Of interest, cells overexpressing Pex7 had reduced growth in methanol medium compared with Pex7 expressed under its endogenous promoter (Figure 2C). This suggests physiological regulation of Pex7 degradation in methanol medium.
Dysfunctional Pex7 activates degradation of Pex7 in oleate
To further characterize the negative correlation between physiological need and Pex7 degradation, we asked whether this degradation pathway could be activated in oleate medium, in which Pex7 is normally stable. To this end, we used the Pex7 (A248R) mutant, which is unable to bind the PTS2 sequence (Elgersma et al., 1998). When grown in methanol, this mutant was degraded in Δpex7 cells but was stabilized in Δpex4 Δpex7 and Δpex7 Δpex8 mutant cells, indicating that it is capable of a full import cycle (Figure 3A). However, in oleate, the Pex7 (A248R) mutant became unstable, unlike wild-type Pex7, but was stabilized in the pex mutant strains (Figure 3B). Similar to wild-type Pex7 in methanol-grown cells, Pex7 (A248R) in oleate-grown cells was mostly degraded in the presence of cycloheximide (Figure 3C), whereas wild-type Pex7 was stable, regardless of cycloheximide treatment. The inability to bind PTS2 proteins renders this mutant nonfunctional and thus physiologically unnecessary, which, in turn, triggers its degradation, via an unknown signaling mechanism. Because thiolase is the main PTS2 cargo protein in P. pastoris, we sought to determine whether the loss of thiolase import in oleate-induced cells was sufficient to activate Pex7 degradation. However, Pex7 was stable in Δpot1 cells (Figure 3D), suggesting that import of all PTS2-containing proteins, such as Pex8 or other unknown PTS2 proteins, may need to be blocked to render Pex7 dispensable.
FIGURE 3:
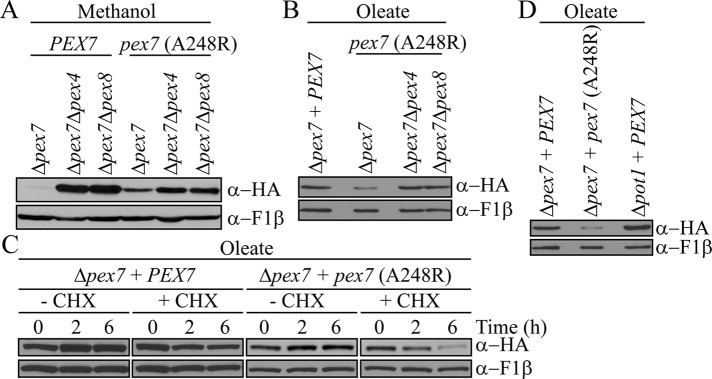
Dysfunctional Pex7 is degraded in oleate. (A, B) Pex7 stability was analyzed in Δpex7, Δpex4 Δpex7, and Δpex7 Δpex8 cells expressing either wild-type Pex7 or the PTS2-binding mutant Pex7 (A248R) grown overnight in methanol (A) or oleate medium (B). (C) Protein turnover of Pex7 and Pex7 (A248R) was compared, after 3 h induction in oleate, by incubation with 5 mg/ml cycloheximide for the indicated amount of time. (D) Pex7 levels were analyzed in Pex7 (A248R) and Δpot1 cells grown overnight in oleate medium.
Pex7 shuttling depends on Pex5 and Pex20
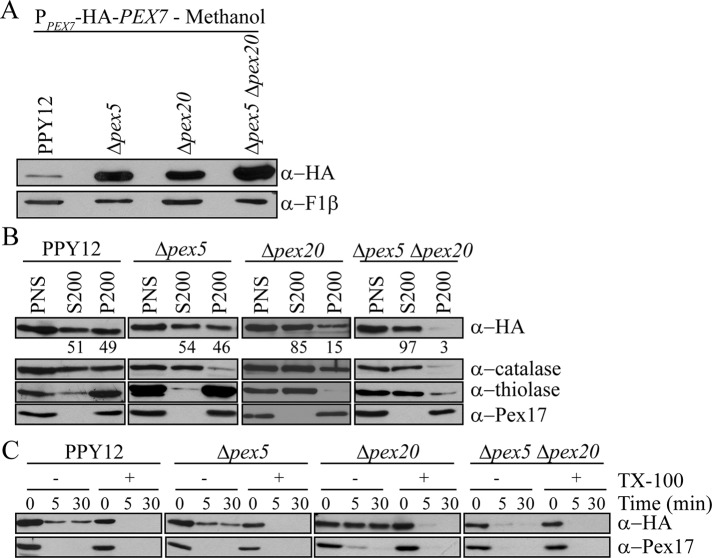
In lower eukaryotes, the PTS1 and PTS2 pathways are believed to be completely independent, whereas in higher eukaryotes, Pex5L has functionally replaced the PTS2 coreceptors, Pex18/21 or Pex20, and is necessary for PTS2 import. Surprisingly, we found that Pex7 accumulated not only in Δpex20 but also in Δpex5 cells after 16-h induction in methanol (Figure 4A). Even greater stabilization of Pex7 was seen in the Δpex5 Δpex20 double mutant compared with each of the single mutants, suggesting synergism between Pex5 and Pex20 in the Pex7 import and degradation process.
FIGURE 4:
Pex7 shuttling depends on Pex5 and Pex20. (A) After overnight growth in methanol medium, Pex7 levels were analyzed in Δpex5, Δpex20, and Δpex5 Δpex20 cells in comparison to wild-type cells. (B) Differential centrifugation of Δpex5, Δpex20, and Δpex5 Δpex20 cells, grown in methanol for 4 h, to determine the subcellular localization of Pex7. The postnuclear supernatant (PNS) was centrifuged at 200,000 × g for 1 h. Equivalent volumes from the resulting supernatant (S200) and pellet (P200) fractions were analyzed by Western blot. Quantification represents the percentage of Pex7 present in each fraction compared with the sum of the amount in the S200 and P200 fractions. (C) Protease protection assay of P200 pellet fractions from B. In B and C, higher exposures are shown for the α-HA blots in wild-type cells due to the instability of Pex7 in this strain.
