Abstract
Significant progress has been made towards our understanding of the mechanism of peroxisome formation, in particular concerning sorting of peroxisomal membrane proteins, matrix protein import and organelle multiplication. Here we evaluate the progress made in recent years. We focus mainly on progress made in yeasts. We indicate the gaps in our knowledge and discuss conflicting models.
Current Opinion in Cell Biology 2014, 29:25–30
This review comes from a themed issue on Cell organelles
Edited by David K Banfield and Will Prinz
For a complete overview see the Issue and the Editorial
Available online 28th March 2014
0955-0674/$ – see front matter, Crown Copyright © 2014 Published by Elsevier Ltd. All rights reserved.
Introduction
Peroxisomes are eukaryotic organelles bound by a single membrane. Their abundance and functions vary between organisms, cell types and environmental conditions. In a seminal review, Lazarow and Fujiki [1] proposed that peroxisomal membrane and matrix proteins are synthesized on free polyribosomes and imported posttranslationally into preexisting peroxisomes. The endoplasmic reticulum was presumed to synthesise the membrane phospholipids of peroxisomes. Thus, they were considered autonomous organelles like mitochondria and chloroplasts. Lazarow and Fujiki stated that one of the implications of this model is that peroxisomes never form de novo [1].
Genetic screens in the late 80s and 90s identified many factors required for import of peroxisomal matrix proteins. The growth and division model was challenged by the discovery that mutants that appear to lack peroxisomal membranes can form peroxisomes de novo from the ER upon complementation. Since then the ER has been central to studies on peroxisome biogenesis. Below we assess the recent literature on peroxisomal matrix protein import and membrane formation.
Posttranslational import of matrix proteins
Protein import into peroxisomes differs from protein import into most other organelles as (1) peroxisomes import folded and even oligomeric proteins and (2) peroxisomal import receptors cycle between a soluble, free form in the cytosol and a cargo-loaded form at the peroxisomal membrane, which is associated with the translocon, and at the end of the cycle is ubiquitinated and released from the membrane in an ATP-dependent process [2]. Peroxisomal matrix proteins contain specific peroxisomal targeting signals (PTS1 or PTS2) that are post-translationally recognized in the cytosol by the import receptors Pex5 and Pex7, respectively [3, 4, 5, 6, 7, 8••].
Receptor-cargo complexes are directed to a docking complex at the peroxisomal membrane (see Figure 1). The PTS1 receptor mediates cargo binding as well as association with the import machinery, whereas the PTS2 receptor binds cargo but requires auxiliary proteins such as PEX5L in mammals and plants, or proteins of the Pex18 family in fungi for membrane association and cargo translocation [2]. The crystal structure of Pex7 in complex with the Pex18 paralog Pex21 and a PTS2 peptide visualizes the cooperative binding mode of the PTS2 pre-import complex [8••].
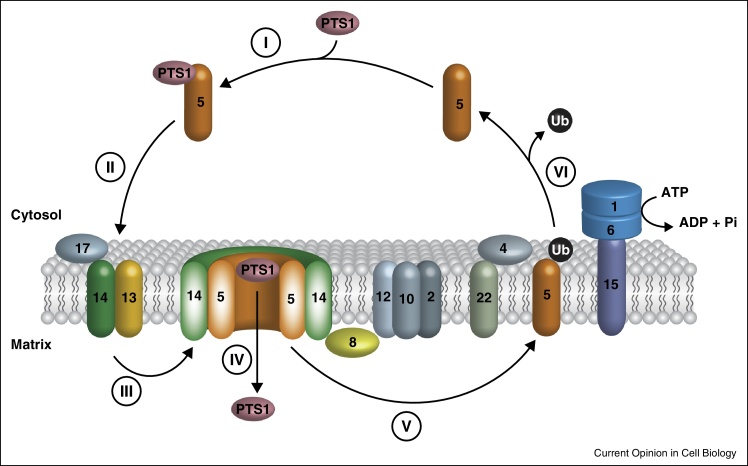
Figure 1.
