Abstract
Breast carcinoma is the leading cause of cancer-related mortality in women worldwide with an estimated 1.38 million new cases and 458,000 deaths in 2008 alone1. This malignancy represents a heterogeneous group of tumours with characteristic molecular features, prognosis, and responses to available therapy2–4. Recurrent somatic alterations in breast cancer have been described including mutations and copy number alterations, notably ERBB2 amplifications, the first successful therapy target defined by a genomic aberration5. Prior DNA sequencing studies of breast cancer genomes have revealed additional candidate mutations and gene rearrangements 6–10. Here we report the whole-exome sequences of DNA from 103 human breast cancers of diverse subtypes from patients in Mexico and Vietnam compared to matched-normal DNA, together with whole-genome sequences of 22 breast cancer/normal pairs. Beyond confirming recurrent somatic mutations in PIK3CA11, TP536, AKT112, GATA313, and MAP3K110, we discovered recurrent mutations in the CBFB transcription factor gene and deletions of its partner RUNX1. Furthermore, we have identified a recurrent MAGI3-AKT3 fusion enriched in triple-negative breast cancer lacking estrogen and progesterone receptors and ERBB2 expression. The Magi3-Akt3 fusion leads to constitutive activation of Akt kinase, which is abolished by treatment with an ATP-competitive Akt small-molecule inhibitor.
Breast cancers are classified according to gene-expression subtypes: Luminal A, Luminal B, Her2-enriched, and basal-like14. Luminal subtypes are associated with expression of estrogen (ER+) and progesterone (PR+) receptors and differentiated luminal epithelial cell markers. The subtypes differ in genomic complexity, key genetic alterations, and clinical prognosis2–4,15. To discover genomic alterations in breast cancers, we performed whole-genome and whole-exome sequencing of 108 primary, treatment naive, breast carcinoma/normal DNA pairs from all major expression subtypes (Table 1; Supplementary Table 1–3), 17 cases by whole-exome and whole-genome sequencing, 5 cases by whole-genome sequencing alone, and 86 cases by whole-exome sequencing alone.
In total, whole-exome sequencing was performed on 103 tumour-normal pairs, 54 from Mexico and 49 from Vietnam, targeting 189,980 exons comprising 33 megabases of the genome and with a median of 85.1% of targeted bases covered at least 30-fold across the sample set. This analysis revealed a total of 4,985 candidate somatic substitutions (see https://confluence.broadinstitute.org/display/CGATools/MuTect for methods and datasets) and insertions/deletions (indels: see https://confluence.broadinstitute.org/display/CGATools/Indelocator for methods) in the target protein-coding regions and the adjacent splice sites, ranging from 14 to 307 putative events in individual samples (Supplementary Table 4). These mutations represented 3,153 missense, 1,157 silent, 242 nonsense, 97 splice site, 194 deletions, 110 insertions and 32 other mutations (Supplementary Table 5). The total mutation rate was 1.66 per Mb (range 0.47–10.5) with a non-silent mutation rate of 1.27 per Mb (range 0.31–8.05), similar to previous reports in breast carcinoma6–9. The mutation rate in breast cancer exceeds that of hematologic malignancies and prostate cancer but is significantly lower than in lung cancer and melanoma10,16–19. The most common mutation events observed are C to T transition events in CpG dinucleotides (Figure 1, Supplementary Figure 4).
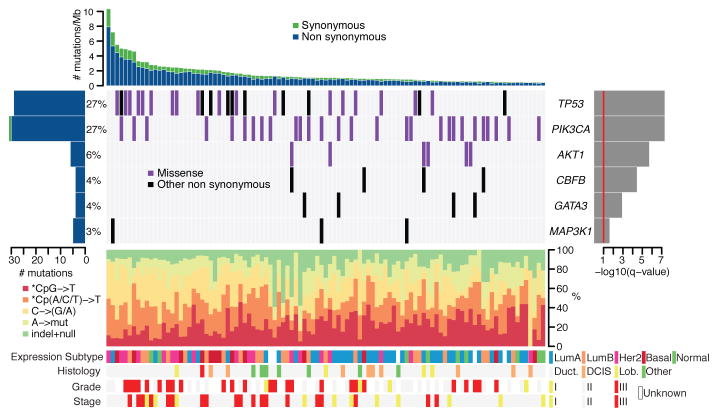
Figure 1. Most significantly-mutated genes in breast cancer as determined by whole exome sequencing (n=103).
