Abstract
Over the past decade the purine riboswitch, and in particular its nucleobase-binding aptamer domain, has emerged as an important model system for exploring various aspects of RNA structure and function. Its relatively small size, structural simplicity and readily observable activity enable application of a wide variety of experimental approaches towards the study of this RNA. These analyses have yielded important insights into small molecule recognition, co-transcriptional folding and secondary structural switching, and conformational dynamics that serve as a paradigm for other RNAs. In this article, the current state of understanding of the purine riboswitch family is examined and how this growing knowledge base is starting to be exploited in the creation of novel RNA devices.
Keywords: riboswitch, purine, RNA structure, co-transcriptional folding, gene expression
1. Introduction.
Over the past decade, structural and biophysical studies of RNA have been heavily influenced by riboswitches, and in particular, purine riboswitches. These regulatory RNA elements are most prevalent in the leader sequences of bacterial mRNAs, controlling expression in a cis-fashion (reviewed in [1]). This activity is imparted through two functional domains: a small molecule binding aptamer (or receptor) domain and a regulatory component (or expression platform) containing a structural switch that most often acts at the level of transcription or translation. Structural and biophysical studies of riboswitches have almost exclusively focused upon the aptamer domain as this domain has both complex tertiary architecture amenable to structure determination and an observable activity—ligand binding. As a consequence, while three-dimensional architectures of almost every major family of riboswitch have been elucidated, only two structures (the SAM-III (or SMK) [2] and hydroxocobalamin riboswitch [3]) have been shown directly and experimentally to encompass all of the sequence elements necessary and sufficient to impart both the ligand binding and gene regulatory activities. While several riboswitch aptamer domains such as SAM-I [4], TPP [5, 6], and preQ1-I [7-9] have been extensively studied by a number of approaches, the aptamer domain of members of the purine riboswitch family [10, 11] have been adopted as as a significant model system for studying these RNAs.
The purine riboswitch was originally discovered as a conserved regulatory feature in leader sequences of mRNAs associated with purine metabolism [12]. This study recognized the two mutually conserved stem-loop structures, a rho-independent transcriptional terminator and an antiterminator, secondary structural features of the mRNA known to be associated with transcriptional attenuation [13]. In addition, repression of transcription was effected by low molecular weight compounds hypoxanthine, guanine and weakly, xanthine. However, a proposed regulatory protein mediating this process, such as those acting on the trp and pyr operons, was not identified. Similar findings with leader sequence elements associated with operons related to S-adenosylmethionine [14], thiamine pyrophosphate (TPP) [15], riboflavin [16] and adenosylcobalamin [17] metabolism led to the very prescient speculation these mRNAs directly bind their small molecule effectors [18]. Only after the discovery of several other conserved RNA elements that directly bind small molecule metabolites [19-23] was it established some purine leader sequences interact with either guanine or adenine [24, 25]. Shortly after this discovery, the Batey laboratory was the first to report the structure of a riboswitch, the B. subtilis xpt-pbuX guanine-responsive aptamer domain, revealing core principles of RNA-small molecule recognition and regulation of gene expression by riboswitches [10]. Later, a third class of riboswitches within the purine riboswitch family was discovered to be regulated by 2′-deoxyguanosine, but this RNA is currently only found in a single organism, Mesoplasma florum [26].
There are several reasons why the purine riboswitch has emerged as a favored model system. First, and likely foremost, it is a structurally simple “complex” RNA—that is a multi-helix structure with side-by-side helical packing. This architecture, described as a three-way junction supported by a distal tertiary interaction (see Section 2), is a highly recurrent theme in RNA biology [27]. In addition to the purine riboswitch aptamer this structural theme is employed to create a functional center in several other riboswitch aptamer domains, the hammerhead ribozyme, signal recognition particle and various substructures of the ribosomal RNA. Further, of the three classes of three-way junctions, the purine riboswitch is a member of the most common class (class C) [28]. Secondly, this RNA has several activities—namely ligand binding and gene regulation—that can be monitored by standard molecular, biochemical, and biophysical techniques. A significant advantage is adenine binding variants also recognize 2-aminopurine (2AP), enabling fluorescence approaches to be used that do not require labeling of the RNA or ligand. Third, RNAs derived from purine riboswitch aptamer domains are remarkably well behaved. While many RNAs tend to misfold, multimerize, or readily degrade, the purine binding aptamer has proven itself easy to work with. In part, this is likely due to the strong selective pressures on rapid and efficient folding present inside the cell as riboswitches may have only a brief timeframe in which to influence the expression machinery (see Section 6). Finally, the purine riboswitch aptamer at ~60 nucleotides in length is one of the smallest RNAs that embodies the above features. The small size makes NMR spectroscopy tractable as well as facilitates the facile incorporation of modifications, such as fluorophores, into the RNA. In this review, we discuss how the purine riboswitch has been used as a model system to study aspects of RNA chemistry and biology.
2. Purine nucleobase recognition
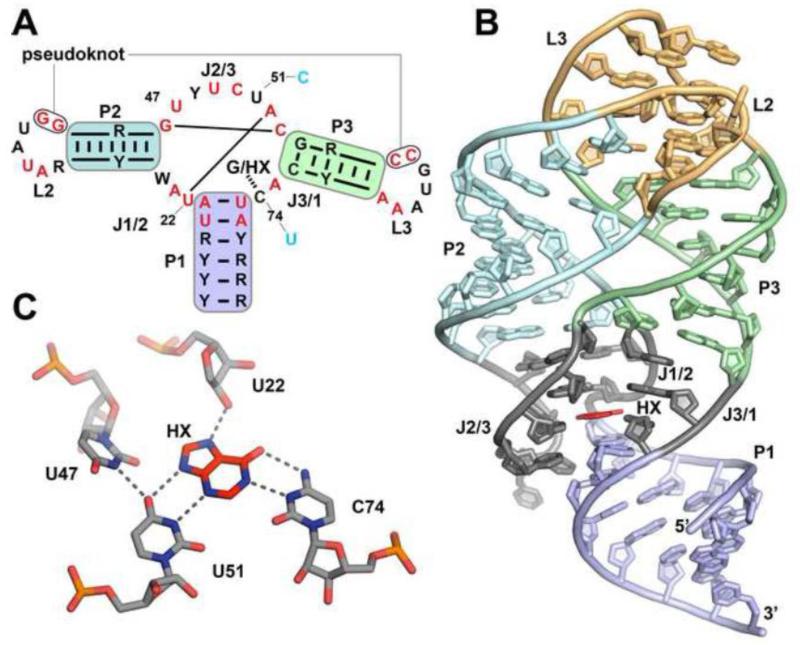
Currently, the aptamer domain structure of four members of the purine riboswitch family have been solved by X-ray crystallography: Bacillus subtilis xpt-pbuX guanine-responsive [10, 11], Vibrio vulnificus add adenine-responsive [11], B. subtilis pbuE adenine-responsive [29], and Mesoplasma florum 1A 2′-deoxyguanosine-responsive [30]. The aptamer domains of these four riboswitches share a common three-dimensional architecture and ligand binding pocket, which are highly conserved throughout the family [24, 26, 31]. The aptamer domain consists of three Watson-Crick paired regions (P1 – P3; Figure 1A, B) organized around a central three-way junction. Sequence conservation within the aptamer is localized to the two terminal loops (L2 and L3) that form a pseudoknot interaction and the three joining regions (J1/2, J2/3 and J3/1) that comprise the junction. Within the junction is a set of nearly invariant nucleotides (highly conserved nucleotides across the purine family [32, 33] are highlighted in red in Figure 1A) organizing three critical bases for hydrogen bonding interactions with the ligand forming essentially a base quartet (nucleotides 47, 51, and 74 in the xpt numbering scheme; for simplicity, features of all members of the purine family in this review will be referenced using this numbering scheme). These bases, along with the 2′-hydroxyl group of U22, create the network of hydrogen bonds that fully recognize the ligand (Figure 1C). Adenine-responsive riboswitches discriminate against guanine primarily through a single change in the pocket (C74 for U74), enabling Watson-Crick pairing to this nucleobase. While in principle U74 could pair to guanine through a wobble pair, the architecture of the junction appears to prevent shifting of nucleotide 74 towards the major groove [34].
Figure 1. The purine riboswitch consensus and binding architecture.
(A) A simplified consensus sequence projected upon the secondary structure of the purine family of riboswitches, derived from the Rfam database [33]. Red nucleotides indicate at least 90 percent conservation. Colored backgrounds represent paired regions corresponding to the overall three-dimensional structure (B) Global architecture of a representative member of the purine family (B. subtilis xpt-pbuX) bound to hypoxanthine (HX). Structure is taken from PDB ID 1U8D. (C) Hydrogen bonding pattern of the two residues that confer ligand specificity (xpt:hypoxanthine U51 and C74) and the critical organizing nucleotide U47.
Recognition of 2′-deoxyguanosine, on the other hand, is achieved primarily through the identity of the nucleotide 51 being cytosine (Figure 1A) [30, 35]. In the bound state, C51 shifts towards 74 relative to U51 in the guanine-bound aptamer, sterically accommodating the 2′-deoxyribose sugar. Accordingly, nucleotide 47 disengages from its interactions with residue 51 and reorients itself out towards the solvent. Thus, it appears that identities of just two nucleotides, 51 and 74, govern discrimination between different purine nucleobases and nucleosides [35]. In principle, the C51/U74 combination would enable recognition of 2′-deoxyadenosine, although this sequence has not yet been observed in biology [26]. Additionally, the M. florum 1A riboswitch has a ~100-fold preference for 2′-deoxyguanosine over guanosine [26]. Comparison of the wild type aptamer bound to both compounds revealed that the ribose sugar of guanosine adopts the C3′-endo conformation that prevents hydrogen bonding of the 3′-hydroxyl group with C48 (C56 in M. florum numbering) [30]. This causes C48 to be flipped out towards solvent, which was speculated to destabilize the conformation of J2/3, leading to lower affinity for guanosine.