Model of peroxisomal matrix protein import. (I) Proteins harboring a peroxisomal targeting signal of type 1 (PTS1) are recognized and bound by the import receptor Pex5 in the cytosol. (II) The cargo-loaded receptor is directed to the peroxisomal membrane and binds to the docking complex (Pex13/Pex14/Pex17). (III) The import receptor assembles with Pex14 to form a transient pore and (IV) cargo proteins are transported into the peroxisomal matrix in an unknown manner. Cargo release might involve the function of Pex8 or Pex14. (V) The import receptor is monoubiquitinated at a conserved cysteine by the E2-enzyme complex Pex4/Pex22 in tandem with E3-ligases of the RING-complex (Pex2, Pex10, Pex12). (VI) The ubiquitinated receptor is released from the peroxisomal membrane in an ATP-dependent manner by the AAA-peroxins Pex1 and Pex6, which are anchored to the peroxisomal membrane via Pex15. As the last step of the cycle, the ubiquitin moiety is removed and the receptor enters a new round of import. The designation is based on the yeast nomenclature.
The cargo-loaded receptor is thought to assemble with components of the docking complex to form the translocon, which allows translocation of folded proteins across the peroxisomal membrane into the matrix. The current opinion is based on the concept of a transient pore that assembles at the peroxisomal membrane and is disassembled after import, with its components being recycled for further rounds of protein import [9]. The major constituents of the dynamic pore for PTS1 import are the PTS1 receptor and the PMP Pex14: this constitutes the minimal functional unit for translocation of matrix proteins in vivo [10], and in electrophysiological studies it displays features of a regulated pore [11]. A major question is whether PTS1 and PTS2 proteins are imported via common or distinct import pores. Also currently debated is whether the cargo-loaded receptor remains associated with the pore (shuttle hypothesis) or whether it is released as a soluble receptor-cargo complex into the peroxisomal matrix (extended shuttle hypothesis). Once the cargo has reached the peroxisomal matrix, it is released from the receptor, which may require the intraperoxisomal peripheral membrane protein Pex8 [2, 12] or Pex14 [13]. It is unknown how folded proteins are translocated through the pore. Moreover, the exact composition of the pore as well as the driving force for cargo translocation remained elusive. During or after dissociation of the receptor-cargo complex, the PTS1 receptor is mono-ubiquitinated at a conserved cysteine, which serves as a signal for ATP-dependent dislocation of the receptor from the membrane to the cytosol [14]. It remains to be investigated why a cysteine and not a lysine is the evolutionarily conserved residue. Interestingly, in the PTS2-pathway, Pex7 but the auxiliary Pex18 family proteins are mono-ubiquitinated, again at a conserved cysteine [15, 16].
Mono-ubiquitination of receptors is performed by the peroxisomal ubiquitin-conjugating enzyme Pex4 [17]. Mono-ubiquitination is thought to prime the receptor for recognition by a complex of the AAA-type ATPases Pex1 and Pex6, which functions as dislocase to release the modified receptor from the membrane to the cytosol [18, 19]. AWP1 has been identified as a cofactor of mammalian Pex6 that binds mono-ubiquitinated Pex5 and is involved in the regulation of Pex5 export [20]. During or shortly after export, the PTS1 receptor is deubiquitinated and thus made available for another round of import [21, 22].
The mechanism of how folded proteins are translocated through the peroxisomal pore and also the driving force for this process remain elusive. The tagging of a substrate by monoubiquitylation or polyubiquitylation and its subsequent recognition and ATP-dependent removal from a membrane by ATPases of the AAA-family of proteins, resembles the mechanism of the membrane release of proteins for ER-associated degradation (ERAD) [23, 24]. As the ATP-dependent release of the ubiquitinated peroxisomal receptor from the membrane is the energy-requiring step of the matrix protein import cascade, the peroxisomal AAA peroxins might induce conformational changes of the receptor in an ATP-dependent manner that allows receptor release and cargo translocation. This link of receptor export and protein import is described by the export-driven import model [23].
