Abstract
Ultraviolet (UV) radiation impairs intracellular functions by directly damaging DNA and by indirectly generating reactive oxygen species (ROS), which induce cell cycle arrest and apoptosis. UV radiation can also alter gene expression profiles, including those of mRNA and microRNA (miRNA). The effects of UV radiation on cellular functions and gene expression have been widely documented in human skin cells such as keratinocytes, melanocytes and dermal fibroblasts, but the effect it has on other types of skin cell such as dermal papilla cells, which are crucial in the induction of hair follicle growth, remains unknown. In the current study, the effect of UV radiation on physiological changes and miRNA-based expression profiles in normal human dermal papilla cells (nHDPs) was investigated. UVB radiation of ≥50 mJ/cm2 displayed high cytotoxicity and apoptosis in a dose-dependent manner. In addition, ROS generation was exhibited in UVB-irradiated nHDPs. Furthermore, using miRNA microarray analysis, it was demonstrated that the expression profiles of 42 miRNAs in UVB-irradiated nHDPs were significantly altered compared with those in the controls (35 upregulated and 7 downregulated). The biological functions of the differentially expressed miRNAs were studied with gene ontology analysis to identify their putative target mRNAs, and were demonstrated to be involved in cell survival- and death-related functions. Overall, the results of the present study provide evidence that miRNA-based cellular mechanisms may be involved in the UVB-induced cellular response in nHDPs.
Keywords: microRNA, ultraviolet B, human dermal papilla cells, microarray
Introduction
Previous studies have established that chronic exposure to solar radiation leads to skin damage (1,2). Ultraviolet (UV) radiation, which can be categorized into UVA, UVB and UVC according to wavelength, has the potential to cause DNA damage, leading to sunburn and skin cancer (3). UVC radiation has the shortest wavelength and thus emits the highest energy levels (4), but the majority of UVC from sunlight is absorbed by the atmosphere, in particular the ozone layer, so is not a threat to health. UVA and UVB, however, reach the skin by penetrating the atmosphere (5), and in hair follicles, androgenetic alopecia is a result of UV-induced photo-aggravated dermatitis (6). Additionally, UV represses growth and cycling of hair follicles and follicular melanogenesis in vitro (7).
microRNAs (miRNAs) are small, non-coding RNAs that regulate mRNA translation (8,9) and have been implicated in the regulation of apoptosis, survival and differentiation (9). UV radiation has been demonstrated to regulate miRNAs in various types of cell (10,11), including melanocytes, in which UV-induced miR-145, miR-148 and miR-25 regulate pigmentation by repressing Myo5a and MITF (12,13). miR-125b and miR-22 promote cell survival by targeting p38α and PTEN following UV irradiation (10,14). Additionally, Pothof et al (11) implicated miRNA-mediated gene interference in the UV-induced DNA damage response. In other studies, miRNA expression in UV-irradiated mouse epidermis and human keratinocytes was profiled via microarray analysis (15,16). However, despite these studies, changes in miRNA expression in response to UV radiation remain unclear in human dermal papilla cells. Therefore, in the present study, global miRNA expression in UVB-irradiated human dermal papilla cells was profiled and bioinformatics were utilized to identify putative miRNA target genes and their associated biological functions. The data from the current study may provide insights into a novel mechanism of UV-dependent damage in human dermal papilla cells.
Materials and methods
Cell culture and UVB irradiation
Normal human dermal papilla cells (nHDPs) were obtained from Cell Engineering For Origin (Seoul, Korea). nHDPs were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA), 5,000 U/ml penicillin G and 5,000 μg/ml streptomycin. The cells were incubated at 37˚C in 5% CO2 humidity.
Cells were exposed to UVB radiation using a G8T5E lamp (Sankyo Denki, Toshima, Japan). Doses were measured with a UV light meter (Lutron UV-340; Lutron Electronic Enterprise Co., Ltd., Taipei, Taiwan). nHDPs were seeded in 60-mm culture dishes and incubated for 24 h, then washed and resuspended in phosphate-buffered saline (PBS) prior to exposure to UVB. The cells were placed in fresh medium following irradiation. Non-exposed control samples were maintained in the dark under the same conditions.
RNA extraction and miRNA microarray
All materials were obtained from Agilent Technologies (Santa Clara, CA, USA) unless otherwise stated. Total RNA was extracted using RiboEx™ (Geneall, Seoul, Korea) according to the manufacturer’s instructions. RNA stability was confirmed using the Bioanalyzer 2100. Nucleic acid purity was calculated from A260/A280 and A260/A230 ratios using the MaestroNano spectrophotometer (Maestrogen, Las Vegas, NV, USA). miRNA expression profiles were analyzed using the SurePrint G3 Human v16.0 miRNA 8x60K Microarray kit (based on miRBase release 19.0), which included 1,368 probes representing 1,205 human miRNAs. Total RNA (100 ng) was dephosphorylated using calf intestine alkaline phosphatase (CIP) and denatured by heat inactivation with dimethyl sulfoxide (DMSO). The dephosphorylated RNA was labeled with pCp-Cy3 using T4 RNA ligase. Unlabeled pCp-Cy3 was removed using the Micro Bio-Spin P-6 column (Bio-Rad, Hercules, CA, USA). Labeled RNA was dried and resuspended in Hi-RPM hybridization buffer prior to hybridization with the microarray at 55˚C and 20 rpm for 20 h in an Agilent Microarray Hybridization Oven (Agilent Technologies, Santa Clara, CA, USA). Following hybridization, microarray slides were washed with wash buffers 1 and 2 and then scanned using an Agilent SureScan Microarray Scanner (Agilent Technologies). The scanned image was quantitated using Agilent Feature Extraction Software (version 10.7; Agilent Technologies). The data were analyzed using GeneSpring GX version 11.5 software.
