Abstract
Bacterial surface components, especially exopolysaccharides, in combination with bacterial Quorum Sensing signals are crucial for the formation of biofilms in most species studied so far. Biofilm formation allows soil bacteria to colonize their surrounding habitat and survive common environmental stresses such as desiccation and nutrient limitation. This mode of life is often essential for survival in bacteria of the genera Mesorhizobium, Sinorhizobium, Bradyrhizobium, and Rhizobium. The role of biofilm formation in symbiosis has been investigated in detail for Sinorhizobium meliloti and Bradyrhizobium japonicum. However, for S. fredii this process has not been studied. In this work we have demonstrated that biofilm formation is crucial for an optimal root colonization and symbiosis between S. fredii SMH12 and Glycine max cv Osumi. In this bacterium, nod-gene inducing flavonoids and the NodD1 protein are required for the transition of the biofilm structure from monolayer to microcolony. Quorum Sensing systems are also required for the full development of both types of biofilms. In fact, both the nodD1 mutant and the lactonase strain (the lactonase enzyme prevents AHL accumulation) are defective in soybean root colonization. The impairment of the lactonase strain in its colonization ability leads to a decrease in the symbiotic parameters. Interestingly, NodD1 together with flavonoids activates certain quorum sensing systems implicit in the development of the symbiotic biofilm. Thus, S. fredii SMH12 by means of a unique key molecule, the flavonoid, efficiently forms biofilm, colonizes the legume roots and activates the synthesis of Nod factors, required for successfully symbiosis.
Introduction
Rhizobia are bacteria that interact with the roots of leguminous plants to develop symbiotic nodules when environmental nitrogen is limited. In these symbiotic structures atmospheric nitrogen is reduced to ammonium making it available to the plant. This process requires the exchange of molecular signals between both members of the symbiosis. Legume roots exude flavonoids which induce, via a mechanism involving the NodD family of transcription activators, the biosynthesis and secretion of strain-specific lipo-chito-oligosaccharides, also known as nodulation factors (Nod factors), which trigger the initiation of nodule organogenesis [1]. Plant flavonoids, besides inducing Nod factor production, attract the bacteria to the legume root [2]; induce type III secretion machinery via NodD1 by activation of the transcriptional regulator TtsI [3], [4]; and activate the rhizobial Quorum Sensing (QS) systems [5].
QS is defined as a regulation mode of bacterial gene expression in response to changes in their population density, which is mediated by small diffusible signal molecules called autoinducers (AI) [6], [7]. QS-regulated genes are involved in adaptive changes in the physiology of the bacterial population, which modify their behaviour when a certain cell density is reached. QS modulates a broad variety of phenotypes, such as biofilm formation, toxin and exopolysaccharide production, virulence, plasmid transfer and motility, which usually are essential for successful establishment of a symbiotic or a pathogenic relationship with the eukaryotic hosts [8], [9], [10], [11], [12]. In plant-associated bacteria QS coordinates the expression of genes involved in virulence, colonization and symbiosis [13].
In all species studied so far, bacterial surface components (flagella, lipopolysaccharides and especially exopolysaccharides) in combination with the presence of bacterial QS signals are crucial for the formation of biofilms [12]. Biofilms are defined as bacterial communities surrounded by a self-produced polymeric matrix, and attached to an inert or a biotic surface [14]. In the course of biofilm formation, the initial reversible attachment to the surface is followed by an irreversible attachment and multiplication of the bacteria forming microcolonies that develop mature communities with a three-dimensional structure, in some cases permeated by channels, which act as the biofilm circulatory system. All these processes are coordinated by bacterial QS systems [14], [15]. Many rhizobia have been described as forming microcolonies or biofilms when they colonize legume roots. These biofilms are mainly composed of water and bacterial cells. However, the three-dimensional structure of the biofilm is due to an extracellular matrix, which is formed by exopolysaccharides (EPS) [16]. In S. meliloti, the biofilm formation is affected and/or regulated by nutritional and environmental conditions [17], [18]; exopolysaccharides and flagella [19]; ExoR with the ExoS–ChvI two-component system [20]; Nod factors synthesized by nod genes [19]; and regulation of exopolysaccharide biosynthesis by means of QS systems [21]. Development of biofilms by rhizobia is crucial to overcoming environmental stresses and in certain species the biofilm formation is clearly an important feature of symbiotic ability [12], [19], [22], [23], [24]. S. fredii SMH12, a wide host-range rhizobium that nodulates soybean and dozens of other legumes, produces at least three AHLs, N-octanoyl homoserine lactone (C8-HSL), 3-oxo N-octanoyl homoserine lactone (3-oxo-C8-HSL) and N-tetradecanoyl homoserine lactone (C14-HSL). In the presence of the flavonoid genistein, an activator of its nodulation genes, the overall production of AHLs is enhanced, being detected the C14-HSL only in bacterial cultures supplemented with inducing flavonoids [5]. SMH12 possesses at least one gene, traI, which is responsiblefor the synthesis of the 3-oxo-C8-HSL [5]; and at least two non-identified luxI-type gene involved in the synthesis of the C8-HSL and the C14-HSL. Furthermore, in this bacterium the AHLs and the QS systems are involved in the biofilm formation on the abiotic surface [25].
In this work, the role and regulation of biofilm formation in S. fredii SMH12 have been studied during symbiosis with Glycine max cv. Osumi. For this purpose, the wild-type strain, a nodD1 mutant and a lactonase overproducing strain of SMH12 were constructed. The lactonase enzyme hydrolyzes the ester bond of the homoserine lactone ring of acylated homoserine lactones preventing these signalling molecules from binding to their target transcriptional regulators. Both nodD1 and lactonase strains showed an impaired ability for colonization of Glycine max roots with respect to the wild-strain, probably due to an abnormal symbiotic biofilm formation which determines a less effective symbiosis. Interestingly, QS-biofilm formation and effective nodulation are connected through flavonoids, since these molecules initiate the molecular dialogue between bacteria and plants and allow the symbiotic biofilm formation. In summary, this report unequivocally demonstrates that the development of biofilm is crucial for successful root colonization and optimal symbiosis in S. fredii SMH12.
Materials and Methods
Strains and media
Bacterial strains and plasmids used in this work are listed in Table 1. S. fredii SMH12 and derivative strains were grown at 28°C in tryptone-yeast extract (TY) medium [26], yeast extract mannitol (YM) medium [27] with a lower mannitol concentration (3 g l−1) and low-phosphate minimal glutamate mannitol (MGM) medium [21], supplemented when necessary with genistein 3.7 µM as inducing flavonoid, umbelliferone 6.2 µM as non-inducing flavonoid, or with commercial AHLs [25]. Agrobacterium tumefaciens NT1 (pZLR4) was grown at 28°C in YM with carbenicillin (100 µg ml−1) and gentamicin (30 µg ml−1). Escherichia coli strains were cultured in Luria-Bertani (LB) medium (28) at 37°C. When required, the media were supplemented with the appropriate antibiotics as described by Lamrabet et al. [29]. Commercial AHLs were dissolved in methanol and used at different concentrations. Flavonoids and AHLs were purchased from Fluka (Sigma-Aldrich, USA).
