Abstract
The aim of the present study was to assess the influence of WWOX gene upregulation on the transcriptome and phenotype of the T98G glioblastoma cell line. The cells with high WWOX expression demonstrated a significantly different transcription profile for approximately 3,000 genes. The main cellular pathways affected were Wnt, TGFβ, Notch and Hedgehog. Moreover, the WWOX-transfected cells proliferated at less than half the rate, exhibited greatly lowered adhesion to ECM, increased apoptosis and impaired 3D culture formation. They also demonstrated an increased ability for crossing the basement membrane. Our results indicate that WWOX, apart from its tumor-suppressor function, appears to be a key regulator of the main cellular functions of the cell cycle and apoptosis. Furthermore, our results showed that WWOX may be involved in controlling metabolism, cytoskeletal structure and differentiation.
Keywords: WWOX, glioblastoma, proliferation, microarrays, adhesion
Introduction
Glioblastoma (GBM) is the most common primary brain tumor in adults. It is also the most aggressive and resistant to therapy and has the worst prognosis. To date, there is no effective treatment for patients suffering from this disease (1). After excision of the tumor, its relapse is inevitable, and the mean survival time of patients rarely exceeds several months (2). The molecular pathology of GBM is diverse, and a number of chromosomal aberrations are known to be hallmarks of glioblastoma carcinogenesis. Among them are the loss of heterozygosity on 10q, EGFR amplification, CDKN2A deletion and mutations of the PTEN and TP53 genes (3). Ineffective treatment, as well as the very high mortality rate and particularly complex molecular background of GBM constitute a strong rationale for research aiming to elucidate the processes underlying the carcinogenesis of neural cells.
The WWOX gene is localized at a common fragile site FRA16D (4). It is known to behave as a tumor suppressor. Unlike most tumor-suppressor genes, the loss of functionality of only one of its alleles is enough to predispose a patient to cancerogenesis - the haploinsufficiency phenomenon (5). Growing evidence indicates that WWOX is not a classical tumor suppressor. Its action is clearly not limited only to cell cycle control or genome integrity maintenance. Although interactions with several transcription factors and signal transduction proteins are well documented (6–9), it seems that only a tiny piece is known of the physiological cellular role of WWOX and its implications for cancerogenesis. Our previous findings on glioblastoma samples showed that the WWOX expression level is correlated with genes important to tumor formation and progression, such as ERBB4, Ki67 and Bcl-2 (10). The present study was conducted on the glioblastoma cell line T98G and aimed to assess the influence of WWOX upregulation on the transcriptome and phenotype of these cells.
Materials and methods
Cell line and culture conditions
T98G cells, derived from a human glioblastoma, were obtained from the European Collection of Cell Cultures (ECACC). The cells were grown according to the manufacturer’s protocol in Minimum Essential Medium (MEM; Gibco) supplemented with 2 mM L-glutamine (Gibco), 0.1 mM NEAA (Gibco), 10% heat inactivated FBS (Gibco), 0.05 mg/ml penicillin (Gibco), 0.05 mg/ml streptomycin (Gibco) and 0.1 mg/ml neomycin (Gibco) in a humidified atmosphere containing 5% CO2 at 37°C.
Stable retroviral transfection
The WWOX gene cDNA was introduced into T98G glioblastoma cells by retroviral transfection. The pLNCX2 retroviral vector with cloned WWOX was produced in the PT67 packaging line. Target cells were grown to 30% confluency and infected with the viruses (~106 colony-forming units/ml) suspended in culture medium with Polybrene as vehicle (8 μg/ml, Sigma-Aldrich). After 24 h, the medium was replaced, and stable transfectants were selected with 400 μg/ml G418 (Sigma-Aldrich) for 3 weeks. A pool of stable transfectants was used for the microarray study of global gene expression and biological experiments. Transfection efficiency was confirmed by real-time RT-PCR and western blot analysis.
Real-time RT-PCR
The real-time RT-PCR procedure was conducted to assess the efficiency of the retroviral transfection and to validate the microarray experiment. Total RNA was isolated using TRIzol reagent (Life Technologies). The cDNA synthesis was performed using 10 μg of total RNA at a volume of 100 μl using ImProm RT-II™ reverse transcriptase (Promega). Reverse transcription was carried out under the following conditions: incubation at 25°C for 5 min and 42°C for 60 min and heating at 70°C for 15 min. cDN A samples were diluted with sterile deionized water to a total volume of 150 μl, and 2 μl was added to the PCR reaction. Real-time RT-PCR was performed using Light Cycler 480 (Roche). PCR products were detected using SYBR® Green I and qPCR Core kit for SYBR® Green I (Eurogentec). All reactions were performed in duplicate. The relative expression levels of the WWOX, BIRC5 and ID3 genes were assessed. The expression levels of the investigated genes were normalized to 3 reference housekeeping genes (RPS17, H3F3A, RPLP0). The relative gene expression was calculated based on the Pfaffl method (11). Universal Human Reference RNA (Stratagene) was used as a calibrator. The primer sequences, PCR reaction conditions and lengths of the obtained products are available upon request.
