We describe drug resistance and levels of susceptibility after first-line virologic failure in patients with several subtypes. High-level nucleoside and nonnucleoside reverse transcriptase inhibitor resistance was noted. Associations between resistance patterns and screening viral load, CD4 cell count, and site were observed.
Keywords: HIV-1 drug resistance, nonsubtype B, first-line failure, resource limited setting
Abstract
Background. The development of drug resistance to nucleoside reverse transcriptase inhibitors (NRTIs) and nonnucleoside reverse transcriptase inhibitors (NNRTIs) has been associated with baseline human immunodeficiency virus (HIV)-1 RNA level (VL), CD4 cell counts (CD4), subtype, and treatment failure duration. This study describes drug resistance and levels of susceptibility after first-line virologic failure in individuals from Thailand, South Africa, India, Malawi, Tanzania.
Methods. CD4 and VL were captured at AIDs Clinical Trial Group (ACTG) A5230 study entry, a study of lopinavir/ritonavir (LPV/r) monotherapy after first-line virologic failure on an NNRTI regimen. HIV drug-resistance mutation associations with subtype, site, study entry VL, and CD4 were evaluated using Fisher exact and Kruskall–Wallis tests.
Results. Of the 207 individuals who were screened for A5230, sequence data were available for 148 individuals. Subtypes observed: subtype C (n = 97, 66%) AE (n = 27, 18%), A1 (n = 12, 8%), and D (n = 10, 7%). Of the 148 individuals, 93% (n = 138) and 96% (n = 142) had at least 1 reverse transcriptase (RT) mutation associated with NRTI and NNRTI resistance, respectively. The number of NRTI mutations was significantly associated with a higher study screening VL and lower study screening CD4 (P < .001). Differences in drug-resistance patterns in both NRTI and NNRTI were observed by site.
Conclusions. The degree of NNRTI and NRTI resistance after first-line virologic failure was associated with higher VL at study entry. Thirty-two percent of individuals remained fully susceptible to etravirine and rilpivirine, protease inhibitor resistance was rare. Some level of susceptibility to NRTI remained; however, VL monitoring and earlier virologic failure detection may result in lower NRTI resistance.
Global access to antiretroviral therapy (ART) has increased rapidly in recent years, with more than 9.7 million infected individuals now receiving ART in resource-limited settings (RLS) [1]. ART can fail as a result of toxicity, transmitted drug resistance, inadequate medication adherence, or incomplete suppression of viral replication, resulting in the emergence of human immunodeficiency virus (HIV)-1 drug resistance [2–6].
RLS treatment programs commonly rely on a nucleoside reverse transcriptase inhibitor (NRTI) backbone of either zidovudine (AZT) or stavudine (d4T) and lamivudine (3TC) or, more recently, tenofovir (TDF) combined with either nevirapine (NVP) or efavirenz (EFV) as first-line treatment. Depending on the country, first-line treatment failure is determined by clinical, immunological, and virological biomarkers or a combination of markers. Of the individuals who fail first-line treatment, between 75% and 90% of individuals who access a failing regimen have 1 or more drug-resistant mutations associated with NRTI and nonnucleoside reverse transcriptase inhibitors (NNRTIs) [7–10]. The pattern of drug-resistance mutations observed are associated with specific drugs in the regimen, levels of adherence, HIV-1 RNA level (VL), CD4 cell counts (CD4), and the duration of treatment failure [7, 11, 12].
There is currently only a partial understanding of patterns of HIV-1 drug resistance that occur with ART failure in developing countries, where VL testing is uncommon and treatment failure may be defined by clinical and immunological indicators only. In contrast to North America and Europe, drug-resistance testing is rarely available and the provision of second-line therapy is based on a public health approach that uses standardized regimens that are based on national guidelines and available drugs. Circulating subtypes are generally nonsubtype B [13], where there is limited information about the activity of second-line boosted protease inhibitors, new second-generation NNRTIs, and TDF usage in either first- or second-line treatment. The optimal strategy for maintaining long-term virologic suppression with limited access to only 2 or at most 3 distinct ART regimens is a challenge to the sustainability of treatment in RLS. Identifying suppressive regimens for those who have failed first-line therapy is essential for maintaining the benefits of ART and the prevention of transmitted drug resistance.
HIV-1 drug-resistance testing at the time of virologic failure is the standard of care in developed countries but is not widely available in RLS. Nevertheless, estimated genotypic susceptibility after sustained failure of first-line regimens may guide the selection of alternative regimens and preserve future treatment options. Here, we present genotypic resistance data from 148 individuals who were failing first-line regimens in a RLS where nonsubtype B viruses, including C, CRF_01AE, A, and D, predominate.
METHODS
Study Design and Participants
AIDs Clinical Trial Group (ACTG) A5230 is a single-arm, open-label, multicenter, pilot study to evaluate the safety and efficacy associated with lopinavir/ritonavir (LPV/r) monotherapy in protease inhibitor–naive individuals failing an initial NNRTI-containing regimen [14] in Thailand (Chiang Mai University ACTG case reporting system [CRS]), South Africa (University of the Witwatersrand HIV CRS), India (Y.R. Gaitonde Centre for AIDS Research and Education VHS CRS), Malawi (Kamuzu Central Hospital, University of North Carolina Lilongwe CRS), and Tanzania (Kilimanjaro Christian Medical Centre CRS). Plasma samples from individuals with suspected failure (confirmed viral load >1000 cpm) of first-line ART screening for A5230 were tested for HIV-1 drug resistance. CD4 cell counts and VLs were available as part of the study at screening.