Table 1. Resistance phenotypes: GmR, KmR, NxR, ApR and TcR, gentamicin, kanamycin, nalidixic acid, ampicillin and tetracycline, respectively.
| Strain or plasmid | Relevant properties | Reference |
| Agrobacterium tumefaciensNT1 (pZRL4) | A.tumefaciens without pTiC58;with pZRL4, which carries thefusion traG::lacZ and the gene traR, GmR | [13] |
| Escherichia coliDH5α | SupE44, ΔlacU169, 5hsdR17,recA1, endA1, gyrA96, thi-1, relA1, NxR | [25] |
| Sinorhizobium fredii SMH12 | Wild-type strain, ApR | [40] |
| SMH12 (pME6000) | SMH12 with the plasmid pME6000, TcR | This work |
| SMH12 (pME6863) | SMH12 with the plasmid pME6863, whichcarries the lactonase gene, TcR | This work |
| SMH12 (pMP2463) | SMH12 with the plasmid pMP2463, whichcarries the gene of the GFP, GmR | This work |
| SMH12 (pMP6000)(pMP2463) | SMH12 with the plasmids pME6000 and pMP2463,which carry the empty plasmid and thegreen fluorescent protein, respectively, TcR GmR | This work |
| SMH12 (pMP6863)(pMP2463) | SMH12 with the plasmids pME6863 and pMP2463,which carry the gene that encode the lactonaseand the green fluorescent protein, respectively, TcR GmR | This work |
| SVQ648 | SMH12 nodD1::lacZ-GmR | This work |
| SVQ648 (pMP2463) | SVQ648 with the plasmid pMP2463, whichcarries the gene of the GFP, GmR | This work |
| pBBR1MCS-5 | Broad-host-range cloning vector, GmR | [41] |
| pK18mobsacB | Cloning vector (suicide in rhizobia), KmR | [42] |
| pME6000 | Broad-host-range cloning vector, TcR | [43] |
| pME6863 | pME6000::aiiA, plasmid carrying thelactonase gene, TcR | [44] |
| pMP2463 | pBBR-MCS-5::egfp-1, plasmid carryingthe green fluorescent protein gene, GmR | [45] |
| pMUS534 | Plasmid pK18mob derivative containingnodD1::lacZ-GmR | [46] |
| pRK2013 | Helper plasmid, KmR | [47] |
Plasmids were transferred from E. coli to SMH12 by conjugation as described by Simon [30] using plasmid pRK2013 as helper. Recombinant DNA techniques were performed according to the general protocols of Sambrook et al. [28]. For hybridization, DNA was blotted to Hybond-N nylon membranes (Amersham, UK), and the DigDNA method of Roche was employed according to the manufacturer’s instructions. PCR amplifications were performed as previously described [31]. Using this methodology the plasmid pMUS534 was employed for the homogenotization of the mutated version of the nodD1 gene in S. fredii SMH12, generating the mutant strain SVQ648. The double recombination event was confirmed by Southern blotting (data not shown).
For the growth curves of the different strains, bacteria were grown in 5 ml of YM medium to early stationary phase and then diluted to OD600 values around 0.03 in fresh YM medium with or without flavonoids. Growth was monitored by measuring the OD600 for at least 46 h. Each experiment was performed two days, eight replicates each day.
RNA isolation, cDNA synthesis and Quantitative RT-PCR
Bacterial RNA was extracted from bacterial cultures from the microtiter plates in the same conditions employed for the biofilm assays and described below. For the RNA extraction the PowerBiofilm RNA Isolation Kit was employed following the manufacturer’s instructions (MO BIO, USA). Three independent RNA extractions were performed.
To quantify the expression of the S. fredii SMH12 traI gene using quantitative RT-PCR, primers rt-traI-F and rt-traI-R described by Pérez-Montaño et al. [5] were used. Expression was calculated relative to bacteria grown without flavonoid. The S. fredii SMH12 RNA 16S gene was used as internal control to normalize gene expression. RNA 16S primers used are described in the same work [5]. The expression data shown are the mean (± standard deviation of the mean) for three biological replicates. The fold change in the target gene, normalized to RNA 16S, and relative to gene expression in the culture without flavonoids was calculated. PCR was conducted on the LightCycler 480 (Roche, Switzerland) with the following conditions: 95°C, 10 min; 95°C, 30 s; 50°C, 30 s; 72°C, 20 s; forty cycles, followed by the melting curve profile from 60 to 95°C to verify the specificity of the reaction. The threshold cycles (Ct) were determined with the iCycler software and the individual values for each sample were generated by averaging three technical replicates that varied less than 0.5 per cycle.
Confocal laser scanning microscopy
The different events of biofilm formation were visualized by confocal laser scanning microscopy using a method described by Russo et al. [32]. Bacterial cultures were placed in 8 well chambered cover glass slides containing a borosilicate glass base 1 µm thick (Thermo Fischer Scientific Inc., USA) for 4 days without shaking. To avoid desiccation, the chambers were incubated in a humid sterilized petri dish. Confocal microscope image capture was carried out with Leica TCS SP2 (Leica Microsystems, Germany). In silico 3D reconstruction analysis was executed using the computer program ImageJ (Java, USA).
Biofilm formation assay
The biofilm formation assay was based on the method described by O′Toole and Kolter [33] with modifications [34]. Cultures were grown in 5 ml of low-phosphate MGM medium, diluted to an OD600 of 0.2, with or without flavonoids and AHLs, and inoculated with 100 µl aliquots and placed on polystyrene microtiter plates, U form (Deltalab S.L., Spain). The plates were inverted and incubated at 28°C for 7 days with gentle rocking. Cell growth was analyzed by measuring OD600 using a microtiter reader Synergy HT (Biotek, USA). The culture in each well was removed carefully; the wells were dried, washed three times with 0.9% NaCl and dried again. Biofilms in each well were stained with 100 µl of 0.1% crystal violet for 20 minutes, then washed with water three times and dried again. Finally, 100 µl of 96% ethanol were added to each well and the OD570 was measured. Every experiment was performed six times with eight replicates each time.
Quantification of overall AHLs
Supernatants of the rhizobial strains grown for 7 days in 96 wells on U shaped polystyrene microtiter plates (Deltalab S.L., Spain) were collected and sterilized by microfiltration. To determine the overall autoinducer production in each condition the method described by Pérez-Montaño et al. was used. [5]. Briefly, 25 µl, 2.5 µl or 0.25 µl of the supernatants were mixed with YM to obtain a final volume of 2.5 ml reaching supernatant concentrations of 1%, 0.1% and 0.01% (v/v). The mixtures were inoculated with approximately 107 cells ml−1 of the A. tumefaciens NT1 (pZLR4), incubated with shaking for 12 h at 28°C and assayed for β-galactosidase activity [35]. As controls, 125 µl of distilled water or 125 µl of N-(3-oxo-hexanoyl)-l-homoserine lactone (3-oxo-C6-HSL) 5.5 µM, were used. To obtain the standard curve with synthetic AHLs, 125 µl of 3-oxo-C8-HSL, C8-HSL or C14-HSL at different concentration were added. The experiments were repeated independently five times with 3 replicates.
Thin Layer Chromatography
For TLC analysis, cultures previously removed from each well of the microtiter plate were extracted with the same volume of dichloromethane, evaporated to dryness and analyzed by thin-layer chromatography as described by Pérez-Montaño et al. [25]. Briefly, 1 µl of each culture extract was loaded on TLC plates (HPTLC plates RP-18 F254s 1.13724 and 1.05559, Merck, Germany) using methanol:water (60∶40 v/v) as eluent, dried and overlaid with a soft agar culture of the biosensor A. tumefaciens NT1 (pZLR4).
Plant assays
Nodulation assays on Glycine max (L.) Merrill cultivar Osumi were performed as described by de Lyra et al. [36]. Plants were inoculated with approximately 5×108 bacteria and were grown in Leonard jars with Fåhraeus nutrient solution [27] for 42 days with a 16 hour-photoperiod at 26°C in light and 18°C in the dark with 70% of humidity. Shoots were dried at 70°C for 48 h and weighed. Experiments were performed three times. For root colonization assays and microscopy, seeds were surface sterilized, germinated and inoculated with a modified method described in Pérez-Montaño et al. [25]. Briefly, Glycine max (L.) Merrill cultivar Osumi seeds were soaked for 30 seconds in 96% ethanol and for 8 minutes in commercial bleach. Then, seeds were washed repeatedly with sterilized distilled water, germinated and checked for sterility in LB medium. Each plant was inoculated with approximately 5×108 bacteria. Plants were grown under hydroponic controlled conditions in Fåhraeus solution for 7 days in the same conditions as those mentioned above.