To characterize the role of Pex5 and Pex20 in the degradation of Pex7, we analyzed the subcellular localization of HA-Pex7 in these mutants by differential centrifugation (Figure 4B) and protease protection assays of the pellet fractions (Figure 4C). To prevent accumulation of alcohol oxidase crystals, which could burst peroxisomes during the experimental handling, cells were induced for 4 h in methanol before cell lysis. By 4 h, Pex7 was stabilized in pex mutant strains at similar levels to 16-h induction (Supplemental Figure S4). Catalase and thiolase were used as PTS1 and PTS2 matrix protein controls, respectively, and Pex17 was used as a peroxisomal membrane protein control. Consistent with its role as a shuttling receptor, in wild-type cells, Pex7 was distributed between the organellar pellet fraction (P200) and the cytosolic fraction (S200; Elgersma et al., 1998). The pool of Pex7 associated with the P200 fraction was protease protected in the absence of the detergent Triton X-100, supporting the extended shuttling receptor model. Pex7 localization was not significantly affected in the absence of Pex5 (Figure 4B); however, because Pex7 is stable, the degradation of Pex7 is somehow still affected. In contrast, loss of Pex20 shifted the majority of Pex7 to the cytosol, suggesting that Pex20 plays a role in the import of Pex7 into peroxisomes (Figure 4B). The small amount of Pex7 still associated with the pellet was protected from protease attack in the absence of detergent, signifying that Pex7 can still enter peroxisomes, although not as efficiently, in the absence of Pex20 (Figure 4C). Furthermore, the deletion of both Pex5 and Pex20 further intensified the cytosolic shift, as almost no Pex7 was found in the pellet (Figure 4B). The small amount of Pex7 associated with the pellet was susceptible to protease treatment in the absence of Triton-X (Figure 4C), suggesting that Pex7 import may be completely abrogated, consistent with the additional stabilization seen in the double mutant (Δpex5 Δpex20). These results suggest that Pex20 is necessary for Pex7 import into peroxisomes, whereas Pex5 is necessary for Pex7 degradation. The somewhat milder effects of the single mutants relative to the double mutant on Pex7 stability and localization suggests Pex5 and Pex20 may work synergistically to allow Pex7 to undergo a complete import cycle.
The recycling of Pex5 and Pex20 is necessary for Pex7 degradation
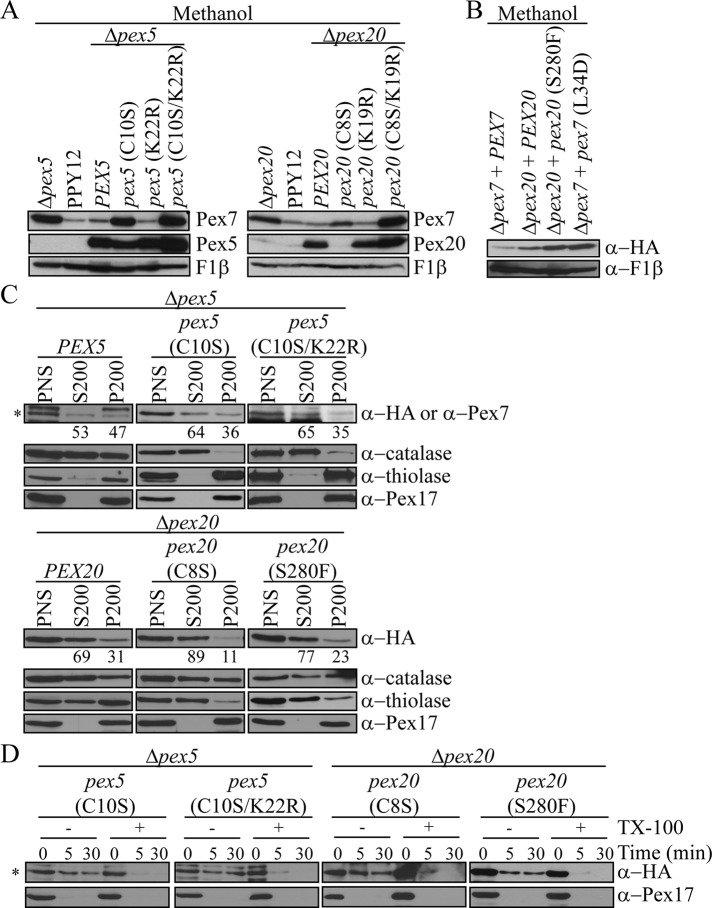
After cargoes dissociate from the PTS receptors in the peroxisome lumen, Pex5 and Pex20 are monoubiquitinated at conserved N-terminal cysteines (C10 and C8, respectively, in P. pastoris) and exported out of peroxisomes to be recycled for another round of import (Léon and Subramani, 2007; Okumoto et al., 2011). However, when these cysteines are mutated or when the receptor recycling machinery is impaired, the receptors are polyubiquitinated at a conserved lysine (K22 on PpPex5 and K19 on PpPex20), resulting in their rapid degradation by the RADAR pathway (Léon et al., 2006; Platta et al., 2007). Mutation of both the cysteine and lysine residues blocks both of these pathways and would result in the stabilization and accumulation of Pex5/20 inside peroxisomes.
We wished to probe further the role of Pex5 and Pex20 in Pex7 shuttling by analyzing Pex7 stability in Pex5 and Pex20 mutants defective in one or both of these export pathways (Figure 5A). Blocking the ability of either Pex5 or Pex20 to recycle resulted in the accumulation of Pex7. However, no accumulation of Pex7 was observed when blocking the degradation pathway of Pex5 or Pex20 alone (Figure 5A). These results signify the importance of the Pex5/Pex20 recycling pathway, but not the degradation pathway, in Pex7 shuttling. Blocking the export of either Pex5 or Pex20 by eliminating both the recycling and degradation pathways for each similarly stabilized Pex7 (Figure 5A). Perhaps Pex7 degradation requires a shared component required for the recycling of both Pex5 and Pex20, and additionally its export and degradation cannot proceed when both recycling and degradation of either Pex5 or Pex20 is blocked.