miRNA target gene prediction and gene ontology (GO) analysis
The target genes of miRNAs that exhibited altered expression levels in response to UVB exposure were predicted using TargetScan (http://www.targetscan.org). The target genes were predicted from the high context score percentile (50–100) in the conserved and nonconserved database. The predicted target genes underwent GO analysis to identify their associated biological functions.
Cell viability
nHDPs were seeded into a 96-well plate at a density of 5x103 cells/well and incubated for 24 h. The cells were irradiated with different doses of UV (0–400 mJ/cm2) and incubated for another 24 h. The cells were then incubated with 0.5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dissolved in DMSO for 1 h. The absorbance at 490 nm was measured with an iMark Microplate Absorbance reader (Bio-Rad).
Cell cycle analysis
nHDPs were plated on 60-mm tissue culture dishes at a density of 2x106 cells/plate and then grown until 70% confluent, prior to irradiation with different doses of UVB (0–400 mJ/cm2). Following the 24-h incubation, the cells were trypsinized and fixed with 70% ethanol for 18 h at 4˚C. The cells were then resuspended in 1 ml propidium iodide (PI) staining solution (50 μg/ml PI, 0.1 μg/ml RNase, and 0.05% Triton X-100 in PBS) for 1 h. The stained cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). A minimum of 10,000 events were collected in each analysis. The various cell cycle populations were determined by ModFit LT (Verity Software House, Topsham, ME, USA).
Reactive oxygen species (ROS) staining
Intercellular ROS levels were measured using 2′,7′-dichlorofluorescein diacetate (DCF-DA) (Sigma-Aldrich) as previously described (17). nHDPs were plated onto 60-mm tissue culture dishes at a density of 2x106 cells/plate. The cells were irradiated with UVB and then incubated for 24 h prior to staining with 20 μM DCF-DA for 30 min. The stained cells were analyzed via flow cytometry using the FACSCalibur flow cytometer.
Statistical analysis
Statistical significance was determined by the Student’s t-test and data were subjected to global normalization. P<0.05 was considered to indicate a statistically significant difference.
Results
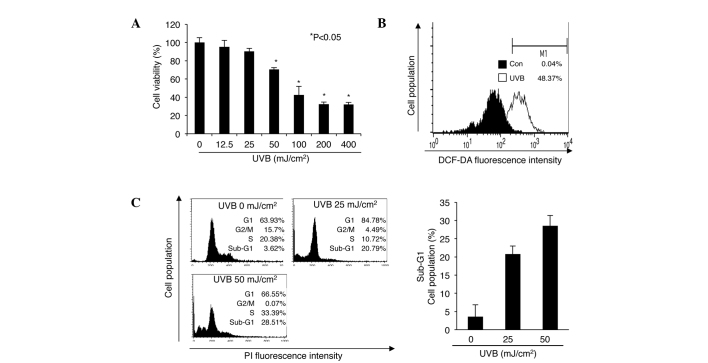
UVB irradiation decreases cell viability by increasing the occurrence of cell cycle arrest or apoptosis in nHDPs
Agents such as UV radiation, that induce DNA damage in mammalian cells, trigger growth arrest, cell cycle arrest, and apoptosis by regulating the ATM-p53 pathway (18,19). However, different types of cell exhibit varied responses to equivalent UV doses (5,20–23). Therefore, in the current study, to determine how nHDPs respond to UV irradiation, these cells were exposed to 0–400 mJ/cm2 of UVB radiation for 24 h, prior to a cell viability assessment (Fig. 1A). Cell viability was significantly reduced to 70.34% following exposure to 50 mJ/cm2 of UVB compared with that following exposure to 0 mJ/cm2. As previous studies have shown that UVB radiation regulates cell viability and apoptosis by increasing ROS production (24,25), intracellular ROS was examined using DCF-DA in the current study. The results demonstrated that ROS production increased in cells exposed to 50 mJ/cm2 UVB compared with those exposed to 0 mJ/cm2 (Fig. 1B). Analysis of cell cycle progression in cells that were exposed to 0, 25 and 50 mJ/cm2 UVB for 24 h revealed that G1-phase cell cycle arrest was induced by 25 mJ/cm2 UVB irradiation (Fig. 1C). Notably, the frequency of sub-G1 cells, which represent apoptotic cells, was increased in nHDPs exposed to 50 mJ/cm2 UVB compared with those exposed to 0 mJ/cm2.