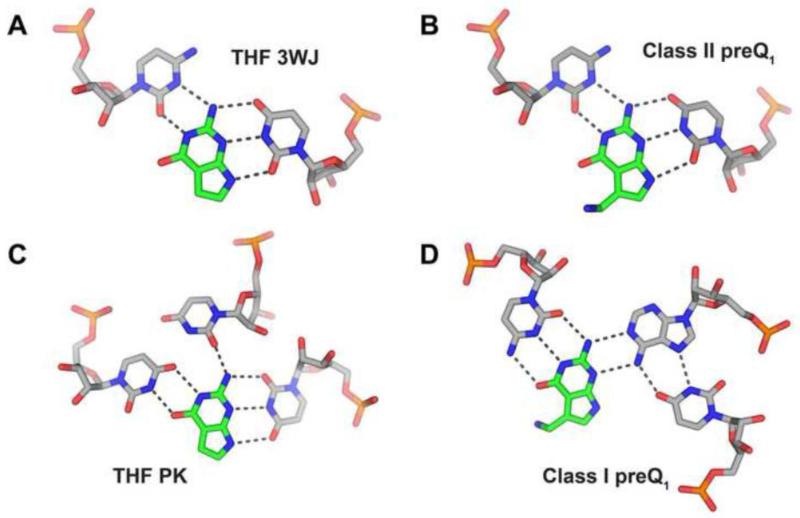
Other riboswitches use similar pyrimidine-rich binding sites to recognize purine nucleobases. For example, the three-way junction (3WJ) site of the THF riboswitch [36] and the preQ1-II riboswitch [37] recognizes guanine through a U-purine-C base triple (Figure 2A, B). The second site in the THF riboswitch, at the pseudoknot (PK), has a slightly different arrangement of pyrimidine residues around the purine base, but still uses a uridine base to recognize the N9/N3 face (note that the N9/N3 face corresponds to the “sugar edge” of purine nucleotides) (Figure 2C) [36]. The only deviation from this theme is found in the preQ1-I riboswitch where the N9/N3 face of the guanine base interacts with the Watson-Crick face of the base of an adenosine residue (Figure 2D) [7-9]. Similarly, the in vitro selected theophylline aptamer uses two pyrimidine residues to hydrogen bond to the Hoogsteen and N3/N9 edges of its ligand [38]. Thus, for most aptamers recognizing purine nucleobases, a recognition strategy involving two pyrimidine residues appears to be the easiest solution. This simple scheme, however, is not reflected in the binding of purines as part of more complex molecules such as S-adenosylmethionine, cyclic-di-GMP or adenosylcobalamin by riboswitches as well as GMP recognition by group I introns. In these cases, recognition of the nucleobase of the ligand is idiosyncratic, more typically using hydrogen bonding interactions with purines in the RNA [39-41].
Figure 2. Comparing the modes of recognition between various purine binding pockets.
A common theme of pyrimidine-rich binding pockets is observed in most purine-binding aptamers, often with a uracil base recognizing the N3/N9 face of the ligand. The binding pocket of the (A) THF three-way junction (PDB ID 4LVW) and (B) class II preQ1 (PDB ID 4JF2) are nearly identical, with recognition of the ligand achieved by the same spatial arrangement. Again in the (C) THF pseudoknot site, guanine’s N3/N9 face is paired to a uracil base, while additional hydrogen bonds are mediated by two ancillary pyrimidines juxtaposed to the Watson-Crick face. The only major departure from this trend is the (D) class I preQ1 riboswitch (PDB ID 3FU2) where the binding pocket is still populated by several pyrimidines, yet recognition is partially achieved by an adenine base structured via auxiliary hydrogen bonding to a uracil.
The purine riboswitches also bind a variety of non-natural purine [24, 31, 34, 42-44] and pyrimidine analogs [45, 46]. Many of these analogs represent trivial solutions to recognition in that they bind with the same hydrogen bonding configuration as the natural effectors and no alteration in the conformation of the RNA. For example, 2-aminopurine (2AP) binds to the adenine riboswitch and the (C74U)xpt-pbuX riboswitch in the same configuration as adenine, enabling it to be widely exploited as a means of investigating the ligand binding properties of purine riboswitches due to its spectral properties [43, 47-49].
2AP recognition by purine riboswitches revealed an interesting aspect of effector discrimination by these RNAs. Several compounds with substitutions at the purine 6-position, such as 2-AP (deletion of N6), 6-chloroguanine (substitution of the guanine O6 with a chlorine atom), and 6-O-methylguanine (methylation of the guanine O6), bind guanine and adenine riboswitches with nearly the same affinity. This is in stark contrast to the nearly 10,000-fold specificity each riboswitch has for its associated purine (guanine vs. adenine) [24, 25]. A comparison of the structures of the wild type and C74U xpt aptamers bound to 2AP, revealed that cytosine 74 is capable of shifting to the minor groove to re-establish a two-hydrogen bonding interaction with the ligand [34]. Presumably, uridine 74 in adenine riboswitches cannot shift towards the major groove, which would enable a G•U wobble pair to form and diminish the discrimination of this riboswitch for adenine over guanine. These observations highlight that in searches for compounds that bind purine riboswitches as potential antimicrobial agents [50], the ability of nucleotides 51 and 74 to reposition themselves within the ligand binding pocket to establish alternative hydrogen bonding patterns must be considered.
3. RNA structural nuances yield functional differences
Global organization of the purine riboswitch family is governed by interactions between L2 and L3 serving to pack P2 against P3 (Figure 1B). Under near physiological magnesium ion concentrations (0.5 - 1 mM), this interaction provides ~4.1 kcal/mol to the ligand binding energy in the xpt/pbuX riboswitch [51] and at least 2.9 kcal/mol in the pbuE adenine riboswitch [52]. Formation of this tertiary interaction is critical for high affinity recognition of ligands, but low affinity binding can be achieved in its absence [51-53]. At the core of this interaction within all members of the purine clan is the formation of two invariant G-C Watson-Crick pairs between L2 and L3 (“pseudoknot”, Figure 1A). Beyond this core element, the guanine, adenine, and 2′-deoxyguanosine riboswitches all have differences contributing to their effector binding properties.
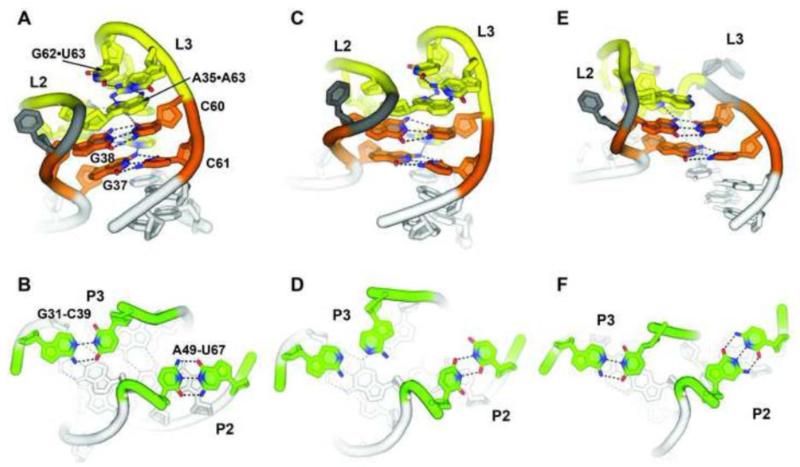
The guanine riboswitches, as exemplified by the B. subtilis xpt-pbuX riboswitch, contain an extremely stable L2-L3 interaction capable of forming in the absence of either magnesium ions or ligand. The overall architecture of this tertiary interaction is a set of four non-canonical base pairs that scaffold two essential Watson-Crick G-C pairs (Figure 3A, orange). The G38-C60 Watson-Crick pair interacts with a reverse Watson-Crick/Hoogsteen A33-A66 pair in its minor groove, while the G37-C61 pair interacts with a reverse Watson-Crick/Hoogsteen U34-A65 pair. It is interesting to note the packing of the two non-canonical base pairs into the minor groove of the G-C pairs is reminiscent of the type-I/type-II A-minor triples that define the interaction of GAAA tetraloops into the minor groove of A-form helices [54, 55]. Distal to these two base quartets are two other pairs: an A35•A63 pair and a side-by-side G62•U63 pair. These pairs make significantly weaker energetic contributions to ligand binding, as reflected in their lower degree of phylogenetic conservation [53]. smFRET [56], NMR [57], and in-line chemical probing studies [24] all show the L2-L3 tertiary interaction is fully formed and stable in the absence of guanine and moderate (1 mM) magnesium ion concentrations. Even in the absence of Mg2+, the “docked” conformation is significantly sampled, indicating that magnesium is not essential for the L2-L3 interaction [56]. Therefore, it is likely that this tertiary structure is tightly formed under physiological conditions such that it promotes preorganization of the binding pocket to enable rapid and efficient ligand binding [58]. Observations by NMR of ligand binding in the absence of magnesium reinforce this observation [11, 59], although the interaction is significantly weakened in the absence of divalent ions [10]. Strikingly, the L2-L3 interaction is observed to form outside the context of the three-way junction or helix 1, further emphasizing its intrinsic stability [57].
Figure 3. Spatial arrangement of the loop-loop tertiary interaction in various purine riboswitch aptamers.