FIGURE 5:
The recycling of Pex5 and Pex20 is necessary for Pex7 degradation. (A) Pex7 levels were analyzed in Pex5 and Pex20 mutants incapable of recycling and/or degrading after overnight induction in methanol medium. (B) Pex7 levels were analyzed in cells expressing Pex20 (S280F) and Pex7 (L34D) mutants, which disrupt the interaction between Pex7 and Pex20 (Léon et al., 2006; Pan et al., 2013). (C) Differential centrifugation of cells grown for 4 h in methanol expressing Pex5 (C10S), Pex5 (C10S/K22R), Pex20 (C8S), or Pex20 (S280F). For detection of HA-Pex7, all samples were immunoblotted with α-HA, except for pex5 (C10S/K22R) cells, which were immunoblotted with α-Pex7. Asterisk indicates HA-Pex7. In PEX5 cells, the other bands are degradation products of Pex5-HA. In pex5 (C10S/K22R) cells, the other bands are nonspecific binding of the Pex7 antibody. Quantification represents the percentage of Pex7 present in each fraction compared with the sum of the amount in the S200 and P200 fractions. (D) Protease protection assay of P200 pellet fractions from C. Asterisk indicates HA-Pex7. The other bands are degradation products of Pex5-HA.
Pex20 interacts with Pex7 via a C-terminal Pex7-binding domain, with the conserved serine in this region (S280 on PpPex20) being required for the interaction (Léon et al., 2006). Similarly, the crystal structure of the S. cerevisiae Pex7/Pex21 complex has defined a conserved leucine near the N-terminus (L34 in S. cerevisiae and P. pastoris) to be indispensable for complex formation (Pan et al., 2013). Disruption of the interaction between Pex7 and Pex20, by mutating either of the conserved residues on Pex20 or Pex7, resulted in stabilization of Pex7 (Figure 5B). Consistent with the stabilization data, the import of Pex7 into peroxisomes was affected in both Pex20 (C8S) and (S280F) mutants, as seen by the shift of distribution of Pex7 to the cytosol compared with wild-type Pex20 cells (Figure 5C). However, just as was seen in the Δpex20 mutant, small amounts of Pex7 were still protease protected, indicating that they are intraperoxisomal and capable of partial import (Figure 5D). Thus loss of interaction between Pex7 and Pex20, either through deletion or mutation of Pex20, causes a defect in the import of Pex7. On the other hand, in both Pex5 (C10S) and (C10S/K22R) mutants, Pex7 localization was mostly unchanged compared with wild-type Pex5 cells. Thus, although in the absence of Pex5 the relative subcellular distribution of Pex7 is not affected, meaning that Pex7 can enter and exit peroxisomes, Pex7 degradation is specifically dependent on the recycling capability and export of Pex5.
The docking complex and receptor recycling machinery are necessary for proper localization of Pex7
To better characterize the dynamics of Pex7 shuttling in methanol medium, we performed subcellular fractionation and protease protection assays with Δpex14, Δpex2, and Δpex4 cells (Figure 6, A and B). In the absence of the docking protein Pex14, Pex7 was localized predominantly in the cytosol with the small amount of peroxisomally associated Pex7 susceptible to protease degradation, suggesting that Pex14 is crucial for the entry of Pex7 into peroxisomes, as expected. Similarly, in the absence of Pex4, Pex7 shifted mostly to the cytosolic fraction, although the small amount of peroxisome-associated Pex7 was partially protease protected, indicating a defect in Pex7 import. This result is not surprising in view of the known instability of both Pex5 and Pex20 in Δpex4 cells and the data given earlier that these two proteins are necessary for Pex7 instability and localization.
FIGURE 6:
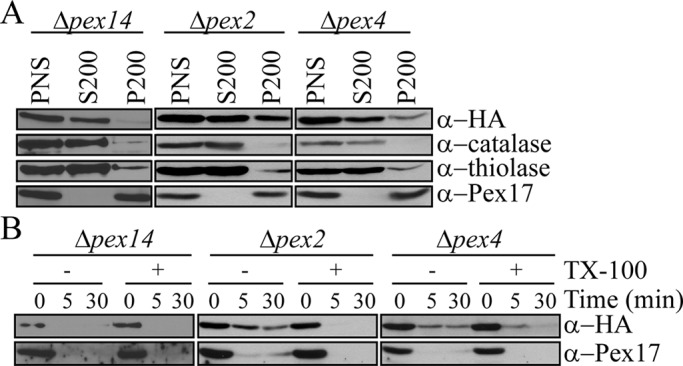
The docking complex and receptor recycling machinery are necessary for proper localization of Pex7. (A) Differential centrifugation of Δpex14, Δpex4, and Δpex2 cells grown in methanol for 4 h. The postnuclear supernatant (PNS) was centrifuged at 200,000 × g for 1 h. Equivalent volumes from the resulting supernatant (S200) and pellet (P200) fractions were analyzed by Western blot. (B) Protease protection assays of P200 pellet fractions from A.
Knockout of any of the RING E3 ligases results in instability of the remaining E3 ligases (Hazra et al., 2002); thus Δpex2 cells were considered as functional deletions of Pex2, Pex10, and Pex12. The peroxisomal E3 ligases are needed for both monoubiquitination and polyubiquitination of both Pex5 and Pex20 in P. pastoris (Platta et al., 2009; Liu and Subramani, 2013). Furthermore, our earlier studies showed that Pex5 and Pex20 are stuck in the peroxisomal lumen in the absence of Pex2 (Léon et al., 2006; Zhang et al., 2006). Taken together, these data suggest that the stabilization seen in the absence of Pex2 (Figure 1C) may be due to the inability for Pex5 and Pex20 to be recycled, which we showed here results in Pex7 stabilization.
Pex4 and Pex2 are not essential for the monoubiquitination of Pex7
Because Pex4 and the peroxisomal RING E3 ligases Pex2, Pex10 and Pex12 have been implicated in the ubiquitination of Pex5 and Pex20 (Williams et al., 2008, 2012; Platta et al., 2009; Liu and Subramani, 2013), we asked whether they are also involved in the monoubiquitination or polyubiquitination of Pex7. In methanol, when Ub (K48R) was overexpressed in Δpex14, Δpex2, and Δpex4 cells, only Δpex14 cells displayed a lack of Pex7 ubiquitination due to the complete blockage of entry into peroxisomes. However, in Δpex2 and Δpex4 cells expressing Ub (K48R) (Figure 7A), monoubiquitinated Pex7 was seen, suggesting that a different set of E2 and E3 ligases is involved in Pex7 monoubiquitination (Figure 7A). Although the other known peroxisomal E3 ligases, Pex10 and Pex12, are unstable in Δpex2 cells, we further confirmed they are also unnecessary for Pex7 monoubiquitination, using the RING domain mutants Pex10 (C313S/C316S) and Pex12 (C339S/C342S; Figure 7B). Because Pex7 is stable in these strains, presence of an ubiquitinated species suggests Pex7 is monoubiquitinated rather than polyubiquitinated; however, it is also possible that Pex7 is polyubiquitinated but degradation is blocked at a later unknown stage.