Figure 1.
Effect of UVB irradiation on cell viability, ROS production and the cell cycle in nHDPs. (A) Effect of UVB radiation on nHDP viability. Cell viability was measured by MTT assay. Results are presented as the mean ± standard error of the percentage of control OD of triplicate samples. *P<0.05 vs. 0 mJ/cm2 UVB irradiation. (B) Effect of UVB on the levels of ROS in nHDPs. Different cell populations are represented by different colors (black, 0 mJ/cm2 UVB; white, 50 mJ/cm2 UVB). (C) Effect of UVB on the cell cycle in nHDPs. The distributions of cells of different populations in the different stages of the cell cycle were analyzed by flow cytometry using PI-stained nHDPs. The frequency of cells in the sub-G1 phase is presented as the mean ± standard error of the percentage of the gated cell population of triplicate samples. UVB, ultraviolet B; DCF-DA, 2′,7′-dichloroflorescein diacetate; PI, propidium iodide; ROS, reactive oxygen species; nHDP, normal human dermal papilla cell; OD, optical density.
miRNA expression is altered by UVB irradiation
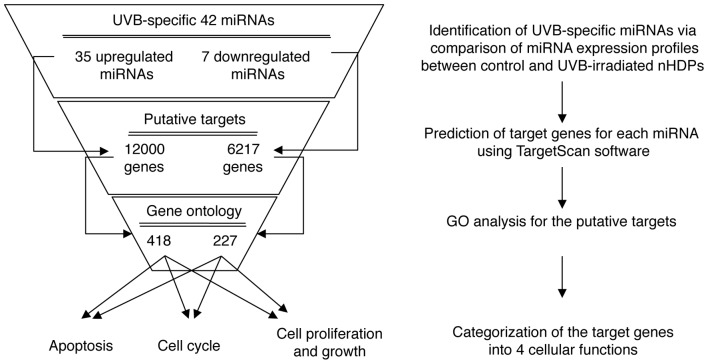
To analyze UVB-dependent changes in miRNA expression in nHDPs, miRNA microarray analysis was performed using arrays containing 1,368 probes representing 1,205 human miRNAs. Total RNA was extracted from non-irradiated and 50 mJ/cm2 UVB-irradiated nHDPs. Data from each sample were subjected to global normalization. To obtain refined data, miRNAs were selected and further considered as a result of flag-present filtration in the data file, indicating that the sensitivity of the selected miRNAs were sufficient for the microarray. A total of 183 miRNAs were selected for further analysis. miRNAs from 50 mJ/cm2 UVB-irradiated nHDPs that were upregulated (n=35) and downregulated (n=7) at least 1.5-fold compared with those of the non-irradiated control nHDPs were identified (Fig. 2), and lists of these 42 miRNAs are presented in Tables I and II.
Figure 2.
UVB radiation alters miRNA expression profiles in nHDPs. (A) Heat-map of miRNAs whose expressions levels were up- and downregulated >1.5-fold in 50 mJ/cm2 UVB-irradiated nHDPs vs. non-irradiated nHDPs. The color bar displaying fluorescence intensity corresponds to miRNA expression levels. Highly expressed miRNAs are red, while those present at low levels are blue. (B) Total number of miRNAs up- and downregulated by 50 mJ/cm2 UVB radiation in nHDPs. UVB, ultraviolet B; miRNA, microRNA; nHDP, normal human dermal papilla cell.
Table I.
miRNAs upregulated at least 1.5-fold in UVB-irradiated nHDPs.
| miRNA | Fold change | Chr |
|---|---|---|
| hsa-miR-1207-5p | 2.00 | chr8 |
| hsa-miR-1225-5p | 1.62 | chr16 |
| hsa-miR-1246 | 1.79 | chr2 |
| hsa-miR-1275 | 1.76 | chr6 |
| hsa-miR-138-5p | 1.80 | chr3 |
| hsa-miR-150-3p | 2.24 | chr19 |
| hsa-miR-181a-5p | 1.56 | chr1 |
| hsa-miR-1915-3p | 2.19 | chr10 |
| hsa-miR-193a-3p | 2.32 | chr17 |
| hsa-miR-1973 | 2.18 | chr4 |
| hsa-miR-222-3p | 2.07 | chrX |
| hsa-miR-26a-5p | 2.08 | chr3 |
| hsa-miR-27a-3p | 1.90 | chr19 |
| hsa-miR-2861 | 2.42 | chr9 |
| hsa-miR-29b-3p | 2.25 | chr1 |
| hsa-miR-30a-5p | 4.03 | chr6 |
| hsa-miR-30d-5p | 2.28 | chr8 |
| hsa-miR-3162-5p | 2.20 | chr11 |
| hsa-miR-3195 | 1.85 | chr20 |
| hsa-miR-337-5p | 1.62 | chr14 |
| hsa-miR-34a-5p | 1.64 | chr1 |
| hsa-miR-34b-5p | 1.76 | chr11 |
| hsa-miR-361-5p | 2.12 | chrX |
| hsa-miR-3651 | 1.55 | chr9 |
| hsa-miR-3663-3p | 2.16 | chr10 |
| hsa-miR-3665 | 2.34 | chr13 |
| hsa-miR-379-5p | 2.09 | chr14 |
| hsa-miR-381-3p | 1.51 | chr14 |
| hsa-miR-423-5p | 1.78 | chr17 |
| hsa-miR-4281 | 2.09 | chr5 |
| hsa-miR-630 | 2.35 | chr15 |
| hsa-miR-638 | 2.55 | chr19 |
| hsa-miR-762 | 1.89 | chr16 |
| hsa-miR-92a-3p | 1.74 | chr13 |
| hsa-miR-99b-5p | 3.28 | chr19 |
miRNA, microRNA; UVB, ultraviolet B; nHDP, normal human dermal papilla cell; Chr, chromosome.
