Abstract
A gram-negative aliphatic hydrocarbon-degrading bacterium SJTD-1 isolated from oil-contaminated soil was identified as Pseudomonas aeruginosa by comparative analyses of the 16S rRNA sequence, phenotype, and physiological features. SJTD-1 could efficiently mineralize medium- and long-chain n-alkanes (C12-C30) as its sole carbon source within seven days, showing the most optimal growth on n-hexadecane, followed by n-octadecane, and n-eicosane. In 36 h, 500 mg/L of tetradecane, hexadecane, and octadecane were transformed completely; and 2 g/L n-hexadecane was degraded to undetectable levels within 72 h. Two putative alkane-degrading genes (gene 3623 and gene 4712) were characterized and our results indicated that their gene products were rate-limiting enzymes involved in the synergetic catabolism of C12–C16 alkanes. On the basis of bioinformatics and transcriptional analysis, two P450 monooxygenases, along with a putative AlmA-like oxygenase, were examined. Genetically defective mutants lacking the characteristic alkane hydroxylase failed to degrade n-octadecane, thereby suggesting a different catalytic mechanism for the microbial transformation of alkanes with chain lengths over C18.
Introduction
Oil pollution poses a severe threat to ecosystems, because of its refractory resistance to environmental degradation. Many remediation technologies have been used for the removal of contaminating oil residues [1]. Bioremediation is considered to be a safe, efficient, and economically viable alternative to physicochemical methods for the elimination of oil contaminants. In the past 30 years, several research groups around the world have tried to build robust biocatalysts for the efficient removal of contaminating oil residues. Although these attempts have not succeeded so far, the constant search for an effective biocatalyst has resulted in the discovery of a large number of microorganisms that could be used for bioremediation [2], [3].
Alkanes are the most abundant hydrocarbons (with an estimated abundance of 20–50%) in crude oil [4]. As alkanes are non-polar molecules with very low chemical activity, their utilization by microorganisms faces significant challenges, owing to known factors, such as low water solubility, high degree of accumulation in cell membranes, and higher activation energies [5], [6]. Nonetheless, at least sixty genera of aerobic bacteria, and five genera of anaerobic bacteria [7], such as Pseudomonas [8], Acinetobacter [9], Rhodococcus [10] and Dietzia [11], have been reported to possess the ability to degrade aliphatic hydrocarbons. Among them, Pseudomonas was found both in soil as well as in aqueous environments [12]. Several Pseudomonas strains are known to utilize aliphatic hydrocarbons as its sole carbon sources [13]–[15]. P. aeruginosa RR1 and P. fluorescens CHA0 degrade n-alkanes ranging from C12 to C34 [16], [17]; and P. aeruginosa DQ8 can grow in the presence of n-tetrodecane, n-docosane, n-triacontane, and n-tetrocontane [8].
Although many strains have been reported to utilize hydrocarbons, most of them can only use a narrow range of substrates. For example, Bacillus stearothermophilus can only grow in media containing hydrocarbons of chain length C15 to C17 [18], whereas A. borkumensis AP1, SK2, and SK7 can only utilize alkanes ranging from C6 to C16 [19]. Indeed, very few strains such as Acinetobacter baylyi ADP1 and Thermus sp. C2 can degrade a wide range of hydrocarbons. However, their genetic characteristics remain elusive and not much is known about the mechanisms through which these microorganisms break down long chain alkanes present in refractory oil residues [20], [21].
In this study, a P. aeruginosa strain SJTD-1 was isolated from oil-contaminated soil; its hydrocarbon utilization capability and n-alkane breakdown efficiency were investigated. Based on its whole-genome DNA sequence [22], several alkane hydroxylases were characterized. These included two AlkB monooxygenases, two P450 monooxygenases, and one AlmA-like monooxygenase. Varying transcriptional expression levels of these genes induced by C12–C24 alkanes indicated the presence of a complex hydrocarbon breakdown mechanism in P. aeruginosa SJTD −1.
Materials and Methods
Chemicals and media
n-Decane (>99% pure) was purchased from Alfa Aesar Organic Co., Inc (Tianjing); n-dodecane, n-tetradecane, n-hexadecane, and n-octadecane (all >99% pure) from Sangon (Shanghai, China); and n-pentadecane, n-eicosane, n-docosane, n-tetracosane, n-triacontane, and n-hexane (of HPLC gradient grade) from Sigma-Aldrich (St. Louis, MO, USA). All other reagents used in this study were of analytical reagent grade.
Luria-Bertani (LB) medium (tryptone 10.0 g, yeast extract 5.0 g, NaCl 10 g/L) and basal salt medium (BSM) (K2HPO4 3.815 g, KH2PO4 0.5 g, (NH4)2HPO4 0.825 g, KNO3 1.2625 g, Na2SO4 0.2 g, CaCl2 0.02 g, FeCl3 0.002 g, and MgCl2 0.02 g/L) were used in this study. To examine the utilization of alkanes by strain SJTD-1, both liquid and solid alkanes, maintained at room temperature, were first dissolved in n-hexane to form 500 mg/mL alkane-hexane solutions. These solutions were then added into the BSM medium to attain various concentrations. The n-hexane was neither toxic to the strain, nor was it utilized by the strain.
Strain Isolation
Samples used for bacterial enrichment were collected from the oil-contaminated soil in the Daqing Oil Field of China (no specific permissions were required and this work did not involve any endangered or protected species). Approximately 5 g of the soil sample was inoculated into a 250 mL flask with 100 mL BSM liquid medium containing 2 g/L hexadecane, and was shaken at 180 rpm for seven days, at 37°C. A 5 mL culture was then inoculated into 100 mL of fresh BSM liquid medium with n-hexadecane, and cultured as above. Enrichment cultures after rounds of enrichment were diluted and plated onto BSM agar plates pre-coated with hexadecane. Bacterial colonies grown on plates with different morphology were tested for their n-alkane utilizing capabilities. One strain, which exhibited a fast growth rate was purified and designated as SJTD-1.
16S rRNA analysis and phylogenic tree construction
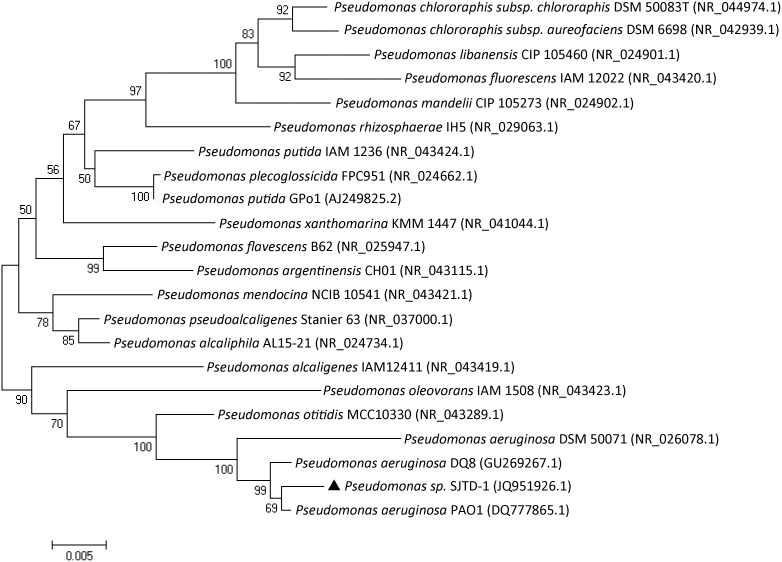
The newly isolated strain SJTD-1 was identified based on its morphological, physiological, and biochemical properties listed in Bergey’s Manual of Determinative Bacteriology [23]. After its genomic DNA was extracted with standard molecular biology techniques [24], the 16S rRNA gene of strain SJTD-1 was amplified using Bacterial 16S rDNA Kit (TaKaRa Biotechnology Co., Ltd. Dalian, China). The PCR program is first denaturized at 94 °C for 5 min, then denaturized at 94 °C for 1 min, annealed at 55 °C for 1 min and elongated at 72 °C for 1.5 min; Repeated this process for 30 times, and then further elongated at 72°C for another 5 min. After the PCR reaction, the fragments were sequenced using the primer pairs 16S-seq-F/R (Table S1), and this 16S rRNA sequence was deposited into GenBank (Accession No. JQ951926.1). A phylogenic tree based on the 16S rRNA sequences of the SJTD-1 strain and some other strains was analyzed with MEGA 5.0, using the Neighbor Joining method with 1,000 replications. The genetic distances were calculated with the Kimura two-parameter distance model.