(A, B) The xpt-pbuX guanine sensing riboswitch contains a stable L2-L3 interaction where four non-canonical base pair interactions (yellow) provide the context for two essential and invariant Watson-Crick G-C pairs (orange) (PDB ID 1U8D). These loops are both closed by Watson-Crick base pairs (green). (C, D) The pbuE riboswitch contains a highly similar tertiary interaction as xpt, (PDB ID 3IVN) but differs in that both loops are closed by non-canonical base pairs. (E, F) The 2′-deoxyguanosine aptamer deviates substantially in the base identities surrounding the G-C pairs containing a deletion of three nucleotides in L3 (PDB ID 3SKI). In all panels, hydrogen bonds are represented as grey dashes, and select oxygen and nitrogen atoms are red and blue, respectively.
A number of adenine riboswitches contain a less stable L2-L3 interaction as compared to the guanine riboswitches. While base specific interactions between L2 and L3 are nearly identical [11], the closing base pairs differ [11, 31]. In the vast majority of guanine riboswitches, the closing pair of L2 and L3 is a Watson-Crick base pair (Figure 3B), whereas in the B. subtilis pbuE adenine riboswitch, L2 is closed by a U•U pair and L3 by an A*A pair (L3 of the V. vulnificus add adenine riboswitch is closed by a G-C Watson-Crick pair) (Figure 3C, D). In the pbuE variant, the absence of magnesium does not promote even transient sampling of the docked configuration [52], which is supported by NMR studies [60]. Instead, at least 50 μM Mg2+ is required to promote the docked state [61]. Differences in the stability of the L2-L3 interaction may have consequences for the regulatory activity of the riboswitch because the tertiary interaction directly influences the conformational ensemble of the unbound junction. Destabilization of this interaction would result in increased disorder and potentially decrease the rate of ligand binding kinetics (kon) and thereby increasing the concentration of ligand required to elicit a regulatory response (see Section 6) [60].
Finally, the class 1A M. florum 2′-deoxyguanosine riboswitch contains a substantial variation in the nature of the L2-L3 interaction. While L2 possesses the same size and pattern of sequence conservation as the rest of the purine riboswitch family, L3 contains a deletion of the middle three nucleotides (Figure 3E, F). While the two core Watson-Crick pairs are maintained, only the equivalent of G37-C69 is engaged by other nucleotides. Distal to these pairs, a single A•A pair between the loops further reinforces the tertiary interaction. Despite the significant difference in tertiary architecture, these novel interactions contribute little to the specificity for 2′-dG over guanine [30, 35]. Instead, sequence differences in P2 adjacent to the three-way junction confer specificity [30, 35].
Within P2 and proximal to the three-way junction is a lightly conserved yet important component called the “P2 tune box” [48]. This sequence element was found through a structure-based sequence alignment of guanine/adenine riboswitches in which only the subset of sequences alignable without insertions and/or deletions were considered. This high-quality alignment yields two sets of nucleotides showing statistically significant co-variation despite being non-interacting: 66/68 in L3 and 24/25 at the interface of J1/2 and P2. Genetic and biochemical analysis revealed a covariation pattern between nucleotide 24, which stacks between nucleotides 72 and 73 at the P3-J3/1 interface and the first two base pairs in P2. Within the purine family there is a marked tendency for the first two base pairs of P2 to be non-canonical pairs. Interestingly, this is a crucial element in the 2′-deoxyguanine aptamer for achieving high-affinity and specific binding of its cognate ligand—more important even than the variant L2-L3 interaction [35]. The structure of this aptamer revealed that nucleotide 24 instead of stacking with P3, forms a triple base pair with the first base pair in P2 [30]. Detailed affinity and kinetic measurements of a series of P2 tune box sequences, both natural and non-natural, reveal that this region has a significant impact upon the association and dissociation rates of ligand binding [48]. Strikingly, while a single point mutation in this box is lethal to the in vivo regulatory activity of the riboswitch, a compensatory mutation to non-interacting nucleotides (i.e., to nucleotides not involve in base pairing) rescue full wild type activity. Like the distal L2 sequence variation affecting the stability of the tertiary interaction, it is hypothesized that the P2 tune box also influences the conformational ensemble of the free state to tune the regulatory activity of the riboswitch to the needs of its associated transcriptional unit.
Another region with clear patterns of conservation exists within the P1 helix that has yet to be investigated. The crystal structure of the guanine and adenine riboswitches reveal that the two base pairs proximal to the ligand binding core (U20-A76 and A21-U75) are involved in ligand-dependent base triples with nucleotides in J2/3 (49 and 50, respectively), and the identity of these nucleotides are almost invariant (Figure 1A). Beyond that, the next three base pairs show a significant preference for the orientation of the purine-pyrimidine pair (for example, the pair below U20-A76 shows a significant tendency for a purine on the 5′-side of the helix). Most likely, these preferences reflect a need to optimize the stability of P1 relative to the competing downstream helix. However, like the rest of the expression platform (see Section 5), the role of specific sequence elements or conservation patterns in the P1 helix remains poorly understood. This is somewhat surprising in light of the central role that this helix plays in communicating the binding state of the aptamer to the downstream elements that direct the expression machinery.
4. The role of metal ions
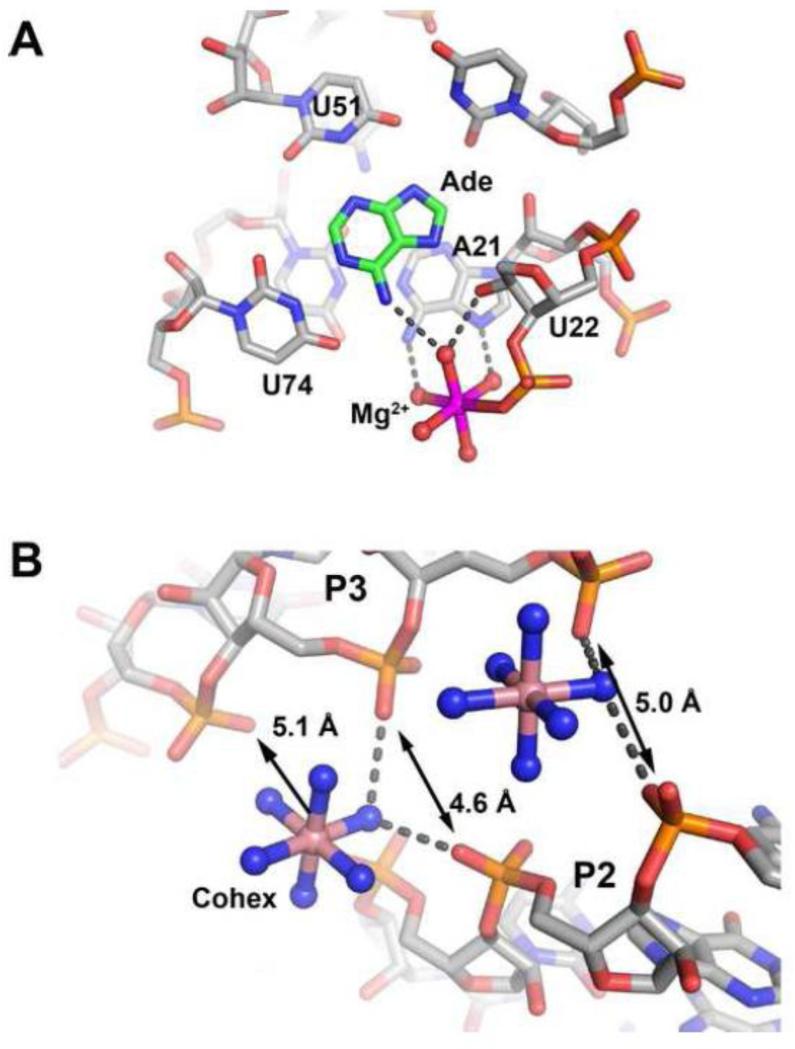
Like almost all RNAs, cations have a strong influence upon the structure and activity of the purine riboswitch (an excellent overview of the role of cations in RNA is given by Williams and colleagues [62]). Multivalent cations interact with RNA in three distinct ways [63], the first being ion specific sites playing specific and critical roles in catalysis or binding. Examples include the TPP [5, 6, 64] and flavin mononucleotide (FMN) [65, 66] aptamer domains in which magnesium mediates interactions between phosphate groups of the ligand and RNA. Quite remarkably, the recent structure of the fluoride riboswitch has revealed how specific magnesium ions completely mediate the halide-RNA interaction [67]. In addition, the magnesium and glycine riboswitch aptamers use specifically bound magnesium cations to stabilize RNA architecture [68-70]. Invariably, these ions form inner sphere contacts with either the RNA or the ligand. Second are divalent ions associating with RNA non-specifically in regions of high electronegative surface potential and may or may not form inner sphere contacts with the RNA. Several examples of such ions are found in the purine riboswitch, such as a magnesium ion that sits adjacent to the ligand binding site in the add-adenine structure [11] (Figure 4A) and cobalt hexamine ions found along P2 and P3 of the xpt-hypoxanthine structure where phosphate groups come into close proximity (Figure 4B). These ions are often observed in X-ray structures, but different monovalent and/or divalent ions can occupy these sites [71]. Finally, “diffuse” ions compose an atmosphere of non well-localized ions around the RNA and their behavior is dictated by long-range electrostatics. From a series of crystallographic and NMR studies of representative purine aptamers, a number of high-occupancy metal ion binding sites dispersed throughout the structure have been observed [10, 11, 72], but none of these appear to be essential for ligand recognition. Instead, the role of divalent metal ions is to increase the affinity of the aptamer for effector by stabilizing global RNA architecture. Thus, for the purine riboswitch, cations appear to play a non-specific role promoting folding and ligand binding.