Table II.
miRNAs downregulated at least 1.5-fold in UVB-irradiated nHDPs.
| miRNA | Fold change | Chr |
|---|---|---|
| hsa-miR-1207-5p | 2.00 | chr8 |
| hsa-miR-1225-5p | 1.62 | chr16 |
| hsa-miR-1246 | 1.79 | chr2 |
| hsa-miR-1275 | 1.76 | chr6 |
| hsa-miR-138-5p | 1.80 | chr3 |
| hsa-miR-150-3p | 2.24 | chr19 |
| hsa-miR-181a-5p | 1.56 | chr1 |
miRNA, microRNA; UVB, ultraviolet B; nHDP, normal human dermal papilla cell; Chr, chromosome.
GO analysis of UVB-specific putative miRNA target genes in nHDPs
To identify the cellular functions associated with UVB-mediated changes in the levels of miRNA expression, a multi-step bioinformatics scheme was used to predict putative miRNA target genes and their biological functions (Fig. 3). TargetScan, a sequence-based miRNA target prediction database, was used to identify 18,217 putative target genes for the up- and downregulated miRNAs. The analysis revealed 12,000 potential targets for the 35 upregulated miRNAs and 6,217 targets for the 7 downregulated miRNAs. Subsequently, the putative target genes involved in UV-mediated cellular damage, including the cell cycle, apoptosis, cell growth and proliferation, were screened. Using a GO web-based tool, AmiGO, the gene lists for the above four GO terms (cell cycle, apoptosis, cell growth and proliferation) were obtained, and these genes were compared with the putative target genes. The overlapping genes in the two groups were then identified (Fig. 3). Among the deregulated 42 miRNAs (Table I and II), the majority of miRNAs were altered >2.0-fold. Thus, the five most upregulated and downregulated miRNAs were selected to obtain more representative biological meanings for the putative genes. The predicted target genes of the top 5 most differentially upregulated and downregulated miRNAs in the UVB-irradiated HDPs are categorized by their functions in Tables III and IV, respectively.
Figure 3.
Scheme of bioinformatic analysis performed. miRNAs whose expression profiles were altered by UVB irradiation were selected by comparing the microarray data of non-irradiated and 50 mJ/cm2 UVB-exposed nHDPs. Predicted targets of the UVB-specific miRNAs were determined using TargetScan software. The target genes were then categorized into three groups, namely apoptosis, cell cycle, and cell growth and proliferation by performing GO analysis. UVB, ultraviolet B; miRNA, microRNA; nHDP, normal human dermal papilla cell; GO, gene ontology.
Table III.
Predicted target genes of the top five most differentially upregulated miRNAs in UVB-irradiated nHDPs.
| Target genes and functions | |||
|---|---|---|---|
|
|
|||
| miRNA | Cell cycle | Apoptosis | Cell growth and proliferation |
| hsa-miR-30a-5p | APBB2, RHOB, EPHB2, NF1 NOTCH2, PNN, TSC1, RECK, CUL3, CUL2, BCL10, MTBP, TBRG1, TBRG1 | ACTC1, CASP3, GJA1, HTT, IL1A, IL2RA, IL17A, MAP3K5, MLL, NAIP, PTGER3, ATXN1, TFDP1, TIA1, UNC5C, TNFSF9, TNFRSF10D, BCL10, ATG12, EBAG9, ARHGEF6, ATG5, BCL2L11, EDAR, TRIM35, SIRT1, TCTN3, CECR2, SH3KBP1, DDIT4, C8orf4, AVEN, BIRC6, TRIB3, NLRP3, RFFL, TICAM1, PRUNE2, RNF144B, BCL2L15 | ADRA1D, ADRA2A, ADRB2, IL7, TLX1, BNC1, DDX11, CAMK2D, JAG2, KRAS, LIFR, LYN, MAFG, PDGFRB, PPP2CA, SOX9, VIPR1, CDC7, CUL3, SOCS1, CREG1, NOV, SOCS3, EBAG9, CFDP1, ENOX2, C19orf10, BIRC6, CDCA7 |
| hsa-miR-99b-5p | MCC, RASSF4 | DFFB, NLRP2, RNF144B | BMPR2, IGF1R, STAT5B |
| hsa-miR-638 | CDKN2B, MCC, MPHOSPH1, XAF1 | CLU, ATN1, TNFRSF11B, PAK2, TNFRSF1B, ATG5, CIDEB, SAP30BP, XAF1, UBE2Z, FAM130A1, RHOT2, RFFL | CD47, CDK2, CLU, CTF1, GAP43, IFNG, IL11, LIF, LIFR, PAK2, TRAF5, VEGFA, HOXB13, BRD8 |
| hsa-miR-2861 | - | - | - |
| hsa-miR-630 | RHOB, CDKN2B, CYLD, NOTCH2, RB1CC1, WWOX, ZAK, C11orf82, LIN9 | XIAP, DOCK1, EP300, GJA1, PAX3, TP63, ATG12, SLK, SMNDC1, NCKAP1, CKAP2, CROP, ZAK, DDIT4, AVEN, ZMAT3, TP53INP1, C11orf82 | BMPR2, KLF5, FOXO1, IGF1R, CYR61, IL7, SSR1, TDGF1, TGFBR2, CDC7, SOCS2, RBM9, CCDC88A, ZMAT3 |
miRNA, microRNA; UVB, ultraviolet B; nHDP, normal human dermal papilla cell.