HIV-1 Drug-Resistance Testing and Subtyping
Population-based HIV drug-resistance testing was performed using the US Food and Drug Administration–approved ViroSeq HIV-1 Genotyping System (v.2.0), per manufacturer's instructions (Celera Diagnostics, Alameda, CA) in batches. Briefly, a 1.7-kb amplicon was generated by reverse transcriptase (RT)-initiated polymerase chain reaction encompassing the entire protease (PR) and partial RT. Sequencing was performed with an ABI Prism 3100-Avant genetic analyzer (Applied Biosystems, USA). Samples that could not be amplified by the ViroSeq kit primers were amplified using a locally developed method [15]. Sequences were analyzed using the ViroSeq v2.7 or Sequencer v4.8 software, and drug-resistance patterns in protease and RT were determined using the Stanford algorithm with the following scoring system: 0–9, susceptible; 10–14, potential low-level resistance; 15–29, low-level resistance; 30–59, intermediate resistance; and >60, high-level resistance. Mutations arising as a consequence of the “reversion” of T215F/Y thymidine analogue mutations (TAMs) were included in the analysis [16].
Statistical Methods
Mutation associations with subtype, VL, CD4, site, and prior NNRTI usage were evaluated using Fisher exact and Kruskall–Wallis tests.
RESULTS
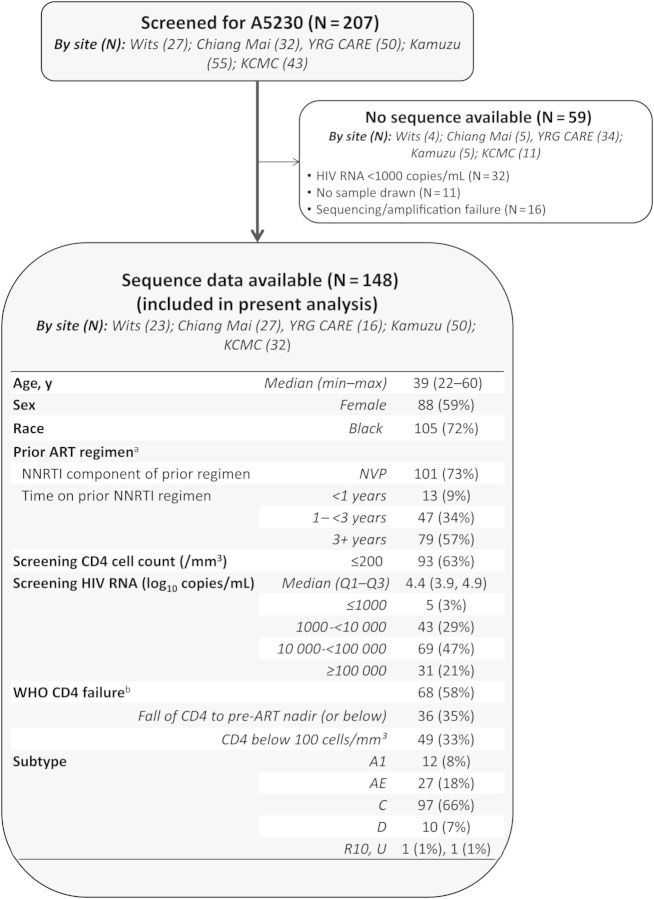
Of the 207 individuals screened for A5230, sequence data were available for 148 (Figure 1). Of the 59 samples that were not sequenced, 32 (64%) had VL <1000 copies/mL, 11 did not have samples available for sequencing, and the remaining 16 failed amplification or sequencing reactions. Most of the individuals without sequence data were from the clinical site in India, likely reflecting distinct prescreening characteristics.
Figure 1.
Study population. aPrior antiretroviral therapy (ART) regimen was not available for 9 individuals (all of whom did not enroll in A5230). bWorld Health Organization (WHO) clinical failure based on a fall in CD4 count to nadir (or below) or a single CD4 level below 100 cells/mm³ at screening; 50% fall from on-treatment peak value and persistent CD4 < 100 cells/mm³ cannot be determined from the data available in the A5230 database. A fall in CD4 counts to pre-ART nadir or below could not be determined for 46 individuals; as a result, 30 individuals for whom WHO CD4 failure criteria could not be evaluated. Abbreviations: Chiang Mai, Thailand (Chiang Mai University ACTG CRS); HIV, human immunodeficiency virus; KCMC, Tanzania (Kilimanjaro Christian Medical Centre CRS); Kumuza, Malawi (Kamuzu Central Hospital, University of North Carolina Lilongwe CRS); NNRTI, nonnucleoside reverse transcriptase inhibitor; Wits, South Africa (University of the Witwatersrand HIV CRS); YRG CARE, India (Y.R. Gaitonde Centre for AIDS Research and Education, VHS CRS).