Growth rate of SJTD-1 in pure n-alkanes
The cell growth rate of strain SJTD-1 in n-alkanes was determined by the Automatic Growth Curve Analyzer (BioScreen Testing Service, Inc., CA, USA). Briefly, a single colony of strain SJTD-1 was inoculated into a 100 mL baffled flask containing 10 mL LB broth, and cultured on a rotary shaker overnight at 37°C. The cells were then harvested by centrifugation at 8,000 rpm for 5 min and washed thrice with sterilized water to exclude the residual organic compounds. The cells were then re-suspended in the BSM medium to OD600≈2.0, to form the inoculum. Subsequently, 10 µL of the cell pellets were inoculated into the 500 µL well of a 10×10 multi-well plate, which contained 190 µL of the BSM medium amended with pure n-alkanes (C10–C24) of different concentrations. The initial cell concentration in each well at OD600 was 0.1. All n-alkanes were prepared as n-hexane solutions. Wells containing cells without n-alkanes and those containing n-alkanes without inoculum were used as blanks. The 10×10 multi-well plates were loaded onto the Automatic Growth Curve Analyzer and incubated at 30°C with constant shaking (180 rpm) for seven days. The cell densities were determined by detecting the OD600 every hour. All experiments were repeated thrice, and the results shown were the average values of three replicates, along with the standard errors.
Degradation analysis of SJTD-1 in the case of pure n-alkanes
The degradation efficiencies of strain SJTD-1 for n-alkanes of different carbon lengths (C14–C16) were determined according to the loss of substrate from each liquid culture. A SJTD-1 cell inoculum was prepared as described above. The cells were distributed in flasks containing 100 mL BSM medium, amended with 500 mg/L of pure C10–C24 n-alkanes (which was the sole carbon source), and cultured for seven days at 30°C in the shaker. The initial OD600 was 0.1. Flasks without cell inoculum were used as blanks to assess the abiotic loss. The cultures were taken out at different time points, to estimate the cell concentrations and the alkane residues. For the residual analysis of alkanes, 3 mL cell cultures were collected into 10 mL-glass centrifuge tubes and extracted with 1 mL n-hexane for the first time. The mixtures were shaken vigorously, centrifuged at 5,000 rpm for 10 min. Then, the organic layer was collected and the aqueous layer was extracted twice, with 1 mL n-hexane each time. The organic extracts were pooled together to a final volume of 3 mL, and dried with anhydrous sodium sulfate. For each sample, n-pentadecane (100 mg/L) was added before the extraction and used as an internal standard. All extractions were performed in triplicate, and the results were expressed as average values with standard errors.
The degradation efficiencies of strain SJTD-1 for n-hexadecane of various concentrations (250, 500, 1,000, 2,000 mg/L) were also analyzed as above. After strain SJTD-1 was cultured for one to seven days, the residual n-hexadecane at different time points were extracted and determined with three replicates.
Analytical methods of n-alkanes concentration
The concentrations of n-alkanes were determined with the gas chromatography-mass spectrometry (GC-MS) technique, using a GC/MS system (7890A GC/5975C MS, Agilent, USA) equipped with a fused DB-5 MS capillary column (0.25 mm×30 m×0.25 µm). The GC program was listed as below. Helium was used as the carrier gas with a constant flow rate of 1.0 mL/min. The split ratio was 10∶1, and the temperatures of the injector and the connector were 270°C and 280°C, respectively. The temperatures of the ion source and the quadrupole were 230°C and 150°C, respectively. The column oven temperature was kept at 150°C for 2 min, and then raised to 200°C at a rate of 5°C/min, followed by an increase to 290°C, at a rate of 30°C/min, and at an isotherm of 290°C. The ionization mode was set as EI+, 70 ev, and the voltage of the detector was 1,388 V. The GC-MS spectra were analyzed with the ChemStation software, and the relative abundance of different hydrocarbon residues was calculated by the ratio of the peak area of each hydrocarbon to the peak area of n-pentadecane in the GC chromatograph. The residue ratio of n-alkanes was calculated with the equation R = [S]/[I], where R, [S], and [I] represent the alkane residue ratio, the residual n-alkane concentrations in the samples, and the concentration before inoculation, respectively. The results were expressed as mean values with standard deviation. The cell-free controls were incubated and analyzed in the same manner.
Deletion of the putative alkane-degradation genes from strain SJTD-1
Based on the whole-genome DNA sequence of strain SJTD-1 [22], two putative alkB genes, gene 3623 and gene 4712, showed great similarities to the reported alkB2 and alkB1 genes, respectively. To determine the function of alkB1 and alkB2, the knockout mutants of the single gene and two genes were constructed using the pRKaraRed homologous recombination system [25]. Briefly, plasmid pRKaraRed was first electro-transformed into strain SJTD-1 to generate the SJTD-1/pRKaraRed competent cells. The in-frame scarless knockout mutant of the single gene was generated by two-step homologous recombination. First, the sacB-bla cassettes were amplified from plasmid pEX18Ap [26] with primer pairs F1 and R1, and then electro-transformed into the SJTD-1/pRKaraRed competent cells. The transformants were screened on LB plates supplemented with 500 µg/mL carbenicillin and 50 µg/mL tetracyclin. Next, the sacB-bla removal cassettes amplified from the genomic DNA of the first-step colonies (SucSCarbR) using the primer pairs F2 and R2, were electro-transformed into the competent cells of the first step to perform the second recombination. The transformants were spread on LB plates with 10% sucrose and 50 µg/mL tetracycline. The transformants were further selected in parallel on LB plates with 10% sucrose and 50 µg/mL tetracycline, and on LB plates with 500 µg/mL carbenicillin and 50 µg/mL tetracycline. A positive genotype (SucRCarbS) was verified by PCR detection with the test primers and DNA sequencing was performed by Invitrogen. The deletion of multiple genes was achieved by repeating this two-step knockout method. Two alkB genes in strain SJTD-1 were knocked out separately or simultaneously, and the mutant strains were named as MutalkB1, MutalkB2, and MutalkB1&2, respectively. Similarly, two putative P450 genes (gene 5482 and 4609), and one AlmA-like monooxygenase gene (gene 3206) were also deleted and the mutant strains were MutP450-1, MutP450-2, and MutAlmA, respectively. All the mutants are listed in Table 1; all the primers for homologous recombination and PCR detection are listed in Table S1. The sequences of the five alkane hydroxylase genes are shown in Text S1.
Table 1. Strains, plasmids and putative genes studied in the work.
| Name | Description | Source/reference |
| Strains | ||
| P. aeruginosa SJTD-1 | Host strain isolated from oil-contaminated soil, Wild Type | This study |
| MutalkB1/MutalkB2/MutalkB1&2 | AlkB-deleted mutants of P. aeruginosa SJTD-1 | This study |
| MutP450-1/MutP450-2/MutP450-1&2 | P450-deleted mutants of P. aeruginosa SJTD-1 | This study |
| MutalmA | AlmA-deleted mutants of P. aeruginosa SJTD-1 | This study |
| Plasmids | ||
| pRKaraRed | Broad-host-range, λ-Red proteins expression vector, TetR | [25] |
| Genes * | ||
| 4712 | alkane-1 monooxygenase, AlkB1 | This study |
| 3623 | alkane-1 monooxygenase, AlkB2 | This study |
| 5482 | putative cytochrome P450 hydroxylase, P450–1 | This study |
| 4609 | putative cytochrome P450 hydroxylase, P450–2 | This study |
| 3206 | monooxygenase, flavin-binding family, AlmA-like protein | This study |
*Gene numbers presented in RAST server.
To study the roles of these alkane hydroxylases in n-alkane utilization, the growth curves of strain SJTD-1 and mutants (with different n-alkanes as the sole carbon sources) were determined by the Automatic Growth Curve Analyzer as described above. Experiments were replicated thrice and the results obtained were the averages values with standard errors.