Western blot analysis
Cells were lysed on ice with RIPA protein extraction buffer containing protease inhibitor cocktail (Sigma-Aldrich). Proteins (60 μg) were resolved on 10% SDS-PAGE and were transferred on PVDF membranes. The membranes were blocked for 1 h in 5% non-fat milk and incubated with a primary antibody for 18 h at 4°C. The antibodies used were goat polyclonal anti-WWOX (cat. no. sc-20529), mouse monoclonal anti-ARK1 (cat. no. sc-56881), goat polyclonal anti-KLF8 (cat. no. sc-69294), rabbit polyclonal anti-JAK1 (cat. no. sc-277) (all from Santa Cruz Biotechnology). Subsequently, the membranes were washed with TBST and incubated with the appropriate secondary antibody conjugated with alkaline phosphatase (Sigma-Aldrich) for 1 h at room temperature (RT). Next, the membranes were washed with TBST and developed with Novex® AP Chromogenic Substrate (Invitrogen). GAPDH was used as a reference protein. The relative protein amount was assessed with ImageJ (NIH) based on the integrated density of the bands.
Microarray transcriptome study
Human OneArray™ (Phalanx Biotech) high-density microarrays were used in flip dye experiments in 4 replicates for each cell variant. Each sample was hybridized against Universal Human Reference RNA (Stratagene) and labelled with ULS™ Labeling Kit (Kreatech Diagnostics). Preparation of the slide for hybridization included prewash in ethanol and pre-hybridization according to the manufacturer’s protocol. Hybridization was performed in a humidity chamber filled with 2 SSPE buffer at 42°C for 16–18 h. Post-hybridization washes were performed with the following buffers: 1 SSPE/0.03% SDS (2 min, 42°C), 1 SSPE (2 min, RT), 0.1 SSPE (rinsed several times, RT). Slide scanning and preliminary normalization were performed with ProScanArray (Perkin-Elmer) and ScanArray Express, respectively.
Further data analysis was performed with the MultiExperiment Viewer (MeV) from the TM4 package provided by The Institute for Genomic Research at http://www.tm4.org/site. For the ontological classification of genes, the PANTHER classification system was used, which allowed a determination to be made of which pathways are susceptible to change depending on the WWOX expression level. The microarray results were validated by real-time RT-PCR and western blot analysis. The data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession no. GSE51481.
Proliferation, redox potential and apoptosis assays
The three assays assessing cell proliferation, redox potential and apoptosis were multiplexed to eliminate population and culture differences. Proliferation was evaluated with 5-bromo-2′-deoxyuridine (BrdU). BrdU incorporated into DNA during replication was detected with an anti-BrdU monoclonal antibody labeled with europium (Perkin-Elmer). The redox potential was measured with the alamarBlue® cell viability reagent (Invitrogen), the redox indicator metabolized in mitochondria. Apoptosis was assessed by TUNEL reaction with the DELFIA DNA fragmentation assay (Perkin-Elmer). Cells were seeded on a white, clear bottom 96-well plate at a density of 10,000 cells/well. All tests were conducted in a starvation medium (without serum).
Adhesion assay
In order to assess the ability of the cells to integrate into the extracellular matrix, a colorimetric CytoSelect™ 48-well adhesion assay (Cell Biolabs, Inc.) was carried out. The assay evaluates the capability of cells to adhere to five ECM proteins: fibronectin, collagen I, collagen I V, laminin and fibrinogen. The cells were seeded on a 48-well plate coated with selected ECM proteins at a density of 150,000 cells/well in starvation medium and were allowed to adhere for 90 min at 37°C. Next, the adherent cells were dyed and their number was analyzed colorimetrically.
Invasion assay
The invasive potential of the investigated cells was evaluated using the colorimetric CytoSelect™ 24-well invasion assay (Cell Biolabs, Inc.). The assay contains a membrane coated with a layer of basement membrane matrix solution and allows for discrimination of invasive cells. The cells were seeded on inserts placed in a 24-well plate at a density of 300,000 cells/well and left to invade for 48 h. Next, the cells that crossed the membrane were dyed and their number was analyzed colorimetrically.