Table IV.
Predicted target genes of the top five most differentially downregulated miRNAs in UVB-irradiated nHDPs.
| Target genes and functions | |||
|---|---|---|---|
|
|
|||
| miRNA | Cell cycle | Apoptosis | Cell growth and proliferation |
| hsa-miR-218-5p | TRIM13, CDK6, CDKN1B, KHDRBS1, CETN2, MAPRE2, RCC1, FOXN3, PYHIN1, DCC, E2F2, SENP5, FANCD2, MAPRE3, SEPT2, SASH1, CLASP1, SPECC1L, SH3BP4, EGFL6, PDCD4, HLA, QB1, HPGD, BIRC5, MCC, NEDD9, ZAK, ANLN, PPP1CB, PPP1CC, RIF1, RCBTB1, FANCI, SMPD3, PCNP, CCND1, SIAH2, BRCA1, TACC1, PTP4A1, MAP9, RASSF5, PARD6B, CCNA2, CCND3, LYK5, RASSF2 | - | NAMPT, CDKN1B, NET1, SOCS4, CTGF, FLT1, KIT, KRT6A, LIF, LIFR, MST1R, NEDD9, NODAL, NRAS, PAK2, C20orf20, BIRC6, PURA, APPA2, BNC1, SHC1, SSR1, TRAF5, YEATS4, CUL3, SOCS3, SOCS6, HTRA3, CD47, SOCS5 |
| hsa-miR-450a-5p | RCC1, EGFR, CD2AP, XAF1 | - | - |
| hsa-miR-299-5p | CDKN1A, MAEA, SMC2, CTCF, SPIN1, POLS, CEP110, FOXN3, GADD45A, AHR, SENP5 , FANCD2, SIRT2, SEP2, CCNDBP1, CD2AP, RABGAP1, KIF11, NF1, NPAT, ZAK, MPHOSPH8, PPP1CB, CEP55, MTUS1, RAD21, RAP1A, AVPI1, SIAH1, PTP4A1, CDC7, PPAPDC1B, MCM8, TBRG1, CDC16, CCNG1, CCNG2, CCNH, AURKB, HDAC4 | - | IGFBP3, IL8RB, NOV, SIRPG, SSR1, TDGF1, CDC7, SOCS6, CD86, SOCS5 |
| hsa-miR-374a-5p | CDK6, SPIN1, POLS, ZWINT, ADCYAP1, ESCO2, SLC5A8, GADD45A, AHR, CCRK, CD2AP, NIPBL, EGFL6, APPL1, GAS1, MTBP, LIN9, ANXA1, HELLS, HPGD, FLJ44060, ING1, LOH11CR2A, MCM2, MCM6, MLH1, NBN, ATM, NEDD9, NEK2, NF1, ZAK, ANLN, ERBB2IP, MAPK1, MAPK6, MAPK7, PCNP, PTEN, SPC25, ARHGAP20, CCND1, NCAPG, PAPD5, BMP2, NEK4, TACC1, TFDP1, TP53BP2, SUV39H2, MAP9, CHAF1B, RECK, MCM8, PARD6B, TBRG1, CDC14A, BCL10, STARD13, CCNE2, KNTC1 | - | NAMPT, DNAJA2, UBE2E3, CHAD, CLU, SOCS4, HES1, IGFBP3, IGFBP7, IL7, CXCL10, KRAS, LIFR, MYC, NEDD9, PAK2, CRIM1, PURA, BCL2, PAPPA2, CXCL5, SLAMF1, BTC, TBX3, TSHR, VEGFA, YEATS4, SOCS6, CIAO1, NTN1, CD47, SOCS5 |
| hsa-miR-495-3p | CDKN1A, MAEA, SMC2, CTCF, SPIN1, POLS, CEP110, FOXN3, GADD45A, AHR, SENP5, FANCD2, SIRT2, SEP2, CCNDBP1, CD2AP, RABGAP1, KIF11, NF1, NPAT, ZAK, MPHOSPH8, PPP1CB, CEP55, MTUS1, RAD21, RAP1A, AVPI1, SIAH1, CD2AP, SH3BP4, CADM1, ANAPC13, GAS1, LIN9, UHRF1, HHEX, TMPRSS11A, ING1, JAG2, LIG4, AD2L1, MCC, MCM2, MCM3, MCM7, NBN, EDD9, PNN, ANLN, XRN1, TXNL4B, TIPIN, PPP1CB, RIF1, RCBTB1, PPP6C, CCAR1, SEP11, BIN3, ERBB2IP, KLK10, PCNP, PTCH1, PTEN, SPC25, ARHGAP20, RAD17, RAP1A, CCND1, NCAPG, PAPD5, SIAH1, BMP2, NEK4, TACC1, BUB1, TFDP1, WEE1, EVI5, HMGA2, CDC7, RASSF5, CUL4B, PARD6G, JUB, CDC14A, RUNX3, PCAF, BCL10, UBA3, CCNT2, PKMYT1, CCNE2, ZNF830, CCPG1, WTAP, MDC1, HDAC4, DLGAP5, MTSS1, RB1CC1, CDC2, TLK1, CDC6, DLEC1, CASP8AP2, SEP7 | - | CDKN1B, NET1, TNFSF13B, MORF4L1, ESM1, FGFR1OP, SOCS4, ADM, ADRB2, DDX11, S1PR3, FGF7, TMEM97, ID4, IFNG, IGF1, IGFBP3, IL6, IL8RB, IL11, LIFR, NAP1L1, NDN, NEDD9, NRAS, CRIM1, FGFRL1, TIPIN, C19orf10, HTRA1, CXCL5, SSR1, KLF5, TGFB2, TRAF5, VEGFA, HMGA2, CAMK2D, CDC7, BRMS1L, CREG1, FGF17, NRP1, SOCS3, SOCS6, CD47, MORF4L2 |