Upper histogram: rates of sample-specific mutations (substitutions and indels), green = synonymous, blue = non-synonymous.
Left histogram: number of mutations per gene and percentage of samples affected (colour coding as in upper histogram).
Central heatmap: Distribution of significant mutations across sequenced samples (“Other non synonymous” mutations = nonsense, indel, splice-site).
Right histogram: -log10 score of MutSig q value. Red line at q = 0.1.
Lower chart: top - rates of non-silent mutations within categories indicated by legend; bottom - key molecular features of samples in each column (Expression subtypes: “Lum” = luminal. Histology: “Duct.” = Infiltrating ductal carcinoma, “DCIS” = Ductal carcinoma in situ, “Lob.” = Infiltrating lobular carcinoma).
We performed validation experiments on 494 candidate mutations (representing all significantly mutated genes and genes in significantly mutated genesets) using a combination of mass-spectrometric genotyping, 454 pyrosequencing, Pacific Biosciences sequencing, and Illumina sequencing of matched formalin-fixed paraffin embedded tissue, and confirmed the presence of 94% of protein-altering point mutations (Supplementary Table 4, Supplementary Figure 5); this validation rate is consistent with previous results that 95% of point mutations can be validated with orthogonal methods16,17. Only 18 of 39 (46%) indels among significantly mutated genes were confirmed.
Six genes were found to be mutated with significant recurrence in the 103 whole exome sequenced samples, by analysis with the MutSig algorithm16,17 (https://confluence.broadinstitute.org/display/CGATools/MutSig) at a False Discovery Rate (FDR) <0.1 after correction for multiple hypothesis testing (Supplementary Table 6A), manual review of reads, and subsequent orthogonal confirmation of somatic events (Figure 1, Supplementary Figure 6). One gene, CBFB is identified for the first time as a significantly mutated gene in breast cancer or any other epithelial cancer, to our knowledge, while the other 5 genes (TP53, PIK3CA, AKT1, GATA3, and MAP3K1) have previously been reported as mutated in breast cancer7,10,13. This significantly mutated genes list, as any list produced by a statistical method, is likely incomplete and reflects the statistical power of our cohort size -- larger sample sets will provide further statistical power.
Somatic mutations in TP53 and PIK3CA were each present in 27% of samples, consistent with published frequencies10,20 (Figure 1). TP53 mutations occur in samples with a higher mutation rate (T-test p = 0.0079 comparing samples with mutation rates greater than or less than the median 1.66 mutations/Mb) and were distributed across the gene in sites reported in COSMIC (http://www.sanger.ac.uk/genetics/CGP/cosmic/). Also, using the ABSOLUTE algorithm for determining allele-specific copy number (Carter et al. manuscript submitted), we observed that 21 of 31 TP53 mutations were homozygous (Supplementary Table 4). PIK3CA mutations were clustered in the helical (amino acids 542/545; 40%) and kinase domains (amino acid 1047; 47%)20. Six samples harboured the AKT1 E17K mutation that alters the pleckstrin-homology (PH) domain and leads to activation of the kinase12. AKT1 and PIK3CA mutations, which activate the phosphatidylinositol-3-kinase (PI3K) pathway, were mutually exclusive in our dataset. MAP3K1, recently reported as mutated in ER+ breast cancers10, harboured 5 mutations in 3 patients with ER+ disease, and followed a pattern consistent with positive selection for recessive inactivation of the gene. In total, 2 frameshift, 2 nonsense, and 1 missense mutation combined with a homozygous deletion spanning the coding region were observed. Although the point mutations appeared to be heterozygous by copy-number analysis, two patients harboured dual mutations, consistent with compound heterozygous inactivation, although confirmatory phasing data were not available. The GATA3 transcription factor gene harboured mutations in 4 patients with luminal tumours, including 3 novel frameshift mutations near the 3′-end of the coding sequence. We also identified one previously described splice-site mutation that disrupts zinc-finger domains in Gata3 required for DNA binding13.