Different mechanisms of PMP sorting
Until recently, two classes of PMPs were distinguished based on their Pex19 dependence for targeting to peroxisomes [25]. Class 1 PMPs contain targeting information that is recognized by Pex19, which binds newly synthesized PMPs in the cytosol and acts as a chaperone/targeting receptor, thus (1) preventing them from aggregating and (2) delivering them to the peroxisomal membrane for docking on Pex3 [26, 27, 28]. Although posttranslational targeting of PMPs to peroxisomes in vivo and in vitro is well documented [25, 28, 29, 30, 31, 32••], the mechanism of insertion remains unclear. The central role of Pex3 and Pex19 in PMP biogenesis is evident from the phenotype of pex3 and pex19 mutants, in which peroxisomal membranes are undetectable and class I PMPs are unstable or mislocalised [33]. On the basis of these observations we proposed that these mutants lack any peroxisomal structures.
Class 2 PMPs, the best-studied example of which is Pex3, contain targeting signals that are not recognized by Pex19. Upon de novo formation of peroxisomes by induction of Pex3 expression in S. cerevisiae pex3 cells, most data support a model whereby Pex3 enters the ER and forms preperoxisomes that develop into peroxisomes [34, 35, 36]. Its targeting and subsequent sorting signals have been described [37, 38], and the ER translocon has been implicated in this process [37]. In vitro budding reactions release vesicles containing Pex3, dependent on Pex19, cytosol and ATP [39, 40]. Budding occurs independently of the machinery required for ER exit of secretory proteins. These vesicles therefore might represent transport vesicles that fuse with pre-existing peroxisomes, thereby delivering Pex3 and lipids to peroxisomes (Figure 2).
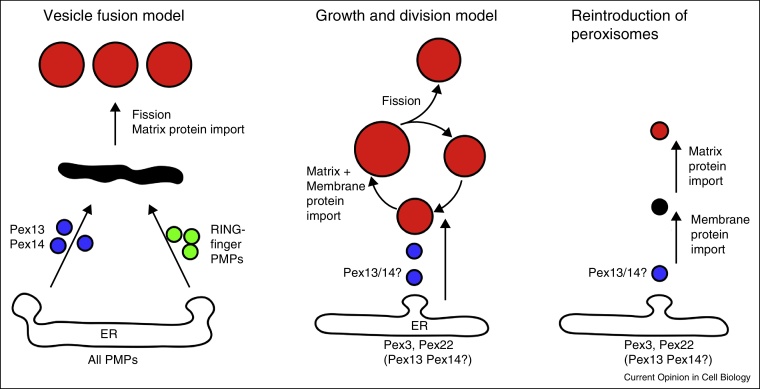
Figure 2.
Schematic representation of models for peroxisome multiplication. The vesicle fusion model proposes that all PMPs enter the ER, where they segregate and exit the ER in distinct vesicles: vesicles containing the docking complex proteins Pex13,14 (blue) fuse with vesicles containing the RING-finger complex proteins Pex2,10,12 (green) forming a preperoxisomal membrane structure (black). As this membrane structure now contains a complete importomer, matrix protein import commences. Subsequent fission results in peroxisomes of final size and membrane protein composition (red). The growth and division model proposes that peroxisomes (red) are derived from existing peroxisomes by fission. A small subset of PMPs (Class 2) insert into the ER and exit it in a transport vesicle (blue) that fuses with existing peroxisomes, where it provides the docking site for Pex19-mediated import of Class 1 PMPs (Class 1). Since Pex3 can only be detected in peroxisomes in wild type cells it cannot be excluded that it inserts directly into peroxisomes. Reintroduction of Peroxisomes. In the absence of pre-existing peroxisomes, the ER-derived Pex3-containing vesicle matures slowly into a peroxisome, with Pex3 again providing a docking site for Pex19/Class I PMPs complexes; this vesicle is thus slowly converted into a membrane structure containing all PMPs (black), finally becoming import-competent for matrix proteins (red). These newly formed peroxisomes will further multiply by growth and division. Whether Pex13 and Pex14 traffic via the ER or insert directly into a membrane structure distinct from the ER is not established.