The median age of the 148 individuals with sequence data included in the present analysis was 39 years (range, 22–60 years). The majority of individuals (59%) were female and black (72%). Most individuals (73%) were on an NVP-based regimen at the time of screening. Although NNRTI use varied across sites, the majority of individuals from South Africa and India were using EFV-based regimens. Seventy-nine of the individuals had received an NNRTI regimen for 3 or more years, 47 individuals for 1–3 years, and 13 individuals for longer than 1 year at time of screening. The median VL at time of screening was 4.4 log10 copies/mL (first–third quartile, 3.9–4.9 copies/mL); 5 individuals with sequence results had VL < 1000 copies/mL. The median CD4 cell count was 155 cells/mm3 (69–256 cells/mm3); 58% met World Health Organization (WHO) treatment failure criteria (among those with nadir CD4 cell count available); however, this varied across sites (P < .001, not shown). Of note, fewer individuals from the South African site met previous WHO CD4 immunologic failure criteria [17] of <250 cells/mm3 (13% compared with at least 45% at the other sites), likely reflecting the routine VL monitoring of the South African national program for failure at that site.
Of the 148 individuals, the most prevalent subtype was subtype C (66%) followed by AE (18%), A1 (8%), and D (7%; Figure 1); these were linked to geographic regions. There was 1 circulating recombinant form (CRF_10) and 1 sequence for which the subtype could not be classified. No associations between number of mutations and level of resistance were observed by subtype (P = .92).
Of the 148 individuals, 93% (n = 138) and 96% (n = 142) had at least 1 RT mutation associated with NRTI and NNRTI resistance, respectively; no differences were observed by site (P = .29 and P = .58 for NRTI and NNRTI, respectively; Table 1A). Only 2 individuals had major protease inhibitor (PI)-associated mutations that were linked to low-level resistance to LPV/r (L90M and V32I/M46L), 1 individual each from Malawi and Tanzania clinical sites.
Table 1.
Stanford Resistance Score for Potential First-line Failures Screened for ACTG A5230
| (A) Prevalence of Resistance-Associated Mutations by Clinical Site | ||||||||
|---|---|---|---|---|---|---|---|---|
| Total (N = 148) | Wits (N = 23) | Chiang Mai (N = 27) | YRG CARE (N = 16) | Kamuzu (N = 50) | KCMC (N = 32) | P Value | ||
| NRTI | ||||||||
| N (%) with ≥1 mutation | 138 (93%) | 21 (91%) | 27 (100%) | 15 (94%) | 44 (88%) | 31 (97%) | .29a | |
| Median no. (1st, 3rd quartile) | [3 (1, 5)] | [1 (1, 2)] | [4 (2, 5)] | [3.5 (1, 6)] | [4 (2, 7)] | [3.5 (2, 5)] | <.001b | |
| NNRTI | ||||||||
| N (%) with ≥1 mutation | 142 (96%) | 21 (91%) | 27 (100%) | 16 (100%) | 47 (94%) | 31 (97%) | .58a | |
| Median no. (1st, 3rd quartile) | [2.5 (2, 3)] | [2 (1, 3)] | [3 (2, 3)] | [2.5 (2, 3)] | [3 (2, 3)] | [2 (2, 3)] | .09b | |
| Lopinavir/ritonavir | ||||||||
| N (%) with ≥1 mutation | 2 (1%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (2%) | 1 (3%) | … | |
| Median no. (1st, 3rd quartile) | [0 (0, 0)] | [0 (0, 0)] | [0 (0, 0)] | [0 (0, 0)] | [0 (0, 0)] | [0 (0, 0)] | ||
| (B) N (%) Individuals With Stanford Resistance Level by Drug | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stanford Resistance Level | NRTI |
NNRTI |
PI |
|||||||||
| Abacavir | Zidovudine | Stavudine | Didanosine | Tenofovir | Lamivudine | Emtricitabine | Efavirenz | Nevirapine | Etravirine | Rilpivirine | Lopinavir | |
| Susceptible | 11 (7%) | 73 (49%) | 61 (41%) | 55 (37%) | 85 (57%) | 10 (7%) | 10 (7%) | 6 (4%) | 6 (4%) | 47 (32%) | 48 (32%) | 146 (99%) |
| Potential low-level resistance | 44 (30%) | 2 (1%) | 5 (3%) | 10 (7%) | 14 (9%) | 1 (1%) | 1 (1%) | 0 (0%) | 0 (0%) | 11 (7%) | 10 (7%) | 0 (0%) |
| Low-level resistance | 25 (17%) | 11 (7%) | 22 (15%) | 22 (15%) | 15 (10%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 20 (14%) | 20 (14%) | 2 (1%) |
| Intermediate resistance | 36 (24%) | 35 (24%) | 35 (24%) | 28 (19%) | 28 (19%) | 7 (5%) | 7 (5%) | 41 (28%) | 0 (0%) | 51 (34%) | 51 (34%) | 0 (0%) |
| High-level resistance | 32 (22%) | 27 (18%) | 25 (17%) | 33 (22%) | 6 (4%) | 130 (88%) | 130 (88%) | 101 (68%) | 142 (96%) | 19 (13%) | 19 (13%) | 0 (0%) |
Abbreviations: Chiang Mai, Thailand (Chiang Mai University ACTG CRS); HIV, human immunodeficiency virus; KCMC, Tanzania (Kilimanjaro Christian Medical Centre CRS); Kumuza, Malawi (Kamuzu Central Hospital, University of North Carolina Lilongwe CRS); NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor; Wits, South Africa (University of the Witwatersrand HIV CRS); YRG CARE, India (Y.R. Gaitonde Centre for AIDS Research and Education, VHS CRS).
a Fisher exact test for differences in the prevalence of resistance mutations at screening for A5230 by site.
b Kruskal–Wallis test for shifts in the distributions of number of resistance mutations present at screening for A5230 by site.