Epifluorescence and scanning electron microscopy
Epifluorescence microscopy assays were carried out with S. fredii SMH12 and derivatives carrying GFP expressing plasmid pME2463. After growth, roots were excised (one centimetre of the proximal area of the main root and the last centimetre of a lateral root) and thoroughly washed with water to eliminate any loosely attached bacteria. Afterwards, roots were soaked for 3 minutes in 0.5% crystal violet to avoid auto-fluorescence, following 3 washes with distilled water. Microscopy analysis was carried out using an Olympus BH2-FRCA microscope with 40x and 100x magnifications (Olympus, Japan). Images of GFP-labelled bacterial cells were obtained by using a filter set consisting of a 400 to 490 nm (BP490) bandpass exciter, a 505 nm dicroic filter and a 530 nm longpass emitter (EO530). In all cases, exposure length of red and green channels was 30 seconds.
For electron scanning microscopy, 7 day-soybean plants were collected and the roots were excised as described above. Sample preparation and visualization were done according to Barahona et al. [37].
Root attachment
Whole plants were carefully taken from the hydroponic solution and roots were excised and thoroughly washed with water to eliminate any loosely attached bacteria. Remaining attached bacteria were recovered from the root by vortexing (one centimetre of the proximal area of the main root or the last centimetre of a lateral root) for 2 min in a tube containing YM medium and plating the appropriate dilutions on YM medium plates. Experiments were performed three times with three plants per assay.
Results
Nodulation Test
In a previous work [5] we described that certain plant flavonoids, besides inducing synthesis of Nod factors, increase the overall production of QS signals in S. fredii SMH12. In many bacteria, these systems regulate a broad variety of phenotypes which are important for the successful establishment of a relationship with the eukaryotic hosts. For this reason, a nodulation test was performed to elucidate if SMH12 QS systems are regulating any important phenotype during the symbiosis with Glycine max. The symbiotic properties of the wild-type strain, SVQ648 (SMH12 nodD1::LacZ::GmR), hereafter nodD1 mutant, SMH12 (pME6863), hereafter lactonase strain (this enzyme prevents AHL accumulation), and SMH12 (pME6000), which carries the empty plasmid, were determined in plant infection tests with soybean cultivar Osumi which are effectively nodulated by the wild-type strain S. fredii SMH12.
As expected, the nodD1 mutant did not induce nodule development due to its inability to produce Nod factors, which are responsible for the formation of these structures in the soybean roots. Consequently, the plant top dry mass was significantly lower (a 60%, where * is the statistical significance of the differences observed using the Mann–Whitney non-parametric test) in plants inoculated with the nodD1 mutant than in those inoculated with the parental strain SMH12. In the cases of the plants inoculated with the lactonase strain, the top dry mass, number of nodules and fresh mass of nodules formed were lower than in the wild-type strain (20%, 40% and 35% respectively). Only in the case of the number of nodules were these differences statistically significant (where * is the statistical significance of the differences observed using the Mann–Whitney non-parametric test) (Table 2). To confirm that differences in this strain are not due to the presence of the plasmid, the symbiotic parameters of SMH12 carrying the empty plasmid (pME6000) were studied. No changes were observed with respect to the wild-type strain. In summary, these results indicate that QS systems could regulate some important phenotypes during the symbiosis with Glycine max.
Table 2. Plant responses to inoculation of Glycine max cv. Osumi with S. fredii SMH12 and derivatives.
| Inoculant | Number of nodules | Fresh mass of nodules (g) | Plant-top dry mass (g) |
| None | 0±0* | 0±0* | 0.48±0.11* |
| SMH12 | 124.30±12.06 | 1.56±0.20 | 1.34±0.18 |
| SVQ648 | 0±0* | 0±0* | 0.46±0.10* |
| SMH12 (pME6863) | 76.20±16.02* | 1.04±0.29 | 1.10±0.30 |
| SMH12 (pME6000) | 131.00±22.68 | 1.60±0.38 | 1.35±0.25 |
Data represent means ± sd of six soybean jars. Each jar contained two soybean plants. Determinations were made six weeks after inoculation. Mutant nodD1 and the lactonase strain parameters were individually compared with the parental strain SMH12 parameters by using the Mann-Whitney non-parametric test. Values tagged by * are significantly different at the level α = 5%.
Plant root colonization
As commented in the introduction, in plant-associated bacteria one of the processes controlled by bacterial QS systems is the root colonization. For this reason three independent assays to study root colonization were carried out: root attachment quantification, observation studies of root by epifluorescence microscopy and electronic barrier microscopy. In these studies, proximal and lateral roots were analyzed separately to test differences in bacterial colonization in the whole root.
Root attachment assays showed a statistically significant reduction in the bacterial number per cm on the proximal area of the main root in the lactonase strain, the detected bacteria being around 10-fold less than in the other two strains. In lateral roots, colonization differences (statistically significant) were even more pronounced, these differences being 100-fold less than in the wild-type strain. A slight reduction in the number of bacteria per cm on the lateral root was also observed in the nodD1 mutant (10-fold less than SMH12). No changes in root attachment were observed in plants inoculated with the SMH12 strain carrying the empty plasmid with respect to those inoculated with the wild-type strain (Table 3). No differences in the bacterial growth-curves were detected among the four strains (data not shown).
Table 3. Glycine max cv. Osumi root attachment.
| Inoculant | CFU/cm of proximal root | CFU/cm of lateral root |
| None | 0±0* | 0±0* |
| SMH12 | 1.68±0.18×105 | 1.22±0.29×104 |
| SVQ648 | 1.10±0.42×105 | 3.83±0.5×103* |
| SMH12 (pME6863) | 5.00±0.10×104* | 8.45±0.46×102* |
| SMH12 (pME6000) | 1.55±0.27×105 | 1.20±0.39×104 |
Soybean plants were inoculated with the test strains, one centimetre of the proximal root and of the lateral roots of each plant were collected after 7 days, and the bacteria present were resuspended and plated. Data are the mean ± SD of at least three independent experiments performed in triplicate. Mutant nodD1 and the lactonase strain attachment were individually compared with the parental strain SMH12 attachment by using the Mann-Whitney non-parametric test. Values tagged by * are significantly different at the level α = 5%.
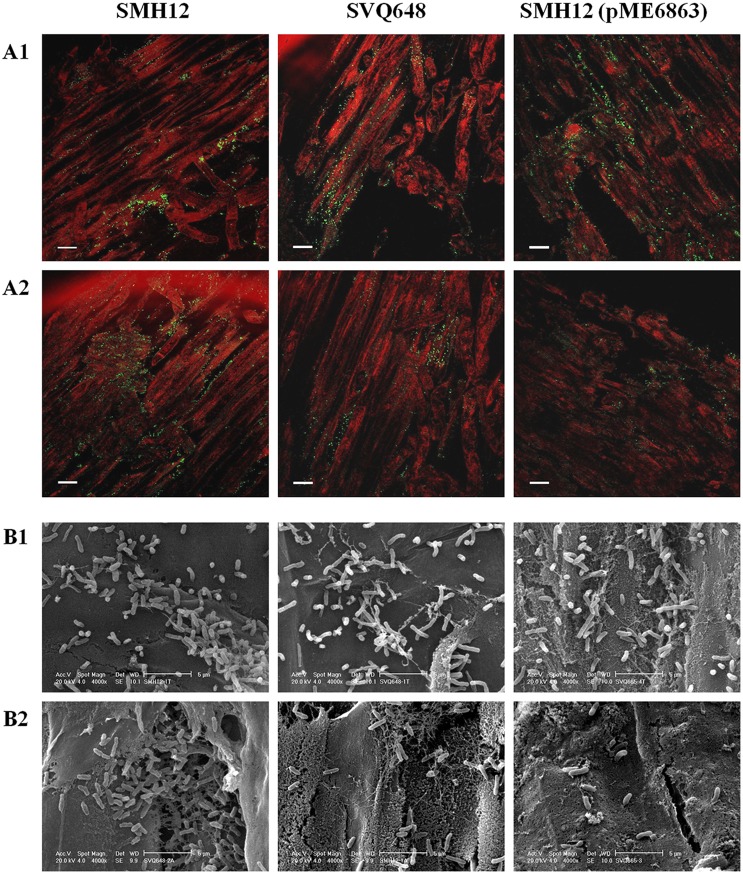
Epifluorescence microscopy and electronic barrier microscopy assays (Fig. 1) showed that in lateral roots of plants inoculated with nodD1 mutant and lactonase strain, the soybean root surface presented lower bacterial density with respect to the wild-type strain. Interestingly, the wild-type strain seems to be also distributed more clustered than the other two strains along the main root and especially in the lateral roots, which could indicate different biofilm structures in these strains. All these results suggest that the nodD1 mutant and especially the lactonase strain are defective in root colonization.