As Pex3 is not observed in the ER of WT cells, one has to propose that its transit through the ER is fast. Thus far, Pex3 has been visualized in the ER only after overexpression, or by using mutant versions appended with large tags in cells blocked in peroxisome formation (pex19 or pex3 cells). It is therefore possible that yeast Pex3 bypasses the ER in wild type cells and is inserted directly into the peroxisomal membrane as has been reported for human Pex3 [41]. In line with this is the study by Knoops et al. [42]. They could detect no ER pool of Pex3 after its reintroduction into pex3 cells and propose that H. polymorpha Pex3 bypasses the ER altogether (see also below). This is an issue that needs further investigation.
Recently, detailed microcopy studies challenged the view that pex3 and pex19 cells are devoid of peroxisomal structures with the finding that the H. polymorpha pex3 and pex19 cells contain pre-peroxisomes to which Pex13 and Pex14 localise [42]. This suggests that Pex13 and Pex14 represent a third class of PMPs that also sort independent of Pex3 and Pex19. Pex13-containing membrane structures have been found in Pichia pastoris pex3 cells [43], and in S. cerevisiae pex3 and pex19 cells [44]. Furthermore, mammalian Pex13 inserts into membranes independent of Pex19 [45]. Also, Pex13 is required for Pex14 sorting [46], underlining that these PMPs behave differently from class I and II PMPs.
Peroxisome multiplication and the contribution of the ER
In wild type yeast cells, peroxisomes receive newly synthesized membrane and matrix proteins and lipids (growth) and multiply by fission [38, 47, 48, 49••, 50]. Peroxisome fission is mediated by dynamin-related proteins and Pex11 (for recent review see [51]). Most peroxisomal membrane lipids are synthesized in the ER. They may be directly transferred from the ER to peroxisomes [59] or reach peroxisomes via vesicular transport. However, it has also been proposed that peroxisomal membranes derive from the ER via budding of vesicles containing PMPs. Tabak and coworkers recently reported that all newly synthesized S. cerevisiae PMPs insert into the ER, and that the Pex13/14 docking complex and the RING-finger complex exit the ER in distinct vesicles [44, 52]. These ‘half importomer’ complexes were proposed to be brought together by heterotypic vesicle fusion dependent on Pex1 and Pex6, forming a pre-peroxisomal membrane structure able to import matrix proteins. Subsequent fission would produce peroxisomes of final size and membrane composition (Figure 2). Besides being responsible for de novo formation of peroxisomes in peroxisome-deficient mutant cells, Tabak and coworkers claim that this mechanism also provides a ‘continuous stream’ of de novo formed peroxisomes in wild type cells [52, 53]. Although elegant, this model is irreconcilable with many of the independent studies described above. This model disregards for instance the well established role of Pex19-dependent posttranslational insertion of newly synthesized PMPs directly into peroxisomes [25, 26, 27, 28, 29, 30, 31, 32••], the mislocalisation of many S. cerevisiae PMPs to the cytosol in pex3 and pex19 cells [33], and that new peroxisomes are derived from preexisting peroxisomes [47, 48, 49••, 50]. Furthermore, Pex25 is required for de novo peroxisome formation, but most pex25 cells contain peroxisomes, which suggests the de novo pathway is not essential for peroxisome maintenance [54••, 55]. Additionally, the studies by Knoops et al. [42] indicated that not the ER, but the Pex13/Pex14-containing structures present in H. polymorpha pex3 cells are the target for reintroduced Pex3. Although the origin of the Pex13/Pex14 preperoxisomal structures is unclear, their formation is independent of Pex3.
The shape of the ER appears to affect peroxisome multiplication. Pex30 is a Class 2 PMP [56] that localises to both peroxisomes and an ER subdomain found in close association with peroxisomes [57, 58••]. It has been postulated that new peroxisomes can form from these ER subdomains. Interestingly, COPI components were found in complex with Pex30, although the functional significance of this is unclear. The ER reticulons Rtn1, Rtn2 and Ypo1 were identified as core components of Pex30 complexes. ER reticulons are important for maintaining the morphology of tubular ER by stabilizing the strongly curved membranes. Interestingly, in cells lacking the ER reticulons or Pex30, de novo formation of peroxisomes is accelerated [58••]. Whether these mutants form peroxisomes de novo all the time is not known.