3D culture growth
For a three dimensional (3D) culture assay, cells were seeded on a 96-well plate at a density of 15,000 cells/well on a solidified 2-mm layer of growth factor-reduced Geltrex™ basement membrane matrix (Gibco). The cells were grown in an assay buffer consisting of full culture medium and 2% Geltrex™. The assay buffer was exchanged every 4 days. The cells were cultured for 12 days.
Statistical analysis of biological assays
The results are presented as means. Statistical significance in all biological tests was assessed with the Student’s t-test. The results were recognized as being statistically significant at a confidence level >95% (p<0.05).
Results
Retroviral transfection and multiclonal selection allows for stable overexpression of the WWOX gene in T98G glioblastoma cells
The level of WWOX expression was assessed by real-time RT-PCR and western blot analysis. The amount of WWOX mRN A in the T98G/WWOX transfectants was >29-fold greater than that noted in the T98G/vec control cells. The higher gene expression resulted in an elevated protein level. The protein level in the T98G/vec control was comparable to that found in the untreated T98G cells (Fig. 1).
Figure 1.
WWOX protein levels in the native and transfected T98G cells.
Transcriptome analysis of the WWOX transfectants
The microarray study was used to assess changes in the expression level of ~29,000 genes. A total of 2,846 genes showed a significant increase or decrease in expression depending on the WWOX level (p<0.05, t-test). For 1,802 of the genes, the change in expression level was 2-fold or greater. All the genes identified in the microarray experiment were ontologically classified using the PANTHER classification system and grouped according to cellular pathways and biological processes. The cellular pathways containing 10 or more genes modulated by WWOX overexpression are presented in Table I. Selected biological processes with the highest number of WWOX-modulated genes are shown in Table II.
Table I.
Ontological analysis of the genes modulated by WWOX overexpression: cellular pathways.
| Cellular pathway | No. of genes |
|---|---|
| Unclassified | 2,041 |
| Inflammation mediated by chemokine and cytokine signaling pathway | 39 |
| Wnt signaling pathway | 38 |
| Heterotrimeric G-protein signaling pathway (Gi α and Gs α mediated) | 32 |
| Integrin signalling pathway | 28 |
| Interleukin signaling pathway | 25 |
| Angiogenesis | 22 |
| PDGF signaling pathway | 21 |
| Huntington disease | 21 |
| Alzheimer disease-presenilin pathway | 20 |
| p53 pathway | 20 |
| Parkinson disease | 20 |
| Heterotrimeric G-protein signaling pathway (Gq α and Go α mediated) | 20 |
| PI3 kinase pathway | 19 |
| TGF-β signaling pathway | 18 |
| EGF receptor signaling pathway | 17 |
| Endothelin signaling pathway | 15 |
| Transcription regulation by bZIP transcription factor | 14 |
| Ionotropic glutamate receptor pathway | 14 |
| FGF signaling pathway | 14 |
| Cytoskeletal regulation by Rho GTPase | 14 |
| Apoptosis signaling pathway | 13 |
| Alzheimer disease-amyloid secretase pathway | 13 |
| Oxidative stress response | 13 |
| Nicotinic acetylcholine receptor signaling pathway | 13 |
| Metabotropic glutamate receptor group III pathway | 13 |
| Cadherin signaling pathway | 13 |
| Angiotensin II-stimulated signaling through G-proteins and β-arrestin | 12 |
| Ras pathway | 11 |
| p38 MAPK pathway | 11 |
| T cell activation | 10 |
| Insulin/IGF pathway-protein kinase B signaling cascade | 10 |
Table II.
Ontological analysis of the genes modulated by WWOX overexpression: biological processes.
