Abstract
The marine bacterium Vibrio parahaemolyticus, a major cause of food-borne gastroenteritis, employs a type VI secretion system 1 (T6SS1), a recently discovered protein secretion system, to combat competing bacteria. Environmental signals such as temperature, salinity, cell density and surface sensing, as well as the quorum-sensing master regulator OpaR, were previously reported to regulate T6SS1 activity and expression. In this work, we set out to identify additional transcription regulators that control the tightly regulated T6SS1 activity. To this end, we determined the effect of deletions in several known virulence regulators and in two regulators encoded within the T6SS1 gene cluster on expression and secretion of the core T6SS component Hcp1 and on T6SS1-mediated anti-bacterial activity. We report that VP1391 and VP1407, transcriptional regulators encoded within the T6SS1 gene cluster, are essential for T6SS1 activity. Moreover, we found that H-NS, a bacterial histone-like nucleoid structuring protein, which mediates transcription silencing of horizontally acquired genes, serves as a repressor of T6SS1. We also show that activation of surface sensing and high salt conditions alleviate the H-NS-mediated repression. Our results shed light on the complex network of environmental signals and transcription regulators that govern the tight regulation over T6SS1 activity.
Introduction
Gram-negative bacteria use the recently discovered type VI secretion system (T6SS) (Ho et al., 2014) in competition against other bacteria or as a virulence determinant against eukaryotic hosts (Russell et al., 2014). The T6SS is a macromolecular protein complex that is analogous to contractile phage tail but in a topologically reversed orientation (Basler et al., 2012; Ho et al., 2014). The central part of the T6SS is composed of a tube made of hexameric rings of Hcp proteins capped with a trimer of VgrG proteins and a PAAR repeat-containing protein at the tip (Basler et al., 2012; Hachani et al., 2011; Ho et al., 2014; Shneider et al., 2013). While inside the cell, this tube, which is decorated with toxic effector proteins (Ho et al., 2014), is surrounded by a sheath structure made of heterodimers of VipA and VipB (also known as TssB and TssC) proteins (Basler et al., 2012). Upon perception of an extracellular signal, the sheath contracts, propelling the Hcp–VgrG–PAAR tube and the effectors out of the cell and into an adjacent recipient cell (Basler et al., 2012; Brunet et al., 2014). Interestingly, up to six different T6SSs can be encoded in the same genome (Boyer et al., 2009), and each of them may be differentially regulated (Salomon et al., 2013; Sana et al., 2013; Schwarz et al., 2010).
Various mechanical and environmental cues were found to activate T6SSs in different bacteria, such as temperature (Hockett et al., 2013; Ishikawa et al., 2012; Pieper et al., 2009; Salomon et al., 2013), salinity (Ishikawa et al., 2012; Salomon et al., 2013), surface sensing (Salomon et al., 2013; Silverman et al., 2011), quorum sensing (Salomon et al., 2013; Sana et al., 2012; Zhang et al., 2011; Zheng et al., 2010) and membrane disruption (Ho et al., 2013). Each bacterium appears to have evolved regulatory mechanisms to activate an appropriate T6SS under the specific conditions in which it requires its activity (Ho et al., 2014). We have previously characterized the environmental conditions required to activate the two T6SSs (T6SS1 and T6SS2) encoded by Vibrio parahaemolyticus, a halophilic marine bacterium that is a major cause of food-borne gastroenteritis (Newton et al., 2012; Zhang & Orth, 2013) and of acute hepatopancreatic necrosis syndrome in shrimp (Tran et al., 2013). The two T6SSs are differentially regulated by temperature, salinity, quorum sensing and surface sensing (Salomon et al., 2013). Remarkably, both T6SSs are inactive at 37 °C, suggesting that they do not play a role during infection of a mammalian host (Salomon et al., 2013). The V. parahaemolyticus T6SS1 (Fig. S1, available in the online Supplementary Material), which plays a role in inter-bacterial competition mediated by at least three identified effectors (Salomon et al., 2014) is active under warm marine-like conditions (in high salt media at 30 °C) and is further induced when surface sensing is activated (Salomon et al., 2013). OpaR, a quorum-sensing master regulator that operates under high cell densities, represses T6SS1 (Gode-Potratz & McCarter, 2011; Ma et al., 2012; McCarter, 1998; Salomon et al., 2013). In addition, we have previously identified two transcription regulators, VP1391 and VP1407, that are encoded within the T6SS1 gene cluster and are required for expression of the T6SS1 Hcp (Hcp1) (Salomon et al., 2013). However, the role of VP1391 and VP1407 in regulation of T6SS1 activity was not determined.