FIGURE 7:
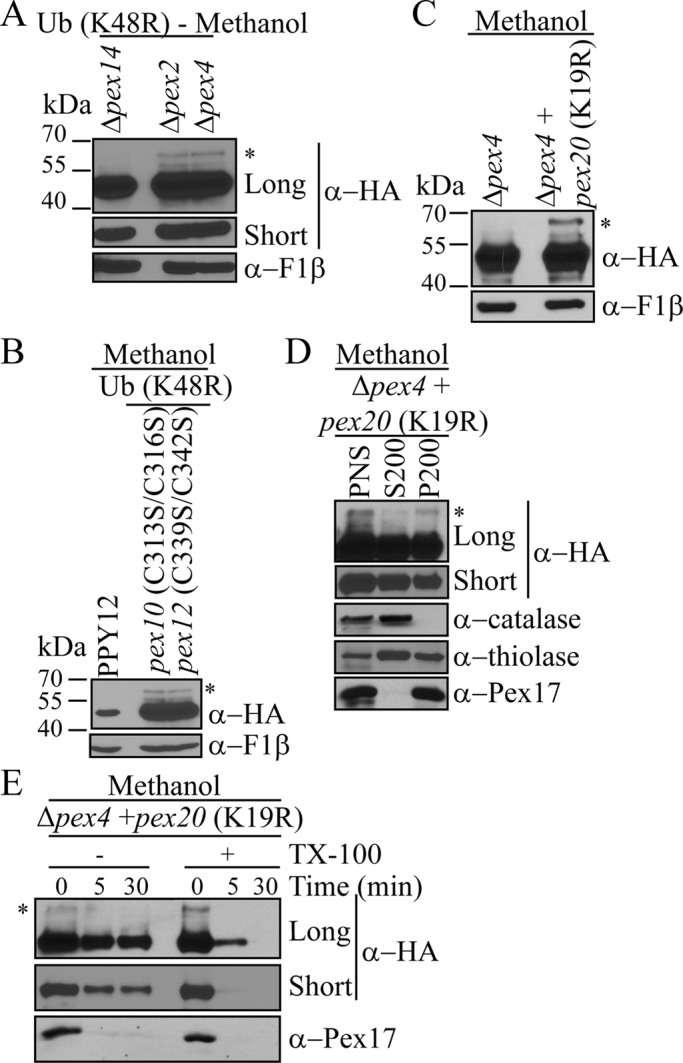
Pex2 and Pex4 are not essential for Pex7 monoubiquitination. (A, B) Strains overexpressing His-myc-Ub (K48R) with the indicated pex deletions (A) or point mutations (B) were induced overnight in methanol. Equal-OD samples were collected and analyzed by immunoblotting for the presence or absence of Pex7 ubiquitination. (C) Pex7 levels were compared in Δpex4 + pex20 (K19R) cells after overnight growth in methanol. (D) Differential centrifugation of Δpex4 + pex20 (K19R) cells grown in methanol for 4 h. (E) Protease protection assay of P200 in (D). In D and E, long and short exposures are shown for α-HA. Asterisks indicate ubiquitinated HA-Pex7.
Because any effects on Pex7 seen in Δpex4 cells could be due to the instability of Pex5 and Pex20, we analyzed Pex7 levels in Δpex4 cells expressing Pex20 (K19R), which cannot be polyubiquitinated and therefore is stabilized. Of interest, endogenous levels of monoubiquitinated Pex7 were detectable after overnight growth in methanol (Figure 7C). As seen by subcellular fractionation and protease protection assays (Figure 7, D and E), stabilizing Pex20 in the absence of Pex4 rescued the Pex7 import defect seen in Δpex4 cells (Figure 6, A and B). In this strain, Pex7 entered peroxisomes wherein it was monoubiquitinated in a Pex4-independent manner. In this sense, monoubiquitination of Pex7 behaves distinctly from Pex5 and Pex20 in its lack of dependence on the E2 and E3 peroxins. Of interest, because the relative distribution of Pex7 in the pellet and supernatant fractions of Δpex2 cells (Figure 6A), as well as in Δpex4 cells expressing Pex20 (K19R; Figure 7D), is essentially similar to that observed in wild-type cells (Figure 4B), the monoubiquitination of Pex7 may be sufficient to shuttle it from peroxisomes to the cytosol, very much like the recycling of Pex5 and Pex20 (Ma et al., 2011).
However, because Pex7 was stabilized in cells either mutated or deleted for the RING E3 ligases or Pex4, we infer that polyubiquitination of Pex7 is not happening in these mutant cells, preventing us from ruling out the possibility that Pex4 and peroxisomal RING E3 ligases may be involved in polyubiquitination of Pex7.