Quantitative reverse transcription-PCR (Q-RT-PCR)
To study the transcription profiles of the five alkane hydroxylases in different conditions, the wild type and mutant SJTD-1 were cultured in the BSM medium with sodium acetate and various alkanes (C12–C24) to the mid-exponential phase. The alkanes were the sole carbon sources. Total RNA was extracted using the FastRNAPro Blue Kit for microbes (MP Biomedicals, Santa Ana, CA) and the RNA extraction was performed according to the manufacturer’s instructions. The RNA yield was estimated using a Nanodrop UV spectrometer (Thermo Scientific, Wilmington, DE, USA). The reverse transcription was achieved using the PrimeScript Reverse Transcriptase Kit (TaKaRa, Dalian, China). Approximately 1 µg RNA and 20 ng random primers were used. The Q-RT-PCR was performed using the IQSYBR Green Super-mix (Bio-Rad Laboratories, Hercules, CA) and gene-specific primers (RT-F and RT-R, Table S1) in the IQTM 5 Multicolor Real-time PCR Detection System (Bio-Rad, CA, USA). The Q-RT-PCR procedure carried out was as follows: 15 min of pre-denaturing, 40 cycles of 95°C for 10 s; 60°C for 30 s; and 72°C for 30 s; followed by a melting curve stage from 60°C to 95°C. The cDNA amplification efficiency of the samples, internal standards (16S rRNA), and calibrators (samples without induction by alkanes) were equivalently modulated, and the relative fold change in mRNA quantity was calculated using the DDCt method [27]. At least three independent Q-RT-PCR experiments were conducted for each RNA sample. The change in the transcriptional levels of strains grown with alkanes was calculated by comparing with the transcriptional level of the strains grown in sodium acetate [17].
Results
Isolation and identification of the oil-degrading strain SJTD-1
Strain SJTD-1 isolated from the enrichment culture of oil-contaminated soil is a rod-shaped, gram-negative bacterium. Its growth pH ranged from 4.0 to 10.0, and optimal growth occurred at pH 7.0. After growing on LB agar at 37°C for 24 h, the cells formed green, round, moist and glossy colonies, approximately 1.0 mm in diameter. Strain SJTD-1 could utilize n-alkanes, from n-dodecane (C12) to n-triacontane (C30), as its (sole) carbon sources. Although SJTD-1 could not grow in the presence of n-decane or other shorter length n-alkanes, its growth was not inhibited by the short-chain n-alkanes like n-hexane. The 16S rRNA gene sequence of SJTD-1 (Genebank Accession No. JQ951926.1) was 99% identical to that of P. aeruginosa PAO1 (Genebank Accession No. DQ777865.1), and the corresponding phylogenetic tree analysis supported a strong relationship between SJTD-1 and members of Pseudomonas sp. (Fig. 1). Therefore, SJTD-1 was classified as a P. aeruginosa strain.
Figure 1. Phylogenetic tree based on 16S rDNA gene sequences indicating the SJTD-1 position.
The Kimura two-parameter distance model was used, and bootstrap analysis was performed with 1,000 repetitions using MEGA 5.0.
Growth curve of strain SJTD-1 and its n-alkane degradation efficiency
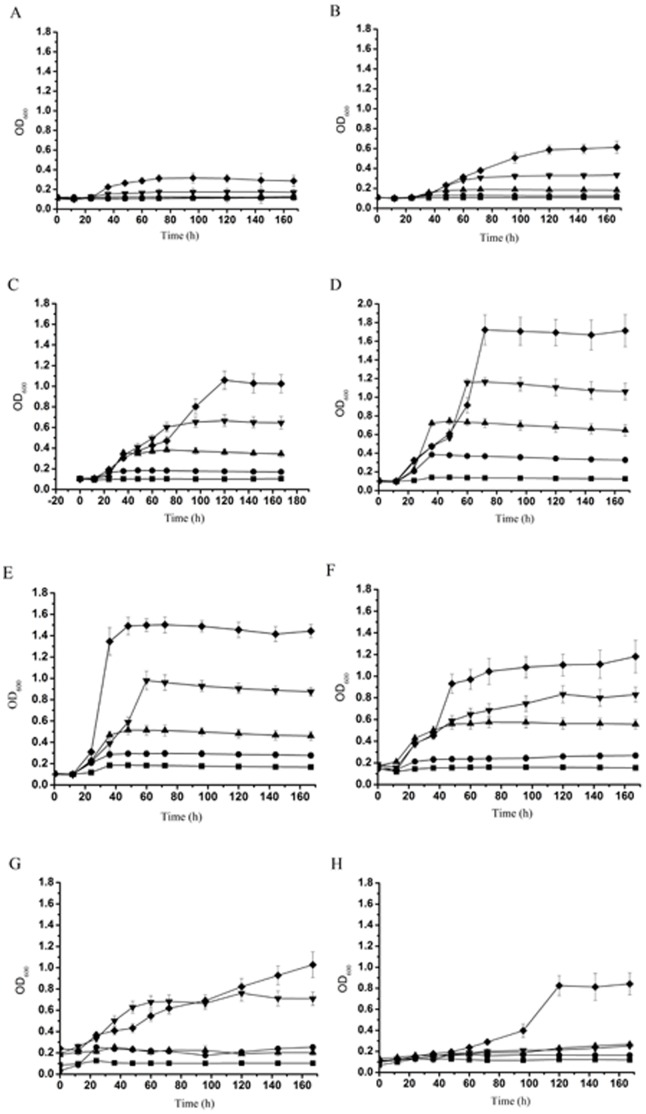
The SJTD-1-mediated breakdown and utilization of n-alkanes was studied by monitoring its cell growth for seven days in a BSM medium containing a variety of n-alkanes (C10–C24) at various concentrations (100 mg/L to 2 g/L) (Fig. 2). Cells started multiplying immediately after incubation and reached the exponential growth phase between day 1 and day 3. Of all the n-alkanes, n-hexadecane caused the highest amount of cell growth, followed by n-octadecane and n-eicosane, thus suggesting that n-hexadecane was probably the best available carbon source for strain SJTD-1 in this set of experiments. At higher concentrations of n-alkanes, strain SJTD-1 accumulated more biomass (as reflected by OD600), and also required more time to reach the stationary phase. The stationary phase was attained after 36 h when 500 mg/L of n-tetradecane, n-hexadecane, n-octadecane, and n-eicosane were used. At 1 g/L of n-dodecane and n-docosane, the stationary phase was reached after 60 h and 36 h, respectively. When 2 g/L n-tetracosane was used, the stationary phase was attained after nearly 100 h (Fig. 2).
Figure 2. Growth curves of P. aeruginosa SJTD-1 in n-alkanes.
Strain SJTD-1 was cultured in BSM supplemented with different concentrations (▪ 100 mg/L, • 250 mg/L, ▴ 500 mg/L, ▾ 1,000 mg/L, and ♦ 2,000 mg/L) of n-alkanes: n-decane (A), n-dodecane (B), n-tetradecane (C), n-hexadecane (D), n-octadecane (E), n-eicosane (F), n-docosane (G), and n-tetracosane (H) for seven days at 30°C. The data are expressed as mean values and the standard deviations are shown.