Figure 4. Coordination of metal ions along the purine riboswitch.
(A) Magnesium ion (magenta) present in the crystal structure of the add riboswitch from B. subtilis (PDB ID 1Y26 that is adjacent to the binding pocket. (B) Two cobalt hexammines observed in the xpt riboswitch structure (PDB ID 1U8D) reveals the interaction of diffuse ions along the riboswitch bridging a region of the structure where the backbone comes into very close proximity to itself (distances between three non-bridging phosphate oxygens shown).
More recently, Leipply and Draper developed a quantitative model describing the effect of magnesium on folding of the V. vulnificus add adenine riboswitch [73]. Structural characterization of this RNA by small angle X-ray scattering (SAXS) and hydroxyl radical probing enabled four states of the RNA to be defined: (1) an unbound and extended form of the aptamer in which L2 and L3 do not interact, (2) a ligand bound form with the L2 and L3 interaction unformed, (3) an unbound form displaying the L2-L3 tertiary interaction and (4) the bound and folded state. These four states are connected in a thermodynamic cycle by three free energy values: (1) formation of the L2-L3 interaction (ΔGdock), (2) binding of ligand (ΔGLBP) and (3) a coupling factor linking the two processes (ΔGw). The magnesium dependence of each of these energies yields information about the mechanism(s) by which divalent cations act on the free energy folding landscape of the RNA. For the adenine riboswitch, magnesium appears to act on the RNA in two ways with nearly equal effectiveness: promoting formation of the L2-L3 interaction and stabilizing the ligand binding pocket. Conversely, coupling of the tertiary interaction and the ligand binding pocket is only weakly magnesium dependent, indicating that their linkage is largely a structural feature of the aptamer [73].
Monovalent cations also influence the stability of the tertiary architecture of the purine riboswitch aptamer domain. In the absence of magnesium, the thermal stability of the V. vulnificus add-DAP complex is strongly influenced by the identity of the monovalent cation [74]. As the radius of the monovalent cation increases down the group I series (Li+, Na+, K+, Rb+, and Cs+), stability of the complex decreases by almost 3 kcal/mol. This effect is rationalized as a result of high charge density in the RNA and small ions have access to sterically restricted regions of high electronegative potential. This effect is counter to the higher desolvation energies of smaller monovalents such that inner sphere ion-RNA interactions are more costly [75, 76]. The lack of a specific monovalent effect, such as observed for GAAA-tetraloop receptor for K+ [74, 77], indicates that monovalent cations interact with purine riboswitch aptamer domains as diffuse ions. In support of this trend, a soak of the M. florum 2′-deoxyguanosine riboswitch with 10 mM CsCl reveals three unique monovalent binding sites; two sitting in the major groove of P2 and a third adjacent to the 2′-hydroxyl of C31 and non-bridging phosphate oxygens of A32 (equivalent to U22, A23 in the xpt numbering system) [30]. None of these sites suggest a significant number of direct RNA-ion interactions, but rather generally reflect regions of high negative potential.
5. Beyond the aptamer domain: the nebulous expression platform
While the aptamer domain, defined as the minimal RNA sequence necessary to achieve high affinity binding of the cognate effector ligand, is structurally well characterized, the structural features of the expression platform are largely unexplored. A serious limitation in the analysis of this region is that most alignments of riboswitches, particularly those found in Rfam [32, 33] generally exclude features other than the easily identifiable and alignable aptamer domain. Structural elements in the expression platform, such as terminator/antiterminator or sequester/antisequester hairpins, are proposed based upon computational secondary structural prediction algorithms. While these programs are good at predicting relatively strong elements of secondary structure, such as rho-independent terminators, the presence of weaker elements that may nonetheless influence the regulatory properties of the RNA remain largely unknown. Additionally, transcriptional termination elements can vary significantly in their structure impeding identification of consensus elements and conserved motifs [78].
Studies of other classes of riboswitches illustrate the need to consider structural elements beyond the aptamer and the generally obvious alternative secondary structural switch in the expression platform. In the expression platform of the B. subtilis lysC lysine riboswitch, there is a small hairpin structure, P6, that lies between the P1 helix and terminator element that does not appear to be directly involved in the secondary structural switch—it is present in the antiterminator and terminator states [79]. The presence of this stem-loop, which would be contiguous with the antiterminator helix, is important for efficient lysine-dependent regulation. Deletion of this hairpin without affecting the antiterminator/terminator structures causes the riboswitch to be constitutively “OFF”, whereas stabilization of this element prohibits formation of the terminator in an in vitro single-turnover transcription assay. Presumably, this hairpin facilitates nucleation of the antiterminator helix at the expense of P1, preventing rho-independent termination. While this element is not conserved in lysine riboswitches, these results suggest that sequence and structure in the expression platform that are not directly involved in alternative structure formation may be important for efficient function [79].
A second instructive example of how inconspicuous structural features adjacent to the aptamer domain can profoundly influence activity is found in the glycine riboswitch. Originally, this riboswitch family was defined by two structurally similar aptamer domains, each binding a single glycine molecule with strong cooperativity [80]. However, just outside of the presumed aptamer domain exists a recently discovered kink-turn motif connecting the two aptamers [81, 82]. This motif and the associated P0 helix promotes interdomain association in the absence of glycine, abrogating cooperative ligand binding under physiological conditions [82, 83]. These studies highlight the need to investigate the sequence and structure of expression platforms to fully define the role the expression platform plays in gene regulation by purine riboswitches.
6. Mechanisms: a tale of two adenine riboswitches
In bacteria, riboswitches typically control gene expression by transcriptional and/or translational attenuation, although a recent study has uncovered further evidence for widespread regulation of gene expression via regulation of antisense RNAs [84]. Also, some riboswitches have been identified that require the Rho termination factor as part of the regulation mechanism [85]. Regulatory control at the transcriptional level has a significant consequence: the riboswitch has only a short timeframe in which to influence RNA polymerase before it escapes beyond the intrinsic terminator. Thus, the riboswitch must do three things rapidly and efficiently: (1) acquire secondary and tertiary structure in the aptamer domain, (2) survey the cellular environment for the cognate ligand, and (3) direct alternative secondary structure formation based on the status of the aptamer domain. An early study of the B. subtilis pbuE adenine riboswitch revealed that adenine binds to the aptamer with slow association and dissociation kinetics [49]. A theoretical consideration of the timeframe required to fully saturate the aptamer at a given intracellular ligand concentration yielded the conclusion that the timeframe of equilibration of the aptamer is substantially longer than what is required for the polymerase to transcribe the expression platform through the terminator. Thus, it was proposed that there is a subset of riboswitches under “kinetic control”. The hallmark of kinetic control is that concentrations of effector required to elicit a half maximal regulatory response (referred to as the EC50 for in vivo and T50 for in vitro) are greater than the concentrations of ligand required to half-saturate the aptamer under full equilibrium (referred to as the KD). Presumably, riboswitches that control gene expression at the translational level do not have a similar temporal constraint, and thus could be under “thermodynamic control” in which the EC50 or T50 is equivalent to the aptamer’s KD.
To understand this potential feature of riboswitches, the Lafontaine and Massé groups performed a comparative study of the B. subtilis pbuE adenine riboswitch (transcriptional regulation) and the V. vulnificus add adenine riboswitch (translational regulation) [86]. It had been previously established that the full length pbuE riboswitch encompassing both the aptamer domain and expression platform cannot efficiently bind adenine, indicating that it is thermodynamically locked into a state where the terminator stem loop has formed at the expense of the aptamer [52, 86, 87]. This implies that the RNA does not act as a reversible switch, but rather as a “fuse” whose ligand-directed folding must occur during the transcription process [88].
The regulatory properties of the pbuE riboswitch are strongly influenced by transcription conditions. First, in a single-turnover transcription assay the T50 showed a significant dependence upon the concentration of NTPs [86], which influences the rate of transcription [89]. As the concentration of NTPs was raised from 20 to 150 μM, the T50 increases from 0.2 to 1.3 μM for 2,6-diaminopurine (an adenine analog). Similar results have been observed for the B. subtilis ribD FMN [88] and lysC lysine [90] riboswitches. Second, a long-lived transcriptional pause located downstream of the pbuE adenine riboswitch aptamer domain was observed at a stretch of uridine residues at positions 114-117 [86]. Uridine tracts are well-known pause sites for bacterial RNAPs, and a similar finding was again observed in the FMN riboswitch [49]. Presumably, the pause stalls the polymerase affording the aptamer more time to interrogate the cellular environment and reach equilibrium with respect to effector concentration. Third, NusA, a promiscuous acting transcription factor that generally affects termination, lowers the T50 by decreasing the rate of transcription. Finally, all of the observed T50 values are greater than that for DAP binding the pbuE riboswitch (KD ~ 25 nM) [86]. Together, these data indicate the riboswitch only acts in the context of transcription and is modulated by factors affecting the rate of RNA synthesis by means of altering the residence time of the polymerase at the riboswitch. While these in vitro data strongly suggest that pbuE is under kinetic control, analogous experiments need to be performed in vivo to firmly establish that in cellular conditions, the same properties are observed.