In this work, we set out to identify additional regulators of the V. parahaemolyticus T6SS1, and to determine whether the regulators VP1391 and VP1407 are required for T6SS1 activity. To this end, we studied the effect on T6SS1 activity of four V. parahaemolyticus master regulators that were previously shown to regulate virulence: AphA, a quorum-sensing master regulator that operates at low cell densities and was previously shown to be required for virulence (Wang et al., 2013a) and to negatively regulate the V. parahaemolyticus T6SS2 in an indirect manner (Wang et al., 2013b); ToxRS, a two-component regulator that is required for toxin induction (Lin et al., 1993) and colonization of a murine model (Whitaker et al., 2012); LafK, a regulator of surface sensing that is required for swarming and virulence (Gode-Potratz et al., 2010; Gode-Potratz et al., 2011; Stewart & McCarter, 2003); and the histone-like nucleoid structuring protein (H-NS), which mediates transcription silencing of horizontally acquired genes (Atlung & Ingmer, 1997; Lucchini et al., 2006) and was shown to regulate the cytotoxic V. parahaemolyticus type III secretion system (T3SS) 1 (Kodama et al., 2010). H-NS was also described as a negative regulator of T6SS gene transcription in several other bacterial strains (Bernard et al., 2010; Eijkelkamp et al., 2013). In addition, we tested the effect of deletions in vp1391 and vp1407 on T6SS1 activity. Our results show that VP1391 and VP1407 are necessary for T6SS1 activity, whereas AphA, ToxRS and LafK do not play a role in T6SS1 regulation. Interestingly, we found that H-NS serves as a negative regulator of T6SS1 under various environmental conditions.
Methods
Bacterial strains and media.
The V. parahaemolyticus RIMD 2210633 derivative strain POR1 [RIMD 2210633 ΔtdhAS (listed in Table S1)] (Park et al., 2004) and its derivatives were routinely cultured in marine Luria–Bertani (MLB) broth [Luria–Bertani broth containing 3 % (w/v) sodium chloride] or grown on marine minimal media agar plates (Eagon, 1962) at 30 °C. Escherichia coli strains DH5α and S17λpir were routinely cultured in 2xYT broth or Luria–Bertani (LB) broth at 37 °C. The medium was supplemented with kanamycin (250 µg ml−1) or chloramphenicol (25 µg ml−1) where necessary.
Plasmids.
Plasmids for expression of VP1391 and VP1407 were described previously (Salomon et al., 2013). For expression of H-NS (VP1133), the gene coding sequence including 1 kb upstream was cloned into the pBAD/Myc–His vector (Invitrogen) harbouring a kanamycin-resistance cassette (Salomon et al., 2013) in-frame with the Myc–His tag. For gene deletions, the 1 kb upstream and 1 kb downstream of aphA (vp2762), lafK (vpa1538), toxRS (vp0819–vp0820) and hns (vp1133) were cloned into the suicide plasmid pDM4 (chloramphenicol resistant, oriR6K). Primers used for creating constructs listed in Table S2.
Construction of deletion and knock-in strains.
To generate in-frame deletions of the entire gene coding region in aphA, lafK, toxRS and hns pDM4 plasmids described above were conjugated into the POR1 strain from E. coli S17λpir and transconjugants were selected on media containing chloramphenicol. Bacteria were then counter-selected by growing on media containing 15 % (w/v) sucrose. Deletions were confirmed by PCR analyses. The construction of Δvp1391 and Δvp1407 deletion strains, as well as of the Hcp1–myc knock-in strain was previously described (Salomon et al., 2013). Strains are listed in Table S1.
Expression and secretion assays.
Hcp1–myc expression and secretion assays were performed as previously described (Salomon et al., 2013). In brief, V. parahaemolyticus cultures were grown overnight in MLB with appropriate antibiotics when necessary to maintain plasmids. Cultures were then normalized to OD600 0.18 in 5 ml of the indicated medium containing appropriate antibiotics and 0.1 % (w/v) arabinose. Cultures were then incubated with agitation at 30 °C or 37 °C for 5 h and OD600 values were determined. For expression fractions (cells in Fig. 1) 1.5 OD600 units were collected and cell pellets were resuspended in 100 µl of 2× protein sample buffer. Supernatants of 10 OD600 units were filtered and precipitated with deoxycholate and TCA. Precipitated proteins were pelleted and washed with acetone, prior to resuspension in 40 µl of 1× protein sample buffer. Expression and secretion samples were resolved on SDS-PAGE, transferred onto PVDF membranes, and immunoblotted using anti c-Myc antibodies (Santa Cruz Biotechnology). Equal loading of total protein lysate was confirmed by analysis of representative bands using Ponceau S staining of the immunoblot membrane. Equal volumes of medium were used from cleared, centrifuged cultures for all TCA precipitations. Experiments were repeated at least twice with similar results. A representative experiment is shown.
Fig. 1.