| Biological process | No. of genes |
|---|---|
| Metabolic process | 1,063 |
| Primary metabolic process | 1,019 |
| Cellular process | 777 |
| Unclassified | 629 |
| Cell communication | 556 |
| Signal transduction | 524 |
| Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 472 |
| Protein metabolic process | 415 |
| Transport | 370 |
| Transcription | 295 |
| Transcription from RNA polymerase II promoter | 293 |
| Developmental process | 288 |
| Cell surface receptor linked signal transduction | 256 |
| Immune system process | 243 |
| Regulation of transcription from RNA polymerase II promoter | 240 |
| System process | 220 |
| Protein transport | 192 |
| Intracellular protein transport | 192 |
| Neurological system process | 176 |
| Protein modification | 174 |
| System development | 174 |
| Cell cycle | 163 |
| Response to stimulus | 156 |
| Intracellular signaling cascade | 148 |
| Proteolysis | 136 |
| Cell adhesion | 131 |
| Cell-cell signaling | 123 |
| Cellular component organization | 122 |
| Ectoderm development | 120 |
| Lipid metabolic process | 118 |
| Vesicle-mediated transport | 117 |
| G-protein coupled receptor protein signaling pathway | 114 |
| Nervous system development | 109 |
| Mesoderm development | 108 |
| Apoptosis | 101 |
Phenotype analysis of the WWOX transfectants
To investigate how the changes in the transcriptome translate into cell phenotype, a number of biological experiments were performed. Proliferation rate, redox potential, apoptosis, ability to adhere to extracellular matrix proteins, invasiveness and 3D culture formation were all evaluated in the transfected cells.
Proliferation, redox potential and apoptosis assays
Multiplexing assays for proliferation, redox potential and apoptosis allowed for elimination of population and culture differences. An analysis of the proliferation rate showed that T98G/WWOX cells proliferated 53% more slowly than the control T98G/vec cells (p<0.01). Simultaneously apoptosis was increased, although without statistical significance (p>0.05). A test with alamarBlue to assess mitochondrial metabolism revealed that cells overexpressing WWOX had a considerably higher rate of metabolizing the substrate, which may signify an intensification of overall mitochondrial redox potential.
Invasion
WWOX-transfected cells were examined to test whether they exhibit changes in invasiveness. T98G/WWOX cells demonstrated a slightly greater ability to cross a membrane coated with basement membrane matrix solution, with 29% more invasive cells. However, the result was not statistically significant (p>0.05).
Adhesion
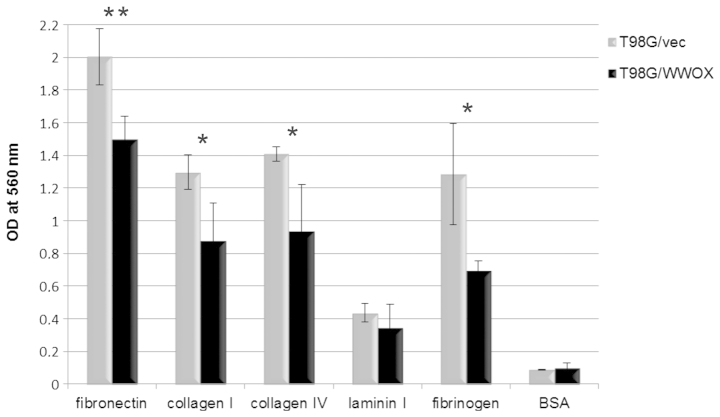
Cells overexpressing WWOX showed a greatly reduced ability to adhere to extracellular matrix proteins. Decreased adhesion was noted to all examined proteins: fibronectin (p<0.01), collagen I (p<0.05), collagen I V (p<0.05), laminin (p>0.05) and fibrinogen (p<0.05). A graphical presentation of changes in adhesion between T98G cells overexpressing WWOX and those with a native WWOX level is provided in Fig. 2.
Figure 2.
Ability for adhesion of the T98G/WWOX and T98G/vec control cells to ECM proteins. *p<0.05, **p<0.01.
3D culture growth
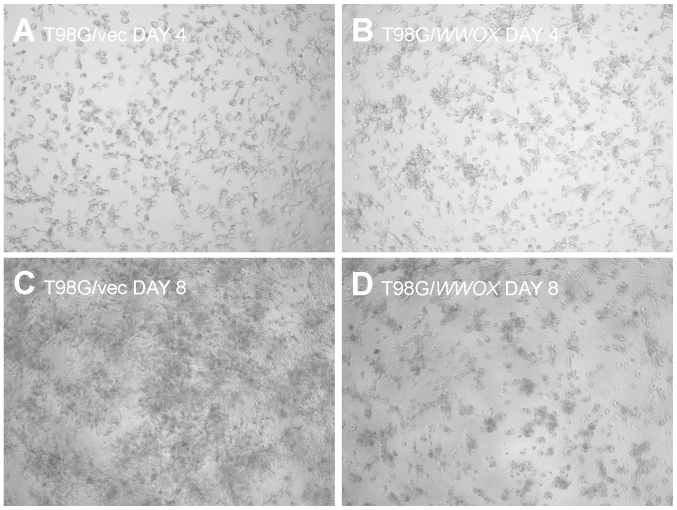
The T98G/WWOX cells exhibited impaired 3D culture formation. The cells seeded in a thick Geltrex® layer after 12 days remained as isolated, single cells that did not proliferate. In contrast, control T98G/vec cells exhibited extensive proliferation and network formation (Fig. 3).