Similar experiments on the V. vulnificus add riboswitch indicate that it exerts translational control in a thermodynamic regime. In contrast to the pbuE riboswitch, the full length add riboswitch binds adenine with only a slight reduction in affinity as compared to the aptamer domain alone [86, 87]. This suggests the alternative P1 stem and sequester hairpin reversibly exchange between the two states. This was confirmed by chemical probing of the native and several mutant sequences that stabilized either the “ON” or “OFF” state in the presence and absence of adenine as well as by single molecule force extension spectroscopy [86, 91]. Further, this riboswitch upregulates β-galactosidase expression in either a coupled or uncoupled in vitro translation system, again indicating the reversibility of the switch [86]. However, in vivo, the riboswitch requires ~500 μM adenine in the extracellular media to elicit a half-maximal regulatory response, substantially higher than the KD (~500 nM). While this might be interpreted as kinetic control in vivo, the authors point out that the cellular uptake of adenine may be very poor or the metabolic turnover high, such that the intracellular concentration of adenine is substantially lower than that in the medium. Further, the pioneer round of translation is physically coupled to transcription in vivo via NusG [92], which also stimulates Rho-dependent transcriptional termination. Thus, translational riboswitches may face temporal constraints via a competition between the rate of ribosome association and Rho-dependent termination. It remains to be determined whether the thermodynamic control of the add riboswitch in vitro also occurs in the cell. While it may be a tempting speculation to associate translational regulation with thermodynamic control [86], this is still an open question.
Another excellent study addressing the question of thermodynamic versus kinetic control of riboswitches investigated the Fusobacterium nucleatum preQ1-1 riboswitch [93]. A critical feature of this riboswitch is that the expression platform is “bistable” in which the antiterminator and terminator hairpins are in reversible equilibrium, where ligand binding to the aptamer significantly shifts the equilibrium towards the terminator state. This nearly unique property enabled a detailed characterization of the ligand-dependent switching properties of the full length riboswitch using both NMR and fluorescence approaches [93]. Kinetic analysis of the terminator/antiterminator rearrangement and ligand binding indicate that, in the absence of polymerase pausing, that the association rate is sufficiently rapid to trap the terminator state at proposed intracellular preQ1 concentrations (30 μM). However, the authors clearly state that without in vivo analysis, the kinetic versus thermodynamic issue in the context of transcription in the cell remains an open question [93].
7. Folding of the riboswitch
Since many riboswitches act co-transcriptionally, the ability to fold rapidly and efficiently is critical for the riboswitch function. First, the RNA must acquire the secondary and tertiary structure that defines the global architecture of the aptamer domain. Second, the binding pocket is formed during this phase and becomes competent for ligand binding. Generally, in the apo-state, riboswitch binding pockets are weakly organized and exhibit a conformational ensemble, of which only a subset are productive for ligand binding, which in turn strongly influences the association rate [51, 66, 94]. Furthermore, since the purine nucleobase is almost completely solvent inaccessible when bound to the aptamer, there must be conformational change in the three-way junction coupled to binding. Finally, since the P1 helix must at least partially form to create the high-affinity aptamer for the ligand, if the riboswitch aptamer remains unbound then an alternative secondary structure must form at its expense. Together, these events illustrate the purine riboswitch is highly dynamic during the course of its function and to fully understand how this RNA transduces intracellular effector concentration into a regulatory response, these processes must be considered.
The folding of the aptamer domain has been investigated using a number of experimental techniques including NMR [57, 95, 96], smFRET [52, 56], chemical probing [51], fast fluorescence spectroscopy [97, 98], single molecule force extension spectroscopy [91, 99, 100], as well as molecular dynamics simulation [101-104]. While all of these techniques observe different aspects of the folding process, together they yield a reasonably consistent model of the folding process. The best models for acquisition of secondary structure comes from single molecule force extension spectroscopy of the pbuE and add adenine riboswitch aptamers [91, 100]. These studies reveal sequential formation of the P2 and P3 hairpins, then the L2-L3 interaction, and finally P1. Since only a few base pairs of P1 are required for moderate-affinity ligand binding [47], it is likely that productive ligand binding starts to occur prior to full formation of the P1 helix. smFRET studies indicate that the L2-L3 interaction is dynamic and forms to variable extents in the apo-state in different purine riboswitch variants (see Section 3) [52, 56]. It should be noted that these experiments investigate the folding of the full length aptamer domain and that the observed folding pathway may be different from a co-transcriptional folding pathway (see Section 7).
One of the most poorly understood aspects of bound state formation is the process of initial ligand docking with the three-way junction and the coupled folding of RNA around the ligand. There is substantial disagreement about the degree of preorganization of the junction and how the aptamer initially recognizes the appropriate ligand. In competing NMR studies of the xpt riboswitch, one group finds that in the absence of ligand the three-way junction lacks any indication of hydrogen bonding that defines the bound state [57], while another argues that the core is moderately preorganized around the junction adjacent to P2/P3 [96]. It should be pointed out that in the latter study, mutations were introduced into the P2 tune box artificially stabilizing this part of the RNA, which may have significantly influenced their observations. A more illuminating study by the Schwalbe group used time-resolve NMR methods to investigate ligand induced folding of the junction using photo caged hypoxanthine to synchronize binding [95]. Two sets of folding rates were observed: a faster group (t1/2 = 19 - 24 s) in the junction and a slower set (t1/2 = 27 - 30 s) in the L2-L3 interaction. Along with other experiments indicating line broadening for non-specific ligands in the core, these data suggest a model in which the ligand forms a low-affinity encounter complex with the core, followed by organization of the core around the specific ligand followed by full stabilization of the L2-L3 interaction that locks P2 and P3 into place.
Using ultrafast multidimensional NMR methods, the Varani group was able to generate an even higher resolution model of adenine/magnesium induced folding of the add aptamer [105]. Their model, generally consistent with those based upon single molecule studies, clearly reveals fast initial docking of adenine with core (~16 s), while the L2-L3 interaction and a region of the P1 helix proximal to the ligand binding site remain unfolded. Kinetically slower folding events include full stabilization of the L2-L3 interaction (~28 - ~58 s) while P1 remains flexible. Finally, the native structure is fully acquired after 2 - 3 minutes. In particular, these data highlight the direct role of ligand in stabilizing the P1 helix, a central feature of the alternative structural switch of many riboswitches.
The most elusive aspect of the folding process is the nature of the initial docking event with between the ligand and the three-way junction. Based upon other kinetic and temperature-dependent chemical protection data, the Batey group proposed a model in which the ligand initially docks with nucleotide 74 in J3/1 and J2/3 folds around the ligand to form the final complex [51]. Various molecular dynamics (MD) simulations disagree on this point, proposing models in which the ligand initially docks with nucleotide 51 [102, 103], while others agree that 74 is the initial docking site [104, 106]. However, all of these studies must be taken with some caution as the force fields used for these calculations are still a work in progress and struggle to accurately describe experimental observations of even simple RNA motifs [107-109]. Nonetheless, these simulations present interesting hypotheses that may become the basis for further experiments that will address this issue regarding both the initial ligand recognition and specificity for the cognate effector.
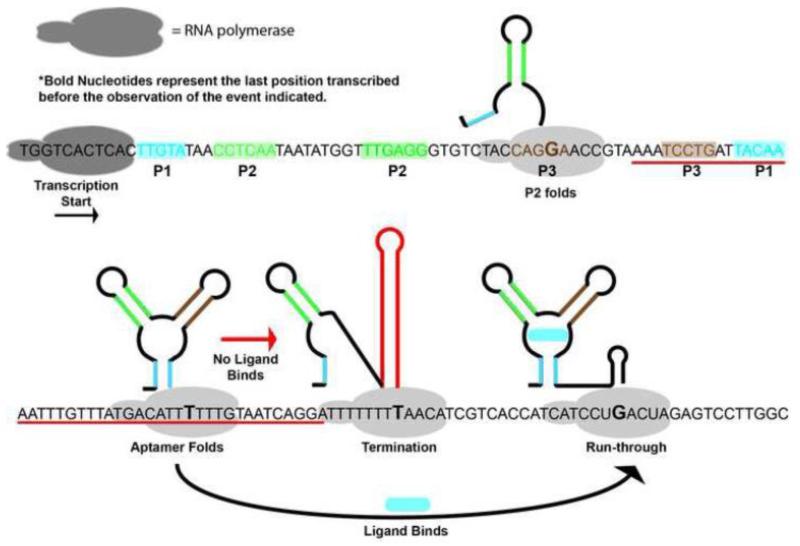
To fully understand the folding pathway of the purine riboswitch and its relationship to regulatory activity, optimal folding needs to be observed in the context of transcription. Currently this is not technically possible using ensemble methods because it would require that the entire population be nearly synchronous and stochastic events such as the entry and exit from programmed pauses frustrate this approach. To address this issue, Frieda and Block used force extension spectroscopy to unfold and refold nascent RNA pbuE riboswitch transcripts in the presence of varying concentrations of adenine [99]. In this experiment, E. coli RNAP stalled downstream of the promoter was attached to a bead in one optical trap, while the 5′-end of the RNA transcript was hybridized to a DNA handle attached to a second trapped bead. Transcription was reinitiated and allowed to proceed to the end of the transcription template where further elongation was blocked by streptavidin bound to the biotinylated DNA (a good review of the tools and experimental methods of force extension spectroscopy as applied to RNA is given by [110]). If transcriptional termination occurs at the Rho-independent terminator, the RNAP disengages and the complex falls apart such that the two beads are no longer tethered. Since these experiments are capable of observing transcriptional events in real time, they yield a complete co-transcriptional folding landscape of an RNA--the first to be achieved (Figure 5). Like previous studies of the aptamer alone, the earliest event is formation of the P2 helix. Shortly after the polymerase cleared the aptamer domain sequence, the aptamer domain obtained its global architecture. If adenine binds to the aptamer, ligand stabilizes this structure against invasion from the terminator sequence long enough to enable RNAP to escape past the polyuridine tract of the intrinsic terminator. If adenine does not bind, then the terminator stem-loop forms rapidly at the expense of the P1 helix once its sequence had been fully transcribed.
Figure 5. Single nucleotide resolution co-transcriptional folding map of the purine riboswitch.