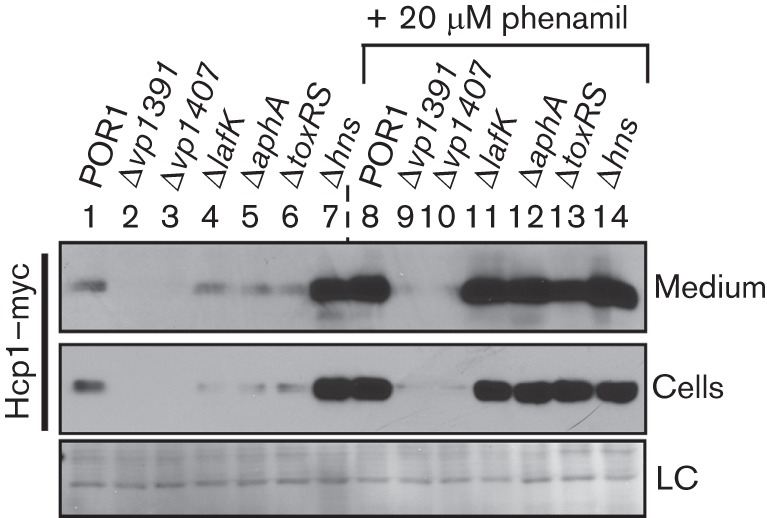
Deletion of hns de-represses Hcp1 expression and secretion. V. parahaemolyticus POR1-derivative deletion strains containing endogenously C-terminal myc-tagged hcp1 were grown in MLB media for 5 h at 30 °C with or without 20 µM phenamil. Expression (cells) and secretion (medium) of Hcp1–myc were detected by immunoblot using anti-myc antibodies. The loading control (LC) is shown for total protein lysate.
Bacterial killing assays.
Bacterial killing assays were performed as previously described (Salomon et al., 2013). In brief, bacteria were grown overnight in MLB (V. parahaemolyticus) or 2xYT (E. coli). Cultures were normalized to OD600 0.5, mixed at a 4 : 1 ratio (V. parahaemolyticus:E. coli) so that the number of prey cells was the same in each co-culture tested, and 25 µl of the co-cultures were spotted on MLB or LB plates in triplicate and incubated at 30 °C or 37 °C for 4 h. Bacterial spots were harvested from plates and the c.f.u. ml−1 was determined by spotting 10-fold serial dilutions on selective media plates. Experiments were repeated at least twice with similar results. A representative experiment is shown.
Bacterial growth assays.
Triplicates of V. parahaemolyticus strains grown overnight in MLB were normalized to OD600 0.1 in MLB or LB, and growth was monitored by measuring OD600 of cultures incubated at 30 °C with agitation over time. Experiments were repeated at least twice with similar results. A representative experiment is shown.
Results
H-NS represses Hcp1 expression and secretion in the absence of surface-sensing activation
To determine the effect of the master regulators AphA, ToxRS, LafK and H-NS, and of VP1391 and VP1407 on the activity of the V. parahaemolyticus T6SS1, we first generated strains with deletions in the corresponding genes and verified that their growth was comparable to the commonly used POR1 parental strain (RIMD 2210633 ΔtdhAS), which is an attenuated strain that was derived from the V. parahaemolyticus clinical isolate RIMD 2210633 (Park et al., 2004) (Fig. S2). We have previously reported that deletions in the genes encoding VP1391 and VP1407 have no effect on growth (Salomon et al., 2013). Next, we tested the effect of these deletions on the expression and secretion of Hcp1, a core component and hallmark secreted protein of T6SS1 (Salomon et al., 2013), under warm marine-like conditions in the presence or absence of phenamil, an inhibitor of the polar flagellar motor that activates surface sensing (Gode-Potratz et al., 2011; Salomon et al., 2013). In agreement with our previous report, VP1391 and VP1407 were required for expression and secretion of Hcp1, even when surface sensing was activated in the presence of phenamil (Fig. 1, lanes 2–3 and 9–10). Whereas deletion of lafK, aphA and toxRS had a mild negative effect on Hcp1 expression in the absence of surface-sensing activation compared to the parental POR1 strain, both expression and secretion of Hcp1 were comparable to the POR1 strain in the presence of phenamil (Fig. 1, compare lanes 4–6 to lane 1, and compare lanes 11–13 to lane 8). Interestingly, deletion of hns resulted in increased expression and secretion of Hcp1 in the absence of surface-sensing activation to levels comparable to those seen in the POR1 parental strain when surface sensing was activated (Fig. 1, compare lane 7 to lanes 1 and 8). No additional increase in Hcp1 expression and secretion were apparent in the Δhns strain when surface sensing was activated (Fig. 1, compare lanes 8 and 14). These results suggest that H-NS is a repressor of Hcp1 expression and secretion in the absence of surface-sensing stimulus. Furthermore, the regulators VP1391 and VP1407 are required for Hcp1 expression and secretion under T6SS1 inducing conditions.