Figure 3.
3D culture formation in a Geltrex basement protein matrix. (A and C) T98G/vec cells; (B and D) T98G/WW OX cells. (A and B) 4-day culture; (C and D) 8-day culture.
Discussion
Our previous report showed that WWOX may be involved in GBM carcinogenesis and/or tumor progression. The study describes the association of WWOX expression with the transcription level of several genes involved in signal transduction and cell cycle control (10). To specify how WWOX may influence cancer cell metabolism, we decided to use the T98G glioblastoma cell line, which has a very low level of expression of endogenous WWOX to ascertain the effect an increase in WWOX expression may have on these cells.
WWOX cDNA was introduced into T98G cells by retroviral transfection. In the stable transfectants, the expression profile and basic biological processes were examined. The microarray study for global gene expression allowed the relevance of WWOX to be investigated, with regard to overall cellular signaling. The experiment identified 2,846 genes whose expression levels were significantly altered as a consequence of WWOX overexpression. The ontological analysis categorized the differentially expressed genes into 121 signaling pathways. The most significant were pathways important both for carcinogenesis and development, such as Wnt, TGFβ, Notch and Hedgehog. The cellular pathways with the highest number of assigned genes regulated by WWOX expression level are presented in Table I. The biological experiments on the WWOX-transfected T98G/WWOX cells revealed a more intensive energetic mitochondrial metabolism, but a lower rate of proliferation and higher apoptosis level than the T98G/vec control cells. Moreover, the T98G/WWOX cells demonstrated a higher migration potential and reduced attachment to extra-cellular matrix proteins. Furthermore, cells overexpressing WWOX failed to grow in an extracellular matrix environment on Geltrex substrate.
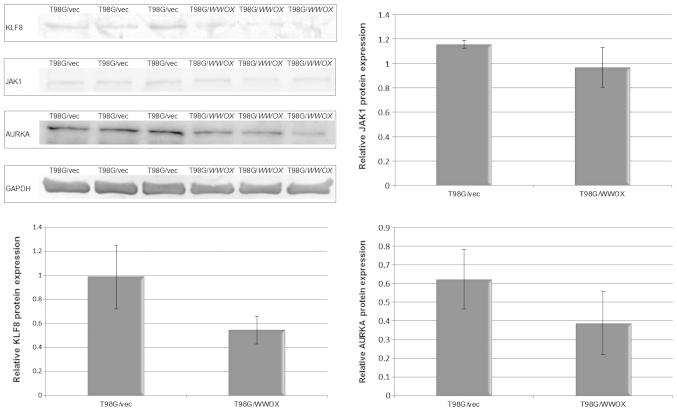
Loss of cell cycle control and intense proliferation are hallmarks of neoplastic cells. In the proliferation assay with BrdU, T98G/WWOX cells demonstrated a 53% lower proliferation rate than the T98G/vec control (p<0.01). An ontological analysis of the microarray data revealed 163 genes to be involved in cell cycle regulation, whose expression was modulated by WWOX overexpression. Among these genes are potent oncogenes linked with brain cancerogenesis: AURKA, KLF8 and JAK1 (12–16). In our microarray experiment, the T98G/WWOX cells demonstrated significantly lower expression of all afore-mentioned genes. Western blot analysis confirmed that the level of these proteins was considerably lower in the cells with high WWOX expression (Fig. 4).
Figure 4.
Western blot analysis of the levels of KLF8, JAK1 and AURKA proteins in relation to the WWOX expression level. The level of GAPDH was used as a loading control.
After enhancement of WWOX expression in the T98G cell line, a tendency for increased apoptosis was observed (p>0.05). Such an effect of WWOX overexpression was found to be cosistant across various cell lines (17–24). While apoptosis seems to proceed through a mitochondrial pathway in breast, prostate and lung cells, Chiang et al showed that in U373MG glioblastoma cells, WWOX overexpression triggers a mitochondrial/caspase-3-independent pathway of apoptosis (25).
WWOX overexpression in T98G cells resulted in an increased number of invasive cells crossing the basement membrane. This effect was also observed in breast and colon cancer cells with ectopic WWOX expression (26,27). It can be hypothesized that elevated cell motility is related to the function of WWOX as a regulator of differentiation. The microarray gene expression analysis revealed a large group of WWOX-regulated genes whose products are engaged in nervous system development (108 genes, classification presented in Table II ). Recently, Abdeen et al reported that WWOX knockdown in MCF-10A normal breast cells resulted in impaired growth in a 3-D culture Matrigel assay and mammary ductal formation (28). This suggests that WWOX expression is required for the proper development of this gland. Contrary to Abdeen’s results, in our experiment, the 3-D culture formation by glioblastoma cells was inhibited by a high level of WWOX.