A multi-step model for peroxisome biogenesis
There is overwhelming evidence in favor of a growth and division model, whereby the insertion machinery for Class1 PMPs is central. As Pex3 is present at steady-state on peroxisomes in wild type cells, Pex19 bound to newly synthesized Class 1 PMPs may dock here to deliver them. Class 2 PMPs, including Pex3, traffic via the ER and vesicular transport to peroxisomes. This route may provide sufficient membrane constituents for continuing growth and division of peroxisomes, although a non-vesicular membrane transfer may contribute as well [59]. In cells forming peroxisomes de novo, Pex3 first inserts in the ER and sorts to preperoxisomes, where it facilitates Class 1 PMP insertion, allowing these membrane structures to import matrix proteins and mature into peroxisomes. Pex13 and Pex14 may follow the Pex3 route in cells synthesizing peroxisomes de novo, or they may bypass the ER and insert directly into preperoxisomes that can form independently of Pex3. Whether Pex13 and Pex14 follow this route in wild type cells is an important question and remains to be determined.
Concluding remarks
The global mechanisms of peroxisome membrane biogenesis and matrix protein import are becoming clear, but many questions remain. To mention but a few: how are ER-inserted PMPs sorted away from the secretory pathway? How do PMPs leave the ER, and how are they delivered specifically to peroxisomes? Do Class1 PMPs insert spontaneously into peroxisomal membranes after targeting, or is a specialised machinery required? How is peroxisome size and number regulated? Which proteins make up the pores for matrix protein import, and how is matrix protein translocation mediated? New approaches will need to be developed to shed light on these questions.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (FOR1905), and a Wellcome Trust Senior Research Fellowship in Basic Biomedical Science awarded to EHH, WT084265MA. The authors would like to thank Alison Motley for discussions.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
References
- 1.Lazarow P.B., Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 2.Liu X., Ma C., Subramani S. Recent advances in peroxisomal matrix protein import. Curr Opin Cell Biol. 2012;24:484–489. doi: 10.1016/j.ceb.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanley W.A., Filipp F.V., Kursula P., Schuller N., Erdmann R., Schliebs W., Sattler M., Wilmanns M. Recognition of a functional peroxisome type 1 target by the dynamic import receptor pex5p. Mol Cell. 2006;24:653–663. doi: 10.1016/j.molcel.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brocard C., Hartig A. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim Biophys Acta. 2006;1763:1565–1573. doi: 10.1016/j.bbamcr.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhary G., Kataya A.R., Lingner T., Reumann S. Non-canonical peroxisome targeting signals: identification of novel PTS1 tripeptides and characterization of enhancer elements by computational permutation analysis. BMC Plant Biol. 2012:12. doi: 10.1186/1471-2229-1112-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petriv O.I., Rachubinski R.A. Lack of peroxisomal catalase causes a progeric phenotype in Caenorhabditis elegans. J Biol Chem. 2004;279:19996–20001. doi: 10.1074/jbc.M400207200. [DOI] [PubMed] [Google Scholar]
- 7.Schliebs W., Kunau W.H. PTS2 co-receptors: diverse proteins with common features. Biochim Biophys Acta. 2006;1763:1605–1612. doi: 10.1016/j.bbamcr.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 8••.Pan D., Nakatsu T., Kato H. Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p. Nat Struct Mol Biol. 2013;20:987–993. doi: 10.1038/nsmb.2618. [DOI] [PubMed] [Google Scholar]; This study describes the crystal structure of Pex7 in complex with its coreceptor.