VP1391 and VP1407 are required for T6SS1 activity
After verifying that VP1391 and VP1407 were required for Hcp1 expression and secretion, we next tested whether VP1391 and VP1407 are required for T6SS1 anti-bacterial activity. To this end, we monitored the bacterial killing activity of the Δvp1391 and Δvp1407 deletion strains compared to the POR1 parental strain. A Δhcp1 strain with an inactive T6SS1 was also used as a control strain (Salomon et al., 2013). In agreement with the lack of Hcp1 expression and secretion seen in Fig. 1, the Δvp1391 and Δvp1407 deletion strains, like the control Δhcp1 strain, were unable to kill E. coli in a co-culture (Fig. 2). Complementation of vp1391 and vp1407 from a plasmid restored bacterial killing activity to levels similar to those of the parental POR1 strain (Fig. 2). These results suggested that vp1391 and vp1407 are indeed required for T6SS1 activity.
Fig. 2.
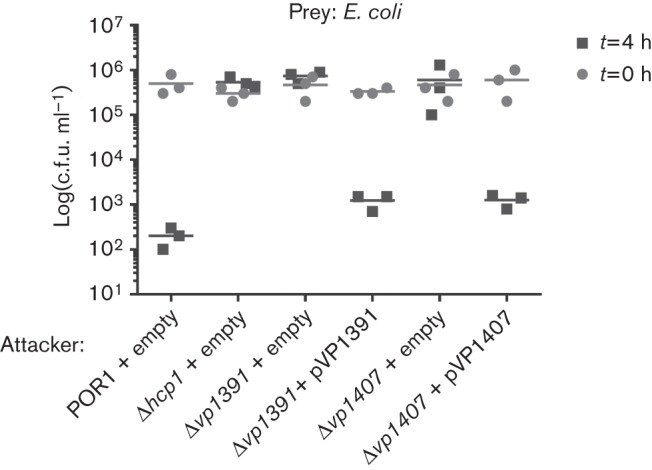
VP1391 and VP1407 are required for T6SS1 anti-bacterial activity. Viability counts of E. coli (prey) before (0 h) and after (4 h) co-culture. E. coli were co-cultured with V. parahaemolyticus (attacker) POR1 or the following POR1 derivative strains, POR1Δhcp1 and POR1Δvp1391, or POR1Δvp1407, carrying an empty vector or a vector encoding the arabinose-inducible expression of vp1391 (pVP1391) or the expression of vp1407 from its endogenous promoter (pVP1407). Mixed cultures at a 4 : 1 OD600 ratio (attacker : prey) were spotted on MLB plates containing 0.1 % (w/v) arabinose and incubated at 30 °C for 4 h.
Even though our results suggested that AphA, ToxRS, LafK and H-NS have no negative effect on Hcp1 expression and secretion under T6SS1-inducing conditions (Fig. 1), we set out to test whether other components of T6SS1 may be controlled by these regulators and affect T6SS1 activity. To this end, we monitored the bacterial killing activity of the ΔlafK, ΔaphA, ΔtoxRS and Δhns strains when co-cultured with E. coli. As shown in Fig. 3, all tested deletion strains were able to kill E. coli, indicating that these regulators are not required for T6SS1 activity. Importantly, all deletion strains grew similarly to the parental POR1 during the co-culture incubation (Fig. 3). Notably, the c.f.u. ml−1 values of E. coli prey and V. parahaemolyticus attacker strains were similar between samples at 0 h (data not shown).
Fig. 3.
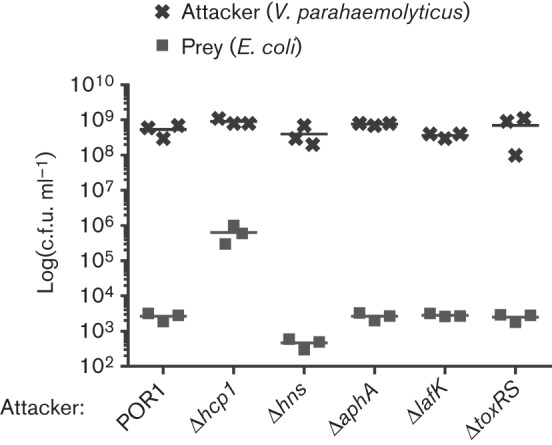
H-NS, AphA, LafK and ToxRS are not required for T6SS1 anti-bacterial activity. Viability counts of E. coli (prey) and V. parahaemolyticus deletion strains (attacker) 4 h after co-culture. Mixed cultures at a 4 : 1 OD600 ratio (attacker : prey) were spotted on MLB plates containing 0.1 % (w/v) arabinose and incubated at 30 °C for 4 h.