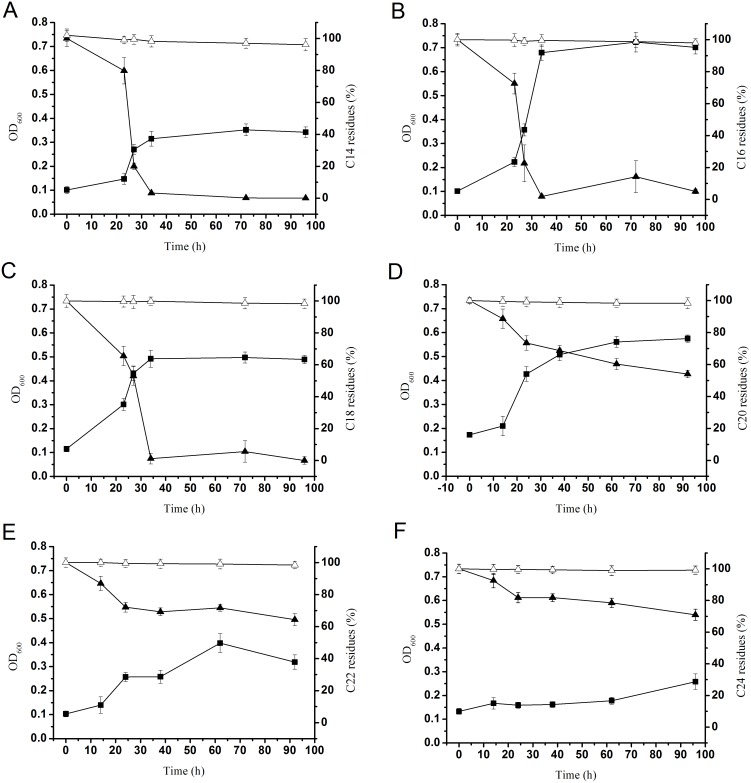
We also determined the n-alkane degradation efficiency of strain SJTD-1 with GC-MS. The corresponding time courses are illustrated in Fig. 3. GC-MS results showed that substrate consumption was closely followed by biomass increase (the loss due to vaporization and extraction was excluded by running two blank controls). As shown in Fig. 3A to 3C, more than 95% of 500 mg/L n-tetrodecane, n-hexadecane, and n-octadecane were degraded within 36 h (from 500 mg/L to about 15 mg/L, 5 mg/L, and 5 mg/L, respectively). Complete degradation occurred in less than 48 h, and the biomass at OD600 of n-tetrodecane, n-hexadecane, and n-octadecane increased to 0.35, 0.72, and 0.50, respectively. It was obvious that the degradation rates of strain SJTD-1 for various lengths of n-alkanes were different. Strain SJTD-1 could degrade n-eicosane, n-docosane, and n-tetracosane completely until the end of the experiments (96 h or 160 h). Only about 50% of the n-eicosane was degraded in 96 h. Although a rapid decline to about 70% residues was observed in the first 24 h, the degradation rate decreased in subsequent hours and the net biomass accumulation corresponded to an OD600≈0.58 (Fig. 3D). Similarly, the utilization efficiencies for n-docosane and n-tetracosane were about 35% and 25%, respectively, and their respective biomass reached an OD600 of approximately 0.39 and 0.26, respectively (Fig. 3E and 3F). All these indicated that strain SJTD-1 degraded the medium-chain n-alkanes more rapidly than the long-chain n-alkanes, probably due to the much higher hydrophobicity of the latter [17], [28], [29].
Figure 3. Growth of strain SJTD-1 in different n-alkanes (500 mg/L) and its consumption of n-alkanes.
▪ indicated as OD600 in the left vertical axis; ▴expressed as n-alkane residues detected by GC-MS with respect to abiotic controls in the right vertical axis); Δ indicated as the n-alkane residues of the abiotic controls. A, B, C, D, E, and F represented the n-alkanes C14, C16, C18, C20, C22, and C24, respectively. Standard errors were calculated from three independent determinations.
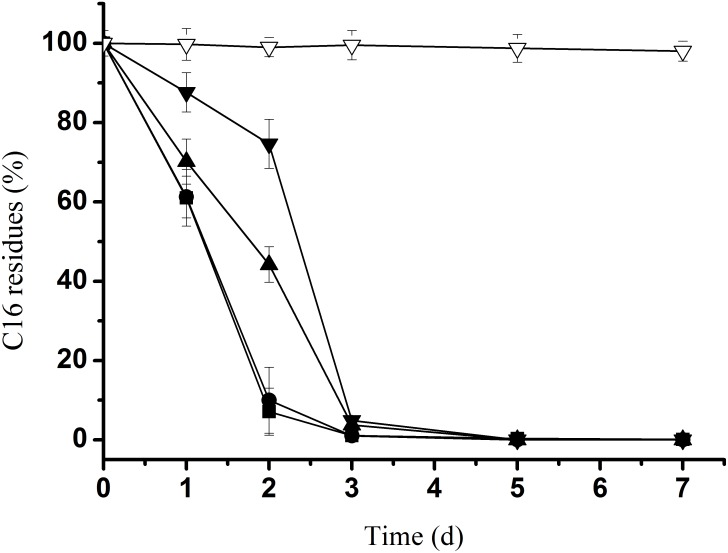
Moreover, to find the highest concentration of n-alkanes that the SJTD-1 strain could tolerate and utilize for growth, we analyzed its degradation efficiency for n-hexadecane (concentration ranging from 250 mg/L to 2.0 g/L) because n-hexadecane was possibly its greatest carbon source. As shown in Fig. 4, all the n-hexadecane could be completely degraded in three days. Additionally, 250 mg/L and 500 mg/L of n-hexadecane were totally degraded within 36 h; as for 1.0 g/L and 2.0 g/L n-hexadecane, approximately one more day was needed for a complete degradation. Therefore, we concluded that SJTD-1 could tolerate and completely biotransform 2.0 g/L n-hexadecane into a relatively large amount of biomass (OD600 = 1.7, Fig. 2). This result reveals its great tolerance ability, fast degradation speed, and high utilization efficiency for n-alkanes.
Figure 4. Quantitative estimation of the SJTD-1-mediated degradation of n-hexadecane.
The strain was cultured in BSM supplemented with 250 mg/L (▪), 500 mg/L (•), 1,000 mg/L (▴), and 2,000 mg/L (▾) of n-hexadecane at 30°C for seven days; Δ indicated as the n-alkane residues of the abiotic controls. Standard errors were calculated from three independent determinations.
Sequence analysis and alignment of the alkane-degradation genes in strain SJTD-1
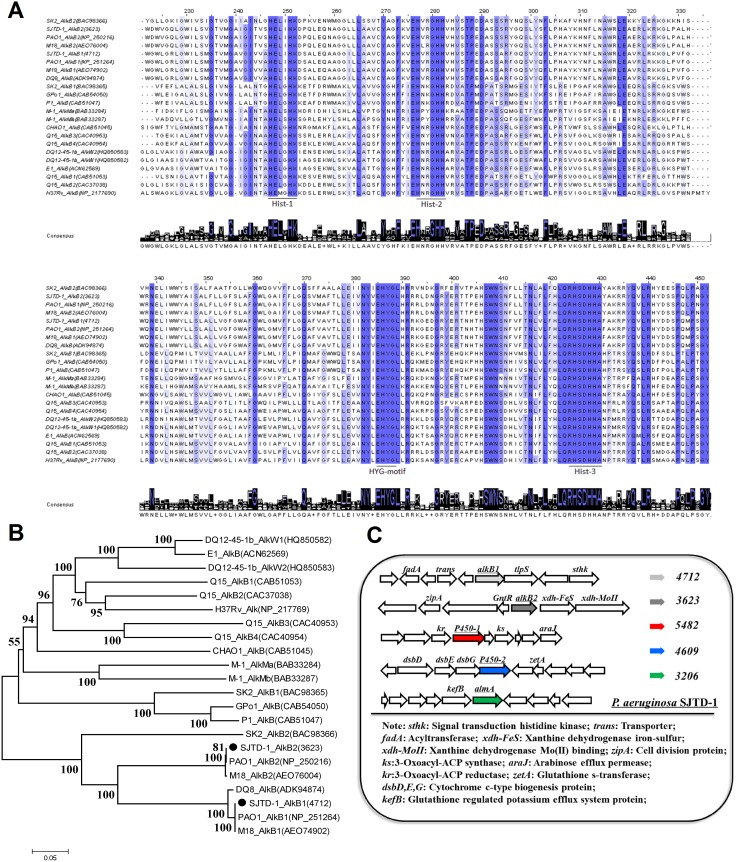
The whole-genome DNA sequence of strain SJTD-1 has been obtained and deposited in GenBank under the accession number AKCM00000000 [23]. It was also annotated by the RAST server. Two putative genes encoding AlkB monooxygenases were found to be involved in the first step of alkane degradation. Gene 4712 (alkB1) encodes a putative AlkB1 monooxygenase, comprising 383 amino acids, and with an estimated molecular weight of 42.1 kDa. The second alkB gene 3623 (alkB2) encodes AlkB2 monooxygenase, comprising 378 amino acids and with an estimated mass of 41.6 kDa. The sequence of alkB1 and alkB2 genes exhibited 78% identity, and their derived proteins exhibited 68% identity. The cross-genera sequence alignments of AlkB1 and AlkB2 with other published AlkB sequences indicated that all of these alkane hydroxylases share one conserved HYG motif and three highly conserved regions of the Hist boxes containing eight histidines [30]–[32] (Fig. 5A).
Figure 5. Sequence alignment of alkane hydroxylases AlkB1 and AlkB2 from several strains and genetic distribution of five putative alkane hydroxylases in P. aeruginosa SJTD-1.