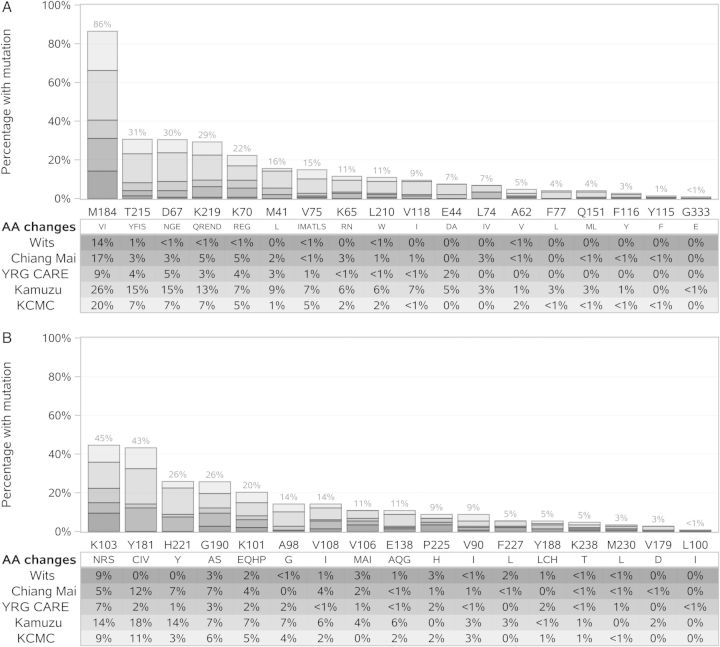
One hundred thirty-eight individuals (93%) had reduced susceptibility to 3TC and FTC (Table 1B). Resistance (defined as low, intermediate, or high) to AZT (49%), d4T (55%), ABC (63%), and didanosine (ddI) (56%) was common. The majority of individuals were fully susceptible to TDF (n = 85, 57%), with only a few demonstrating full resistance by the Stanford algorithm (n = 6, 4%); however, this was increased to 10% when the presence of K65R was interpreted as TDF resistant. The most frequent NRTI mutation was M184V/I (n = 128, 86%), followed by TAMs at codons 215 (31%), 67 (30%), 219 (29%), 70 (22%), and 41 (16%) and mutations at codon 65 (11%; Figure 2A). Rare deletions in RT codons 66, 69, and 70 were identified in 12 individuals; these individuals had virus with subtypes A1, AE, C, D, and U.
Figure 2.
Mutation frequency by enrolling site. Bar chart shows the proportion of individuals with amino acid changes associated with resistance to (A) nucleoside reverse transcriptase inhibitor and (B) nonnucleoside reverse transcriptase inhibitor agents. The height of the bar shows the proportion of participants with any significant amino acid changes at a given codon broken down by enrolling site; details of the proportions with specific mutations are provided in the text. Abbreviations: AA, Amino acids; Chiang Mai, Thailand (Chiang Mai University ACTG CRS); KCMC, Tanzania (Kilimanjaro Christian Medical Centre CRS); Kumuza, Malawi (Kamuzu Central Hospital, University of North Carolina Lilongwe CRS); Wits, South Africa (University of the Witwatersrand HIV CRS); YRG CARE, India (Y.R. Gaitonde Centre for AIDS Research and Education, VHS CRS).
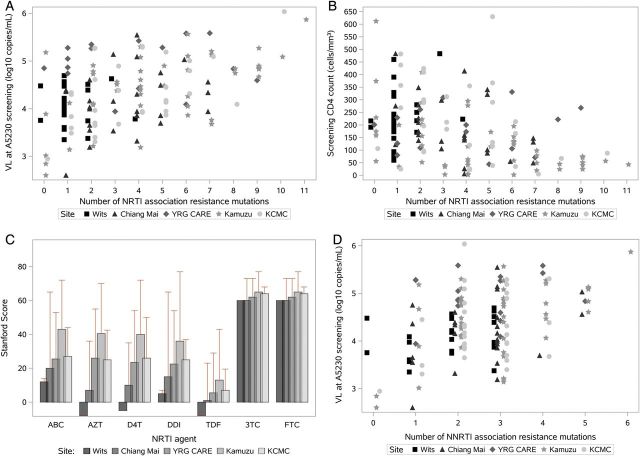
The number of NRTI mutations was significantly associated with a higher VL (P < .001) and lower CD4 cell count (P < .001; Figure 3A and 3B). Samples with a VL >5 log10 copies/mL at study enrollment had a median of 5 NRTI-associated mutations compared with a median of 2 for those with VL < 4 log10 copies/mL and 3 for those with VL between 4 and 5 log10 copies/mL. The distribution of the number of NRTI-associated resistance mutations differed across the sites (P < .001); the South African clinical site, where VL are routinely performed, had lower VL and fewer NRTI mutations (Figure 3A). Moreover, the NRTI resistance scores differed across sites (AZT, d4T, TDF, ddI, ABC, P < .001; 3TC, FTC, P = .06). Again, the highest NRTI susceptibility scores were observed at the South African clinical site (Figure 3C). Conversely, more complex resistance patterns (TAM, K65R, and Q151M) were observed at the 4 sites that do not routinely perform VL testing.