Figure 1. Visualization of the soybean root colonization.
A. Epifluorescence microscopy analysis of the colonization of the soybean rhizosphere by gfp-tagged bacteria [SMH12, SVQ648, SMH12 (pME6863)]. Roots were visualized 7 days after inoculation. 1. Proximal root. 2. Lateral roots. Bar, 100 µm. B. Scanning microscopy analysis of the colonization of the soybean rhizosphere by SMH12, SVQ648 and SMH12 (pME6863). Roots were visualized 7 days after inoculation. 1. Proximal root. 2. Lateral roots. Bar, 5 µm. SMH12: wild-type, SVQ648: nodD1 mutant, SMH12 (pME6863): lactonase strain.
Influence of inducing flavonoids on biofilm structure
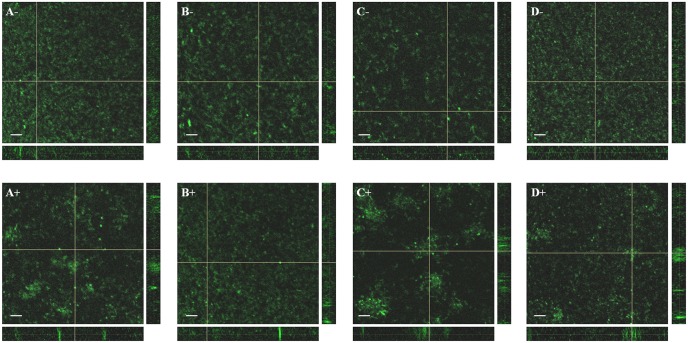
When rhizobia colonize the legume root surface microcolonies or biofilms are formed. Could the observed colonization differences be a consequence of abnormal biofilm formation in the lactonase strain and in the nodD1 mutant? To study the role of the biofilm formation during symbiosis, the biofilm structures both in the presence/absence of nod-gen inducing flavonoid (genistein) were observed using confocal microscopy experiments. Biofilm images of the wild-type, the nodD1 mutant, the lactonase and the control strain carrying the empty plasmid, all labelled with GFP, were obtained (Fig. 2 and Fig. 3). Results showed that after 4 days of growth without flavonoid, the formed biofilm consisted of a monolayer in all the studied strains being the surface coverage statistically lower (about 50%) in the lactonase strain biofilm (Fig. 2c). In the presence of genistein, the formed biofilm changed from monolayer to microcolony type in SMH12, being the coverage statistically lower (almost 45%) (Fig. 2a). In the case of the nodD1 mutant, no change was observed in the biofilm three-dimensional structure or surface coverage with genistein (Fig. 2b). Moreover, the lactonase strain showed a less surface-coverage (43%) without genistein with respect to the wild-type, but interestingly, in the presence of genistein the biofilm development underwent the transition to microcolony but no reduction in the coverage was obtained with respect to the lactonase strain without flavonoid (Fig. 2c). The control strain carrying the empty plasmid showed the same phenotype as the wild-type strain (Fig. 2d). These results suggest that, in S. fredii SMH12, the nodulation gene inducing flavonoids via NodD1 protein provoke a decrease in the bacterial attachment to an abiotic surface, since the monolayer biofilm structure (covering the whole surface) developed in the absence of flavonoid changed to microcolony-type (covering only some parts of the surface). Furthermore, these data indicate that QS signals must to be involved at least in the full formation of the monolayer-type biofilm on the glass surface.
Figure 2. Biofilm structure of S. fredii SMH12 and derivatives on glass surfaces: reconstruction of the Z-stacks and measure of the surface coverage.
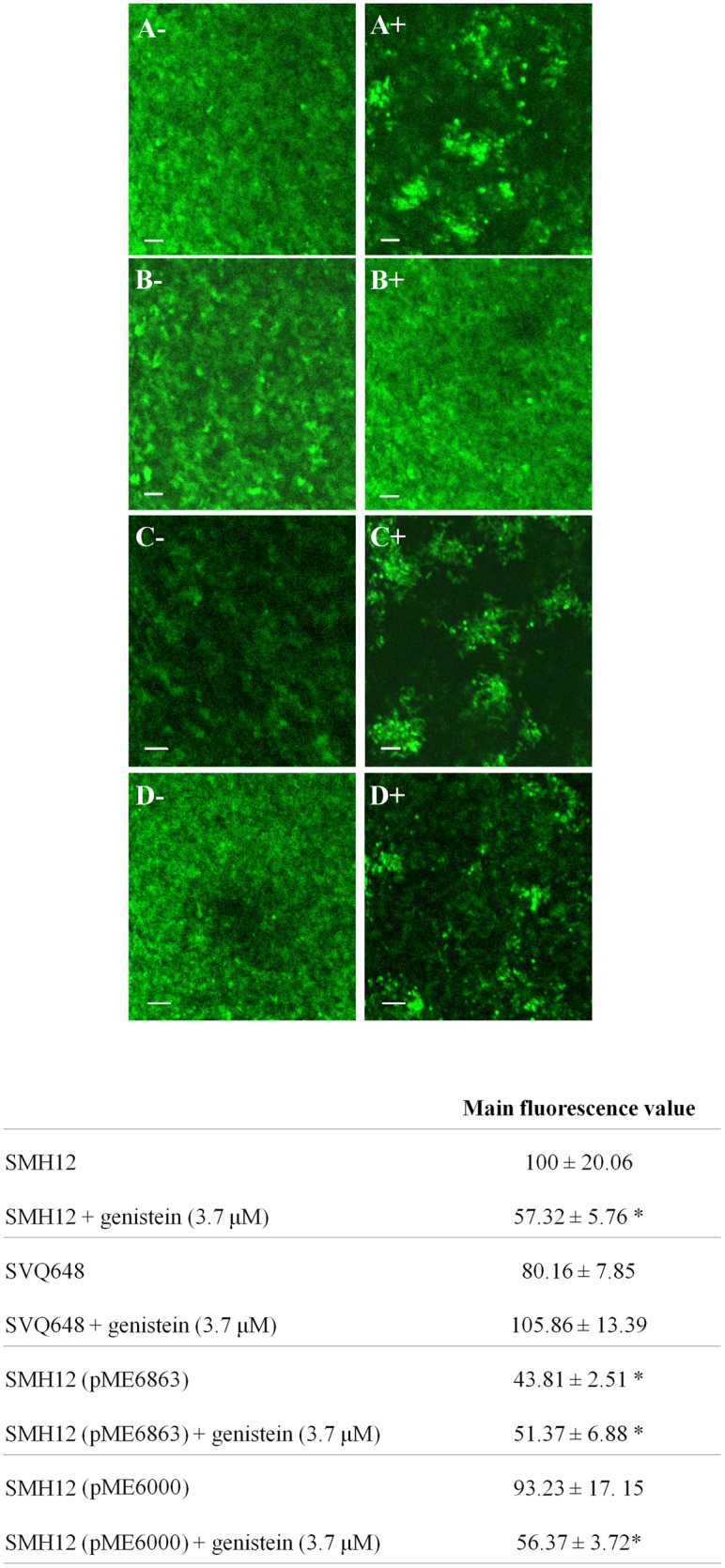
Main fluorescence value of the wild-type strain was arbitrarily given a value of 100. Averages and standard deviations of five randomized optical fields per strain corresponding to two independent experiments are shown. The asterisks indicate a significant different at the level α = 5% with respect to wild-type strain by using the Mann-Whitney non-parametrical test. Left side corresponds to cultures without flavonoids. Right side corresponds to cultures with inducing flavonoid. A. SMH12. B. SVQ648. C. SMH12 (pME6863). D. SMH12 (pME6000). Bar, 20 µm. SMH12: wild-type, SVQ648: nodD1 mutant, SMH12 (pME6863): lactonase strain. SMH12 (pME6000): carrying the empty plasmid.