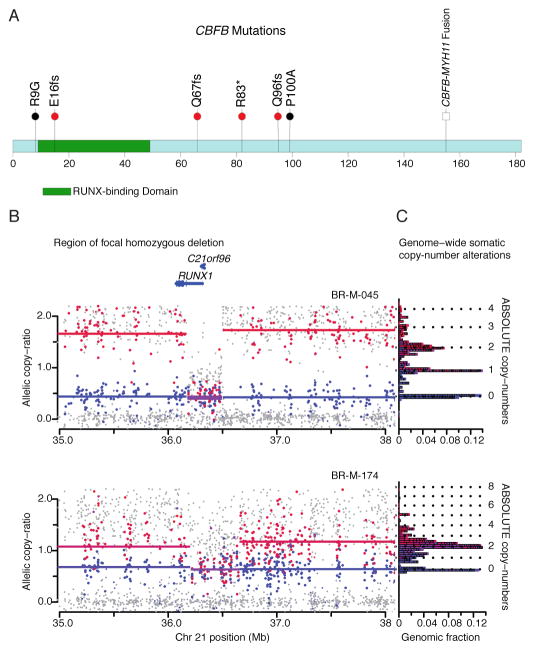
CBFB, encoding the core-binding-factor beta subunit, was mutated in 4 ER+ samples, with one nonsense mutation and three truncating frameshift mutations (Figure 2A). CBFB somatic mutations have been noted in isolated cases of breast cancer6,10. This is the first report of these mutations recurring at a significant rate above background; the sample size is not sufficient to determine whether these mutations are specific for ER+ subtypes. CBFB encodes the non-DNA binding component of a heterodimeric protein complex, together with the DNA-binding RUNX proteins encoded by RUNX1, RUNX2, and RUNX3. Copy-number analysis, using the ABSOLUTE algorithm (Carter et al., manuscript submitted), provides further evidence for loss of function of the Runx1/Cbfb complex in breast cancer: the cases with CBFB mutations appear to have hemizygous deletions of one parental allele while two additional cases harbour homozygous deletions of RUNX1 (Figure 2B, 2C, Supplementary Figures 7,8). Oncogenic rearrangements of RUNX1 or CBFB are common in acute myeloid leukemia21,22 (including the CBFB-MYH11 translocation believed to have dominant negative function21). This is to our knowledge the first report of inactivation of this transcription factor complex in epithelial cancers.
Figure 2. CBFB mutations and RUNX1 deletions.
A. CBFB coding region diagram: RUNX binding domain in green. Mutations identified in this study (red bullets), previously identified mutations6,10 (black bullets), and known CBFB-MYH11 fusion indicated.
B. Allelic copy ratios for the 3 Mb region surrounding RUNX1 in samples BR-M-045 and BR-M-174. Dots indicate copy-ratios for individual SNP alleles: Red = higher copy-ratio allele for informative SNPs that are heterozygous in matched normal DNA; Blue = lower-copy ratio SNPs; Grey = uninformative SNPs (homozygous in matched normal).
Lines indicate inferred segmental copy-ratios. Red = higher copy segment; Blue = lower copy segment; Purple = equal copy segment.
C. Histogram depicting bins of segmented copy number (y-axis), with inferred integral copies shown by dotted lines; the length of each horizontal block corresponds to the fraction of the haploid genome at the copy number level, or “genomic fraction” (x-axis).
Significance analysis restricted to somatic mutations in genes reported in COSMIC revealed 3 significantly mutated genes, including PIK3CA, TP53, and ERBB2, the latter below the significance threshold in the complete analysis (Supplementary Table 7). ERBB2 contained somatic mutations in three samples, with two being identical S310F mutations (these two samples are distinct based on their germline and somatic genotypes.) The S310F mutation can activate ERBB2 and is transforming in vitro (personal communication from H. Greulich). Neither sample with the S310F activating mutation has ERBB2 amplification (Supplementary Figure 9). The two samples belong to the Her2-enriched and Luminal B subtypes, which typically have ERBB2 amplification; this supports the notion that the observed mutations have a driving role in these tumours10,23.
To identify candidate genomic rearrangements, we applied the dRanger algorithm16,17 to the 22 cases with paired tumour/normal whole-genome sequencing data (Supplementary Table 8). The rate of rearrangements ranged from a median of 30 rearrangements per sample in the Luminal A subtype (range 0–218) to the basal-like and Her2-enriched subtypes with a median of 237 and 246 rearrangements, respectively (Supplementary Figure 10); the rates are similar to a recent report15. We performed PCR amplification on a subset of the candidate rearrangements (Supplementary Methods) and confirmed 89 of 165 events (54%). No rearrangement was seen in more than one sample (Supplementary Table 8). In addition, we did not identify rearrangements previously observed by DNA sequencing15 nor by cDNA-sequencing, including MAST and Notch family-gene fusions24.