Figure 3.
Frequency of nucleoside reverse transcriptase inhibitor and nonnucleoside reverse transcriptase inhibitor mutations by human immunodeficiency virus-1 RNA level, CD4 cell count, and clinical site. Abbreviations: Chiang Mai, Thailand (Chiang Mai University ACTG CRS); KCMC, Tanzania (Kilimanjaro Christian Medical Centre CRS); Kumuza, Malawi (Kamuzu Central Hospital, University of North Carolina Lilongwe CRS); NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; VL, human immunodeficiency virus-1 RNA level; Wits, South Africa (University of the Witwatersrand HIV CRS); YRG CARE, India (Y.R. Gaitonde Centre for AIDS Research and Education, VHS CRS).
The median number of NNRTI mutations was 2.5 (first–third quartile, 2–3), with no differences observed by site (P = .09; Table 1). The presence of NNRTI mutations was significantly associated with a higher VL (P = .003; Figure 3D). However, 96% of the individuals were resistant to either EFV or NVP, and 32% were fully susceptible to etravirine (ETR) or rilpivirine (RPV). The most frequent mutations associated with reduced NNRTI susceptibility were at codon 181 (n = 64; 43%) and codon 103 (n = 66; 45%; Figure 2B). ETR and RPV resistance was associated more frequently with previous NVP usage compared with EFV usage (P < .001; 60% vs 10%, with at least intermediate-level resistance; Table 2). Median (first–third quartile) resistance scores for EFV, NVP, ETR, and RPV were 65 (50–90), 90 (65–120), 25 (5–35), and 25 (0–35), respectively (Table 2). Y181C was observed more frequently in NVP-exposed individuals (53% [NVP], 8% [EFV]); K103N was observed more frequently in EFV-exposed individuals (32% [NVP], 63% [EFV]). A median of 2 mutations associated with ETR resistance and 1 associated with RPV resistance were observed. Sixty percent of individuals were classified as resistant (low- to high-level resistance) and 32% as fully susceptible to both ETR and RPV.
Table 2.
Nonnucleoside Reverse Transcriptase Inhibitor (NNRTI) Resistance by Prior NNRTI Use
| Total (N = 148) | Prior Nonnucleoside Reverse Transcriptase Inhibitor |
P Value | ||
|---|---|---|---|---|
| NVP (N = 101) | EFV (N = 38) | |||
| EFV: Stanford resistance score | ||||
| Median (Q1, Q3) | 65 (50, 90) | 60 (40, 85) | 90 (60, 100) | <.001a |
| Susceptible | 5 (4%) | 3 (3%) | 2 (5%) | <.001b |
| Intermediate resistance | 36 (26%) | 34 (34%) | 2 (5%) | |
| High-level resistance | 98 (71%) | 64 (63%) | 34 (89%) | |
| No. of mutations, median (Q1, Q3) | 2 (2, 3) | 3 (2, 3) | 2 (1, 3) | .12a |
| NVP: Stanford resistance score | ||||
| Median (Q1, Q3) | 90 (65, 120) | 90 (65, 120) | 78 (60, 120) | .55a |
| Susceptible | 6 (4%) | 3 (3%) | 2 (5%) | .61b |
| High-level resistance | 142 (96%) | 98 (97%) | 36 (95%) | |
| No. of mutations, median (Q1, Q3) | 2 (2, 3) | 3 (2, 3) | 2 (1, 3) | .10a |
| ETR: Stanford resistance score | ||||
| Median (Q1, Q3) | 25 (5, 35) | 30 (10, 40) | 5 (0, 15) | <.001a |
| Susceptible | 47 (32%) | 22 (22%) | 22 (58%) | <.001b |
| Potential low-level resistance | 11 (7%) | 5 (5%) | 6 (16%) | |
| Low-level resistance | 20 (14%) | 14 (14%) | 6 (16%) | |
| Intermediate resistance | 51 (34%) | 44 (44%) | 2 (5%) | |
| High-level resistance | 19 (13%) | 16 (16%) | 2 (5%) | |
| No. of mutations, median (Q1, Q3) | 2 (1, 2) | 2 (1, 3) | 1 (0, 2) | <.001a |
| RPV: Stanford resistance score | ||||
| Median (Q1, Q3) | 25 (0, 35) | 30 (10, 40) | 0 (0, 15) | <.001a |
| Susceptible | 48 (32%) | 23 (23%) | 22 (58%) | <.001b |
| Potential low-level resistance | 10 (7%) | 4 (4%) | 6 (16%) | |
| Low-level resistance | 20 (14%) | 14 (14%) | 6 (16%) | |
| Intermediate resistance | 51 (34%) | 44 (44%) | 2 (5%) | |
| High-level resistance | 19 (13%) | 16 (16%) | 2 (5%) | |
| No. of mutations, median (Q1, Q3) | 1 (0, 2) | 2 (1, 3) | 0 (0, 2) | <.001a |
Nine individuals for whom prior nonnucleoside reverse transcriptase inhibitor usage was not available are excluded.
Abbreviations: EFV, efavirenz; ETR, etravirine; NVP, nevirapine; RPV, rilpivirine.
a Wilcoxon test.
b Fisher exact test.