The increase in mitochondrial redox activity in T98G/WWOX cells is intriguing. One of the genes whose expression was elevated in our microarray experiment was DLD, a gene encoding dihydrolipoamide dehydrogenase, diaphorase which catalyzes the reduction of resazurin (alamarBlue®) into resofurin (29). The DLD expression in T98G/WWOX was >3-fold higher than that in the T98/vec control cells. This may explain the higher redox potential observed in the T98G/WWOX cells in the alamarBlue assay. O’Keefe et al showed in a Drosophila model that WWOX takes part in aerobic metabolism and the generation of ROS (30). In their recent study, Dayan et al claimed that WWOX not only influences metabolism, but its mRN A level is also related to the metabolic state of the cell. They reported that switching the metabolism from glycolysis to oxidative phosphorylation causes a stable increase in the amount of WWOX mRNA. Consequently, hypoxia, a state where cells rely on glycolysis, causes its decrease (31). GBM cells are known to switch their metabolism to glycolysis in response to hypoxic conditions inside the tumor and the demand for the accelerated production of substrates needed by the rapidly proliferating cells (32).
The T98G cells overexpressing WWOX exhibited a significantly lowered ability to adhere to fibronectin, collagen I, collagen IV and fibrinogen. The decrease in adhesion to laminin, was also noted, although without statistical significance. A similar result of WWOX influence on decreased adhesion to fibronectin was observed by Gourley et al in ovarian cell lines (33). Adhesion to the extracellular matrix is fundamental for cancer cell behavior. Cell adhesion-mediated drug resistance (CAM-DR) is a phenomenon of apoptosis resistance caused by close interactions between cancer cells and ECM proteins (34). Furthermore, one of the main GBM hallmarks is ability for invasion and infiltration of surrounding tissue; this is the reason for the almost inevitable relapse of the disease. The invasion of GBM corresponds with increased adhesion to ECM proteins and its proteolytic degradation (35). In the light of the present knowledge in regards to adhesion in the progression of GBM, the observed reduction in the attachment of cells to ECM proteins caused by WWOX overexpression can be interpreted as decreasing cell malignancy.
The decrease in adhesion of the cells to ECM proteins and their reduced ability to proliferate and form 3D structures in the extracellular matrix indicate that WWOX overexpression causes substantial changes in cytoskeleton organization and the structure of membrane proteins such as integrins. Indeed, the global gene expression examination showed a large group of WWOX-regulated genes acting in the integrin and cadherin signaling pathways (28 and 13 genes, respectively). Moreover, an ontological classification of the genes affected by WWOX overexpression with respect to the cellular component revealed that the largest group of genes was connected with the cytoskeleton. The organization of the cytoskeleton regulates such processes as cell adhesion and motility (36,37).
Additionally, besides the cellular functions discussed above, cell cycle control, metabolism, invasiveness, apoptosis and adhesion, several other notable aspects of cell behavior which could be affected by WWOX emerged from the microarray study. In the group of genes whose expression was altered by WWOX overexpression were genes involved in angiogenesis, development and intracellular transport. The precise mechanism and the nature of WWOX acting on the numerous cellular functions indicated here is yet to be determined.
In conclusion, the observed reduction in ECM adhesion, lowering of the proliferation rate and the increase in apoptosis can be regarded as a decrease in cell malignancy due to WWOX overexpression. The most striking evidence of the WWOX tumor suppressor effect is the inhibition of the growth of glioblastoma cells in an extracellular matrix environment. The higher migration rates may be linked with the presumed role played by WWOX in differentiation and the regulation of development.
The obtained results indicate that WWOX may be a key regulator of the main cellular processes: cell cycle, apoptosis, metabolism, cytoskeleton structure and differentiation. The increase in WWOX expression influences the phenotype of T98G cells, reducing their malignancy.
Present knowledge of WWOX function suggests that, contrary to initial assumptions, it does not act as a classical tumor suppressor. It appears that the role of WWOX is not limited to that of cell cycle control or genome integrity protection, but its influence on cell function is more global. It is probable that it is one of the pivotal regulators of genes involved in cell differentiation, responsible for maintaining tissue structure.
Acknowledgements
The present study was funded by the Medical University of Lodz, grants 503/0-078-02/503-01 and 502-03/0-078-02/502-04-020.