Figure 3. Biofilm structure of S. fredii SMH12 and derivatives on glass surfaces: reconstruction of the XY-axis, XZ-axis and YZ-axis. The top corresponds to cultures without flavonoids.
The bottom corresponds to cultures with inducing flavonoid. A. SMH12. B. SVQ648. C. SMH12 (pME6863). D. SMH12 (pME6000). Bar, 20 µm. SMH12: wild-type, SVQ648: nodD1 mutant, SMH12 (pME6863): lactonase strain. SMH12 (pME6000): carrying the empty plasmid.
Influence of inducing flavonoids on the QS systems
Our earlier results demonstrated that nod gene inducing flavonoids increase the overall production of QS signals in S. fredii SMH12 [5]. To investigate if the activation of the QS systems takes place during the biofilm formation a quantification of the overall AI production was carried out in each condition and strain during the biofilm formation experiments on microtiter plates using the A. tumefaciens NT1 (pZRL4) biosensor. Firstly, the optimal concentration of bacterial supernatant required to obtain the wider differences between each condition without reaching saturation were determined adding different concentrations of bacterial supernatants (1%, 0.1% and 0.01%). The optimal concentration for these assays was 1% of the total volume of the biosensor culture (data not shown). Under these experimental conditions, results showed that the presence of genistein provoked a statistically significant higher production of AIs only in SMH12 (17% more), indicating that nod gene inducing flavonoids enhanced the QS signals accumulation via NodD1. Furthermore, β-galactosidase activity only in the presence of these flavonoids was similar to that obtained in the negative control (Table 4). As expected, a strong decrease in AI accumulation was detected in the supernatants from the lactonase strain, with or without genistein (Table 4). A supplementary TLC with extracts of the supernatants of biofilm experiments in microtiter plates showed a complete degradation of all AHLs in the lactonase strain. In the SMH12 strain carrying the empty plasmid no changes in the AHL profile were observed with respect to the wild-type strain (data not shown).
Table 4. β-galactosidase activity obtained using an adapted assay with A. tumefaciens NT1 (pZLR4) as bioreporter and grown in the presence of supernatants from biofilm cultures (1% v/v).
| Miller units | n (%) | |
| Control (YM) | 143.5±6.4 | |
| Genistein (37 nM) | 122.1±25.6 | |
| Umbelliferone (62 nM) | 131.2±7.2 | |
| 3-oxo-C6-HSL (5.5 µM) | 824.7±21.2 | |
| SMH12 | 717.6±23.6 | 100 |
| SMH12+genistein (3.7 µM) | 843.9±24.6 | 117* |
| SVQ648 | 702.7±28.4 | 97 |
| SVQ648+genistein (3.7 µM) | 745.8±39.2 | 104 |
| SMH12 (pME6863) | 150.2±53.8 | 21* |
| SMH12 (pME6863)+genistein (3.7 µM) | 152.2±9.71 | 21* |
Data are the mean ± SD of three independent experiments performed in triplicate.
n: percentage of induction of each supernatant with respect to SMH12 without flavonoids, defined as 100%.
Each β-galactosidase activity using biofilm supernatant was individually compared to that obtained in SMH12 without flavonoids by using the Mann-Whitney non-parametrical test. Numbers on the percentage of induction column followed by * are significantly different at the level α = 5%.
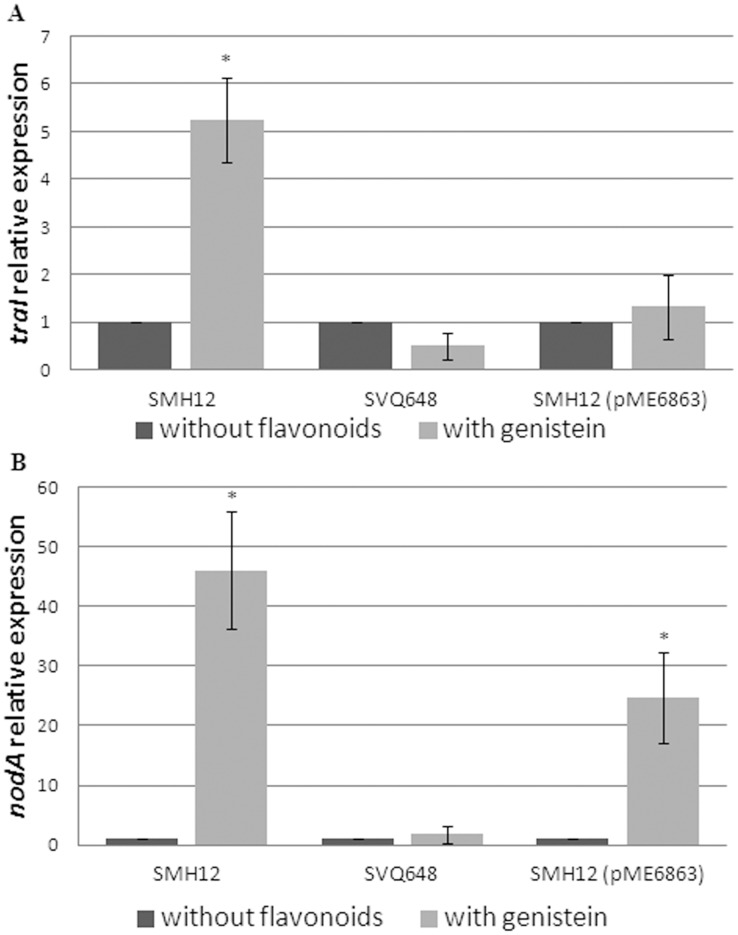
Finally, to ascertain if the increase in AHL production with flavonoids in biofilm culture assays was correlated with an increase of gene transcription, a quantitative real time RT-PCR assays was carried out. Results showed that the expression of traI, an AHL-synthesis gene from SMH12 [5], significantly increased (5-fold) in the presence of genistein compared to the control without flavonoids. No changes in the traI expression were observed in the presence of flavonoid in the case of the nodD1 mutant or in the lactonase strain, suggesting that the induction of the transcription takes place via NodD1 and that QS systems are necessary for the gene expression enhancement probably due to the typical positive feedback of the QS systems at high cellular density (Fig. 4a). As control, the expression of nodA, a gene positively regulated via NodD1 and flavonoids, was measured. As expected, a statistically significant increase was observed in nodA gene expression (46-fold and 24-fold, respectively) in both SMH12 and lactonase strains in the presence of genistein, but not in the nodD1 mutant. Interestingly, the nodA gene expression in the lactonase strain in the presence of genistein was significantly lower than in the wild-type (Fig. 4b), which could explain both the reduced soybean nodulation and the less rizosphere attachment capacities of the lactonase strain, connecting the Nod factor production with these capacities (Table 2, Table 3, Fig. 1).
Figure 4. Quantitative RT-PCR analysis of the expression of traI and nodA from S. fredii SMH12 and derivatives from biofilm cultures.
Expression data shown are the mean (± standard deviation of the mean) for three biological replicates. Expression was calculated relative to the expression without flavonoids of the wild-type strain by using the Mann-Whitney non-parametrical test. The asterisks indicate a significant different at the level α = 5%. White bars: biofilm cultures without flavonoids. Gray bars: biofilm cultures with genistein. A. traI relative expression. B. nodA relative expression. SMH12: wild-type, SVQ648: nodD1 mutant, SMH12 (pME6863): lactonase strain.
All these observations suggest that SMH12 nod gene inducing flavonoids enhance the transcription of certain AHL synthesis genes and the overall AHL production in biofilm experiments via NodD1 when the bacteria have reached the necessary cell density threshold.