H-NS is not a temperature-dependent repressor of Hcp1 expression
Regulation of gene expression by H-NS was previously shown to be controlled by temperature and osmolarity in various bacteria (Hurme & Rhen, 1998; Porter & Dorman, 1994). Since our results suggested that H-NS represses Hcp1 expression in the absence of surface-sensing activation (Fig. 1, lane 7), we next set out to characterize additional environmental conditions that possibly control H-NS-mediated repression of Hcp1 expression. To this end, we determined the expression levels of Hcp1 in low or high salt media (LB and MLB, respectively) and at 30 °C or 37 °C, in the presence or absence of phenamil. Aside from de-repressing Hcp1 expression in MLB at 30 °C in the absence of phenamil (Fig. 4, compare lanes 5 and 6), deletion of hns also resulted in an increase of Hcp1 expression under T6SS1 non-inducing low salt conditions when bacteria were grown in LB medium (Fig. 4, compare lanes 3 and 4; and Fig. S3). Because we previously found that the V. parahaemolyticus T6SS1 is inactive when cells are grown at 37 °C, we hypothesized that H-NS could also be the repressor that prevents T6SS1 activation at this temperature. Surprisingly, however, deletion of hns did not result in high level expression of Hcp1 when cells were grown at 37 °C either in high salt (Fig. 4, lanes 1 and 2) or low salt (Fig. S3, lanes 4 and 5). This is in contrast to the de-repressing effect of the hns deletion on Hcp1 expression in absence of surface-sensing activation at 30 °C. Nevertheless, low expression of Hcp1 was detected when the Δhns strain was grown at 37 °C in the presence of phenamil, an inducer of surface sensing, but only in high salt (Fig. 4, lanes 8 and 9; and Fig. S3, lanes 10 and 11). Importantly, ectopic complementation of H-NS from a plasmid resulted in a decrease of Hcp1 expression (Fig. 4, lanes 7 and 14; and Fig. S3, lanes 3 and 9). Thus, these results indicate that H-NS is a repressor of Hcp1 expression in the absence of surface-sensing activation, whereas repression of Hcp1 expression at 37 °C is not dependent on H-NS.
Fig. 4.
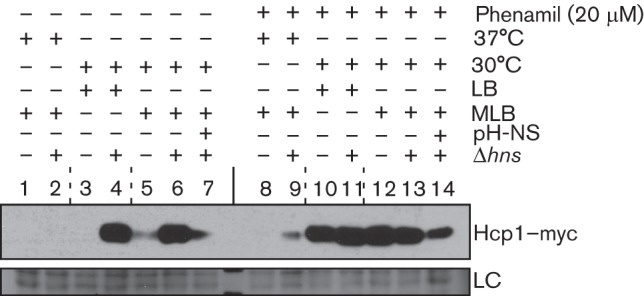
H-NS does not mediate repression of Hcp1 expression at 37 °C. Expression of endogenously C-terminal myc-tagged Hcp1 in V. parahaemolyticus POR1 and the Δhns (POR1 derivative) strains harbouring an empty vector or a vector for the expression of hns driven by its native promoter (pH-NS). Cultures were grown in different media (LB or MLB) and at different temperatures (30 °C or 37 °C) and in the absence or presence of phenamil as indicated. Expression of Hcp1–myc was detected by immunoblot using anti-myc antibodies. The loading control (LC) is shown for total protein lysate.
H-NS is a salinity-dependent regulator of T6SS1
After identifying the environmental conditions under which H-NS represses Hcp1 expression, we next asked whether H-NS represses the bacterial killing activity of T6SS1 under non-inducing conditions. To test this, we monitored the bacterial killing activity of a Δhns strain under low salt conditions or at 37 °C, conditions under which the T6SS1 is not normally induced (Salomon et al., 2013). Because the bacterial killing assays are performed on solid media surface sensing is activated, and therefore, we were unable to test the effect of H-NS on T6SS1 activity in the absence of surface-sensing activation. In agreement with our observations that the deletion of hns did not affect the repression of Hcp1 at 37 °C, the Δhns strain was unable to kill E. coli when co-cultured at 37 °C (Fig. 5a). Interestingly, when we tested the T6SS1-mediated bacterial killing of E. coli in low salt (LB) media at 30 °C, the POR1 parental strain was less efficient in killing E. coli compared to its killing ability in high salt (MLB) media (Fig. 5a), in agreement with our previous report that Hcp1 requires high salt for induction (Salomon et al., 2013). In contrast, the Δhns strain was able to kill E. coli similarly in both high and low salt conditions, at a level similar to that of the parental POR1 strain-mediated killing under inducing high salt conditions (Fig. 5). Importantly, complementation of H-NS from a plasmid resulted in a decrease in killing ability (Fig. 5b). Notably, the growth of the Δhns strain was comparable to that of the parental POR1 strain in both high and low salt media (Fig. S2). Thus, our results indicate that H-NS represses T6SS1 activity under low salt conditions, but not at 37 °C.
Fig. 5.