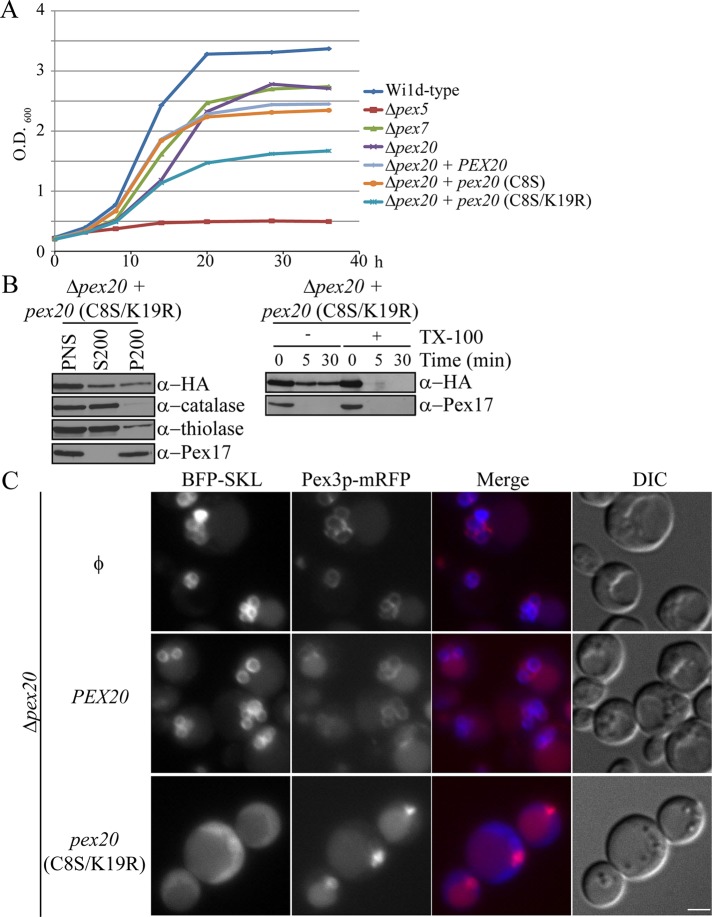
Pex20 (C8S/K19R) blocks both the PTS1 and PTS2 pathways
Unexpectedly, we noticed that the Pex20 (C8S/K19R) strain had a partial growth defect when cells were induced in methanol medium (Figure 8A). Because the PTS2 pathway is not required in methanol medium, the biogenesis defect caused by Pex20 (C8S/K19R) most likely represents a defect in PTS1 import. This was indeed the case, as catalase was mislocalized to the cytosolic fraction (Figure 8B). To further prove that Pex20 (C8S/K19R) blocks the PTS1 pathway, we expressed the PTS1 cargo protein BFP-SKL in Δpex20 cells transformed with wild-type or mutant Pex20 (Figure 8C). In Δpex20 cells and those expressing wild-type Pex20, BFP-SKL colocalized with Pex3-RFP. However, in cells expressing Pex20 (C8S/K19R), BFP-SKL was mislocalized in a diffuse pattern in the cytosol. The nonfunctional Pex20 (C8S/K19R) mutant, which is incapable of export from peroxisomes, accumulates at the peroxisome membrane (Léon and Subramani, 2007) and may block the export of other proteins as well. Indeed, in this mutant, Pex7 remained peroxisomally associated and protease protected in this mutant, suggesting that stabilization may be due to a late-stage defect in the degradation process (Figure 8B).
FIGURE 8:
Pex20 (C8S/K19R) blocks both the PTS1 and PTS2 pathways. (A) Growth curve of the indicated deletion or mutant strains induced in methanol medium. (B) Differential centrifugation and protease protection assay of cells harboring the Pex20 (C8S/K19R) mutation and grown in methanol for 4 h. (C) Fluorescence microscopy of Δpex20 cells untransformed (Φ) or transformed with wild-type or mutant Pex20 (C8S/K19R) grown overnight in methanol medium. Pex3-mRFP was used to label peroxisomes, and BFP-SKL was used to track PTS1 protein import. DIC; differential interference contrast. Scale bar, 2 μm.
DISCUSSION
Pex7 shuttling to and from peroxisomes requires Pex5 and Pex20
In this study, the shuttling pathway of Pex7 has been elucidated. Unlike Pex5 and Pex20, which have conserved cysteine and lysine residues near their N-termini that can be monoubiquitinated or polyubiquitinated for recycling and degradation, respectively (Léon et al., 2006; Platta et al., 2007, 2009; Liu and Subramani, 2013). Pex7 cannot shuttle between the cytosol and the peroxisome lumen by itself. Pex7 entry into peroxisomes depends on Pex20, and to a lesser extent on Pex5, because in Δpex20 cells, Pex7 is mostly cytosolic (Figure 4B), compared with the roughly equal peroxisomal and cytosolic localization seen in wild-type cells (Figure 4B), although the small pellet-associated fraction is protease protected (Figure 4C). Supporting this conclusion is the observation that in Δpex5 cells, Pex7 distribution and protease protection are largely unaffected (Figure 4, B and C). Pex7 entry and exit from peroxisomes does not require a recycling Pex5 as long as Pex20 is present, as reflected by the insignificant change in the relative subcellular locations of Pex7 in the cytosol versus the peroxisome pellet (Figure 5C) and the protease-protected nature of the Pex7 associated with the pellet fractions in the Pex5 (C10S) and Pex5 (C10S/K22R) mutants (Figure 5D). Consistent with this idea, the import of Pex7 was completely abolished only when both Pex5 and Pex20 were eliminated (Figure 4, B and C). Together these data suggest that Pex5 and Pex20 may be acting synergistically, as both are required for efficient import of Pex7.
Pex7 was significantly stabilized when the recycling of either Pex5 or Pex20 was compromised using the Pex5 (C10S) or Pex20 (C8S) cells, respectively (Figure 5A), or the respective double mutants. However, inhibition of Pex20 recycling, using Pex20 (C8S), or prevention of Pex7-Pex20 interaction, using Pex20 (S280F), appeared to have a more dramatic effect on Pex7 import and association with the pellet fraction than did the Pex5 (C10S) mutant (Figure 5B). Because of the greater instability of Pex20 (C8S) compared with Pex5 (C10S) (Figure 5A), it is not surprising that the Pex20 (C8S) mutant behaves essentially like Δpex20 cells with respect to Pex7 stability. The instability of Pex20 (C8S) and the necessity of Pex20 interaction with Pex7 for Pex7 import make it hard to tease out whether Pex20 also has a significant effect on Pex7 export from peroxisomes. However, Pex5 (and likely Pex20) recycling definitely appears to be necessary for Pex7 instability, suggesting that Pex7 degradation by the UPS may be coupled to the recycling of Pex5 (and Pex20). Further work needs to be done to define the precise roles these peroxins play in the different aspects of Pex7 shuttling.
We found that Pex7 can be monoubiquitinated (in oleate, where it is necessary for growth and not degraded, or in mutants of E2 and E3 peroxins) and polyubiquitinated (in methanol-grown cells, where it is dispensable). Although the use of Ub (K48R) impeded direct detection of polyubiquitinated Pex7, our data suggest that differential monoubiquitination versus polyubiquitination of Pex7 may dictate its stability, as is the case for Pex5 and Pex20 (Ma et al., 2011). Our work suggests that monoubiquitinated Pex7 serves as a signal for its shuttling and equilibration roughly equally between the organelle pellet and cytosolic fractions, as seen in wild-type cells. In contrast, the polyubiquitination of Pex7 targets it for proteasomal degradation after export with recycled Pex5 or Pex20.