Strains list: SJTD-1, P. aeruginosa SJTD-1; PAO1, P. aeruginosa PAO1; DQ8: P. aeruginosa DQ8; GPo1, P. putida Gpo1; P1, P. putida P1; CHAO: P. fluorescens CHAO; SK2, Alcanivorax borkumensis SK2; M-1, Acinetobacter sp. M-1; Q15, Rohodococcus erythropolis Q15; H37Rv, Mycobacterium tuberculosis H37Rv; DQ12-45-1b, Dietzia sp. DQ12-45-1b; E1, Dietzia sp. E1. (A) The conserved amino acid residues in all the sequences are indicated with a blue background, and the dark degree is in direct proportion to the degree of conservation. The three conserved histidine boxes (Hist-1, Hist-2, and Hist-3), and one HYG motif are underlined. The conserved amino acid associated with each position is shown below the alignment (created by Clustal Origin). (B) Phylogenetic relationship based on the complete amino acid sequences of AlkB1 and AlkB2 from P. aeruginosa SJTD-1 and other published AlkB amino acid sequences. The phylogenetic tree was determined using the Kimura two-parameter distance model, and bootstrap analysis was performed with 1,000 repetitions using MEGA 5.0. (C) Schematic profile of the genomic organization of AHs homologues in P. aeruginosa SJTD-1. alkB1 and alkB2, alkane-1 monooxygenases (lawngreen and grey, separately); P450-1 and P450-2, cytochrome P450s (red and blue, respectively); almA, AlmA-like monooxygenase (green); tlpS, methyl-accepting chemotaxis protein.
To further understand the relationship between AlkB proteins, phylogenetic analysis was performed based on the characterized AlkB monooxygenases from Pseudomonas, Alcanivorax, Acinetobacter, Mycobacteria, and Rhodococci (Fig. 5B). In the phylogenetic tree, the two AlkB proteins from P. aeruginosa PAO1 and M18 were clustered as alkane monooxygenase 1, and these were close to the alkane monooxygenases from other gram-negative bacteria, such as Alcanivorax borkumensis SK2 and Acinetobacter sp M-1. In fact, the AlkBs of strain SJTD-1 showed very high sequence identity with other Pseudomonas AlkBs. For instance, the identity of SJTD-1 AlkB1 to P. aeruginosa PAO1 and M18 AlkB1 was 100%, and the AlkB2 identity was 100% and 99%, respectively. Another reported Pseudomonas strain DQ8, that could easily break down alkanes and polycyclic aromatic hydrocarbons harbored only one AlkB homologue, which was mapped to SJTD-1 AlkB2 with 100% identity [8]. In addition, two cytochrome P450 monooxygenase homologues (gene 5482 and 4609) and one AlmA-like monooxygenase homologue (gene 3206) were also found in the genome of strain SJTD-1. Sequence analysis showed that genes P450-1 (gene 5482) and P450-2 (gene 4609) encoded a 418-amino-acid-protein and a 444-amino-acid-protein, respectively; they shared 99% amino acid sequence identity to those of P. aeruginosa PAO1 and 26% to those of Alcanivorax dieselolei B-5 [33]. As for the putative almA-like gene, its protein (499 aa) displayed 56% identity with the putative flavin-containing monooxygenase of A. dieselolei B-5, and only 50% identity to the AlmA of Acinetobacter sp. DSM 17874 [34].
As shown in Fig. 5C, the genomic organization of the five alkane hydoxylases, none of which are located within the gene cluster encoding the genes responsible for alkane catabolism like those seen in P. putida GPo1 and other hydrocarbonoclastic bacteria [19], [33], [35]. It should be noted that the surrounding open reading frames of the alkB genes in strain SJTD-1 were different from those in other strains, while the rubredoxin (Rd)-encoding genes, the rubredoxin reductase-encoding genes, and the transcriptional regulatory protein-encoding genes were often located immediately downstream of the alkB gene. No rubredoxin reductase-encoding gene was sought out; and no fusion Rd domain was found near the alkB genes, as opposed to that observed in Dietzia sp. DQ12-45-1b [31]. However, some important clues were found despite the decentralized distributions over the chromosome [36]. One exception was tlpS, a gene coding for a methyl-accepting chemotaxis protein that is potentially involved in chemotaxis towards long-chain n-alkanes. It was found located downstream of alkB2 [37]. A putative transcriptional regulator of the GntR family was located upstream of AlkB2 and the intergenic region between alkB2 and gntR was only 181 bp. Therefore, it is conceivable that in strain SJTD-1, n-alkanes may be utilized through a more diversified pattern than that in other reported hydrocarbonoclastic bacteria.
Phenotypic study of the putative alkane hydroxylation genes in strain SJTD-1
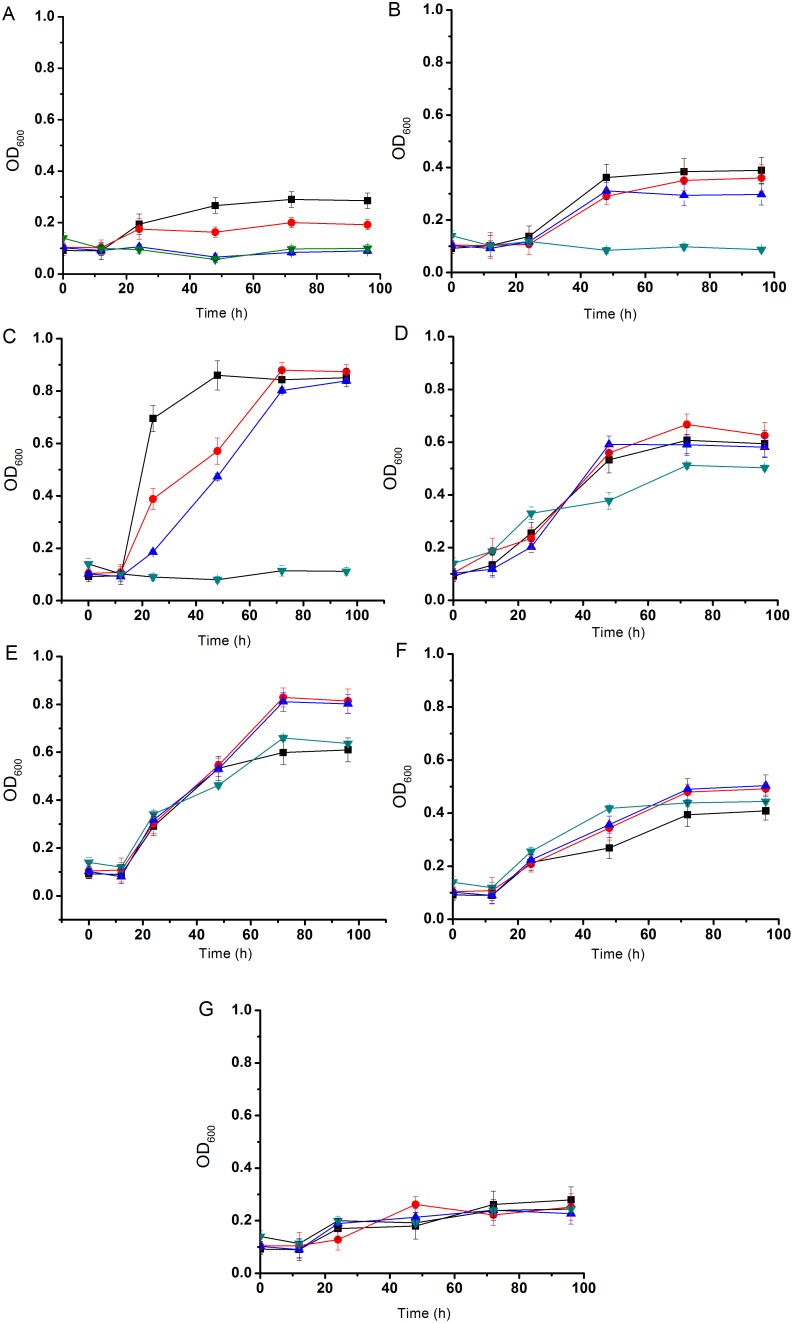
In order to determine the functions of the two alkB genes in the n-alkane oxidative process and in order to understand their preferences to n-alkanes, we knocked out the two genes separately and simultaneously, and then monitored the utilization capability of the three mutants (MutalkB1, MutalkB2, and MutalkB1&2) for the C12–C24 n-alkanes. The mutants were verified using PCR detection (Fig. S1). For the medium-chain n-alkanes (C12–C16), single-knockout mutants showed an obvious delay as compared to the wild type SJTD-1, and double-knockout mutants could not utilize the n-alkanes at all (Fig. 6A–6C). It meant that these two genes were responsible for the degradation of C12–C16 n-alkanes and that both of them were involved in the oxidative process. The alkB2 defect showed a more pronounced effect on the cell growth than did the alkB1 defect, thereby suggesting that AlkB2 played a more major role in the oxidation of medium-chain alkanes (Fig. 6A–6C and Fig. 7). Unexpectedly, the poor viability of MutalkB1&2 recovered to a normal state when C18–C24 alkanes were used as the sole source of carbon. No obvious growth decrease was found in the single-knockout mutants (Fig. 6D–6G). This implied that there were some hitherto unknown genes involved in the utilization of longer-chain n-alkanes.