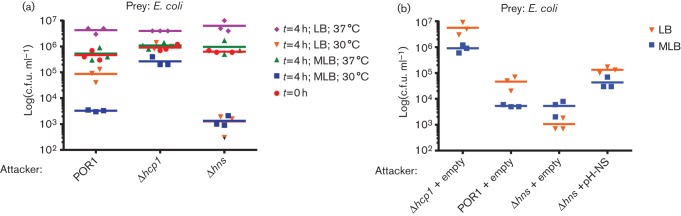
H-NS represses T6SS1 anti-bacterial activity under low salt conditions. (a) Viability counts of E. coli (prey) before (0 h) and after (4 h) co-culture. E. coli were co-cultured with V. parahaemolyticus (attacker) POR1 or the POR1 derivative strains, Δhcp1, or Δhns. Mixed cultures at a 4 : 1 OD600 ratio (attacker : prey) were spotted on LB or MLB plates and incubated at 30 °C or 37 °C for 4 h. (b) Viability counts of E. coli (prey) 4 h after co-culture. E. coli were co-cultured with V. parahaemolyticus (attacker) POR1 or the POR1 derivative strains, Δhcp1, or Δhns, harbouring an empty vector or a vector for the expression of hns driven by its native promoter (pH-NS). Mixed cultures at a 4 : 1 OD600 ratio (attacker : prey) were spotted on LB or MLB plates and incubated at 30 °C for 4 h.
Discussion
T6SSs are tightly regulated protein secretion systems that are often used by Gram-negative bacteria to combat bacterial competitors (Ho et al., 2014). In this work, we determined the effect of several virulence regulators on the activity of the V. parahaemolyticus anti-bacterial T6SS1. We found that T6SS1 activity is positively regulated by VP1391 and VP1407, two transcription regulators encoded within the T6SS1 gene cluster. In addition, we identified H-NS as a negative regulator of T6SS1 under conditions of low salt and in the absence of surface-sensing activation.
We have previously reported that VP1391 and VP1407 are required for Hcp1 expression (Salomon et al., 2013). Here we extended our analysis and showed that these two regulators are also required for Hcp1 secretion, even when surface sensing was activated to induce T6SS1. As Hcp1 is a core component of the T6SS, it was not surprising that we also found VP1391 and VP1407 to be absolutely required for the anti-bacterial activity mediated by T6SS1. Our results demonstrated that the T6SS1 gene cluster encodes its own positive regulators, which are, at least in part, responsible for its tight regulation.
In addition, we tested the effects of the known eukaryotic virulence regulators AphA, LafK, ToxRS and H-NS on Hcp1 expression and secretion, as well as on T6SS1 anti-bacterial activity. Our results demonstrated that AphA, LafK and ToxRS have a mild effect on Hcp1 expression under non-optimal inducing conditions (i.e. in the absence of surface-sensing activation), but have no apparent effect on Hcp1 expression and secretion or on T6SS1 anti-bacterial activity when surface sensing is activated. Thus, it appears that virulence against eukaryotes, which is mediated by secreted toxins and T3SSs, is differentially regulated from the anti-bacterial toxicity of V. parahaemolyticus, which is mediated by T6SS1.
We anticipated that AphA, a quorum-sensing master regulator that is active at low cell densities (Wang et al., 2013a), would have a regulatory effect on T6SS1 as we have previously reported that quorum sensing, and specifically the master regulator OpaR that is active at high cell densities, regulates T6SS1 activity (Salomon et al., 2013). However, AphA, which was also previously shown to negatively regulate the V. parahaemolyticus T6SS2 in an indirect manner (Wang et al., 2013b), had no effect on T6SS1 activity. These results further support our previous observations that V. parahaemolyticus T6SS1 and T6SS2 are differentially regulated, and also suggest that this differential regulation involves different transcription regulators. Moreover, we found that LafK, a central regulator of surface sensing (Gode-Potratz et al., 2010; Gode-Potratz et al., 2011; Stewart & McCarter, 2003), did not affect T6SS1 activity and did not appear to be involved in T6SS1 regulation. This result was quite surprising as we have previously shown that surface-sensing activates T6SS1 (Salomon et al., 2013). Thus, it is possible that another pathway is responsible for T6SS1 activation when V. parahaemolyticus is grown on a surface, and that this pathway differs from the one that regulates the activation of the lateral flagella, which requires LafK (Gode-Potratz et al., 2010; Gode-Potratz et al., 2011; Stewart & McCarter, 2003).
Remarkably, deletion of hns, a regulator that mediates transcriptional silencing of horizontally acquired genes (Lucchini et al., 2006), de-repressed Hcp1 expression and secretion under non-optimal T6SS1-inducing conditions to levels comparable to T6SS1 activation under optimal inducing conditions when surface sensing is activated. We showed that a Δhns strain was able to kill E. coli under non-optimal inducing conditions (in low salt media) as if the bacterial co-culture was incubated under optimal T6SS1-inducing conditions (in high salt media). Thus, our results demonstrate that H-NS serves as a negative regulator of T6SS1 and plays a major role in the tight regulation of the V. parahaemolyticus T6SS1. Moreover, as activation of surface sensing did not result in further increase in Hcp1 expression and secretion in the Δhns strain, we suggest that H-NS is the major regulator that controls the surface sensing and salinity mediated activation of T6SS1, and that both surface sensing and high salt conditions are required to alleviate the H-NS-mediated repression.