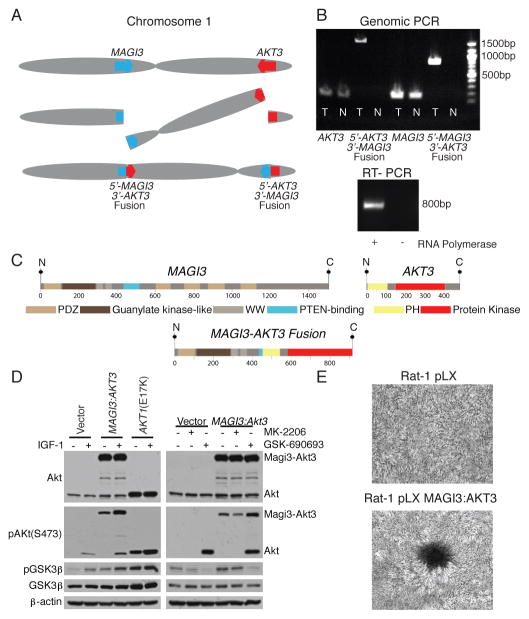
The discovery of recurrent driver rearrangements in other epithelial cancers25,26 led to a closer examination of the list of confirmed rearrangements. In a triple-negative, basal-like subtype tumour, we observed a rearrangement between the genes MAGI3 (membrane associated guanylate kinase, WW and PDZ domain containing 3) on chromosome 1p and AKT3 (v-akt murine thymoma viral oncogene homolog 3) on chromosome 1q, resulting in a balanced translocation from intron 9 in MAGI3 to intron 1 of AKT3 (Figure 3A). The novel fusion genes were confirmed in tumour DNA by sequencing the product of PCR amplification (Figure 3B). The MAGI3 disruption is complemented by a hemizygous deletion of the other allele (Supplementary Figure 11A). The expression levels of individual exons of MAGI3 and AKT3 correspond to the predicted 5′-MAGI3-AKT3-3′ fusion (Supplementary Figure 11B), with this sample having the highest AKT3 expression in the dataset. Expression of the fusion gene was confirmed in the tumour sample by PCR amplification of the cDNA (Figure 3B).
Figure 3. MAGI3-AKT3 fusion gene.
A. Diagram of balanced translocation between MAGI3 and AKT3.
B. (top) Genomic DNA PCR for AKT3, MAGI3, and both fusion products in tumour (T) and normal (N). (bottom) cDNA PCR of fusion gene in tumour.
C. (above) MAGI3 and AKT3 protein domains; (below) putative fusion protein.
D. Immunoblots of lysates from ZR-75 cells transfected with vector, MAGI3-AKT3 fusion, or AKT1 E17K mutant, grown in low-serum media, for the indicated antibodies. (Left) infected cells with and without insulin growth factor 1 (IGF-1) stimulation; (right) treatment of vector or MAGI3-AKT3 overexpressing cells with Akt inhibitors MK-2206 and GSK-690693.
E. Focus formation assays with Rat-1 cells expressing pLX control or MAGI3-AKT3, and stained with crystal violet.
The rearrangement produces an in-frame fusion gene with a predicted Magi3-Akt3 fusion protein that combines Magi3 lacking the second PDZ domain, reported to bind to Pten and be required for Pten’s inhibitory effect on the PI3K pathway27, together with an Akt3 region that retains an intact kinase domain but has a disruption of the pleckstrin homology domain prior to the glutamate at position 17 (Figure 3C). AKT3 shares significant homology to AKT1 and is reported to be the dominant AKT family member expressed in hormone receptor negative breast cancers28. Together, the MAGI3-AKT3 translocation and deletion of MAGI3 could result in the combined loss of function of a tumour suppressor gene (PTEN) and activation of an oncogene (AKT3).