Figure 6. Growth curves of P. aeruginasa SJTD-1 (▪), MutalkB1 (•), MutalkB2 (▴), and MutalkB1&2 (▾) in n-alkanes.
The strains were inoculated in BSM supplemented with 500 mg/L n-alkanes: n-dodecane (A), n-tetradecane (B), n-hexadecane (C), n-octadecane (D), n-eicosane (E), n-docosane (F), and n-tetracosane (G) and cultured for seven days at 30°C.
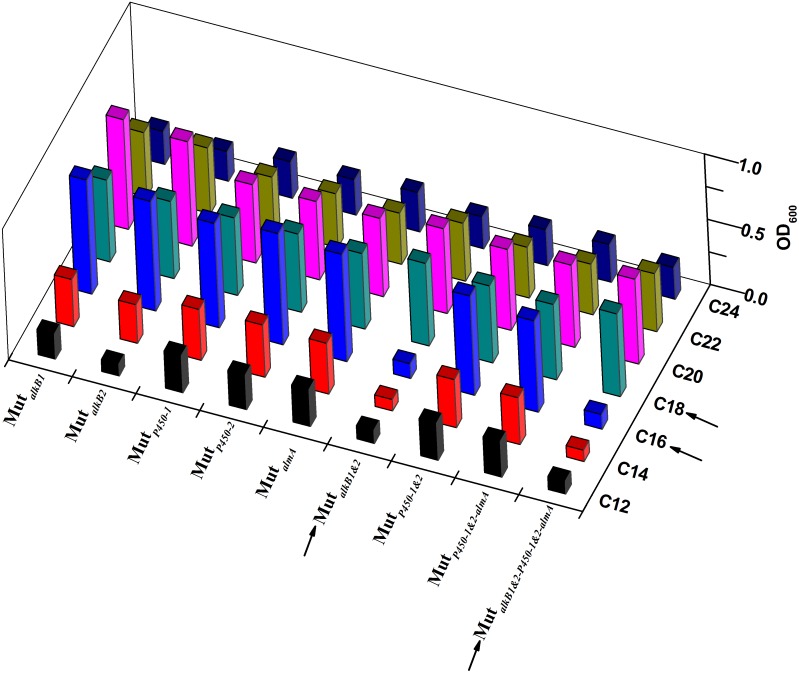
Figure 7. 3D-bar growth representations of all the defective mutants with C12–C24 n-alkanes.
Values on the horizontal axis profile the genotype of the mutants with single or multiple deletions of alkane hydroxylases; The Y-axis specifies the tested n-alkanes, coloring from black to dark blue black. The Z-axis of bar altitudes represents the OD600 for the viability of each mutant. Sharp contrasts were symbolized using arrows when both alkB1 and alkB2 were knocked out, and cultivated in C16 and C18 alkane media, respectively.
We further studied three more potential alkane hydroxylases (AH) genes (two putative cytochrome P450 genes and one almA monooxygenase gene) listed in Table 1. In order to judge the physiological role of each putative AH, single-knockout, double-knockout or multi-knockout mutants were obtained, and their viabilities were evaluated in the BSM media supplemented with n-alkanes (Fig. 7). On the whole, the divergence of the mutants’ viabilities in Fig. 7 resulted from the dissimilar enzymatic preferences of AHs within the range of the substrate spectrum. Strains with absence of two AlkBs almost failed to grow on C12–C16 alkanes; whereas P450 and AlmA made a minor contribution to the degradation of these substrates, as their defective strains still grew to a comparable level relative to the parent. However, it could be concluded that the recovery of viability originated from the transcription of P450 and AlmA oxygenases, as mutant MutalkB1&2-P450-1&2-almA showed a similar growth level as that of the wild type for C18–C24. Therefore, an affirmative hypothesis for the contribution of other undefined AHs to microbial catabolism of long-chain alkanes (C18–C24) was made. So far, these findings explain the metabolic roles of identified AHs within their preferred substrate range and further provided clues for exploring the uncharacterized alkane hydroxylases.
Transcriptional expression profiles of the alkB1, alkB2, p450 and almA genes in strain SJTD-1
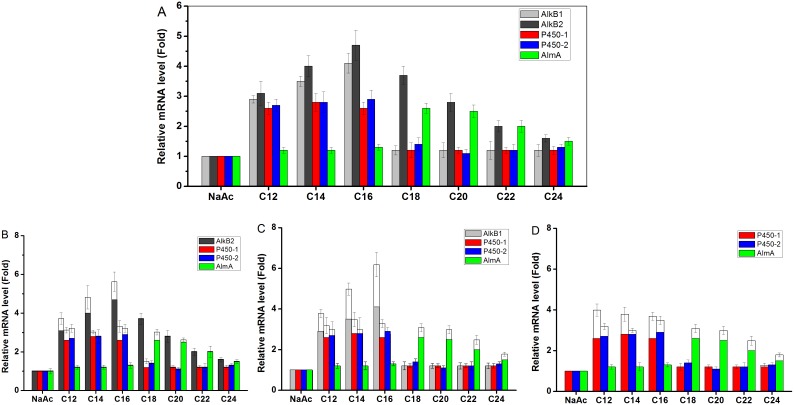
Five putative AHs in strain SJTD-1 were revealed as expected, and the diversity of AHs allowed for a rapid adaptation of SJTD-1 to the changing environments of various alkanes. To articulate the contribution of each AH to the utilization of alkanes, their transcriptional profiles were investigated. Fig. 8A shows that C12–C24 alkanes rendering the viability of SJTD-1 rested with a universal expression of AHs in any case. AlkB1 showed a relatively narrow inducer range of C12–C16 than did AlkB2, which could be induced by C12–C24 alkanes, thereby eliciting more than a two-fold change on its transcriptional level (Fig. 8A). Two P450 monooxygenases probably contributed to the mineralization of C12–C16 alkanes, because of the 2.5-fold change with those substrates. However, when C18–C24 alkanes were used, their transcriptional changes fell down sharply (Fig. 8A). The AlmA-like oxidase was expressed at a low level (1.5–3 fold change), only when the C18–C24 n-alkanes were used (Fig. 8A). So far, it was facile to explain that the abundant transcription of AHs in the hexadecane-induced cells lead to the most significant utilization of n-hexadecane during SJTD-1 growth in Fig. 2.
Figure 8. Transcriptional levels of five putative AHs genes in wild type SJTD-1 and in three knockouts.
Relative expression levels were determined with real-time RT-PCR. 16S rRNA gene was used as the reference gene. The expression levels of genes in strain SJTD-1 (A), strain MutalkB1 (B), strain MutalkB2 (C), and strain MutalkB1&2 (D) were shown. All strains were grown in different n-alkanes (C12 to C24 alkanes as the sole carbon source), and cells grown with sodium acetate were used as controls. Standard errors were calculated from three independent determinations.
As the single deletion of the five AHs made little impact on the cell growth, the transcription levels of the targeted AHs were analyzed in the alkB1-defective, alkB2-defective, and double-deletion mutants. A functional complementation of AHs was observed during the cultivation period. As illustrated in Fig. 8B and 8C, the transcriptional level of an alternative AlkB hydroxylase was enhanced and it was thus responsible to terminally oxidize n-alkanes, particularly when one AlkB was absent. This terminal oxidation of n-alkanes was limited as indicated by the overlapping area of both AlkB substrate spectrums, i.e. C12–C16 alkanes. Among these substrates, two P450s also displayed changes in expression and contributed to the complementary effects of AlkBs. Interestingly, the AlmA-like oxygenase increased its transcription to some extent (within the range of C18–C24 alkanes) when alkB2 was removed rather than alkB1 (Fig. 8B–8D). Therefore, the less attenuated viability of strain SJTD-1 on alkanes even when any alkB gene was inactivated can be attributed to the upregulation of functionally-related AHs and their metabolic contribution.