Role of the QS signals on biofilm formation
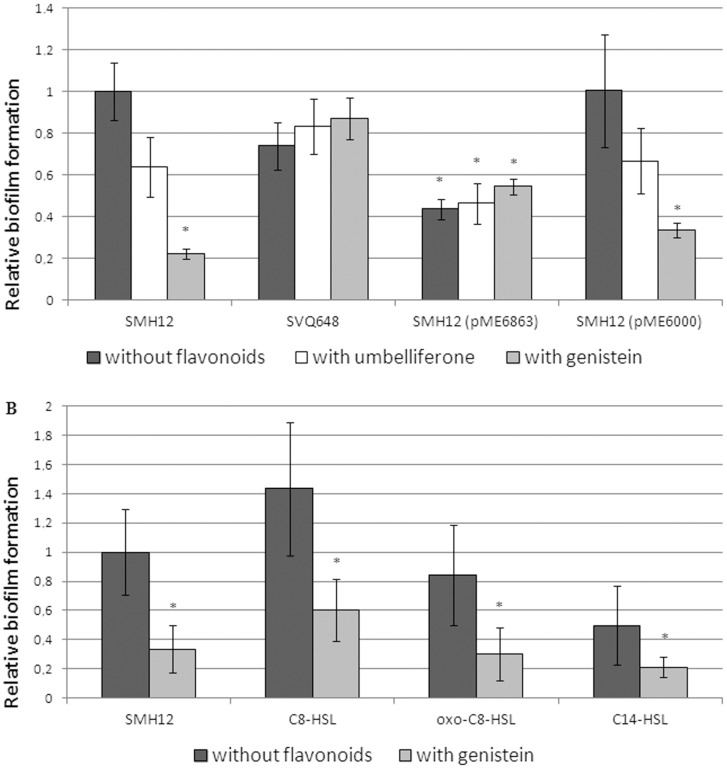
Finally, for a more exhaustive analysis of the role of each QS signal in the biofilm formation, bacterial adhesion to polystyrene surface (biofilm formation experiments) was studied after 7 days of growing in low-phosphate MGM medium (Fig. 5). Firstly, it was confirmed that the glass coverage results obtained in confocal microscopy experiments are correlated with the bacterial adhesion values obtained in biofilm formation in polystyrene microtiter plate experiments. Thus, we could use this experimental approach to study the different biofilm structures. As shown in Figure 5, in the presence of genistein, SMH12 showed a significant reduction in the adhesion value to the abiotic surface (80%, where * is the statistical significance of the differences observed using the Mann–Whitney non-parametric test). Only a slight reduction was obtained using umbelliferone, a non-inducing flavonoid [5]. In the case of nodD1 mutant, the adhesion values with or without flavonoids were similar to those obtained in the case of SMH12 without flavonoid. However, in the lactonase strain a lower adhesion was observed to polystyrene surfaces (50%) in all cases with respect to the wild-type strain without flavonoids, indicating that QS signals must be implied in the full differentiation of both types of biofilm on polystyrene surface since neither the high values without flavonoids nor the low values with inducing flavonoids are reached. To confirm that the differences observed in this strain were not due to the presence of the plasmid, the adhesion values were studied using SMH12 carrying the empty plasmid (pME6000). The experiment confirmed that this strain behaved like the wild-type strain (Fig. 5a). The absorbance at 600 nm after bacterial growth was similar in all strains and conditions (data not shown). Thus, the adhesion values to polystyrene surface in the wild type strain have a correlation with the values of surface coverage obtained on a glass surface with confocal microscopy experiments (Fig. 2).
Figure 5. Adhesion of S. fredii SMH12 and derivatives on polystyrene surfaces.
Biofilms were measured as the amount of crystal violet absorbed by the biofilm formed on multi-well plates and determined by absorbance at 570 nm after de-staining with ethanol (see methods). Absorbance of the wild-type strain was arbitrarily given a value of 1. Averages and standard deviations of eight replicas per strain corresponding to five independent experiments are shown. The asterisks indicate a significant different at the level α = 5% with respect to wild-type strain by using the Mann-Whitney non-parametrical test. A. Dark gray bars correspond to experiments performed without flavonoids, white to experiments with umbelliferone and light grey bars to experiments with genistein. B. Dark gray bars correspond to experiments performed without flavonoids and light grey bars to experiments with genistein. 3-oxo-C8-HSL and C8-HSL are used at 5.5 µM. C14-HSL is used at 55 µM. SMH12: wild-type, SVQ648: nodD1 mutant, SMH12 (pME6863): lactonase strain. SMH12 (pME6000): carrying the empty plasmid.
Once this method was validated as a reporter of the formation of the different biofilm types (monolayer with high values of coverage/adhesion and microcolony with low values of coverage/adhesion), the influence of QS signals in the biofilm formation of SMH12 with or without inducing flavonoids was studied. For this purpose, the cognate AHLs of SMH12, 3-oxo-C8-HSL, C8-HSL and C14-HSL [5], were added to wild-type bacterial cultures at physiological concentrations (data not shown) in biofilm formation experiments on microtiter plates (Fig. 5b). A slight increase or decrease in the adhesion to the polystyrene surface was observed when the medium was supplemented with C8-HSL or C14-HSL, respectively. However, these differences were not statistically significant (Fig. 5b). The absorbance at 600 nm after bacterial growth was similar in all bacteria and conditions (data not shown).
Discussion
During the first stages of root colonization, leguminous plants exudate molecules which chemotactically attract rhizobia. Once in the rhizosphere, the rhizobial population colonizes the root surface, forming a bacterial community surrounded by a matrix produced by its own bacteria, the biofilm. In the family Rhizobiaceae, the bacterial intrinsic factors that are required for the biofilm formation are mainly EPS and bacterial QS systems as well as in the case of S. meliloti the Nod factor production [12]. A fundamental question is whether the process of biofilm formation significantly affects nodulation in the symbiosis S. fredii SMH12-G. max cv. Osumi. To answer this question, the significance of QS systems and the nod gene inducing flavonoids on the biofilm formation, the colonization and the symbiosis with soybean were studied by means of both a nodD1 mutant and a lactonase strain of S. fredii SMH12.
As expected, nodulation tests with soybean showed that the symbiotic phenotype of the nodD1 mutant is similar to that obtained in non-inoculated plants, since this bacterium is unable to detect via NodD1 the flavonoids exuded by plants. Consequently the Nod factor production is blocked. Interestingly, results indicate that SMH12 QS systems could regulate some important process during the symbiosis, since the lactonase strain showed reductions in both the plant top dry mass and the fresh mass of nodules formed with respect to the wild-type strain but only in the number of nodules the reduction was statistically significant (Table 2). As described in the introduction, QS regulates a broad variety of phenotypes, including the biofilm formation. In the review of Rinaudi and Giordano [12] the mechanisms involved in both the rhizobial biofilm formation and the attachment to the plant roots are summarized. Taking into consideration the previous reports, they concluded that the biofilm lifestyle allows rhizobia to survive under unfavourable conditions and in certain species the biofilm formation is clearly an important feature of symbiotic ability [19], [22], [23], [24]. However, the role and regulation of biofilm formation during the symbiosis S. fredii SMH12-soybean has not been reported. Our results suggest that there is a correlation between QS systems, biofilm formation, root colonization and symbiosis. Firstly, three independent experiments of bacterial root colonization showed a significant reduction in bacterial root colonization in the nodD1 mutant and especially in the lactonase strain. These differences were more visible in the lateral root colonization (Table 3). Interestingly, in addition to the differences in the bacterial number, these two strains showed more of a spread surface distribution on roots compared to the wild-type, in which the bacteria appeared clustered (Fig. 1). These observations would indicate that the nodD1 mutant and specially the lactonase strain do not colonize the root surface optimally due to an alteration in their biofilm formation ability. The impaired biofilm formation in the case of the lactonase strain would explain its less nodulation capacity in the soybean test (Table 2).
To corroborate this hypothesis, the biofilm structure was analyzed by confocal microscopy. Two types of biofilms were observed, the monolayer-type in the absence of flavonoid (bacterial population cover the entire surface homogeneously), and the microcolony-type in the presence of genistein (bacterial population is clustered) (Fig. 2a and Fig. 3a). These observations would indicate that on the surface of the legume root, with a high concentration of flavonoids, the bacterial biofilm could undergo a transition from monolayer to microcolony. Perhaps this biofilm structure allows the bacterial colonization of specific root areas (those with high flavonoid exudation), which would be the optimal for the symbiosis initiation (i.e. root hairs).