- 9.Erdmann R., Schliebs W. Peroxisomal matrix protein import: the transient pore model. Nat Rev Mol Cell Biol. 2005;6:738–742. doi: 10.1038/nrm1710. [DOI] [PubMed] [Google Scholar]
- 10.Ma C., Subramani S. Peroxisome matrix and membrane protein biogenesis. IUBMB Life. 2009;61:713–722. doi: 10.1002/iub.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meinecke M., Cizmowski C., Schliebs W., Kruger V., Beck S., Wagner R., Erdmann R. The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol. 2010;12:273–277. doi: 10.1038/ncb2027. [DOI] [PubMed] [Google Scholar]
- 12.Wang D., Visser N.V., Veenhuis M., Van Der Klei I.J. Physical interactions of the peroxisomal targeting signal 1-receptor, Pex5p, studied by fluorescence correlation spectroscopy. J Biol Chem. 2003;278:43340–43345. doi: 10.1074/jbc.M307789200. [DOI] [PubMed] [Google Scholar]
- 13.Freitas M.O., Francisco T., Rodrigues T.A., Alencastre I.S., Pinto M.P., Grou C.P., Carvalho A.F., Fransen M., Sa-Miranda C., Azevedo J.E. PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. J Biol Chem. 2011;286:40509–40519. doi: 10.1074/jbc.M111.287201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platta H.W., Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17:474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Leon S., Zhang L., McDonald W.H., Yates, Cregg J.M., Subramani S. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J Cell Biol. 2006;172:67–78. doi: 10.1083/jcb.200508096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensel A., Beck S., El Magraoui F., Platta H.W., Girzalsky W., Erdmann R. Cysteine-dependent ubiquitination of Pex18p is linked to cargo translocation across the peroxisomal membrane. J Biol Chem. 2011;286:43495–43505. doi: 10.1074/jbc.M111.286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platta H.W., Girzalsky W., Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem J. 2004;384:37–45. doi: 10.1042/BJ20040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Platta H.W., Grunau S., Rosenkranz K., Girzalsky W., Erdmann R. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat Cell Biol. 2005;7:817–822. doi: 10.1038/ncb1281. [DOI] [PubMed] [Google Scholar]
- 19.Miyata N., Fujiki Y. Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol Cell Biol. 2005;25:10822–10832. doi: 10.1128/MCB.25.24.10822-10832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyata N., Okumoto K., Mukai S., Noguchi M., Fujiki Y. AWP1/ZFAND6 functions in Pex5 export by interacting with cys-monoubiquitinated Pex5 and Pex6 AAA ATPase. Traffic. 2012;13:168–183. doi: 10.1111/j.1600-0854.2011.01298.x. [DOI] [PubMed] [Google Scholar]
- 21.Debelyy M.O., Platta H.W., Saffian D., Hensel A., Thoms S., Meyer H.E., Warscheid B., Girzalsky W., Erdmann R. Ubp15p, an ubiquitin hydrolase associated with the peroxisomal export machinery. J Biol Chem. 2011;286:28223–28234. doi: 10.1074/jbc.M111.238600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grou C.P., Carvalho A.F., Pinto M.P., Huybrechts S.J., Sa-Miranda C., Fransen M., Azevedo J.E. Properties of the ubiquitin-Pex5p thiol ester conjugate. J Biol Chem. 2009;284:10504–10513. doi: 10.1074/jbc.M808978200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schliebs W., Girzalsky W., Erdmann R. Peroxisomal protein import and ERAD: variations on a common theme. Nat Rev Mol Cell Biol. 2010;11:885–890. doi: 10.1038/nrm3008. [DOI] [PubMed] [Google Scholar]
- 24.Platta H.W., Hagen S., Erdmann R. The exportomer: the peroxisomal receptor export machinery. Cell Mol Life Sci. 2013;70:1393–1411. doi: 10.1007/s00018-012-1136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones J.M., Morrell J.C., Gould S.J. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J Cell Biol. 2004;164:57–67. doi: 10.1083/jcb.200304111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sacksteder K.A., Jones J.M., South S.T., Li X., Liu Y., Gould S.J. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol. 2000;148:931–944. doi: 10.1083/jcb.148.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rottensteiner H., Kramer A., Lorenzen S., Stein K., Landgraf C., Volkmer-Engert R., Erdmann R. Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol Biol Cell. 2004;15:3406–3417. doi: 10.1091/mbc.E04-03-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto M.P., Grou C.P., Alencastre I.S., Oliveira M.E., Sa-Miranda C., Fransen M., Azevedo J.E. The import competence of a peroxisomal membrane protein is determined by Pex19p before the docking step. J Biol Chem. 2006;281:34492–34502. doi: 10.1074/jbc.M607183200. [DOI] [PubMed] [Google Scholar]