Two individuals had low-level resistance to LPV/r, and the remaining individuals (99%) were fully susceptible to LPV/r. There were no associations between the LPV/r-associated mutations and polymorphisms observed between subtype (P = .07), site (P = .59), VL (P = .35), or CD4 cell count (P = .58).
DISCUSSION
This study describes the HIV-1 drug-resistance patterns associated with first-line regimen failures in 148 individuals screened for a study of LPR/r monotherapy after first-line treatment failure [14]. HIV-1 drug-resistance analysis showed that 138 individuals (93%) had at least 1 NRTI mutation and 142 individuals (96%) had at least 1 NNRTI mutation. The mutation profiles are similar to those that have been reported in similar populations with subtype A1, C, and D [18], with more complex resistance profiles observed at sites that do not routinely use VL monitoring.
The most frequent mutation observed was M184V/I followed by K103N and Y181C, which is similar to published resistance data from first-line regimen failures [7–10]. Most individuals were highly resistant to many of the NRTI, presenting with more than 3 TAMs, K65R, or Q151M; this would have an impact on second-line nucleoside analog therapy options. However, the predicted NRTI susceptibility supports TDF as an NRTI for second-line therapy, with only 23% demonstrating mutations consistent with resistance (high and intermediate).
Candidates for enrollment at sites that did not routinely use VL monitoring had lower CD4 cell counts and higher VL at screening for A5230. Furthermore, at sites that did not routinely perform VL testing, the resistance patterns were more complex. This finding is similar to those reported in Bangkok, where it was found that individuals who had failure that was determined based on immunological rather than virological criteria had higher viral loads and a higher frequency of the multidrug-resistant Q151M complex [19]. This accumulation of mutations was associated with the extent to which individuals remained on a failing drug regimen. The introduction of VL monitoring in RLS could facilitate faster switching if second-line regimens are available; a decrease in NRTI- and NNRTI-associated mutations; and an increase in the number of future ARV options for salvage therapy [20], supporting the global drive to improve access to viral load testing [21].
Ten percent of individuals had the K65R mutation, decreasing their susceptibility to TDF. No difference in K65R prevalence was observed among subtypes, although many participants were failing on a d4T-containing regimen. The observed frequency of K65R is thus consistent with other studies in RLS with infrequent monitoring [10, 22, 23] but lower than what was found in 1 study among subtype C individuals who were failing TDF (70%) [12]. In this cohort, more than 90% of individuals were no longer susceptible to cytidine analogue drugs, 3TC/FTC, commonly used in first-line regimens, and fewer than 50% of individuals were susceptible to NRTI commonly used in second-line regimens (AZT, 49%; ddI, 37%). However, because of the low frequency of K65R, TDF retained activity in 57%–77% of individuals.
Only 32% of individuals genotypes predicted full susceptibility to ETR and RPV. High-level ETR and RPV resistance was associated with Y181C in 42% of individuals. Studies have shown that NVP selects for the Y181C and G190A mutations; both lead to reduced ETR or RPV susceptibility [24, 25]. The K103N mutation is more likely to be associated with EFV failure and has little effect on ETR susceptibility. In this study, NVP was more commonly used in RLS, except in South Africa where EFV was used. A study in South Africa, where monitoring for both EFV and VL was used, found that only 10% of first-line failures were fully resistant to ETR [26], similar to the results presented here. Furthermore, ETR resistance was more frequent in individuals who received NVP without VL monitoring for treatment failure.
If only 1 in 3 individuals who fail first-line regimens in this setting are fully susceptible to ETR and RPV, the utility of these agents as part of second- or third-line regimens is limited. The ACTG has a third-line study underway to determine the effectiveness of using ETR in future treatment regimens in RLS. If ETR and RPV are introduced into third-line or salvage regimens, it may be preferable to use EFV rather than NVP in first-line regimens. However, the genotypic resistance data for ETR and RPV presented here were determined using the scoring system from the DUET I and II studies [27], which were predominately based on HIV-1 subtype B. Although there was a high level of ETR- and RPV-associated mutations, the effect of subtype-specific polymorphisms or changes in the RNase H and connection domains of these samples was not examined; therefore, phenotypic studies on these samples are planned.
Protease inhibitor–associated resistance mutations, particularly to LPV/r, were only observed in 2 individuals and thus classified as potential or low-level resistance. It is unlikely that these mutations, which were associated with low-level LPV/r resistance (L90M, M46L, and V32I), resulted from transmitted or acquired drug resistance. There is limited use of protease inhibitors in RLS and no evidence for LPV/r resistance, apart from a report of M46I, which is a subtype-associated polymorphism [28]. However, these 3 mutations have not been observed as naturally occurring polymorphisms in subtype C (where 1 L90M was observed) or A1 (where M46L and V32I were observed), and they are classified as transmitted drug-resistance mutations by the WHO [29]. Of note, the individual with L90M at screening rapidly failed LPV/r, with development of multiple PI mutations.