Furthermore, analysing these results we infer that QS systems is involved at least in the formation of the monolayer type, since the lactonase strain developed incomplete biofilm phenotypes without inducing flavonoids (Fig. 2c and Fig. 3c). In fact, most rhizobial biofilms investigated so far are regulated directly or indirectly by QS systems [12]. Interestingly, the transition to microcolony-type biofilm requires Nod factor production since this biofilm was not developed in the presence of genistein in the nodD1 mutant (Fig. 2b and Fig. 3b). Fujishige et al. [19] reported that nodD1ABC of S. meliloti, involved in the synthesis of Nod factors, are necessary for the three-dimensional architecture of the biofilm and, in fact, these molecules are present in the biofilm matrix. The mutation of any of these genes generates a monolayer-type biofilm. However, in contrast with our results, the presence of inducing flavonoids is not necessary for the development of three dimensional structures in the biofilm of S. meliloti [19].
Interestingly, our findings also suggest that flavonoids, beyond inducing the synthesis of Nod factors (which integrate the symbiotic biofilm matrix), are enhancing the QS systems in biofilm formation experiments (Table 4 and Fig. 4a). In the presence of inducing flavonoids, an increase in the traI expression and in the overall AI production was detected. These effects were not observed in the lactonase strain due to the positive feed-back regulation of the QS systems which only occurs when the threshold in cellular density is reached [6] (Fig. 4a and Table 4). Furthermore, it is clear that the enhancement of the traI gene expression takes place via NodD1, because in the nodD1 mutant this over expression was not observed. He et al. [38] studied the QS systems of Rhizobium sp. NGR234, a Sinorhizobium fredii related bacterium, but they only unequivocally identified the tra system. This QS system is homologous to the one responsible for the plasmid Ti transfer in A. tumefaciens. This transference occurs only in the presence of opines, compounds that are produced by plants. Control of the Ti plasmid transference is modulated by TraM, a small protein that binds to and inactivates TraR, which senses the AI concentration. The induction in the presence of opines allows the synthesis of TraR at levels that overcome the inhibitory activity of TraM, activating the QS systems and the plasmid transfer [39]. In NGR234, the over expression of TraR activates not only the tra system but also other QS systems of the bacteria, since an increase in the overall AI production was detected [34]. However, so far, no natural compounds exuded by plants (similar to opines for A. tumefaciens QS systems) have been identified as inductor molecules for NGR234. Interestingly, results obtained in this paper and those reported in our previous work [5] indicate that the nod gene inducing flavonoids could be these inducing compounds for S. fredii SMH12.
In summary, experiments show that SMH12 QS systems must be involved at least in the full differentiation of monolayer-type biofilm and, moreover, in the presence of inducing flavonoids, these systems increase the AHL production and the biofilm undergoes a transition to microcolony-type (Fig. 4a, Table 4 and Fig. 2). Logically, this expression increase in the presence of flavonoids could be related to the developement of the microcolony-type biofilm, which would only occur in the rhizosphere. This fact would allow, through a unique key molecule, the efficient colonization of the legume root and the activation of the synthesis of Nod factors, both required for successful symbiosis. Interestingly, the study of the role of QS signals on biofilm formation experiments (Fig. 5) showed no statistically differences in adhesion values with or without flavonoid after addition of the wild-type cognate AHLs (Fig. 5b). However, in the case of the lactonase strain, the decrease in the adhesion values in the presence of inducing flavonoids did not reach the values obtained in the wild-type strain. This result suggests that, despite the transition to microcolony-type biofilm is regulated directly by NodD1 protein and inducing flavonoids (Fig. 2), the QS signals could also be involved in the full differentiation of this biofilm on the polystyrene surface (Fig. 5a).
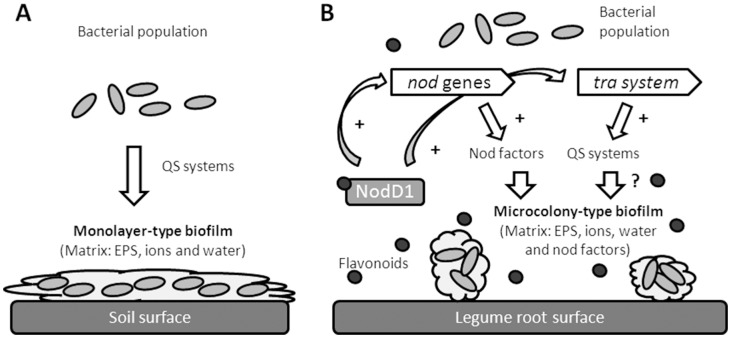
Thus, taking into consideration all the results, we propose the following model (Fig. 6): S. fredii SMH12 forms a QS-regulated monolayer-type biofilm (whose matrix would be mainly composed of EPS, water and ions) when bacterium colonizes soil. In the rhizosphere, the exudation of inducing flavonoids, besides inducing the synthesis of Nod factors which are necessary for the microcolony-type biofilm, potentiate via NodD1 the tra QS system, which leads to an overproduction of the QS signals. This accumulation and the synthesis of Nod factors are required for the full development of the microcolony-type biofilm, whose matrix should be composed of EPS, water, ions and Nod factors. The complete formation of this symbiotic biofilm is necessary for a successful root colonization and symbiosis between rhizobia and legume.
Figure 6. Model of biofilm formation in S. fredii SMH12.
A. In soil. B. In rizosphere. S. fredii SMH12 forms monolayer-type biofilm when colonize soil surfaces. When the bacterium colonizes the legume root (in presence of inducing flavonoids), it forms microcolony-type biofilm, which is necessary for a successful root colonization and symbiosis between rhizobia and legume.
Despite demonstrating in this work that the rhizobial biofilm formation is important for colonization and symbiosis of S. fredii SMH12 with the soybean, there are still many aspects that must to be studied in the future to further clarify this process.