The sequence of the B. subtilis pbuE adenine responsive riboswitch used for single molecule force extension spectroscopy with secondary structural elements in the aptamer highlighted as shaded boxes. Secondary structure acquisition and ligand binding was observed along the transcription trajectory as indicated by the cartoons. For individual experiments that read-through was observed, binding of adenine was observed to occur only once, supporting the idea that this riboswitch is kinetically driven. Figure adapted from (92).
Further, these experiments clearly demonstrate kinetic control of the pbuE riboswitch. In the vast majority of observations, the aptamer folds and binds adenine only once before the polymerase escapes past the terminator. Therefore, the aptamer does not have sufficient time to fully equilibrate (requiring multiple samplings of the bound/unbound states on average) during the transcription process. Finally, in these experiments the polymerase was not observed to pause as expected at the two poly-uridine tracts. The absence of polymerase stalling was interpreted as differences in one of several experimental parameters and merits further investigation since the Lafontaine group uncovered a clear influence on pausing in the pbuE riboswitch [86].
8. Why riboswitches?
Given the pervasiveness of riboswitches across bacteria, the diversity of small molecules recognized, and their regulation of essential metabolite biosynthesis [111], it is clear that these RNAs confer a significant advantage in the maintenance of normal cellular homeostasis. With their numbers outpacing that of known protein repressors in some Firmicutes [1], these RNAs must possess some advantage over their protein counterparts. One reason might be that the RNA has an ability to integrate multiple sensory inputs into a single, appropriate regulatory response. Put another way, the regulatory response is a complex function of not only the effector concentration, but also other intracellular conditions. As discussed above, the intracellular NTP concentration affects the transcription rate, which for kinetically controlled riboswitches alters their T50. A second factor which can significantly fluctuate in some bacteria is magnesium ion concentration, though it is typically tightly regulated [112]. Another class of RNA regulatory devices called “thermosensors” exploits the thermal stability of RNA secondary structure [113-115]. Even more recently, an RNA pH sensor was characterized that represses expression of the alx locus at elevated extracellular pH [116]. Thus, the regulatory landscape of a specific riboswitch can be very complex allowing the RNA to respond in real time to variety of intra- and extracellular cues [90].
An example of how RNA modulates regulation in response to an environmental factor was recently described by the Schwalbe group [117]. They correctly note that a strictly two-state RNA would have difficulty maintaining a strong regulatory response under varying temperature conditions. At low temperatures, the RNA’s affinity for ligand is greater than at high temperatures such that a regulatory RNA would only be efficient at low temperature. However, binding is not a two state process, as has been shown in a number of studies. An earlier study of the xpt-pbuX guanine riboswitch aptamer showed that over a broad temperature range (5 – 60 °C) the free energy of binding (ΔG°) is fairly constant as measured by isothermal titration calorimetry [43]. But these same experiments revealed a marked temperature dependence upon the heat capacity (∂ΔH°/∂T), which is generally interpreted as temperature-dependent changes in the ensemble of conformers of the apo-state. Thus, the population of RNA competent to bind ligand changes as a function of temperature, which in turn is likely to influence its regulatory properties. Similar observations were made for an SAH-binding riboswitch [118]. As many other riboswitch aptamers experience multiple apo-state conformations, this is likely a general feature [94].
In the study by the Schwalbe group of a nearly full length add riboswitch (a small truncation of seven nucleotides was used at the 5′-end), the structure of two distinct apo-form conformers were elucidated by NMR. The first is a native-like state that is competent to bind adenine, while the second has an alternative secondary structure that destroys the P1 helix. Importantly, these two states are bistable such that they interconvert in response to temperature changes. Using a combination of binding analysis and an in vitro transcription/translation assays, they demonstrate that the equilibrium population of the apo-forms changes between 10 and 37 °C in such a way that enables the regulatory response to remain robust. This has important consequences for Vibrio vulnificus as it experiences both temperatures depending upon whether it is living freely in a marine environment (10 °C) or infecting a human host (37 °C), similar to a previously characterized RNA thermosensor in Listeria monocytogenes [114]. Given the diversity of environments and how rapidly conditions can change, it is not surprising that RNA-mediated regulatory mechanisms which are natively predisposed to be exquisitely sensitive to these factors are extensively utilized to regulate gene expression. Strikingly, examples of temperature-dependent riboregulation of virulence factors were recently described in Yersinia species [119] and Neisseria meningitidis [120].
9. Application of purine riboswitches
RNA-based sensors are increasingly seen as important tools for synthetic biology, of which riboswitches represent Nature’s solution to the implementation of such devices [121-124]. One important application where riboswitches can make a significant contribution is the development of new regulatory elements for inducible gene expression. While protein-based regulation is typically used for this task, a number of specific needs have arisen in synthetic biology that demand new regulatory elements for which RNA is particularly well suited. These include differential induction of multiple genes in a pathway or in multi-protein complexes and minimizing cross-talk between regulatory elements.
To address these issues, the Micklefield group has developed orthogonal riboswitches responding to non-natural small molecules using the add adenine-responsive riboswitch as a scaffold [125]. To find variants of the add riboswitch responsive to alternative compounds, all possible nucleotide combinations of positions 47 and 51 (Figure 1C) were cloned upstream of the CAT gene and each screened against a chemical library of ~80 heterocycles for conferring chloramphenicol resistance. One variant (U47C,U51C) was found to specifically bind ammeline (4,6-diamino-2-hydroxy-1,3,5-triazine) and resulted in a regulatory element capable of robustly inducing varying levels of protein expression in response to its effector. This pioneering study suggests reengineering of naturally occurring riboswitch aptamer domains is a viable solution to the creation of genetic control elements that function in a variety of contexts.
To demonstrate how orthogonal riboswitches can assist in the regulatory control of multiple genes, the authors created dual and single promoter operons in which the natural add and the M6 translational “ON” riboswitches regulate the DSRed and eGFP genes, respectively [126]. In both examples, each gene was specifically induced by its cognate effector (2-aminopurine for add and ammeline for M6). For the dual promoter system, there was no cross-talk between the riboswitches despite their sequence differing by only two point mutations in the ligand binding core. In contrast, under a single promoter in which the M6 riboswitch is in the leader sequence and add is an intercistronic control element of the DsRed:eGFP cassette, add exhibits a moderate dependence on both riboswitch effector ligands for maximal expression. The downside to this approach is that the fold induction for these riboswitches (10-25 fold) is lower than some protein systems.
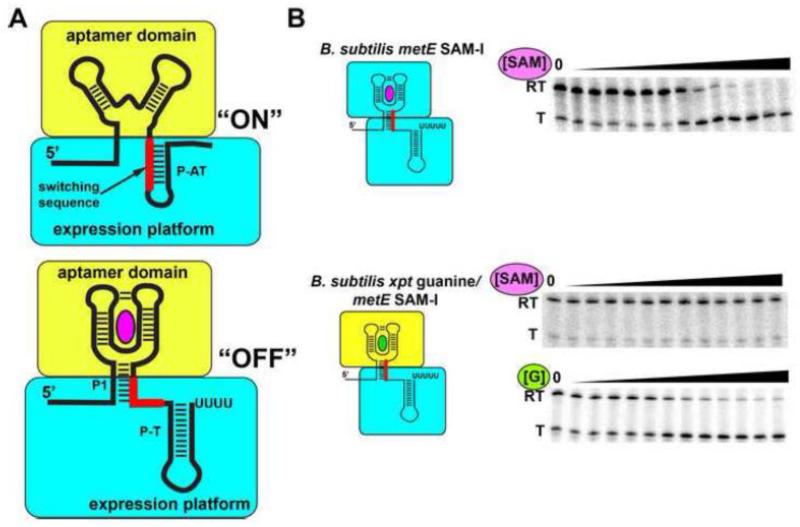
Synthetic riboswitches have also been created through modification of the expression platform. It is well appreciated that aptamer domains of riboswitches, like their SELEX-derived counterparts, are highly composable such that they can function in variety of contexts including synthetic riboswitches, aptazymes (aptamer inducible ribozymes), and fluorescent biosensors [123]. However, modularity of expression platforms had not been demonstrated until recently [127, 128]. In some riboswitches, the sequence required for high affinity ligand binding to the aptamer domain does not overlap with the alternative secondary structural switch, particularly those in the P1 helix considered to be central to interdomain communication. An example of such an arrangement is the B. subtilis metE transcriptional “OFF” riboswitch, in which the two base pairs essential for S-adenosylmethionine (SAM) binding to the aptamer are localized to the 3′-side of the P1 helix while nucleotides that participate in the P1/P-AT switch are localized to the 5′-side (Figure 6A) [127]. By defining the boundary of the two domains as the region of P1 where these elements meet, the aptamer can be replaced by a number of alternative sensors, both natural and in vitro selected with the resultant chimeras functional both in vitro (Figure 6B) and in vivo.
Figure 6. Synthesis of chimeric riboswitches from biological parts.
(A) In order for the expression platform to be used as a modular part, all of the sequences required for the alternative secondary structural switch must be distinct from the sequence that defines the minimal aptamer domain (yellow). In many riboswitches, the key element is the “switching sequence” (red), the region of the expression platform (cyan) shared between the two alternative structures. (B) An example of a functional chimeric riboswitch. The top shows the wild type B. subtilis metE SAM-responsive riboswitch undergoing transcriptional attenuation as a function of increasing ligand concentration in a single-turnover transcription assay (RT, read through product; T, terminated product). The middle shows that a chimera of the B. subtilis xpt-pbuX guanine binding aptamer domain and the B. subtilis metE expression platform is no longer SAM-responsive, but is strongly responsive to guanine (bottom). Figure adapted from (118).