Other pex mutants, such as Δpex14 and Δpex4, affect the stability of Pex7 and its entry into peroxisomes, as shown by subcellular localization and protease protection assays. However, these peroxins may have an indirect role in the shuttling of Pex7, since they are directly involved in the shuttling of Pex5 and Pex20 (Léon et al., 2006; Zhang et al., 2006). Stabilization of Pex20 in the absence of Pex4 leads to accumulation of endogenous monoubiquitinated Pex7 (Figure 7C), demonstrating that Pex4 is not required for Pex7 monoubiquitination. However, because we were only able to detect monoubiquitination of Pex7, either in this strain or in strains overexpressing Ub (K48R), we cannot exclude the possibility that Pex4 is required for extension of the ubiquitin chain, as reported for Pex20 ubiquitination (Liu and Subramani, 2013). Recently a proteomic analysis in transgenic Arabidopsis expressing green fluorescent protein (GFP)-tagged PEX7 revealed that the GTPase RabE1c binds PEX7 and facilitates its proteasomal degradation when GFP-PEX7 accumulated at the peroxisome membrane, but since this behavior was uncovered via use of a GFP fusion, its physiological role is unclear (Cui et al., 2013).
Pex7 is constitutively degraded when it is not needed
The PTS2 import pathway is absent in Caenorhabditis elegans and the model diatom Phaeodactylum tricornutum, suggesting an evolutional selection pressure over the PTS2-related components (Motley et al., 2000; Gonzalez et al., 2011). In P. pastoris, the PTS2 pathway is essential only when cells are grown in oleate medium but not in methanol medium (Elgersma et al., 1998; Léon et al., 2006). We found that Pex7 is polyubiquitinated and degraded constitutively via the UPS when P. pastoris cells are grown in methanol medium. Furthermore, the Pex7 degradation pathway can be activated when cells are transferred from oleate medium to methanol medium. Degradation of Pex7 in methanol medium is probably important not only for removing this unnecessary protein, but also for suppressing the PTS2 pathway to allow for the peroxisomal importomer to become dedicated to the import of essential PTS1 proteins, such as AOX, which can constitute up to 30% of the cellular proteins, which are needed for survival in methanol medium (Lin-Cereghino et al., 2006). Moreover, we found that damaged or nonfunctional Pex7, which can be mimicked by the Pex7 PTS2-binding mutant, can be selectively recognized for degradation in oleate media. Pex18 similarly has been found to be degraded constitutively, but it is degraded in oleate medium, a condition in which the PTS2 pathway is essential (Purdue and Lazarow, 2001; Hensel et al., 2011). It is not known whether the degradation of Pex18 has any physiological effect, especially because, in S. cerevisiae, Pex21 serves a redundant function with Pex18 (Purdue and Lazarow, 2001), so perhaps Pex18 is degraded constitutively because it is dispensable. To some extent, the degradation of Pex7 found in methanol medium in P. pastoris is similar to the peroxisome-associated matrix protein degradation observed in Arabidopsis seedlings, where the obsolete proteins of the glyoxylate cycle, malate synthase and isocitrate lyase, are degraded and replaced by photorespiration enzymes (Lingard et al., 2009). Our study illustrates how the PTS2 receptor, Pex7, is subject to environmentally regulated remodeling and how damaged Pex7 undergoes turnover.
Inherent interconnection between the PTS1 and PTS2 pathways
In mammals and plants, the PTS2 pathway has merged with the PTS1 pathway, since the longer isoform of Pex5 (Pex5L), which is orthologous to Pex18/21 or Pex20, interacts with Pex7 (Dodt et al., 2001). However, it has been suggested for a long time that the PTS1 and PTS2 pathways are completely separate in lower eukaryotic cells. Our data suggest that P. pastoris Pex5 has an unexpected role in Pex7 shuttling, suggesting the PTS1 and PTS2 pathways are interconnected, even in fungi. Consistent with this, in the yeast Hansenula polymorpha, Pex5 and Pex20 have been shown to form a complex detectable by electron microscopy (Moscicka et al., 2007). Although this complex is capable of binding catalase, its function and physiological significance are unknown. Furthermore, we found that the accumulation of Pex20 (C8S/K19R) in peroxisomes not only blocks the degradation of Pex7 and the import of PTS2 proteins, but also has a deleterious effect on the targeting of PTS1 proteins, probably by blocking the export of Pex5. To the best of our knowledge, these data suggest for the first time that the PTS1 and PTS2 receptors may use the same export machineries after releasing cargo in the peroxisome lumen.
MATERIALS AND METHODS
Yeast strains and culture conditions
P. pastoris strains used in this study are listed in Supplemental Table S1, and all represent genomic integrations. Yeast cells were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose) overnight at 30°C. Cultured cells were diluted in YPD and grown for 6–7 h. Cells were washed in sterile distilled water and transferred to either methanol (YNM: 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 0.05% yeast extract, 0.79 g/l complete amino acids, 0.5% methanol) or oleate medium (YNO: 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 0.05% yeast extract, 0.79 g/l complete amino acids, 0.2% oleic acid, 0.02% Tween 40). Unless otherwise noted, cells were grown for ∼16 h at 30°C. For analysis of protein levels by Western blot, equal-OD samples were removed and trichloroacetic acid precipitated.
Plasmid construction
The PpPEX7 promoter was cloned into the pIB1 vector at XmaI/SpeI sites, creating pCM93. A fragment containing a tandem HA at the N-terminal of PpPex7 was amplified using pHA7 (a gift of William Snyder; lab stock) as the template and cloned downstream of the PpPEX7 promoter in pCM93 at SpeI/HindIII sites, creating pCM95. A Zeocin cassette was amplified from pMY69 (a gift of Mingda Yan; lab stock) and cloned upstream of the PpPEX7 promoter in pCM95 at the SmaI site, generating pCM255. The pex7 mutants (L34D and A248R) were created using site-directed mutagenesis based on pCM95.