Notably, H-NS-mediated regulation was previously reported as being dependent on external environmental signals such as osmolarity and temperature (Hurme & Rhen, 1998). Indeed, we showed that deletion of hns alleviates T6SS1 repression under low osmolarity conditions (i.e. low salt), but we did not observe a significant role for H-NS in temperature-dependent repression of T6SS1 activity (i.e. when cells were grown at 37 °C). The inability of V. parahaemolyticus to kill E. coli when co-cultured at 37 °C even in the absence of H-NS-mediated repression further demonstrates the complex regulation network of the T6SS1 and indicates that another repressor, yet to be identified, plays a role in regulating temperature-mediated T6SS1 activity. Moreover, our results are in agreement with previous reports suggesting that H-NS regulates T6SSs in various bacteria (Bernard et al., 2010; Eijkelkamp et al., 2013), which could be expected given recent observations that H-NS negatively regulates horizontally acquired genes (Lucchini et al., 2006) such as T6SS gene clusters.
Acknowledgements
K. O., D. S. and J. A. K. are supported by grants from NIH-Allergy and Infectious Disease (R01-AI056404) and the Welch Foundation (I-1561). D. S. is a Chilton Foundation Fellow. K. O. is a Burroughs Wellcome Investigator in Pathogenesis of Infectious Disease, a W.W. Caruth Jr. Biomedical Scholar and has an Earl A. Forsythe Chair in Biomedical Science.
Abbreviations:
- H-NS
histone-like nucleoid structuring
- MLB
marine Luria–Bertani
- T3SS
type III secretion system
- T6SS
type VI secretion system
Footnotes
Three supplementary figures and two supplementary tables are available with the online version of this paper.
References
- Atlung T., Ingmer H. (1997). H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol 24, 7–17. 10.1046/j.1365-2958.1997.3151679.x [DOI] [PubMed] [Google Scholar]
- Basler M., Pilhofer M., Henderson G. P., Jensen G. J., Mekalanos J. J. (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. 10.1038/nature10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C. S., Brunet Y. R., Gueguen E., Cascales E. (2010). Nooks and crannies in type VI secretion regulation. J Bacteriol 192, 3850–3860. 10.1128/JB.00370-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F., Fichant G., Berthod J., Vandenbrouck Y., Attree I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10, 104. 10.1186/1471-2164-10-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet Y. R., Hénin J., Celia H., Cascales E. (2014). Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep 15, 315–321. 10.1002/embr.201337936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagon R. G. (1962). Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes. J Bacteriol 83, 736–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelkamp B. A., Stroeher U. H., Hassan K. A., Elbourne L. D., Paulsen I. T., Brown M. H. (2013). H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect Immun 81, 2574–2583. 10.1128/IAI.00065-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz C. J., McCarter L. L. (2011). Quorum sensing and silencing in Vibrio parahaemolyticus. J Bacteriol 193, 4224–4237. 10.1128/JB.00432-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz C. J., Chodur D. M., McCarter L. L. (2010). Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus. J Bacteriol 192, 6025–6038. 10.1128/JB.00654-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz C. J., Kustusch R. J., Breheny P. J., Weiss D. S., McCarter L. L. (2011). Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol 79, 240–263. 10.1111/j.1365-2958.2010.07445.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachani A., Lossi N. S., Hamilton A., Jones C., Bleves S., Albesa-Jové D., Filloux A. (2011). Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J Biol Chem 286, 12317–12327. 10.1074/jbc.M110.193045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B. T., Basler M., Mekalanos J. J. (2013). Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science 342, 250–253. 10.1126/science.1243745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B. T., Dong T. G., Mekalanos J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21. 10.1016/j.chom.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockett K. L., Burch A. Y., Lindow S. E. (2013). Thermo-regulation of genes mediating motility and plant interactions in Pseudomonas syringae. PLoS ONE 8, e59850. 10.1371/journal.pone.0059850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurme R., Rhen M. (1998). Temperature sensing in bacterial gene regulation–what it all boils down to. Mol Microbiol 30, 1–6. 10.1046/j.1365-2958.1998.01049.x [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Sabharwal D., Bröms J., Milton D. L., Sjöstedt A., Uhlin B. E., Wai S. N. (2012). Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun 80, 575–584. 10.1128/IAI.05510-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T., Yamazaki C., Park K. S., Akeda Y., Iida T., Honda T. (2010). Transcription of Vibrio parahaemolyticus T3SS1 genes is regulated by a dual regulation system consisting of the ExsACDE regulatory cascade and H-NS. FEMS Microbiol Lett 311, 10–17. 10.1111/j.1574-6968.2010.02066.x [DOI] [PubMed] [Google Scholar]