From a mechanistic perspective, the adaptability of select expression platforms suggests these RNAs exploit an “encoded co-transcriptional” folding process [129]. In this model of RNA folding, the secondary structure adopted by the RNA is a function of the relative thermodynamic stability of competing helical elements as well as their 5′-to-3′ polarity. This principle was first demonstrated in folding of small model RNAs that have isoenergetic, mutually exclusive structures [129]. For these RNAs, the folding landscape is directed by the sequential ordering of the helical elements. In transcriptional “OFF” expression platforms ordering of the P1/P-AT elements remain fixed, but the relative stabilities of the helical elements are variable [130]. In the absence of ligand, P-AT is the more stable element, enabling its formation to dominate over P1. Conversely, ligand to a site in the aptamer domain that is typically adjacent to the 3′-side of the P1, increases the relative stability of P1 over P-AT, leading to formation of the downstream intrinsic terminator. Thus, the expression platform does not necessarily care about the particulars of the ligand-aptamer complex—only that it alters the stability of P1, which is why many distinct aptamer domains can efficiently direct ligand-dependent transcriptional termination of the metE expression platform [127]. More importantly, since SELEX-derived aptamers can be incorporated into functional chimeric riboswitches, there is nothing special about biological aptamers that enables them to interface with an RNA switch. Instead, in most cases, the aptamer merely has to position the ligand binding site (or a coupled conformational change in the RNA) to influence P1 stability.
A more useful modular switch is one that can induce gene expression in response to ligand binding, like the M6 riboswitch [125, 126]. However, most of the “ON” switches surveyed for potential modularity have significant sequence overlap between the two domains, preventing their simple decoupling at a defined boundary like the metE riboswitch [130]. Instead, the well-characterized adenine-responsive pbuE riboswitch from B. subtilis was re-engineered to enable its coupling to different aptamers [130]. In the natural pbuE riboswitch, the terminator (P-T) invades into L3, such that the alternative secondary structural switch includes sequence elements of the aptamer necessary for both ligand binding and tertiary structure formation. To decouple the two domains, a short sequence was added to the 3′-side of the P1 helix that prevented the invasion of P-T into the aptamer, mimicking the arrangement observed in metE. This engineered variant of the pbuE expression platform is able to host a variety of aptamers to turn on gene expression. These studies have only explored a small number of expression platforms, and it may be that even more useful regulatory modules can be created from translational attenuators or even eukaryotic riboswitches that regulate splicing or mRNA stability.
10. Conclusion
The purine riboswitch, and particularly its aptamer domain, has become an invaluable model system for exploring facets of RNA folding, small molecule recognition (reviewed more extensively in [40]), and structure. From this extensive body of work, we are beginning to elucidate the foundational principles of these RNAs function to efficiently regulate gene expression across a highly diverse set of organisms that experience a broad spectrum of environmental conditions. The door is now opening to engineering naturally based sensors that are modular, adaptable, and flexible in their application with the benefits stemming from the vast amount of knowledge gained by studies of the purine riboswitch. Nonetheless, there are still many aspects of these RNAs that remain to be elucidated, in particular a level of understanding of the expression platform that rivals the aptamer domain as well as a better appreciation of how these RNAs function in the cellular context.
Highlights.
Purine riboswitch aptamers are a widely used RNA model system
Common features of purine nucleobase recognition between different riboswitch classes
Efficient co-transcriptional folding is a key feature of riboswitch function
Novel RNA devices can be constructed from purine riboswitches
Acknowledgments
Funding for this work was provided by a grant from the National Institutes of Health to R.T.B. (GM 073850).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lu C, Smith AM, Fuch RT, Ding F, Rajashankar K, Henkin TM, Ke A. Crystal structure of the SAM-III/SAM(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struc Mol Biol. 2008;15:1076–1083. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Johnson JE, Jr., Reyes FE, Polaski JT, Batey RT. B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012;492:133–137. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- [5].Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- [6].Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Klein DJ, Edwards TE, Ferre-D’Amare AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kang M, Peterson R, Feigon J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Molecular cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- [9].Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J Biol Chem. 2009;284:11012–11016. doi: 10.1074/jbc.C900024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- [11].Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chemistry & biology. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Christiansen LC, Schou S, Nygaard P, Saxild HH. Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J Bacteriol. 1997;179:2540–2550. doi: 10.1128/jb.179.8.2540-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Landick R, Yanofsky C. Stability of an Rna Secondary Structure Affects Invitro Transcription Pausing in the Trp Operon Leader Region. Journal of Biological Chemistry. 1984;259:1550–1555. [PubMed] [Google Scholar]
- [14].Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- [15].Miranda-Rios J, Navarro M, Soberon M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- [17].Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stormo GD, Ji Y. Do mRNAs act as direct sensors of small molecules to control their expression? Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9465–9467. doi: 10.1073/pnas.181334498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- [22].Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chemistry & biology. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- [23].Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- [25].Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- [26].Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].de la Pena M, Dufour D, Gallego J. Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA. 2009;15:1949–1964. doi: 10.1261/rna.1889509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Delfosse V, Bouchard P, Bonneau E, Dagenais P, Lemay JF, Lafontaine DA, Legault P. Riboswitch structure: an internal residue mimicking the purine ligand. Nucleic Acids Res. 2010;38:2057–2068. doi: 10.1093/nar/gkp1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pikovskaya O, Polonskaia A, Patel DJ, Serganov A. Structural principles of nucleoside selectivity in a 2′-deoxyguanosine riboswitch. Nat Chem Biol. 2011;7:748–755. doi: 10.1038/nchembio.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- [32].Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17:857–868. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Edwards AL, Batey RT. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J Mol Biol. 2009;385:938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Trausch JJ, Batey RT. A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch. Chemistry & biology. 2013 doi: 10.1016/j.chembiol.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liberman JA, Salim M, Krucinska J, Wedekind JE. Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold. Nat Chem Biol. 2013;9:353–355. doi: 10.1038/nchembio.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat Struct Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
- [39].Batey RT. Recognition of S-adenosylmethionine by riboswitches. Wiley Interdiscip Rev RNA. 2011;2:299–311. doi: 10.1002/wrna.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Batey RT. Structure and mechanism of purine-binding riboswitches. Q Rev Biophys. 2012;45:345–381. doi: 10.1017/S0033583512000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- [42].Daldrop P, Reyes FE, Robinson DA, Hammond CM, Lilley DM, Batey RT, Brenk R. Novel ligands for a purine riboswitch discovered by RNA-ligand docking. Chemistry & biology. 2011;18:324–335. doi: 10.1016/j.chembiol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- [44].Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR. Design and antimicrobial action of purine analogues that bind Guanine riboswitches. ACS Chem Biol. 2009;4:915–927. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. J Am Chem Soc. 2006;128:14214–14215. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- [46].Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 2010;6:e1000865. doi: 10.1371/journal.ppat.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lemay JF, Lafontaine DA. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA. 2007;13:339–350. doi: 10.1261/rna.142007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Stoddard CD, Widmann J, Trausch JJ, Marcano-Velazquez JG, Knight R, Batey RT. Nucleotides adjacent to the ligand-binding pocket are linked to activity tuning in the purine riboswitch. J Mol Biol. 2013;425:1596–1611. doi: 10.1016/j.jmb.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- [50].Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- [51].Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chemistry & biology. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- [53].Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat Struct Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- [56].Brenner MD, Scanlan MS, Nahas MK, Ha T, Silverman SK. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry. 2010;49:1596–1605. doi: 10.1021/bi9019912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gilbert SD, Batey RT. Riboswitches: fold and function. Chemistry & biology. 2006;13:805–807. doi: 10.1016/j.chembiol.2006.08.002. [DOI] [PubMed] [Google Scholar]
- [59].Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Noeske J, Schwalbe H, Wohnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lemay JF, Lafontaine DA. The adenine riboswitch: a new gene regulation mechanism. Med Sci (Paris) 2006;22:1053–1059. doi: 10.1051/medsci/200622121053. [DOI] [PubMed] [Google Scholar]
- [62].Bowman JC, Lenz TK, Hud NV, Williams LD. Cations in charge: magnesium ions in RNA folding and catalysis. Curr Opin Struct Biol. 2012;22:262–272. doi: 10.1016/j.sbi.2012.04.006. [DOI] [PubMed] [Google Scholar]
- [63].Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Edwards TE, Ferre-D’Amare AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- [65].Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Vicens Q, Mondragon E, Batey RT. Molecular sensing by the aptamer domain of the FMN riboswitch: a general model for ligand binding by conformational selection. Nucleic Acids Res. 2011;39:8586–8598. doi: 10.1093/nar/gkr565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ren A, Rajashankar KR, Patel DJ. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486:85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Butler EB, Xiong Y, Wang J, Strobel SA. Structural basis of cooperative ligand binding by the glycine riboswitch. Chemistry & biology. 2011;18:293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- [70].Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Molecular cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Batey RT, Doudna JA. Structural and energetic analysis of metal ions essential to SRP signal recognition domain assembly. Biochemistry. 2002;41:11703–11710. doi: 10.1021/bi026163c. [DOI] [PubMed] [Google Scholar]