Cycloheximide assay
Yeast cells were grown in YPD to exponential growth and transferred to methanol or oleate medium at a starting OD600 of 1.0. After 3 h of growth at 30°C, cycloheximide (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 5 mg/ml and, when indicated, MG-132 (UBPBio, Aurora, CO) to a final concentration of 1 mM. Cells continued to grow at 30°C for 6 h, during which equal-OD samples were taken at the indicated time points to be analyzed by Western blot.
Immunoprecipitation
Cells were grown in YPD to exponential growth and transferred to methanol medium at a starting OD600 of 1 for growth overnight. Immunoprecipitations were performed as previously described (Farré et al., 2010), except that the input supernatant was incubated overnight with 40 μl of EZ View Red Anti-HA Affinity Gel (Sigma-Aldrich), followed by three washes with 1 ml of IP lysis buffer (50 mM HEPES-NaOH [pH 8], 0.15 M NaCl, 0.5% Triton X-100, 10% glycerol, 5 mM NaF, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail) supplemented with 100 mM N-ethylmaleimide and 100 μM MG-132 for 1 min.
Subcellular fractionation and protease protection
Cells were grown in YPD overnight to an OD600 of ∼2.0. Cells were washed with sterile distilled water twice and transferred to methanol medium. After 4 h of growth at 30°C, cells were harvested by centrifugation at 3000 × g for 10 min and washed with distilled water twice. To create spheroplasts, 0.5 mg/g cells Zymolyase 100T (Nacalai Tesque, San Diego, CA) was added, and cells were incubated for 30 min at 30°C with gentle rotation. Spheroplasts were harvested by centrifugation at 2000 × g for 10 min at 4°C. The pellet was resuspended in homogenization buffer (5 mM 2-(N-morpholino)ethanesulfonic acid/KOH, pH 5.5, 1 M sorbitol, 12.5 μg/ml leupeptin, 5 μg/ml aprotinin, yeast protease inhibitor cocktail, 5 mM NaF). Cells were broken with 20 strokes in a Dounce homogenizer. The homogenate was spun at 2000 × g for 10 min at 4°C to pellet unbroken cells. The postnuclear supernatant was subject to centrifugation for 1 h at 200,000 × g at 4°C. Equivalent volumes of the resulting supernatant (S200) and pellet (P200) were analyzed by Western blot.
Protease protection assays performed on the pellet fraction obtained were done as previously described (Ma et al., 2009).
Fluorescence microscopy
YPD grown cells were transferred to methanol and grown overnight at 30°C. Images were taken using a Zeiss AxioSkop fluorescence microscope (AxioSkop 2 Plus, motorized) coupled to a cooled charge-coupled device monochrome camera (AxioCam MRM; Zeiss) and analyzed using AxioVision 4 software.
Quantification
Densitometry analysis was performed using ImageJ (National Institutes of Health, Bethesda, MD).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant DK41737 to S.S. We thank former Subramani lab members William Snyder, Xueqian Liu, and Mingda Yan for plasmids and strains.
Abbreviations used:
- ACO
acyl-CoA oxidase
- AOX
alcohol oxidase
- PTS
peroxisomal targeting signal
- RADAR
receptor accumulation and degradation in the absence of recycling
- Ub
ubiquitin
- UPS
ubiquitin-proteasome system
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-12-0716) on July 9, 2014.
*These authors contributed equally to this work.
REFERENCES
- Collins CS, Kalish JE, Morrell JC, McCaffery JM, Gould SJ. The peroxisome biogenesis factors Pex4p, Pex22p, Pex1p, and Pex6p act in the terminal steps of peroxisomal matrix protein import. Mol Cell Biol. 2000;20:7516–7526. doi: 10.1128/mcb.20.20.7516-7526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Fukao Y, Mano S, Yamada K, Hayashi M, Nishimura M. Proteomic analysis reveals that the Rab GTPase RabE1c is involved in the degradation of the peroxisomal protein receptor PEX7 (peroxin 7) J Biol Chem. 2013;288:6014–6023. doi: 10.1074/jbc.M112.438143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debelyy MO, Platta HW, Saffian D, Hensel A, Thoms S, Meyer HE, Warscheid B, Girzalsky W, Erdmann R. Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J Biol Chem. 2011;286:28223–28234. doi: 10.1074/jbc.M111.238600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G, Warren D, Becker E, Rehling P, Gould SJ. Domain mapping of human PEX5 reveals functional and structural similarities to Saccharomyces cerevisiae Pex18p and Pex21p. J Biol Chem. 2001;276:41769–41781. doi: 10.1074/jbc.M106932200. [DOI] [PubMed] [Google Scholar]
- Elgersma Y, Elgersma-Hooisma M, Wenzel T, McCaffery JM, Farquhar MG, Subramani S. A mobile PTS2 receptor for peroxisomal protein import in Pichia pastoris. J Cell Biol. 1998;140:807–820. doi: 10.1083/jcb.140.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Magraoui F, Brinkmeier R, Schrötter A, Girzalsky W, Müller T, Marcus K, Meyer HE, Erdmann R, Platta HW. Distinct ubiquitination cascades act on the peroxisomal targeting signal type 2 co-receptor Pex18p. Traffic. 2013;12:1290–301. doi: 10.1111/tra.12120. [DOI] [PubMed] [Google Scholar]
- Facciotti F, Ramanjaneyulu GS, Lepore M, Sansano S, Cavallari M, Kistowska M, Forss-Petter S, Ni G, Colone A, Singhal A, et al. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat Immunol. 2012;13:474–480. doi: 10.1038/ni.2245. [DOI] [PubMed] [Google Scholar]
- Farré JC, Mathewson RD, Manjithaya R, Subramani S. Roles of Pichia pastoris Uvrag in vacuolar protein sorting and the phosphatidylinositol 3-kinase complex in phagophore elongation in autophagy pathways. Autophagy. 2010;6:86–99. doi: 10.4161/auto.6.1.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez NH, Felsner G, Schramm FD, Klingl A, Maier UG, Bolte K. A single peroxisomal targeting signal mediates matrix protein import in diatoms. PLoS One. 2011;6:e25316. doi: 10.1371/journal.pone.0025316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, McCollum D, Spong AP, Heyman JA, Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- Grou CP, Francisco T, Rodrigues TA, Freitas MO, Pinto MP, Carvalho AF, Domingues P, Wood SA, Rodríguez-Borges JE, Sá-Miranda C, et al. Identification of ubiquitin-specific protease 9X (USP9X) as a deubiquitinase acting on ubiquitin-peroxin 5 (PEX5) thioester conjugate. J Biol Chem. 2012;287:12815–12827. doi: 10.1074/jbc.M112.340158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra PP, Suriapranata I, Snyder WB, Subramani S. Peroxisome remnants in pex3D cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic. 2002;3:560–574. doi: 10.1034/j.1600-0854.2002.30806.x. [DOI] [PubMed] [Google Scholar]