To evaluate oncogenic activity of the MAGI3-AKT3 fusion, we expressed the fusion gene ectopically in ZR-75 cells. The Magi3-Akt3 fusion protein is constitutively phosphorylated at serine 473 in the Akt3 kinase domain (numbered according to the wild-type protein) in the absence of growth factors (Figure 3D); ectopically expressed Akt1 with an engineered E17K mutation is likewise constitutively phosphorylated (Figure 3D), as previously reported12. Constitutive activation of the Magi3-Akt3 kinase in turn activates downstream pathways as demonstrated by phosphorylation of GSK3β, an Akt substrate (Figure 3D). Phosphorylation of GSK3β by the MAGI3-AKT3 fusion can be inhibited with an ATP-competitive small molecule Akt inhibitor, GSK-690693, but not with an allosteric Akt inhibitor, MK-2206, that interacts with the PH domain of Akt (Figure 3D). Over-expression of the MAGI3-AKT3 fusion gene in Rat-1 fibroblast cell lines led to loss of contact inhibition and focus formation (Figure 3E).
We screened 235 additional breast cancer samples for the presence of the 5′-MAGI3-AKT3-3′ fusion event by RT-PCR of cDNA followed by Sanger sequencing of breakpoints. The fusion was present in 8 of the 235 samples, including 5 out of 72 triple negative (ER−/PR−/Her2−) samples, (Supplementary Figure 12).
The power provided by whole-genome and whole-exome sequencing of a relatively large and diverse breast cancer sample set has enabled several significant discoveries including the identification of recurrent inactivating mutations in CBFB and of a recurrent translocation of MAGI3-AKT3. The mutations in CBFB, RUNX1 and GATA3 suggest the importance of understanding epithelial cell differentiation and its regulatory transcription factors in breast cancer pathogenesis. The recurrent genomic fusion involving AKT3 suggests that the use of ATP-competitive Akt inhibitors should be evaluated in clinical trials for the treatment of fusion-positive triple-negative breast cancers, a subtype where limited therapeutic options exist beyond systemic cytotoxic chemotherapy.
Methods Summary
All samples were obtained under institutional IRB approval and with documented informed consent. Breast cancer specimens from Mexico were paired with peripheral blood normal DNA while the Vietnamese samples were paired with DNA from normal adjacent breast tissue. Tumour RNA for each case was analysed on exon arrays to determine breast cancer expression subtype using the PAM50 classification method, while tumour/normal DNA pairs were analysed for copy number, allelic imbalance, and ancestry using SNP arrays. A total of 108 samples, 17 both WGS and WES, 86 WES only, and 5 WGS only, passed initial qualification metrics, library construction, and successfully achieved desired sequencing depth (100X WES; 30X WGS) on the Illumina sequencing platform (Supplementary Figures 1–3; Supplementary Tables 2 and 3). Tumour-specific point mutations, small insertions/deletions (indels), and rearrangements were detected by comparing tumour DNA to its paired normal DNA and using a series of algorithms to identify somatic events (Supplementary Figure 2)16,17. Additional mutation calling was performed separately on tumour and normal DNA to identify germline mutation events that may confer susceptibility to breast carcinoma. Allele-specific copy number of each gene/mutation was determined using the HAPSEG and ABSOLUTE analysis methods. Confirmation of point mutations and indels was performed using mass spectrometry based genotyping and orthogonal next-generation sequencing methods, whereas putative in-frame genomic rearrangements were PCR amplified from DNA to confirm the presence of the event.
A complete description of the materials and methods is provided in the Supplementary Information. Access to the data and computational algorithms used in this study can be found at https://confluence.broadinstitute.org/display/CGATools/Home.
Supplementary Material
Acknowledgments
We would like to thank all patients who contributed samples to this study. This study was a collaboration of the Broad Institute in Cambridge, MA and the National Institute of Genomic Medicine (INMEGEN) in Mexico City, Mexico. The work was conducted as part of the Slim Initiative for Genomic Medicine, a project funded by the Carlos Slim Health Institute in Mexico. This work is part of a global effort in collaboration with the International Cancer Genome Consortium (ICGC). The authors would also like to acknowledge Jordi Barretina and Heidi Greulich for their critical review of the manuscript. In addition, we would like to acknowledge the technical expertise and data generation efforts of The Broad Institute Biological Samples, Genome Sequencing, and Genetic Analysis Platforms. S.B. has received fellowship support cosponsored by CancerCare Manitoba and the University of Manitoba. K.K.B is a recipient of the John Gavin Post-doctoral Fellowship, Genesis Oncology Trust of New Zealand. R.R.B. and S.L.R.C. received a scholarship from the Mexican Council of Science and Technology (CONACyT). R.B. is a V Foundation Scholar. A.T. is funded by NIH grant CA122099. This work was partially supported by the Dana-Farber/Harvard SPORE in breast cancer under NCI grant reference CA089393.