- 29.Diestelkotter P., Just W.W. In vitro insertion of the 22-kD peroxisomal membrane protein into isolated rat liver peroxisomes. J Cell Biol. 1993;123:1717–1725. doi: 10.1083/jcb.123.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imanaka T., Shiina Y., Takano T., Hashimoto T., Osumi T. Insertion of the 70-kDa peroxisomal membrane protein into peroxisomal membranes in vivo and in vitro. J Biol Chem. 1996;271:3706–3713. doi: 10.1074/jbc.271.7.3706. [DOI] [PubMed] [Google Scholar]
- 31.Fang Y., Morrell J.C., Jones J.M., Gould S.J. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J Cell Biol. 2004;164:863–875. doi: 10.1083/jcb.200311131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Yagita Y., Hiromasa T., Fujiki Y. Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J Cell Biol. 2013;200:651–666. doi: 10.1083/jcb.201211077. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes the post translational import of a tail-anchored protein directly into peroxisomes.
- 33.Hettema E.H., Girzalsky W., van Den Berg M., Erdmann R., Distel B. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 2000;19:223–233. doi: 10.1093/emboj/19.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoepfner D., Schildknegt D., Braakman I., Philippsen P., Tabak H.F. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 35.Tam Y.Y., Fagarasanu A., Fagarasanu M., Rachubinski R.A. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J Biol Chem. 2005;280:34933–34939. doi: 10.1074/jbc.M506208200. [DOI] [PubMed] [Google Scholar]
- 36.Kragt A., Voorn-Brouwer T., van den Berg M., Distel B. Endoplasmic reticulum-directed Pex3p routes to peroxisomes and restores peroxisome formation in a Saccharomyces cerevisiae pex3Delta strain. J Biol Chem. 2005;280:34350–34357. doi: 10.1074/jbc.M505432200. [DOI] [PubMed] [Google Scholar]
- 37.Thoms S., Harms I., Kalies K.U., Gartner J. Peroxisome formation requires the endoplasmic reticulum channel protein Sec61. Traffic. 2012 doi: 10.1111/j.1600-0854.2011.01324.x. [DOI] [PubMed] [Google Scholar]
- 38.Fakieh M.H., Drake P.J., Lacey J., Munck J.M., Motley A.M., Hettema E.H. Intra-ER sorting of the peroxisomal membrane protein Pex3 relies on its luminal domain. Biol Open. 2013;2:829–837. doi: 10.1242/bio.20134788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam S.K., Yoda N., Schekman R. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1013397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agrawal G., Joshi S., Subramani S. Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2011;108:9113–9118. doi: 10.1073/pnas.1018749108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuzaki T., Fujiki Y. The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J Cell Biol. 2008;183:1275–1286. doi: 10.1083/jcb.200806062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knoops K., Manivannan S., Cepinska M.N., Krikken A.M., Kram A.M., Veenhuis M., van der Klei I.J. Preperoxisomal vesicles can form in the absence of pex3. J. Cell Biology. 2014;204:659–668. doi: 10.1083/jcb.201310148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hazra P.P., Suriapranata I., Snyder W.B., Subramani S. Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic. 2002;3:560–574. doi: 10.1034/j.1600-0854.2002.30806.x. [DOI] [PubMed] [Google Scholar]
- 44.van der Zand A., Braakman I., Tabak H.F. Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol Biol Cell. 2010;21:2057–2065. doi: 10.1091/mbc.E10-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fransen M., Vastiau I., Brees C., Brys V., Mannaerts G.P., Van Veldhoven P.P. Potential role for Pex19p in assembly of PTS-receptor docking complexes. J Biol Chem. 2004;279:12615–12624. doi: 10.1074/jbc.M304941200. [DOI] [PubMed] [Google Scholar]
- 46.Girzalsky W., Rehling P., Stein K., Kipper J., Blank L., Kunau W.H., Erdmann R. Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J Cell Biol. 1999;144:1151–1162. doi: 10.1083/jcb.144.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuravi K., Nagotu S., Krikken A.M., Sjollema K., Deckers M., Erdmann R., Veenhuis M., van der Klei I.J. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci. 2006;119:3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- 48.Motley A.M., Hettema E.H. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Menendez-Benito V., van Deventer S.J., Jimenez-Garcia V., Roy-Luzarraga M., van Leeuwen F., Neefjes J. Spatiotemporal analysis of organelle and macromolecular complex inheritance. Proc Natl Acad Sci U S A. 2013;110:175–180. doi: 10.1073/pnas.1207424110. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that existing peroxisomes receive newly synthesised Pex3.