- Lin Z., Kumagai K., Baba K., Mekalanos J. J., Nishibuchi M. (1993). Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol 175, 3844–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S., Rowley G., Goldberg M. D., Hurd D., Harrison M., Hinton J. C. (2006). H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog 2, e81. 10.1371/journal.ppat.0020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Zhang Y., Yan X., Guo L., Wang L., Qiu J., Yang R., Zhou D. (2012). Expression of the type VI secretion system 1 component Hcp1 is indirectly repressed by OpaR in Vibrio parahaemolyticus. ScientificWorldJournal 2012, 1–7. 10.1100/2012/982140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. L. (1998). OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J Bacteriol 180, 3166–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A., Kendall M., Vugia D. J., Henao O. L., Mahon B. E. (2012). Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin Infect Dis 54 (Suppl 5), S391–S395. 10.1093/cid/cis243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K. S., Ono T., Rokuda M., Jang M. H., Okada K., Iida T., Honda T. (2004). Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun 72, 6659–6665. 10.1128/IAI.72.11.6659-6665.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper R., Huang S. T., Robinson J. M., Clark D. J., Alami H., Parmar P. P., Perry R. D., Fleischmann R. D., Peterson S. N. (2009). Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology 155, 498–512. 10.1099/mic.0.022160-0 [DOI] [PubMed] [Google Scholar]
- Porter M. E., Dorman C. J. (1994). A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J Bacteriol 176, 4187–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. B., Peterson S. B., Mougous J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12, 137–148. 10.1038/nrmicro3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D., Gonzalez H., Updegraff B. L., Orth K. (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE 8, e61086. 10.1371/journal.pone.0061086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D., Kinch L. N., Trudgian D. C., Guo X., Klimko J. A., Grishin N. V., Mirzaei H., Orth K. (2014). Marker for type VI secretion system effectors. Proc Natl Acad Sci U S A 111, 9271–9276. 10.1073/pnas.1406110111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana T. G., Hachani A., Bucior I., Soscia C., Garvis S., Termine E., Engel J., Filloux A., Bleves S. (2012). The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem 287, 27095–27105. 10.1074/jbc.M112.376368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana T. G., Soscia C., Tonglet C. M., Garvis S., Bleves S. (2013). Divergent control of two type VI secretion systems by RpoN in Pseudomonas aeruginosa. PLoS ONE 8, e76030. 10.1371/journal.pone.0076030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S., West T. E., Boyer F., Chiang W. C., Carl M. A., Hood R. D., Rohmer L., Tolker-Nielsen T., Skerrett S. J., Mougous J. D. (2010). Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6, e1001068. 10.1371/journal.ppat.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneider M. M., Buth S. A., Ho B. T., Basler M., Mekalanos J. J., Leiman P. G. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353. 10.1038/nature12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J. M., Austin L. S., Hsu F., Hicks K. G., Hood R. D., Mougous J. D. (2011). Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol 82, 1277–1290. 10.1111/j.1365-2958.2011.07889.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart B. J., McCarter L. L. (2003). Lateral flagellar gene system of Vibrio parahaemolyticus. J Bacteriol 185, 4508–4518. 10.1128/JB.185.15.4508-4518.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L., Nunan L., Redman R. M., Mohney L. L., Pantoja C. R., Fitzsimmons K., Lightner D. V. (2013). Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis Aquat Organ 105, 45–55. 10.3354/dao02621 [DOI] [PubMed] [Google Scholar]
- Wang L., Ling Y., Jiang H., Qiu Y., Qiu J., Chen H., Yang R., Zhou D. (2013a). AphA is required for biofilm formation, motility, and virulence in pandemic Vibrio parahaemolyticus. Int J Food Microbiol 160, 245–251. 10.1016/j.ijfoodmicro.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhou D., Mao P., Zhang Y., Hou J., Hu Y., Li J., Hou S., Yang R. & other authors (2013b). Cell density- and quorum sensing-dependent expression of type VI secretion system 2 in Vibrio parahaemolyticus. PLoS ONE 8, e73363. 10.1371/journal.pone.0073363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker W. B., Parent M. A., Boyd A., Richards G. P., Boyd E. F. (2012). The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect Immun 80, 1834–1845. 10.1128/IAI.06284-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Orth K. (2013). Virulence determinants for Vibrio parahaemolyticus infection. Curr Opin Microbiol 16, 70–77. 10.1016/j.mib.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Zhang W., Xu S., Li J., Shen X., Wang Y., Yuan Z. (2011). Modulation of a thermoregulated type VI secretion system by AHL-dependent quorum sensing in Yersinia pseudotuberculosis. Arch Microbiol 193, 351–363. [DOI] [PubMed] [Google Scholar]
- Zheng J., Shin O. S., Cameron D. E., Mekalanos J. J. (2010). Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci U S A 107, 21128–21133. 10.1073/pnas.1014998107 [DOI] [PMC free article] [PubMed] [Google Scholar]