- [72].Buck J, Noeske J, Wohnert J, Schwalbe H. Dissecting the influence of Mg2+ on 3D architecture and ligand-binding of the guanine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2010;38:4143–4153. doi: 10.1093/nar/gkq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Leipply D, Draper DE. Effects of Mg2+ on the free energy landscape for folding a purine riboswitch RNA. Biochemistry. 2011;50:2790–2799. doi: 10.1021/bi101948k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lambert D, Leipply D, Shiman R, Draper DE. The influence of monovalent cation size on the stability of RNA tertiary structures. J Mol Biol. 2009;390:791–804. doi: 10.1016/j.jmb.2009.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Shiman R, Draper DE. Stabilization of RNA tertiary structure by monovalent cations. J Mol Biol. 2000;302:79–91. doi: 10.1006/jmbi.2000.4031. [DOI] [PubMed] [Google Scholar]
- [76].Draper DE, Misra VK. RNA shows its metal. Nat Struct Biol. 1998;5:927–930. doi: 10.1038/2901. [DOI] [PubMed] [Google Scholar]
- [77].Basu S, Rambo RP, Strauss-Soukup J, Cate JH, Ferre-D’Amare AR, Strobel SA, Doudna JA. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat Struct Biol. 1998;5:986–992. doi: 10.1038/2960. [DOI] [PubMed] [Google Scholar]
- [78].Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker RR. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Blouin S, Chinnappan R, Lafontaine DA. Folding of the lysine riboswitch: importance of peripheral elements for transcriptional regulation. Nucleic Acids Res. 2011;39:3373–3387. doi: 10.1093/nar/gkq1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- [81].Sherman EM, Esquiaqui J, Elsayed G, Ye JD. An energetically beneficial leader-linker interaction abolishes ligand-binding cooperativity in glycine riboswitches. RNA. 2012;18:496–507. doi: 10.1261/rna.031286.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kladwang W, Chou FC, Das R. Automated RNA structure prediction uncovers a kink-turn linker in double glycine riboswitches. J Am Chem Soc. 2012;134:1404–1407. doi: 10.1021/ja2093508. [DOI] [PubMed] [Google Scholar]
- [83].Baird NJ, Ferre-D’Amare AR. Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches. RNA. 2013;19:167–176. doi: 10.1261/rna.036269.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Mellin JR, Tiensuu T, Becavin C, Gouin E, Johansson J, Cossart P. A riboswitch-regulated antisense RNA in Listeria monocytogenes. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13132–13137. doi: 10.1073/pnas.1304795110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch control of Rho-dependent transcription termination. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lemay JF, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Masse E, Lafontaine DA. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- [88].Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Molecular cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- [89].McDowell JC, Roberts JW, Jin DJ, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- [90].Garst AD, Porter EB, Batey RT. Insights into the regulatory landscape of the lysine riboswitch. J Mol Biol. 2012;423:17–33. doi: 10.1016/j.jmb.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Neupane K, Yu H, Foster DA, Wang F, Woodside MT. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 2011;39:7677–7687. doi: 10.1093/nar/gkr305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].McGary K, Nudler E. RNA polymerase and the ribosome: the close relationship. Curr Opin Microbiol. 2013;16:112–117. doi: 10.1016/j.mib.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Rieder U, Kreutz C, Micura R. Folding of a transcriptionally acting preQ1 riboswitch. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10804–10809. doi: 10.1073/pnas.0914925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wedekind JE. The apo riboswitch as a molecular hydra. Structure. 2010;18:757–758. doi: 10.1016/j.str.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Buck J, Furtig B, Noeske J, Wohnert J, Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ottink OM, Rampersad SM, Tessari M, Zaman GJ, Heus HA, Wijmenga SS. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA. 2007;13:2202–2212. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Eskandari S, Prychyna O, Leung J, Avdic D, O’Neill MA. Ligand-directed dynamics of adenine riboswitch conformers. J Am Chem Soc. 2007;129:11308–11309. doi: 10.1021/ja073159l. [DOI] [PubMed] [Google Scholar]
- [98].Prychyna O, Dahabieh MS, Chao J, O’Neill MA. Sequence-dependent folding and unfolding of ligand-bound purine riboswitches. Biopolymers. 2009;91:953–965. doi: 10.1002/bip.21283. [DOI] [PubMed] [Google Scholar]
- [99].Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338:397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Nguyen PH, Derreumaux P, Stock G. Energy flow and long-range correlations in guanine-binding riboswitch: a nonequilibrium molecular dynamics study. J Phys Chem B. 2009;113:9340–9347. doi: 10.1021/jp902013s. [DOI] [PubMed] [Google Scholar]
- [102].Villa A, Wohnert J, Stock G. Molecular dynamics simulation study of the binding of purine bases to the aptamer domain of the guanine sensing riboswitch. Nucleic Acids Res. 2009;37:4774–4786. doi: 10.1093/nar/gkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Sharma M, Bulusu G, Mitra A. MD simulations of ligand-bound and ligand-free aptamer: molecular level insights into the binding and switching mechanism of the add A-riboswitch. RNA. 2009;15:1673–1692. doi: 10.1261/rna.1675809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Gong Z, Zhao Y, Chen C, Xiao Y. Role of ligand binding in structural organization of add A-riboswitch aptamer: a molecular dynamics. J Biomol Struct Dyn. 2011;29:403–416. doi: 10.1080/07391102.2011.10507394. [DOI] [PubMed] [Google Scholar]
- [105].Lee MK, Gal M, Frydman L, Varani G. Real-time multidimensional NMR follows RNA folding with second resolution. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9192–9197. doi: 10.1073/pnas.1001195107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Priyakumar UD, MacKerell AD., Jr. Role of the adenine ligand on the stabilization of the secondary and tertiary interactions in the adenine riboswitch. J Mol Biol. 2010;396:1422–1438. doi: 10.1016/j.jmb.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Banáš P, Hollas D, Jurečka P, Orozco M, Cheatham TE, III, Šponer J.i., Otyepka M. Performance of molecular mechanics force fields for RNA simulations: Stability of UUCG and GNRA hairpins. Journal of Chemical Theory and Computation. 2010;6:3836–3849. doi: 10.1021/ct100481h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Ditzler MA, Otyepka M, Sponer J, Walter NG. Molecular dynamics and quantum mechanics of RNA: conformational and chemical change we can believe in. Acc Chem Res. 2010;43:40–47. doi: 10.1021/ar900093g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].McDowell SE, a. Špačková N, Šponer J, Walter NG. Molecular dynamics simulations of RNA: An in silico single molecule approach. Biopolymers. 2007;85:169–184. doi: 10.1002/bip.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Woodside MT, Garcia-Garcia C, Block SM. Folding and unfolding single RNA molecules under tension. Curr Opin Chem Biol. 2008;12:640–646. doi: 10.1016/j.cbpa.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Heldal M, Norland S, Erichsen ES, Sandaa RA, Larsen A, Thingstad F, Bratbak G. Mg2+ as an indicator of nutritional status in marine bacteria. ISME J. 2012;6:524–530. doi: 10.1038/ismej.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Meyer M, Plass M, Perez-Valle J, Eyras E, Vilardell J. Deciphering 3′ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Molecular cell. 2011;43:1033–1039. doi: 10.1016/j.molcel.2011.07.030. [DOI] [PubMed] [Google Scholar]
- [114].Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- [115].Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T. Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes & development. 1999;13:655–665. doi: 10.1101/gad.13.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes & development. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Reining A, Nozinovic S, Schlepckow K, Buhr F, Furtig B, Schwalbe H. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature. 2013;499:355–359. doi: 10.1038/nature12378. [DOI] [PubMed] [Google Scholar]
- [118].Edwards AL, Reyes FE, Heroux A, Batey RT. Structural basis for recognition of S-adenosylhomocysteine by riboswitches. RNA. 2010;16:2144–2155. doi: 10.1261/rna.2341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Bohme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, Pisano F, Thiermann T, Wolf-Watz H, Narberhaus F, Dersch P. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 2012;8:e1002518. doi: 10.1371/journal.ppat.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Loh E, Kugelberg E, Tracy A, Zhang Q, Gollan B, Ewles H, Chalmers R, Pelicic V, Tang CM. Temperature triggers immune evasion by Neisseria meningitidis. Nature. 2013;502:237–240. doi: 10.1038/nature12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Benenson Y. Synthetic biology with RNA: progress report. Curr Opin Chem Biol. 2012;16:278–284. doi: 10.1016/j.cbpa.2012.05.192. [DOI] [PubMed] [Google Scholar]
- [122].Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- [123].Wittmann A, Suess B. Engineered riboswitches: Expanding researchers’ toolbox with synthetic RNA regulators. FEBS Lett. 2012;586:2076–2083. doi: 10.1016/j.febslet.2012.02.038. [DOI] [PubMed] [Google Scholar]
- [124].Topp S, Gallivan JP. Emerging applications of riboswitches in chemical biology. ACS Chem Biol. 2010;5:139–148. doi: 10.1021/cb900278x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Dixon N, Duncan JN, Geerlings T, Dunstan MS, McCarthy JE, Leys D, Micklefield J. Reengineering orthogonally selective riboswitches. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2830–2835. doi: 10.1073/pnas.0911209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Dixon N, Robinson CJ, Geerlings T, Duncan JN, Drummond SP, Micklefield J. Orthogonal riboswitches for tuneable coexpression in bacteria. Angew Chem Int Ed Engl. 2012;51:3620–3624. doi: 10.1002/anie.201109106. [DOI] [PubMed] [Google Scholar]
- [127].Ceres P, Garst AD, Marcano-Velazquez JG, Batey RT. Modularity of select riboswitch expression platforms enables facile engineering of novel genetic regulatory devices. ACS Synth Biol. 2013;2:463–472. doi: 10.1021/sb4000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Trausch JJ, Ceres P, Reyes FE, Batey RT. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Xayaphoummine A, Viasnoff V, Harlepp S, Isambert H. Encoding folding paths of RNA switches. Nucleic acids research. 2007;35:614–622. doi: 10.1093/nar/gkl1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Ceres P, Trausch JJ, Batey RT. Engineering modular ‘ON’ RNA switches using biological components. Nucleic Acids Res. 2013;41:10449–10461. doi: 10.1093/nar/gkt787. [DOI] [PMC free article] [PubMed] [Google Scholar]