- 50.Nagotu S., Krikken A.M., Otzen M., Kiel J.A., Veenhuis M., van der Klei I.J. Peroxisome fission in Hansenula polymorpha requires Mdv1 and Fis1, two proteins also involved in mitochondrial fission. Traffic. 2008;9:1471–1484. doi: 10.1111/j.1600-0854.2008.00772.x. [DOI] [PubMed] [Google Scholar]
- 51.Schrader M., Bonekamp N.A., Islinger M. Fission and proliferation of peroxisomes. Biochim Biophys Acta. 2012;1822:1343–1357. doi: 10.1016/j.bbadis.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 52.van der Zand A., Gent J., Braakman I., Tabak H.F. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell. 2012;149:397–409. doi: 10.1016/j.cell.2012.01.054. [DOI] [PubMed] [Google Scholar]
- 53.van der Zand A., Tabak H.F. Peroxisomes: offshoots of the ER. Curr Opin Cell Biol. 2013;25:449–454. doi: 10.1016/j.ceb.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 54••.Saraya R., Krikken A.M., Veenhuis M., van der Klei I.J. Peroxisome reintroduction in Hansenula polymorpha requires Pex25 and Rho1. J Cell Biol. 2011;193:885–900. doi: 10.1083/jcb.201012083. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]; This paper shows for the first time that distinct proteins are required for de novo peroxisome formation versus peroxisome maintenance by growth and division.
- 55.Huber A., Koch J., Kragler F., Brocard C., Hartig A. A subtle interplay between three Pex11 proteins shapes de novo formation and fission of peroxisomes. Traffic. 2012;13:157–167. doi: 10.1111/j.1600-0854.2011.01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vizeacoumar F.J., Vreden W.N., Aitchison J.D., Rachubinski R.A. Pex19p binds Pex30p and Pex32p at regions required for their peroxisomal localization but separate from their peroxisomal targeting signals. J Biol Chem 2006. 2006;281:14805–14812. doi: 10.1074/jbc.M601808200. [DOI] [PubMed] [Google Scholar]
- 57.Yan M., Rachubinski D.A., Joshi S., Rachubinski R.A., Subramani S. Dysferlin domain-containing proteins. Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol Biol Cell. 2008;19:885–898. doi: 10.1091/mbc.E07-10-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that ER morphology influences peroxisome reintroduction. Furthermore, Pex30 was found to associate with reticulons and coatomer.
- 58••.David C., Koch J., Oeljeklaus S., Laernsack A., Melchior S., Wiese S., Schummer A., Erdmann R., Warscheid B., Brocard C. A combined approach of quantitative interaction proteomics and live-cell imaging reveals a regulatory role for endoplasmic reticulum (ER) reticulon homology proteins in peroxisome biogenesis. Mol Cell Proteomics. 2013;12:2408–2425. doi: 10.1074/mcp.M112.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raychaudhuri S., Prinz W.A. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2008;105:15785–15790. doi: 10.1073/pnas.0808321105. [DOI] [PMC free article] [PubMed] [Google Scholar]