miRNA, microRNA; UVB, ultraviolet B; nHDP, normal human dermal papilla cell.
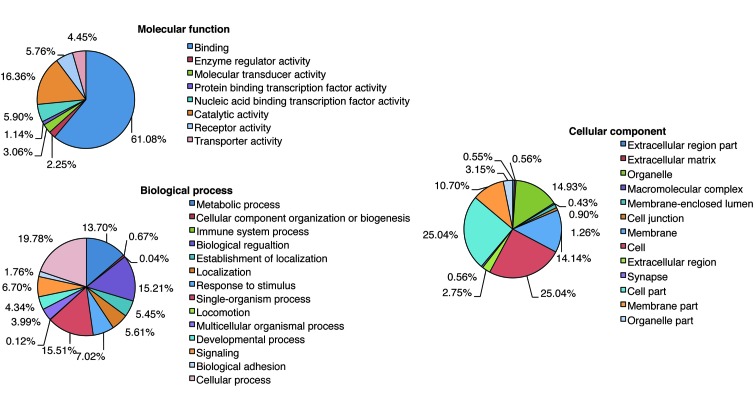
Each putative target gene was subjected to GO analysis to reveal its cellular function. GO was analyzed using three significant categories as follows: Molecular function, biological process and cellular component. The analysis revealed a wide distribution of cellular functions, which are presented in Fig. 4. Cellular process, which comprised the largest percentage of the biological process category in Fig. 4, was analyzed in greater detail. The highest percentage of gene functions were in the cellular process category, which encompasses the processes analyzed in Fig. 5. In particular, the target genes of miRNAs up- and downregulated by UVB radiation are associated with cellular responses to stimuli and cell communication. These results suggest that UVB-induced growth arrest and apoptosis may be mediated by miRNAs involved in cellular stimulation and communication in nHDPs.
Figure 4.
GO analysis of predicted target genes. miRNA target genes were predicted using web-based TargetScan software. Biological roles of each target were analyzed by GO. A diagrammatic representation of the GO analysis was produced using GeneSpring GX. GO, gene ontology; miRNA, microRNA.
Figure 5.

Detailed GO analysis of cellular process, which was the highest regulated subsection of the biological process category.
Discussion
Sunlight is a multi-wavelength light that modulates cellular signaling in the skin (3). UV radiation from sunlight is known to induce DNA damage in types of skin cell such as keratinocytes and fibroblasts (26), but the UV-mediated cellular effects in nHDPs have not yet been reported. In the current study it was determined that UVB radiation represses nHDP growth via cell cycle arrest and apoptosis, and that it can induce ROS generation. miRNAs have been demonstrated to have a major influence over the control of UVB-induced growth repression in a previous study (27). Therefore, in the present study, the miRNA expression profile in nHDPs following UVB-irradiation was analyzed. A total of 42 miRNAs whose expression in nHDPs changed at least 1.5-fold following UVB irradiation were identified. One of the upregulated miRNAs identified in the current study was miR-30a-5p, which has been reported to modulate cell growth by targeting the denticleless protein homolog, a gene implicated in S phase and UVB-induced growth arrest (28). miR-34a-5p and miR-34b-5p, which were upregulated 1.6- and 1.8-fold in the current study, respectively, are induced by DNA damage in various types of cell (29,30), and by p53, which is activated by DNA break-induced ataxia telangiectasia mutated activation (29). Overall, the results of the current study indicate that, in nHDPs, UVB regulates specific miRNAs in order to regulate cell growth and death.