Acknowledgments
We would like to thank the Laboratorio de Microscopía de Barrido y Análisis por Energía Dispersiva de Rayos X from the Universidad Autónoma de Madrid for electronic barrier microscopy assays. Thanks to Servicio General de Biología of the CITIUS from the University of Seville for allowing us to use their laboratory equipment. Thanks to Diane Haun for English style supervision. Special thanks to Miguel Cámara, for his comments on an earlier version of the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the following grants: AGL2012-38831 from the Spanish Ministerio de Economía y Competitividad, http://www.idi.mineco.gob.es/portal/site/MICINN/ P10-AGR-5821 and P11-CVI-7050 from the Junta de Andalucía, Consejería de Innovación, Ciencia y Empresas. http://www.juntadeandalucia.es/organismos/economiainnovacioncienciayempleo.html. Dr. Pérez-Montaño’s work was supported by an FPU fellowship from the Spanish Ministerio de Ciencia y Tecnología and a contract from the University of Seville. http://www.idi.mineco.gob.es/portal/site/MICINN/http://www.us.es/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Murray JD (2011) Invasion by invitation: rhizobial infection in legumes. Mol Plant-Microbe Interact 24: 631–639. [DOI] [PubMed] [Google Scholar]
- 2. Somers E, Vanderleyden J, Srinivasan M (2004) Rhizosphere bacterial signalling: a love parade beneath our feet. Crit Rev Microbiol 30: 205–240. [DOI] [PubMed] [Google Scholar]
- 3. Krause A, Doerfel A, Göttfert M (2002) Mutational and Transcriptional Analysis of the Type III Secretion System of Bradyrhizobium japonicum. . Mol Plant Microbe Interact 12: 1228–1235. [DOI] [PubMed] [Google Scholar]
- 4. López-Baena FJ, Vinardell JM, Pérez-Montano F, Crespo-Rivas JC, Bellogín RA, et al. (2008) Regulation and symbiotic significance of nodulation outer proteins secretion in Sinorhizobium fredii HH103. Microbiology 154: 1825–1836. [DOI] [PubMed] [Google Scholar]
- 5. Pérez-Montaño F, Guasch-Vidal B, González-Barroso S, López-Baena FJ, Cubo T, et al. (2011) Nodulation-gene-inducing flavonoids increase overall production of autoinducers and expression of N-acyl homoserine lactone synthesis genes in rhizobia. Res Microbiol 162: 715–723. [DOI] [PubMed] [Google Scholar]
- 6. Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miller MB, Bassler BL (2001) Quorum sensing in bacteria. Annu Rev Microbiol 55: 165–199. [DOI] [PubMed] [Google Scholar]
- 8. Marketon MM, Glenn SA, Eberhard A, Gonzalez JE (2003) Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti . J Bacteriol 185: 25–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ohtani K, Hayashi H, Shimizu T (2002) The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens . Mol Microbiol 44: 171–179. [DOI] [PubMed] [Google Scholar]
- 10. Quiñones B, Dulla G, Lindow SE (2005) Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae . Mol Plant-Microbe Interact 18: 682–693. [DOI] [PubMed] [Google Scholar]
- 11. Rice SA, Koh KS, Queck SY, Labbate M, Lam KW, et al. (2005) Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J Bacteriol 187: 3477–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rinaudi LV, Giordano W (2010) An integrated view of biofilm formation in rhizobia. FEMS Microbiol Lett 304: 1–11. [DOI] [PubMed] [Google Scholar]
- 13. Cha C, Gao P, Chen YC, Shaw PD, Farrand SK (1998) Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol Plant-Microbe Interact 11: 1119–1129. [DOI] [PubMed] [Google Scholar]
- 14. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (1995) Microbial biofilm. Annu Rev Microbiol 49: 711–745. [DOI] [PubMed] [Google Scholar]
- 15. Stanley NR, Lazazzera BA (2004) Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol 52: 917–924. [DOI] [PubMed] [Google Scholar]
- 16. Sutherland IW (2001) Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147: 3–9. [DOI] [PubMed] [Google Scholar]
- 17. Rinaudi L, Fujishige NA, Hirsch AM, Banchio E, Zorreguieta A, et al. (2006) Effects of nutritional and environmental conditions on Sinorhizobium meliloti biofilm formation. Res Microbiol 157: 867–875. [DOI] [PubMed] [Google Scholar]
- 18. Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM (2006) Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol 56: 195–206. [DOI] [PubMed] [Google Scholar]
- 19. Fujishige NA, Lum MR, De Hoff PL, Whitelegge JP, Faull KF, et al. (2008) Rhizobium common nod genes are required for biofilm formation. Mol Microbiol 67: 504–515. [DOI] [PubMed] [Google Scholar]
- 20. Wells DH, Chen EJ, Fisher RF, Long SR (2007) ExoR is genetically coupled to the ExoS-ChvI two-component system and located in the periplasm of Sinorhizobium meliloti . Mol Microbiol 64: 647–664. [DOI] [PubMed] [Google Scholar]
- 21. Rinaudi LV, González JE (2009) The low-molecular-weight fraction of exopolysaccharide II from Sinorhizobium meliloti is a crucial determinant of biofilm formation. J Bacteriol 191: 7216–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. González JE, Marketon MM (2003) Quorum sensing in nitrogen-fixing rhizobia. Microbiol Mol Biol Rev 67: 574–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loh JT, Yuen-Tsai JP, Stacey MG, Lohar D, Welborn A, et al. (2001) Population density-dependent regulation of the Bradyrhizobium japonicum nodulation genes. Mol Microbiol 42: 37–46. [DOI] [PubMed] [Google Scholar]
- 24. Jitacksorn S1, Sadowsky MJ (2008) Nodulation gene regulation and quorum sensing control density-dependent suppression and restriction of nodulation in the Bradyrhizobium japonicum-soybean symbiosis. Appl Environ Microbiol 74: 3749–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pérez-Montaño F, Jiménez-Guerrero I, Sánchez-Matamoros RC, López-Baena FJ, Ollero FJ, et al. (2013) Rice and bean AHL-mimic quorum-sensing signals specifically interfere with the capacity to form biofilms by plant-associated bacteria. Res Microbiol 164: 749–760. [DOI] [PubMed] [Google Scholar]
- 26. Beringer JE (1974) R factor transfer in Rhizobium leguminosarum . J Gen Microbiol 84: 188–198. [DOI] [PubMed] [Google Scholar]
- 27.Vincent JM (1970) The modified Fahraeus slide technique. In A manual for the practical study of root nodule bacteria, 144–145. Edited by J. M. Vincent. Oxford, UK: Blackwell Scientific Publications.
- 28.Sambrook J, Fritsch EF, Maniatis T(1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor NY: Cold Spring Harbor Laboratory.
- 29. Lamrabet Y, Bellogín RA, Cubo T, Espuny R, Gil A, et al. (1999) Mutation in GDP-fucose synthesis genes of Sinorhizobium fredii alters Nod factors and significantly decreases competitiveness to nodulate soybeans. Mol Plant-Microbe Interact 12: 207–217. [DOI] [PubMed] [Google Scholar]
- 30. Simon R (1984) High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed TnS-Mob transposon. Mol Gen Genet 196: 413–420. [DOI] [PubMed] [Google Scholar]
- 31. López-Baena FJ, Monreal JA, Pérez-Montano F, Guasch-Vidal B, Bellogín RA, et al. (2009) The absence of Nops secretion in Sinorhizobium fredii HH103 increases GmPR1 expression in Williams soybean. Mol Plant-Microbe Interact 22: 1445–1454. [DOI] [PubMed] [Google Scholar]
- 32. Russo DM, Williams A, Edwards A, Posadas DM, Finnie C, et al. (2006) Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum . J Bacteriol 188: 4474–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O’Toole GA, Kolter R (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28: 449–461. [DOI] [PubMed] [Google Scholar]
- 34. Mueller K, González JE (2011) Complex regulation of symbiotic functions is coordinated by MucR and quorum sensing in Sinorhizobium meliloti . J Bacteriol 193: 485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller JH. (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York (USA).
- 36. de Lyra, MCCP, López-Baena FJ, Madinabeitia N, Vinardell JM, Espuny MR, et al. (2006) Inactivation of the Sinorhizobium fredii HH103 rhcJ gene abolishes nodulation outer proteins (Nops) secretion and decreases the symbiotic capacity with soybean. Int Microbiol 9: 125–133. [PubMed] [Google Scholar]
- 37. Barahona E, Navazo A, Yousef-Coronado F, Aguirre de Cárcer D, Martínez-Granero F, et al. (2010) Efficient rhizosphere colonization by Pseudomonas fluorescens f113 mutants unable to form biofilms on abiotic surfaces. Environ Microbiol 12: 3185–3195. [DOI] [PubMed] [Google Scholar]
- 38. He X, Chang W, Pierce DL, Seib LO, Wagner J, et al. (2003) Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J Bacteriol 185: 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Piper KR, Farrand SK (2000) Quorum sensing but not autoinduction of Ti plasmid conjugal transfer requires control by the opine regulon and the antiactivator TraM. J Bacteriol 182: 1080–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, et al. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176. [DOI] [PubMed] [Google Scholar]
- 42. Schäfer A, Tauch A, Jager W, Kalinowski J, Thierbach G, et al. (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene 145: 69–73. [DOI] [PubMed] [Google Scholar]
- 43. Maurhofer M, Reimmann C, Schmidli-Sacherer P, Heeb S, Haas D, et al. (1998) Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 88: 678–684. [DOI] [PubMed] [Google Scholar]
- 44. Reimmann C, Ginet N, Michel L, Keel C, Michaux P, et al. (2002) Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology 148: 923–932. [DOI] [PubMed] [Google Scholar]
- 45. Stuurman N, Pacios-Bras C, Schlaman HR, Wijfjes AH, Bloemberg G, et al. (2000) Use of green fluorescent protein color variants expressed on stable broad-host-range vectors to visualize rhizobia interacting with plants. Mol Plant Microbe-Interact 13: 1163–1169. [DOI] [PubMed] [Google Scholar]
- 46. Vinardell JM, Ollero FJ, Hidalgo A, López-Baena FJ, Medina C, et al. (2004) NolR regulates diverse symbiotic signals of Sinorhizobium fredii HH103. Mol Plant Microbe-Interact 17: 676. [DOI] [PubMed] [Google Scholar]
- 47. Figurski DH, Helinski DR (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA 76: 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.