As mutants lacking both the AlkB hydroxylases showed poor propagation on C12–C16 alkanes, but did not affect the duplication rate on the C18 alkane, the transcription of two P450 and AlmA-like oxygenases was surveyed as depicted in Fig. 8D. However, no obvious alteration was found for these AHs in the single or double alkB mutants. Mutants with a consecutive deletion of five AHs still survived on n-octadecane and also duplicated to a normal level of OD600 (Fig. 7). This strongly suggested that the biodegradation of C18–C24 alkanes differed from that of the C12–C16 medium chain alkanes, and that a cluster of unidentified AHs may participate in this process, by transforming the long chain alkanes into lower hydrocarbons.
Discussion
P. aeruginosa strain SJTD-1, a new soil isolate, was determined as a hydrocarbonoclastic consumer of medium- and long-chain alkanes (C12–C30). Based on the 16S rDNA sequence alignments and phenotype analysis, strain SJTD-1 showed close homogeneity to the well-documented P. aeruginosa PAO1 and P. aeruginosa DQ8. However, strain SJTD-1 exhibited no human pathogenicity like PAO1, and it degraded polycyclic aromatic hydrocarbons only to some extent, unlike DQ8. In fact, a relatively narrower substrate range of C12–C24 supported a sustainable growth of strain SJTD-1. For this range of n-alkanes, strain SJTD-1 showed outstanding degradation efficiency and utilization speed. n-Hexadecane followed by n-tetradecane and n-octadecane were assimilated as preferential carbon sources for this strain as these hydrocarbons could be rapidly biodegraded by SJTD-1. n-Tetradecane, n-hexadecane, and n-octadecane (at a concentration of 500 mg/L) were completely degraded within 36 h. SJTD-1 could also tolerate and degrade 2.0 g/L n-hexadecane within three days. As compared to other alkane-utilizing strains, strain SJTD-1 degrades the same type and same amount of n-alkanes more efficiently and more quickly. Most of the alkane-consuming strains mineralize equivalent amounts of alkanes at a much slower rate and require a much longer degradation time (Table 2). When 500 mg/L of n-octadecane was depleted by strain SJTD-1 in 36 h, Alcanivorax sp. 2B5 simply consumed 16.07% of this substrate at the beginning of 48 h. In contrast to P. aeruginosa DQ8, strain SJTD-1 showed a comparable transformation velocity but the initial concentration of n-tetradecane was 5-fold higher in this case.
Table 2. Comparison of alkane degradation by different strains.
| Strain | Substrates | Conservation rate | Sources/Reference |
| P. aeruginosa SJTD-1 | C14, C16, C18 | >99% in 36 h | This study |
| 500 mg/mL | |||
| P. aeruginosa DQ8 | C14, 100 mg/L | >99% in 24 h* | [8] |
| Alcanivorax sp. 2B5 | C18, 500 mg/L | 16.07% in 48 h | [32] |
| >99% in 96 h | |||
| Acinetobacter | C16, 400 mg/L | >99% in 60 h | [39] |
| Dietzia DQ12-45-1b | C16, 2.5 mg/mL | 31% in 21 d | [11] |
*with 0.005% yeast extract in the medium.
In order to decipher the mechanism by which SJTD-1 breaks down the n-alkanes, the initial but rate-limiting step deserves special attention. This step marks the beginning of the terminal oxidation catalyzed by alkane hydroxylases. The genome sequencing results indicated that at least five alkane hydroxylases in strain SJTD-1 were involved in the oxidization process. Through transcriptional analysis, two alkB genes were found to be greatly induced by n-alkanes, with AlkB2 sharing a broader substrate range. The two P450 monooxygenases were induced only when medium-length alkanes were used, whereas the C18–C24 alkanes were able to regulate the expression of the AlmA oxygenase, which is known to be involved in the oxidation of super long-chain alkanes (>C30) [34]. The fact that n-hexadecane showed a prominent growing effect for strain SJTD-1 was evident from the topmost transcription of AHs during hexadecane-induced culture, as compared to the other n-alkanes. A transcriptional enhancement of two AlkBs was evident when either of the AlkBs was inactivated, strongly suggesting their overlapping substrate spectrum and functional identities.
In order to specify the roles of AHs in the alkane degradation, some important defective strains were deliberately constructed and their phenotypic profiles were represented under the given range of C12–C24 alkanes. For medium chain alkanes (C12–C16), AlkB hydroxylases showed a dramatic performance, and bacterial growth was seriously influenced when the two alkB genes were deleted. Moreover, the defective strain in absence of AlkB2 grew at a slower rate than the AlkB1 defective mutant did. It was therefore speculated that AlkB2 played a central role in the catabolism of C12–C16 alkanes, which was more important than the role played by AlkB1. The other three AHs including P450 and AlmA oxygenases had little effect on n-alkane degradation, although their transcription would somewhat offset the deletion of the alkB genes. This implies a heavy redundancy of AHs for the microbial degradation of n-alkanes. However when C18–C24 n- alkanes were used, strain SJTD-1 utilized another set of alkane hydroxylases to metabolize these n-alkanes. Therefore, it is important to understand how strain SJTD-1, in the absence of all known AHs, still manages to survive on the long-chain alkanes.
The key factors involved in the degradation of alkanes, such as AlkB, rubredoxin, and other related alkane hydroxylases, showed a scattered distribution in the genome of strain SJTD-1, without the occurrence of gene clusters as represented in Pseudomonas putida cells. This was also observed among other P. aeruginosa, such as P. aeruginosa PAO1 and P. aeruginosa RR1. However, their sporadic distribution did not compromise the bacterial utilization of n-alkanes. This indicates that a compensatory mechanism was responsible for the regulation and coordination of these multi-step catalytic reactions. For instance, it has been proposed that genes encoding two significant electron transfer proteins, viz. rubredoxin and rubredoxin reductase, were distant from the inducible alkB genes and constitutively expressed in any case [38]. To better understand the mechanism behind this disordered genetic organization, we need to resort to a comprehensive approach, such as an iTRAQ-based proteomics study, which would systematically characterize the global response of strain SJTD-1 to various types of medium- and long-chain n-alkanes.
Supporting Information
PCR verification of the mutant strains. The genomic DNA of all the mutants as well as the wild type P. aeruginosa SJTD-1 was extracted and amplified with the test primers listed in Table S1. The colonies with shorter PCR products (400–500 bp) represented successful deletions. (A) The PCR products of MutalkB1, MutalkB1&2, and wild type SJTD-1; (B) The PCR products of MutalmA and wild type SJTD-1; (C)The PCR products of MutP450-1, MutP450-2, MutP450-1&2, and wild type SJTD-1.
(TIF)
List of primers used in this work. All primers are listed from 5′ to 3′. The F and R primers represent forward and reverse primers, respectively.
(DOCX)
Nucleotide and amino acid sequences of five Alkane Hydroxylases (AHs) in P. aeruginosa SJTD-1. The five AHs are AlkB1, AlkB2, P450-1, P450-2 and AlmA-like. Gene numbers presented here are the numbers annotated in RAST server.
(DOCX)
Acknowledgments
Thanks Jian He of Nanjing Agricultural University for his support in the isolation of this strain, and thanks Wenjuan Yu of Instrumental Analysis Center of SJTU for her support in the GC-MS detection of n-alkanes.
Funding Statement
This work was sponsored by the National Science Foundation of China (Grant No. 31370152/21135004), Shanghai Pujiang Program and Chen Xing Grant of Shanghai Jiaotong University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mohanty S, Jasmine J, Mukherji S (2013) Practical considerations and challenges involved in surfactant enhanced bioremediation of oil. BioMed. Res. Int. 2013: 328608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Camilli R, Reddy CM, Yoerger DR, Van Mooy BA, Jakuba MV, et al. (2010) Tracking hydrocarbon plume transport and biodegradation at Deepwater Horizon. Science 330: 201–204. [DOI] [PubMed] [Google Scholar]
- 3. Valentine DL, Kessler JD, Redmond MC, Mendes SD, Heintz MB, et al. (2010) Propane respiration jump-starts microbial response to a deep oil spill. Science 330: 208–211. [DOI] [PubMed] [Google Scholar]
- 4. Rehm H, Reiff I (1981) Mechanisms and occurrence of microbial oxidation of long-chain alkanes. Adv. Biochem. Eng. 19: 175–215. [Google Scholar]
- 5. Labinger JA, Bercaw JE (2002) Understanding and exploiting C-H bond activation. Nature 417: 507–514. [DOI] [PubMed] [Google Scholar]
- 6. Rojo F (2009) Degradation of alkanes by bacteria. Environ. Microbiol. 11: 2477–2490. [DOI] [PubMed] [Google Scholar]
- 7.Prince RC (2005) ASM Press, Washington, DC. In Ollivier, B., Magot, M. (Eds.), Petroleum Microbiology. The microbiology of marine oil spill bioremediation. 317–336.