Footnotes
Author Contributions: S.B., K.C., C.R.E., L.Z., M.S.L., A.Y.S, A.H.R., N.S., G.G., A.H.M., and M.M. performed sequencing data analysis. S.B, A.M.F, and J.F. planned and performed fusion gene PCR and cloning. K.B. and A.T. planned and performed fusion gene biochemical and transformation experiments. F.D. and J.J. performed qPCR experiments. C.R.E. and J.C.F.L. performed ancestry analysis. C.R.E., J.C.F.L, L.Z., and S.P. performed expression analysis. S.L.C. and G.G. performed ABSOLUTE analysis. R.R.V., L.U.F., S.E.S., and R.B. performed experimental work on SNP arrays and DNA copy number analysis. K.C., R.C.O., and M.P. performed mutation and rearrangement validation analysis. C.S., M.L.C., K.G.A., D.A., V.B.P, V.Q.J., and S.L.R.C collected data, processed and analysed genetic material, and provided data management. S.B., K.A., D.A., R.R.V, S.L.R.C, A.M.A., S.R.C., K.T., D.C.S., A.L.R. and A.H.M. collected patient materials. S.B., K.C., C.R.E., S.L.C., L.Z., A.M.F., J.C.F.L., N.P., and M.M. contributed text and figures including Supplementary Information. All authors contributed to the final manuscript. K.P., D.C.S., A.L.R., and E.S.L. provided critical review of the manuscript. J.B., G.J.S., E.S.L., S.B.G., L.A.G., T.R.G., J.M.Z., G.G., A.H.M., and M.M. provided leadership for the project.
Author Information: Sequence data have been deposited in the dbGaP repository (http://www.ncbi.nlm.nih.gov/gap) under accession number phs000369.v1.p1. E.S.L., L.A.G., T.R.G., and M.M. have financial interests in Foundation Medicine, which operates in the field of cancer diagnosis, but has no connection or rights to the work described in this study.
They wish to declare this interest, although it does not appear to be a competing interest.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011 doi: 10.3322/caac.20107. [DOI] [Google Scholar]
- 2.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Gatza ML, et al. A pathway-based classification of human breast cancer. Proceedings of the National Academy of Sciences. 2010;107:6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King CR, Kraus MH, Aaronson SA. Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science. 1985;229:974–976. doi: 10.1126/science.2992089. [DOI] [PubMed] [Google Scholar]
- 6.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 7.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 8.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 9.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 11.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 12.Carpten JD, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 13.Usary J, et al. Mutation of GATA3 in human breast tumors. Oncogene. 2004;23:7669–7678. doi: 10.1038/sj.onc.1207966. [DOI] [PubMed] [Google Scholar]
- 14.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachman KE, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 21.Cameron ER, Neil JC. The Runx genes: lineage-specific oncogenes and tumor suppressors. Oncogene. 2004;23:4308–4314. doi: 10.1038/sj.onc.1207130. [DOI] [PubMed] [Google Scholar]
- 22.Shigesada K, van de Sluis B, Liu PP. Mechanism of leukemogenesis by the inv(16) chimeric gene CBFB/PEBP2B-MHY11. Oncogene. 2004;23:4297–4307. doi: 10.1038/sj.onc.1207748. [DOI] [PubMed] [Google Scholar]
- 23.Stephens P, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 24.Robinson DR, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nature medicine. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 26.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y. Interaction of the Tumor Suppressor PTEN/MMAC with a PDZ Domain of MAGI3, a Novel Membrane-associated Guanylate Kinase. Journal of Biological Chemistry. 2000;275:21477–21485. doi: 10.1074/jbc.M909741199. [DOI] [PubMed] [Google Scholar]
- 28.Nakatani K, et al. Up-regulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. J Biol Chem. 1999;274:21528–21532. doi: 10.1074/jbc.274.31.21528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.