- Hensel A, Beck S, El Magraoui F, Platta HW, Girzalsky W, Erdmann R. Cysteine-dependent ubiquitination of Pex18p is linked to cargo translocation across the peroxisomal membrane. J Biol Chem. 2011;286:43495–43505. doi: 10.1074/jbc.M111.286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Agrawal G, Subramani S. Phosphorylation-dependent Pex11p and Fis1p interaction regulates peroxisome division. Mol Biol Cell. 2012;23:1307–1315. doi: 10.1091/mbc.E11-09-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S, Subramani S. A conserved cysteine residue of Pichia pastoris Pex20p is essential for its recycling from the peroxisome to the cytosol. J Biol Chem. 2007:282, 7424–7430. doi: 10.1074/jbc.M611627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S, Zhang L, McDonald WH, Yates J, Cregg JM, Subramani S. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J Cell Biol. 2006;172:67–78. doi: 10.1083/jcb.200508096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Cereghino GP, Godfrey L, de la Cruz BJ, Johnson S, Khuongsathiene S, Tolstorukov I, Yan M, Lin-Cereghino J, Veenhuis M, Subramani S. Mxr1p, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris. Mol Cell Biol. 2006;26:883–897. doi: 10.1128/MCB.26.3.883-897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Monroe-Augustus M, Bartel B. Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:4561–4566. doi: 10.1073/pnas.0811329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Subramani S. Unique requirements for mono- and polyubiquitination of the peroxisomal targeting signal co-receptor, Pex20. J Biol Chem. 2013;288:7230–7240. doi: 10.1074/jbc.M112.424911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Agrawal G, Subramani S. Peroxisome assembly: matrix and membrane protein biogenesis. J Cell Biol. 2011;193:7–16. doi: 10.1083/jcb.201010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Schumann U, Rayapuram N, Subramani S. The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol Biol Cell. 2009;20:3680–3689. doi: 10.1091/mbc.E09-01-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata N, Okumoto K, Mukai S, Noguchi M, Fujiki Y. AWP1/ZFAND6 functions in Pex5 export by interacting with cys-monoubiquitinated Pex5 and Pex6 AAA ATPase. Traffic. 2012;13:168–183. doi: 10.1111/j.1600-0854.2011.01298.x. [DOI] [PubMed] [Google Scholar]
- Moscicka KB, Klompmaker SH, Wang D, van der Klei IJ, Boekema EJ. The Hansenula polymorpha peroxisomal targeting signal 1 receptor, Pex5p, functions as a tetramer. FEBS Lett. 2007;581:1758–1762. doi: 10.1016/j.febslet.2007.03.061. [DOI] [PubMed] [Google Scholar]
- Motley AM, Hettema EH, Ketting R, Plasterk R, Tabak HF. Caenorhabditis elegans has a single pathway to target matrix proteins to peroxisomes. EMBO Rep. 2000;1:40–46. doi: 10.1093/embo-reports/kvd010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair DM, Purdue PE, Lazarow PB. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J Cell Biol. 2004;167:599–604. doi: 10.1083/jcb.200407119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K, Misono S, Miyata N, Matsumoto Y, Mukai S, Fujiki Y. Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic. 2011;12:1067–1083. doi: 10.1111/j.1600-0854.2011.01217.x. [DOI] [PubMed] [Google Scholar]
- Otzen M, Wang D, Lunenborg MG, van der Klei IJ. Hansenula polymorpha Pex20p is an oligomer that binds the peroxisomal targeting signal 2 (PTS2) J Cell Sci. 2005;118:3409–3418. doi: 10.1242/jcs.02463. [DOI] [PubMed] [Google Scholar]
- Pan D, Nakatsu T, Kato H. Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p. Nat Struct Mol Biol. 2013;20:987–993. doi: 10.1038/nsmb.2618. [DOI] [PubMed] [Google Scholar]
- Platta HW, El Magraoui F, Bäumer BE, Schlee D, Girzalsky W, Erdmann R. Pex2 and Pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol. 2009;29:5505–5516. doi: 10.1128/MCB.00388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, El Magraoui F, Schlee D, Grunau S, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol. 2007;177:197–204. doi: 10.1083/jcb.200611012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB. Pex18p is constitutively degraded during peroxisome biogenesis. J Biol Chem. 2001;276:47684–47689. doi: 10.1074/jbc.M106823200. [DOI] [PubMed] [Google Scholar]
- Purdue PE, Yang X, Lazarow PB. Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J Cell Biol. 1998;143:1859–1869. doi: 10.1083/jcb.143.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Smith JJ, Szilard RK, Rachubinski RA. Pex20p of the yeast Yarrowia lipolytica is required for the oligomerization of thiolase in the cytosol and for its targeting to the peroxisome. J Cell Biol. 1998;142:403–420. doi: 10.1083/jcb.142.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, van den Berg M, Geers E, Distel B. Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun. 2008;374:620–624. doi: 10.1016/j.bbrc.2008.07.054. [DOI] [PubMed] [Google Scholar]
- Williams C, van den Berg M, Panjikar S, Stanley WA, Distel B, Wilmanns M. Insights into ubiquitin-conjugating enzyme/co-activator interactions from the structure of the Pex4p:Pex22p complex. EMBO J. 2012;31:391–402. doi: 10.1038/emboj.2011.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell. 2005;16:573–583. doi: 10.1091/mbc.E04-05-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, Tee AR, Tait-Mulder J, Di Nardo A, Han JM, et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat Cell Biol 15, 1186–1196. 2013 doi: 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Léon S, Subramani S. Two independent pathways traffic the intraperoxisomal peroxin PpPex8p into peroxisomes: mechanism and evolutionary implications. Mol Biol Cell. 2006;17:690–699. doi: 10.1091/mbc.E05-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.