In conclusion, widely available RTI-based ART regimens have resulted in unprecedented access to care in RLS. However, the low genetic barrier and high-level resistance of NNRTI-anchored regimens in the absence of frequent VL monitoring results in rapid accumulation of drug resistance mutations to NNRTI and NRTI drugs, which may limit future treatment options. The resistance mutations in nonsubtype B viruses are not dissimilar from those observed in subtype B, with some distinct features, including the prevalence of V106M and K65R and a higher frequency of the TAM-2 (D67N, K70R, T215F, T219Q/E) vs the TAM-1 pathway (M41L, L210W, T215Y). More frequent VL monitoring was associated with fewer RTI mutations. Implementation of VL monitoring could reduce the duration of exposure to a failing regimen and therefore retain enhanced options for future lines of therapy. A5230 study results indicate that LPV/r treatment alone successfully suppressed viremia for up to 48 weeks in nonsubtype B–infected individuals [14]. The continued susceptibility to TDF, despite the accumulation of NRTI mutations, provides additional options for intensification of boosted protease inhibitor treatment with TDF and TDF/FTC.
Notes
Acknowledgments. The authors thank all ACTG A5230 study individuals for their participation. In addition, the authors thank the clinical research site members for their participation and for their supporting grants (listed below) at the 5 sites that conducted ACTG A5230: Drs Agnes Moses and Albert Mwafongo, UNC Project Lilongwe; Drs Venance Maro and John Crump, KCMC; Professor Thira Sirisanthana and Ms Patchanee Samutarlai, Chiang Mai University; Ms Petronilla Borain and Vuyokazi Jezile, University of Witswatersrand; and Dr Poongulali and Beulah Faith, YRG CARE.
Financial support. This work was supported by an award (U01AI068636) from the National Institute for Allergy and Infectious Diseases and was further supported by the National Institute of Mental Health and the National Institute of Dental and Craniofacial Research. Reagents were supplied by Abbott/Abbvie. Drug supplies were provided by Abbott/Abbvie and Gilead Sciences Inc. Site supporting grants included the following: UNC Project Lilongwe, U01AI069518; KCMC, U01069484; University of Witswatersrand, U01AI38858; YRG CARE, U01AI069432; and Statistical Data Analysis Center grant (AI068634).
Potential conflicts of interest. C. W. has received honorarium from Abbott and AbbVie and is a consultant for Celera Diagnostics. H. R. holds several National Institutes of Health-funded grants. M. N. was previously employed by Abbott and is a current employee of AbbVie and may hold stock in both AbbVie and Abbott. W. S. has received honorarium from UNITAID; is a consultant for WHO, the Clinton Foundation, and the Gates Foundation; and holds several research grants. D. K. holds a grant from Gilead. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.UNAIDS. UNAIDS World AIDS Day Report. 2012. Available at: http://www.unaids.org/en/media/unaids/contentassets/documents/epidemiology/2012/gr2012/JC2434_WorldAIDSday_results_en.pdf . Accessed 16 May 2014.
- 2.Boulle A, Orrel C, Kaplan R, et al. Substitutions due to antiretroviral toxicity or contraindication in the first 3 years of antiretroviral therapy in a large South African cohort. Antivir Ther. 2007;12:753–60. doi: 10.1177/135965350701200508. [DOI] [PubMed] [Google Scholar]
- 3.Carr A, Cooper DA. Adverse effects of antiretroviral therapy. Lancet. 2000;356:1423–30. doi: 10.1016/S0140-6736(00)02854-3. [DOI] [PubMed] [Google Scholar]
- 4.Haas DW, Ribaudo HJ, Kim RB, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18:2391–400. [PubMed] [Google Scholar]
- 5.Haas DW, Wu H, Li H, et al. MDR1 gene polymorphisms and phase 1 viral decay during HIV-1 infection: an adult AIDS Clinical Trials Group study. J Acquir Immune Defic Syndr. 2003;34:295–8. doi: 10.1097/00126334-200311010-00006. [DOI] [PubMed] [Google Scholar]
- 6.Johnson VA, Calvez V, Gunthard HF, et al. Update of the drug resistance mutations in HIV-1: March 2013. Top Antivir Med. 2013;21:6–14. [PMC free article] [PubMed] [Google Scholar]
- 7.Hosseinipour MC, van Oosterhout JJ, Weigel R, et al. The public health approach to identify antiretroviral therapy failure: high-level nucleoside reverse transcriptase inhibitor resistance among Malawians failing first-line antiretroviral therapy. AIDS. 2009;23:1127–34. doi: 10.1097/QAD.0b013e32832ac34e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marconi VC, Sunpath H, Lu Z, et al. Prevalence of HIV-1 drug resistance after failure of a first highly active antiretroviral therapy regimen in KwaZulu Natal, South Africa. Clin Infect Dis. 2008;46:1589–97. doi: 10.1086/587109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orrell C, Walensky RP, Losina E, Pitt J, Freedberg KA, Wood R. HIV type-1 clade C resistance genotypes in treatment-naive patients and after first virological failure in a large community antiretroviral therapy programme. Antivir Ther. 2009;14:523–31. [PMC free article] [PubMed] [Google Scholar]