- 8. Zhang Z, Hou Z, Yang C, Ma C, Tao F, et al. (2011) Degradation of n-alkanes and polycyclic aromatic hydrocarbons in petroleum by a newly isolated Pseudomonas aeruginosa DQ8. Bioresour. Technol. 102: 4111–4116. [DOI] [PubMed] [Google Scholar]
- 9. Maeng JH, Sakai Y, Tani Y, Kato N (1996) Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J. Bacteriol. 178: 3695–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Hamme JD, Ward OP (2001) Physical and metabolic interactions of Pseudomonas sp. strain JA5-B45 and Rhodococcus sp. strain F9-D79 during growth on crude oil and effect of a chemical surfactant on them. Appl. Environ. Microbiol. 67: 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang XB, Chi C-Q, Nie Y, Tang YQ, Tan Y, et al. (2011) Degradation of petroleum hydrocarbons (C6–C40) and crude oil by a novel Dietzia strain. Bioresour. Technol. 102: 7755–7761. [DOI] [PubMed] [Google Scholar]
- 12. Spiers AJ, Buckling A, Rainey PB (2000) The causes of Pseudomonas diversity. Microbiology-UK 146: 2345–2350. [DOI] [PubMed] [Google Scholar]
- 13. Alonso A, Rojo F, Martínez JL (1999) Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1: 421–430. [DOI] [PubMed] [Google Scholar]
- 14. van Beilen JB, Wubbolts MG, Witholt B (1994) Genetics of alkane oxidation by Pseudomonas oleovorans . Biodegradation 5: 161–174. [DOI] [PubMed] [Google Scholar]
- 15.van Beilen JB, Veenhoff L, Witholt B (1998) Elsevier Science B.V., Amsterdam, Netherlands. In K. Kieslich, C. P. van der Beek, J. A. M. de Bont, and W. J. J. van den Tweel (ed.), New frontiers in screening for microbial biocatalysts. Alkane hydroxylase systems in Pseudomonas aeruginosa strains able to grow on n-octane, 211–215.
- 16. Yuste L, Corbella ME, Turiégano MaJ, Karlson U, Puyet A, et al. (2000) Characterization of bacterial strains able to grow on high molecular mass residues from crude oil processing. FEMS Microbiol. Ecol. 32: 69–75. [DOI] [PubMed] [Google Scholar]
- 17. Smits TH, Balada SB, Witholt B, van Beilen JB (2002) Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184: 1733–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sorkhoh N, Ibrahim A, Ghannoum M, Radwan S (1993) High-temperature hydrocarbon degradation by Bacillus stearothermophilus from oil-polluted Kuwaiti desert. Appl. Microbiol. Biotechnol. 39: 123–126. [Google Scholar]
- 19. Van Beilen JB, Marin MM, Smits TH, Röthlisberger M, Franchini AG, et al. (2004) Characterization of two alkane hydroxylase genes from the marine hydrocarbonoclastic bacterium Alcanivorax borkumensis. Environ. Microbiol. 6: 264–273. [DOI] [PubMed] [Google Scholar]
- 20. Santos PM, Benndorf D, Sa-Correia I (2004) Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics. Proteomics 4: 2640–2652. [DOI] [PubMed] [Google Scholar]
- 21. Hao R, Lu A, Wang G (2004) Crude-oil-degrading thermophilic bacterium isolated from an oil field. Can. J. Microbiol 50: 175–182. [DOI] [PubMed] [Google Scholar]
- 22. Liu H, Liang R, Tao F, Ma C, Liu Y, et al. (2012) Genome sequence of Pseudomonas aeruginosa strain SJTD-1, a bacterium capable of degrading long-chain alkanes and crude oil. J. Bacteriol. 194: 4783–4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holt J, Krieg N, Sneath P, Staley J, Williams S (1994) Williams & Wilkins, Baltimore. International edition: Bergey's manual of determinative bacteriology.
- 24.Joseph S, David W (2001) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. Molecular cloning: a laboratory manual.
- 25. Liang R, Liu J (2010) Scarless and sequential gene modification in Pseudomonas using PCR product flanked by short homology regions. BMC Microbiol. 10: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP (1998) A broad-host-range Flp- FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212: 77–86. [DOI] [PubMed] [Google Scholar]
- 27. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 28. Feng L, Wang W, Cheng J, Ren Y, Zhao G, et al. (2007) Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Nat. Acad. Sci. USA 104: 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Throne-Holst M, Markussen S, Winnberg A, Ellingsen TE, Kotlar H-K, et al. (2006) Utilization of n-alkanes by a newly isolated strain of Acinetobacter venetianus: the role of two AlkB-type alkane hydroxylases. Appl. Microbiol. Biotechnol. 72: 353–360. [DOI] [PubMed] [Google Scholar]
- 30. Whyte L, Smits T, Labbe D, Witholt B, Greer C, et al. (2002) Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl. Environ. Microbiol. 68: 5933–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nie Y, Liang J, Fang H, Tang YQ, Wu XL (2011) Two novel alkane hydroxylase-rubredoxin fusion genes isolated from a Dietzia bacterium and the functions of fused rubredoxin domains in long-chain n-alkane degradation. Appl Environ. Microbiol. 77: 7279–7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu YC, Li LZ, Wu Y, Tian W, Zhang LP, et al. (2010) Isolation of an alkane-degrading Alcanivorax sp. strain 2B5 and cloning of the alkB gene. Bioresour. Technol. 101: 310–316. [DOI] [PubMed] [Google Scholar]
- 33. Liu C, Wang W, Wu Y, Zhou Z, Lai Q, et al. (2011) Multiple alkane hydroxylase systems in a marine alkane degrader, Alcanivorax dieselolei B-5. Environ. Microbiol. 13: 1168–1178. [DOI] [PubMed] [Google Scholar]
- 34. Throne-Holst M, Wentzel A, Ellingsen TE, Kotlar H-K, Zotchev SB (2007) Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl. Environ. Microbiol. 73: 3327–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Beilen JB, Panke S, Lucchini S, Franchini AG, Röthlisberger M, et al. (2001) Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology 147: 1621–1630. [DOI] [PubMed] [Google Scholar]
- 36. Stover C, Pham X, Erwin A, Mizoguchi S, Warrener P, et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964. [DOI] [PubMed] [Google Scholar]
- 37. Smits TH, Witholt B, van Beilen JB (2003) Functional characterization of genes involved in alkane oxidation by Pseudomonas aeruginosa. Antonie Van Leeuwenhoek. 84: 193–200. [DOI] [PubMed] [Google Scholar]
- 38. Marín MM, Yuste L, Rojo F (2003) Differential expression of the components of the two alkane hydroxylases from pseudomonas aeruginosa. J. Bacteriol. 185: 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun JQ, Xu L, Tang YQ, Chen FM, Wu XL (2012) Simultaneous degradation of phenol and n-hexadecane by Acinetobacter strains. Bioresour. Technol. 123: 664–668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR verification of the mutant strains. The genomic DNA of all the mutants as well as the wild type P. aeruginosa SJTD-1 was extracted and amplified with the test primers listed in Table S1. The colonies with shorter PCR products (400–500 bp) represented successful deletions. (A) The PCR products of MutalkB1, MutalkB1&2, and wild type SJTD-1; (B) The PCR products of MutalmA and wild type SJTD-1; (C)The PCR products of MutP450-1, MutP450-2, MutP450-1&2, and wild type SJTD-1.
(TIF)
List of primers used in this work. All primers are listed from 5′ to 3′. The F and R primers represent forward and reverse primers, respectively.
(DOCX)
Nucleotide and amino acid sequences of five Alkane Hydroxylases (AHs) in P. aeruginosa SJTD-1. The five AHs are AlkB1, AlkB2, P450-1, P450-2 and AlmA-like. Gene numbers presented here are the numbers annotated in RAST server.
(DOCX)