References
- 1.Chen J, Xu T. Recent therapeutic advances and insights of recurrent glioblastoma multiforme. Front Biosci (Landmark Ed) 2013;18:676–684. doi: 10.2741/4130. [DOI] [PubMed] [Google Scholar]
- 2.Lwin Z, MacFadden D, Al-Zahrani A, et al. Glioblastoma management in the temozolomide era: have we improved outcome? J Neurooncol. 2013;115:303–310. doi: 10.1007/s11060-013-1230-3. [DOI] [PubMed] [Google Scholar]
- 3.Kanu OO, Hughes B, Di C, et al. Glioblastoma multiforme oncogenomics and signaling pathways. Clin Med Oncol. 2009;3:39–52. doi: 10.4137/cmo.s1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3–24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 5.Aqeilan RI, Trapasso F, Hussain S, et al. Targeted deletion of Wwox reveals a tumor suppressor function. Proc Natl Acad Sci USA. 2007;104:3949–3954. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aqeilan RI, Palamarchuk A, Weigel RJ, Herrero JJ, Pekarsky Y, Croce CM. Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res. 2004;64:8256–8261. doi: 10.1158/0008-5472.CAN-04-2055. [DOI] [PubMed] [Google Scholar]
- 7.Aqeilan RI, Pekarsky Y, Herrero JJ, et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci USA. 2004;101:4401–4406. doi: 10.1073/pnas.0400805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aqeilan RI, Donati V, Palamarchuk A, et al. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;65:6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 9.Matteucci E, Bendinelli P, Desiderio MA. Nuclear localization of active HGF receptor Met in aggressive MDA-MB231 breast carcinoma cells. Carcinogenesis. 2009;30:937–945. doi: 10.1093/carcin/bgp080. [DOI] [PubMed] [Google Scholar]
- 10.Kosla K, Pluciennik E, Kurzyk A, et al. Molecular analysis of WWOX expression correlation with proliferation and apoptosis in glioblastoma multiforme. J Neurooncol. 2011;101:207–213. doi: 10.1007/s11060-010-0254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borges KS, Castro-Gamero AM, Moreno DA, et al. Inhibition of Aurora kinases enhances chemosensitivity to temozolomide and causes radiosensitization in glioblastoma cells. J Cancer Res Clin Oncol. 2012;138:405–414. doi: 10.1007/s00432-011-1111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehman NL, O’Donnell JP, Whiteley LJ, et al. Aurora A is differentially expressed in gliomas, is associated with patient survival in glioblastoma and is a potential chemotherapeutic target in gliomas. Cell Cycle. 2012;11:489–502. doi: 10.4161/cc.11.3.18996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samaras V, Stamatelli A, Samaras E, et al. Comparative immunohistochemical analysis of aurora-A and aurora-B expression in human glioblastomas. Associations with proliferative activity and clinicopathological features. Pathol Res Pract. 2009;205:765–773. doi: 10.1016/j.prp.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Schnell O, Romagna A, Jaehnert I, et al. Kruppel-like factor 8 (KLF8) is expressed in gliomas of different WHO grades and is essential for tumor cell proliferation. PLoS One. 2012;7:e30429. doi: 10.1371/journal.pone.0030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu Y, Zhong Y, Fu J, et al. Activation of JAK/STAT signal pathway predicts poor prognosis of patients with gliomas. Med Oncol. 2011;28:15–23. doi: 10.1007/s12032-010-9435-1. [DOI] [PubMed] [Google Scholar]
- 17.Aderca I, Moser CD, Veerasamy M, et al. The JNK inhibitor SP600129 enhances apoptosis of HCC cells induced by the tumor suppressor WWOX. J Hepatol. 2008;49:373–383. doi: 10.1016/j.jhep.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabbri M, Iliopoulos D, Trapasso F, et al. WWOX gene restoration prevents lung cancer growth in vitro and in vivo. Proc Natl Acad Sci USA. 2005;102:15611–15616. doi: 10.1073/pnas.0505485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu BS, Tan JW, Zhu GH, Wang DF, Zhou X, Sun ZQ. WWOX induces apoptosis and inhibits proliferation of human hepatoma cell line SMMC-7721. World J Gastroenterol. 2012;18:3020–3026. doi: 10.3748/wjg.v18.i23.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iliopoulos D, Fabbri M, Druck T, Qin HR, Han SY, Huebner K. Inhibition of breast cancer cell growth in vitro and in vivo: effect of restoration of Wwox expression. Clin Cancer Res. 2007;13:268–274. doi: 10.1158/1078-0432.CCR-06-2038. [DOI] [PubMed] [Google Scholar]