In addition, the present study identified putative target genes of the up- and downregulated miRNAs, and categorized their reported biological functions by GO into cell cycle, apoptosis and cell growth and proliferation categories. UV irradiation of various types of cell, including keratinocytes, melanocytes and dermal fibroblasts, can regulate cell fate via the intrinsic apoptosis pathway (26,31,32). Consistent with this, the results of the present study demonstrated that the miRNAs that were upregulated by UVB irradiation targeted intrinsic apoptosis pathway-related genes, including NAIP (targeted by miR-30a-5p), XAF1 (targeted by miR-638) and XIAP (targeted by miR-630). In addition, miRNAs induced by UVB exposure were demonstrated to regulate core cell cycle regulators including cyclins, cyclin dependent kinases (CDKs) and CDK inhibitors. For example, upregulated miRNAs targeted CDC7 (targeted by miR-30a-5p and miR-630) and CDK2 (targeted by miR-638), while downregulated miRNAs targeted CDKN1A (targeted by miR-299-5p), CDKN1B (targeted by miR-218-5p and miR-495-3p) and CCNA2 (targeted by miR-218-5p).
Overall, data of the current study demonstrated that miRNA expression in nHDPs was altered in response to UVB exposure. This furthers the current understanding of the cellular mechanisms mediating the response to UVB exposure, whilst providing insight into the processes involved in sunlight-induced hair and skin aging.
Acknowledgements
This study was supported by a grant from the Korean Health Technology R&D Project (grant no. HN13C0075), Ministry of Health & Welfare, Republic of Korea. Dr Seunghee Bae was supported by the KU Research Professor Program of Konkuk University.
References
- 1.Schreiber MM, Moon TE, Bozzo PD. Chronic solar ultraviolet damage associated with malignant melanoma of the skin. J Am Acad Dermatol. 1984;10:755–759. doi: 10.1016/s0190-9622(84)70090-9. [DOI] [PubMed] [Google Scholar]
- 2.Helfrich YR, Sachs DL, Voorhees JJ. Overview of skin aging and photoaging. Dermatol Nurs. 2008;20:177–184. [PubMed] [Google Scholar]
- 3.Muller HK, Woods GM. Ultraviolet radiation effects on the proteome of skin cells. Adv Exp Med Biol. 2013;990:111–119. doi: 10.1007/978-94-007-5896-4_8. [DOI] [PubMed] [Google Scholar]
- 4.Lee YK, Cha HJ, Hong M, Yoon Y, Lee H, An S. Role of NF-κB-p53 crosstalk in ultraviolet A-induced cell death and G1 arrest in human dermal fibroblasts. Arch Dermatol Res. 2012;304:73–79. doi: 10.1007/s00403-011-1176-2. [DOI] [PubMed] [Google Scholar]
- 5.Xu H, Yan Y, Li L, Peng S, Qu T, Wang B. Ultraviolet B-induced apoptosis of human skin fibroblasts involves activation of caspase-8 and -3 with increased expression of vimentin. Photodermatol Photoimmunol Photomed. 2010;26:198–204. doi: 10.1111/j.1600-0781.2010.00522.x. [DOI] [PubMed] [Google Scholar]
- 6.Trüeb RM. Is androgenetic alopecia a photoaggravated dermatosis? Dermatology. 2003;207:343–348. doi: 10.1159/000074111. [DOI] [PubMed] [Google Scholar]
- 7.Lu Z, Fischer TW, Hasse S, et al. Profiling the response of human hair follicles to ultraviolet radiation. J Invest Dermatol. 2009;129:1790–1804. doi: 10.1038/jid.2008.418. [DOI] [PubMed] [Google Scholar]
- 8.Cha HJ, Shin S, Yoo H, et al. Identification of ionizing radiation-responsive microRNAs in the IM9 human B lymphoblastic cell line. Int J Oncol. 2009;34:1661–1668. doi: 10.3892/ijo_00000297. [DOI] [PubMed] [Google Scholar]
- 9.Wan G, Mathur R, Hu X, Zhang X, Lu X. miRNA response to DNA damage. Trends Biochem Sci. 2011;36:478–484. doi: 10.1016/j.tibs.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan G, Shi Y, Wu ZH. MicroRNA-22 promotes cell survival upon UV radiation by repressing PTEN. Biochem Biophys Res Commun. 2012;417:546–551. doi: 10.1016/j.bbrc.2011.11.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pothof J, Verkaik NS, van IJcken W, et al. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009;28:2090–2099. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dynoodt P, Mestdagh P, Van Peer G, et al. Identification of miR-145 as a key regulator of the pigmentary process. J Invest Dermatol. 2013;133:201–209. doi: 10.1038/jid.2012.266. [DOI] [PubMed] [Google Scholar]