- 10.Wallis CL, Mellors JW, Venter WD, Sanne I, Stevens W. Varied patterns of HIV-1 drug resistance on failing first-line antiretroviral therapy in South Africa. J Acquir Immune Defic Syndr. 2010;53:480–4. doi: 10.1097/QAI.0b013e3181bc478b. [DOI] [PubMed] [Google Scholar]
- 11.Sigaloff KC, Ramatsebe T, Viana R, de Wit TF, Wallis CL, Stevens WS. Accumulation of HIV drug resistance mutations in patients failing first-line antiretroviral treatment in South Africa. AIDS Res Hum Retroviruses. 2012;28:171–5. doi: 10.1089/aid.2011.0136. [DOI] [PubMed] [Google Scholar]
- 12.Sunpath H, Wu B, Gordon M, et al. High rate of K65R for antiretroviral therapy-naive patients with subtype C HIV infection failing a tenofovir-containing first-line regimen. AIDS. 2012;26:1679–84. doi: 10.1097/QAD.0b013e328356886d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papathanasopoulos MA, Hunt GM, Tiemessen CT. Evolution and diversity of HIV-1 in Africa—a review. Virus Genes. 2003;26:151–63. doi: 10.1023/a:1023435429841. [DOI] [PubMed] [Google Scholar]
- 14.Bartlett JA, Ribaudo HJ, Wallis CL, et al. Lopinavir/ritonavir monotherapy after virologic failure of first-line antiretroviral therapy in resource-limited settings. AIDS. 2012;26:1345–54. doi: 10.1097/QAD.0b013e328353b066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallis CL, Papathanasopoulos MA, Lakhi S, et al. Affordable in-house antiretroviral drug resistance assay with good performance in non-subtype B HIV-1. J Virol Methods. 2010;163:505–8. doi: 10.1016/j.jviromet.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yahi N, Fantini J, Henry M, Tourres C, Tamalet C. Structural analysis of reverse transcriptase mutations at codon 215 explains the predominance of T215Y over T215F in HIV-1 variants selected under antiretroviral therapy. J Biomed Sci. 2005;12:701–10. doi: 10.1007/s11373-005-9011-4. [DOI] [PubMed] [Google Scholar]
- 17.WHO Guidelines Approved by the Guidelines Review Committee. Antiretroviral Therapy for HIV Infection in Adults and Adolescents: recommendations for a public health approach. Geneva: World Health Organization; 2006. [PubMed] [Google Scholar]
- 18.Gupta RK, Hill A, Sawyer AW, et al. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. Lancet Infect Dis. 2009;9:409–17. doi: 10.1016/S1473-3099(09)70136-7. [DOI] [PubMed] [Google Scholar]
- 19.Sungkanuparph S, Win MM, Kiertiburanakul S, Phonrat B, Maek-a-nantawat W. HIV-1 drug resistance at virological failure versus immunological failure among patients failing first-line antiretroviral therapy in a resource-limited setting. Int J STD AIDS. 2012;23:316–8. doi: 10.1258/ijsa.2011.011337. [DOI] [PubMed] [Google Scholar]
- 20.Jiang H, Deeks SG, Kuritzkes DR, et al. Assessing resistance costs of antiretroviral therapies via measures of future drug options. J Infect Dis. 2003;188:1001–8. doi: 10.1086/378355. [DOI] [PubMed] [Google Scholar]
- 21.WHO Guidelines Approved by the Guidelines Review Committee. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Geneva: World Health Organization; 2013. [PubMed] [Google Scholar]
- 22.Nouhin J, Madec Y, Ngo-Giang-Huong N, Ferradini L, Nerrienet E. Increased risk of Q151M and K65R mutations in patients failing stavudine-containing first-line antiretroviral therapy in Cambodia. PloS One. 2013;8:e73744. doi: 10.1371/journal.pone.0073744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang MW, Rhee SY, Bertagnolio S, et al. Nucleoside reverse transcriptase inhibitor resistance mutations associated with first-line stavudine-containing antiretroviral therapy: programmatic implications for countries phasing out stavudine. J Infect Dis. 2013;207(Suppl 2):S70–7. doi: 10.1093/infdis/jit114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tambuyzer L, Nijs S, Daems B, Picchio G, Vingerhoets J. Effect of mutations at position E138 in HIV-1 reverse transcriptase on phenotypic susceptibility and virologic response to etravirine. J Acquir Immune Defic Syndr. 2011;58:18–22. doi: 10.1097/QAI.0b013e3182237f74. [DOI] [PubMed] [Google Scholar]
- 25.Vingerhoets J, Nijs S, Tambuyzer L, Hoogstoel A, Anderson D, Picchio G. Similar predictions of etravirine sensitivity regardless of genotypic testing method used: comparison of available scoring systems. Antivir Ther. 2012;17:1571–9. doi: 10.3851/IMP2275. [DOI] [PubMed] [Google Scholar]
- 26.Stevens WS, Wallis CL, Sanne I, Venter F. Will etravirine work in patients failing nonnucleoside reverse transcriptase inhibitor-based treatment in southern Africa? J Acquir Immune Defic Syndr. 2009;52:655–6. doi: 10.1097/QAI.0b013e3181ba1b00. [DOI] [PubMed] [Google Scholar]
- 27.Vingerhoets J, Azijn H, Tambuyzer L, et al. Short communication: activity of etravirine on different HIV type 1 subtypes: in vitro susceptibility in treatment-naive patients and week 48 pooled DUET study data. AIDS Res Hum Retroviruses. 2010;26:621–4. doi: 10.1089/aid.2009.0239. [DOI] [PubMed] [Google Scholar]
- 28.Hunt GM, Ledwaba J, Basson AE, et al. Surveillance of transmitted HIV-1 drug resistance in Gauteng and KwaZulu-Natal Provinces, South Africa, 2005–2009. Clin Infect Dis. 2012;54(Suppl 4):S334–8. doi: 10.1093/cid/cir1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett DE, Camacho RJ, Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PloS One. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]