- 21.Kuroki T, Yendamuri S, Trapasso F, et al. The tumor suppressor gene WWOX at FRA16D is involved in pancreatic carcinogenesis. Clin Cancer Res. 2004;10:2459–2465. doi: 10.1158/1078-0432.ccr-03-0096. [DOI] [PubMed] [Google Scholar]
- 22.Qin HR, Iliopoulos D, Semba S, et al. A role for the WWOX gene in prostate cancer. Cancer Res. 2006;66:6477–6481. doi: 10.1158/0008-5472.CAN-06-0956. [DOI] [PubMed] [Google Scholar]
- 23.Xiong Z, Hu S, Wang Z. Cloning of WWOX gene and its growth-inhibiting effects on ovarian cancer cells. J Huazhong Univ Sci Technolog Med Sci. 2010;30:365–369. doi: 10.1007/s11596-010-0358-z. [DOI] [PubMed] [Google Scholar]
- 24.Zhang P, Jia R, Ying L, et al. WWOX-mediated apoptosis in A549 cells mainly involves the mitochondrial pathway. Mol Med Rep. 2012;6:121–124. doi: 10.3892/mmr.2012.860. [DOI] [PubMed] [Google Scholar]
- 25.Chiang MF, Yeh ST, Liao HF, Chang NS, Chen YJ. Overexpression of WW domain-containing oxidoreductase WOX1 preferentially induces apoptosis in human glioblastoma cells harboring mutant p53. Biomed Pharmacother. 2012;66:433–438. doi: 10.1016/j.biopha.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Lewandowska U, Zelazowski M, Seta K, Byczewska M, Pluciennik E, Bednarek AK. WWOX, the tumour suppressor gene affected in multiple cancers. J Physiol Pharmacol. 2009;60:47–56. [PubMed] [Google Scholar]
- 27.Nowakowska M, Pospiech K, Lewandowska U, Piastowska-Ciesielska AW, Bednarek AK. Diverse effect of WWOX overexpression in HT29 and SW480 colon cancer cell lines. Tumour Biol. 2014 doi: 10.1007/s13277-014-2196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdeen SK, Salah Z, Khawaled S, Aqeilan RI. Characterization of WWOX inactivation in murine mammary gland development. J Cell Physiol. 2013;228:1391–1396. doi: 10.1002/jcp.24310. [DOI] [PubMed] [Google Scholar]
- 29.O’Brien J, Wilson I, Orton T, Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem. 2000;267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 30.O’Keefe LV, Colella A, Dayan S, et al. Drosophila orthologue of WWOX, the chromosomal fragile site FRA16D tumour suppressor gene, functions in aerobic metabolism and regulates reactive oxygen species. Hum Mol Genet. 2011;20:497–509. doi: 10.1093/hmg/ddq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dayan S, O’Keefe LV, Choo A, Richards RI. Common chromosomal fragile site FRA16D tumor suppressor WWOX gene expression and metabolic reprograming in cells. Genes Chromosomes Cancer. 2013;52:823–831. doi: 10.1002/gcc.22078. [DOI] [PubMed] [Google Scholar]
- 32.Wolf A, Agnihotri S, Guha A. Targeting metabolic remodeling in glioblastoma multiforme. Oncotarget. 2010;1:552–562. doi: 10.18632/oncotarget.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gourley C, Paige AJ, Taylor KJ, et al. WWOX gene expression abolishes ovarian cancer tumorigenicity in vivo and decreases attachment to fibronectin via integrin alpha3. Cancer Res. 2009;69:4835–4842. doi: 10.1158/0008-5472.CAN-08-2974. [DOI] [PubMed] [Google Scholar]
- 34.Morin PJ. Drug resistance and the microenvironment: nature and nurture. Drug Resist Updat. 2003;6:169–172. doi: 10.1016/s1368-7646(03)00059-1. [DOI] [PubMed] [Google Scholar]
- 35.Giese A, Laube B, Zapf S, Mangold U, Westphal M. Glioma cell adhesion and migration on human brain sections. Anticancer Res. 1998;18:2435–2447. [PubMed] [Google Scholar]
- 36.Brieher WM, Yap AS. Cadherin junctions and their cytoskeleton(s) Curr Opin Cell Biol. 2013;25:39–46. doi: 10.1016/j.ceb.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Vitriol EA, Zheng JQ. Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron. 2012;73:1068–1081. doi: 10.1016/j.neuron.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]