- 13.Zhu Z, He J, Jia X, et al. MicroRNA-25 functions in regulation of pigmentation by targeting the transcription factor MITF in Alpaca (Lama pacos) skin melanocytes. Domest Anim Endocrinol. 2010;38:200–209. doi: 10.1016/j.domaniend.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Tan G, Niu J, Shi Y, Ouyang H, Wu ZH. NF-κB-dependent microRNA-125b up-regulation promotes cell survival by targeting p38α upon ultraviolet radiation. J Biol Chem. 2012;287:33036–33047. doi: 10.1074/jbc.M112.383273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou BR, Xu Y, Luo D. Effect of UVB irradiation on microRNA expression in mouse epidermis. Oncol Lett. 2012;3:560–564. doi: 10.3892/ol.2012.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou BR, Xu Y, Permatasari F, et al. Characterization of the miRNA profile in UVB-irradiated normal human keratinocytes. Exp Dermatol. 2012;21:317–319. doi: 10.1111/j.1600-0625.2012.01465.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim YJ, Cha HJ, Nam KH, Yoon Y, Lee H, An S. Centella asiatica extracts modulate hydrogen peroxide-induced senescence in human dermal fibroblasts. Exp Dermatol. 2011;20:998–1003. doi: 10.1111/j.1600-0625.2011.01388.x. [DOI] [PubMed] [Google Scholar]
- 18.Smith ML, Ford JM, Hollander MC, et al. p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol Cell Biol. 2000;20:3705–3714. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meek DW. The p53 response to DNA damage. DNA Repair (Amst) 2004;3:1049–1056. doi: 10.1016/j.dnarep.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 20.Lewis DA, Yi Q, Travers JB, Spandau DF. UVB-induced senescence in human keratinocytes requires a functional insulin-like growth factor-1 receptor and p53. Mol Biol Cell. 2008;19:1346–1353. doi: 10.1091/mbc.E07-10-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Abaseri TB, Putta S, Hansen LA. Ultraviolet irradiation induces keratinocyte proliferation and epidermal hyperplasia through the activation of the epidermal growth factor receptor. Carcinogenesis. 2006;27:225–231. doi: 10.1093/carcin/bgi220. [DOI] [PubMed] [Google Scholar]
- 22.Takasawa R, Nakamura H, Mori T, Tanuma S. Differential apoptotic pathways in human keratinocyte HaCaT cells exposed to UVB and UVC. Apoptosis. 2005;10:1121–1130. doi: 10.1007/s10495-005-0901-8. [DOI] [PubMed] [Google Scholar]
- 23.Chainiaux F, Magalhaes JP, Eliaers F, Remacle J, Toussaint O. UVB-induced premature senescence of human diploid skin fibroblasts. Int J Biochem Cell Biol. 2002;34:1331–1339. doi: 10.1016/s1357-2725(02)00022-5. [DOI] [PubMed] [Google Scholar]
- 24.Bossi O, Gartsbein M, Leitges M, Kuroki T, Grossman S, Tennenbaum T. UV irradiation increases ROS production via PKCdelta signaling in primary murine fibroblasts. J Cell Biochem. 2008;105:194–207. doi: 10.1002/jcb.21817. [DOI] [PubMed] [Google Scholar]
- 25.Scharffetter-Kochanek K, Wlaschek M, Brenneisen P, Schauen M, Blaudschun R, Wenk J. UV-induced reactive oxygen species in photocarcinogenesis and photoaging. Biol Chem. 1997;378:1247–1257. [PubMed] [Google Scholar]
- 26.Pustisek N, Situm M. UV-radiation, apoptosis and skin. Coll Antropol. 2011;35(Suppl 2):339–341. [PubMed] [Google Scholar]
- 27.Schneider MR. MicroRNAs as novel players in skin development, homeostasis and disease. Br J Dermatol. 2012;166:22–28. doi: 10.1111/j.1365-2133.2011.10568.x. [DOI] [PubMed] [Google Scholar]
- 28.Baraniskin A, Birkenkamp-Demtroder K, Maghnouj A, et al. MiR-30a-5p suppresses tumor growth in colon carcinoma by targeting DTL. Carcinogenesis. 2012;33:732–739. doi: 10.1093/carcin/bgs020. [DOI] [PubMed] [Google Scholar]
- 29.Hu H, Gatti RA. MicroRNAs: new players in the DNA damage response. J Mol Cell Biol. 2011;3:151–158. doi: 10.1093/jmcb/mjq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato M, Paranjape T, Müller RU, et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamauchi T, Adachi S, Yasuda I, et al. Ultra-violet irradiation induces apoptosis via mitochondrial pathway in pancreatic cancer cells. Int J Oncol. 2011;39:1375–1380. doi: 10.3892/ijo.2011.1188. [DOI] [PubMed] [Google Scholar]
- 32.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]