Abstract
African horse sickness (AHS) is an equine disease with a mortality of up to 90% for susceptible horses. The causative agent AHS virus (AHSV) is transmitted by species of Culicoides. AHSV serogroup within the genus Orbivirus of the Reoviridae family consists of nine serotypes that show no or very limited cross-neutralization. Of the seven structural proteins (VP1-VP7) of AHSV, VP2 is the serotype specific protein, and the major target for neutralizing antibodies. In this report, recombinant VP2 proteins of all nine serotypes were expressed individually by the baculovirus expression system and the immunogenicity of each was studied by immunization of guinea pigs with single VP2 as well as with cocktails of VP2 proteins. Homologous neutralizing antibodies measured by 50% plaque reduction assay showed varying degrees (from 37 to 1365) of titers for different VP2 proteins. A low cross-neutralizing antibody titer was found for genetically related AHSV serotypes. Immunization with VP2 cocktails containing equal amounts of each of the VP2 proteins also triggered neutralizing antibodies albeit to lower titers (4-117) to each of the serotypes in the cocktail. This study is a first step to develop a VP2 subunit vaccine for AHS and our results indicate that VP2 subunit vaccines are feasible individually or in a multi-serotype cocktail.
Keywords: African horse sickness, Recombinant protein, Capsid protein VP2, Subunit vaccine
1. Introduction
African horse sickness virus (AHSV) is the causative agent of African horse sickness (AHS) which is lethal for up to 90% of infected domestic horses [1]. AHSV infections of zebras and donkeys are less severe and mostly cause mild clinical symptoms or an asymptomatic infection. These equids are carriers of AHSV, which is transmitted by Culicoides midges, in particular by C. imicola in endemic areas [1], [2]. It is believed that the distribution of AHSV is associated with the presence of these competent vectors. Currently, AHSV is endemic in tropical and sub-Saharan Africa, but sporadic cases and short-term epidemics in North Africa and Middle-East have been reported in the mid-20th century. In 1987, an outbreak of AHSV-4 on the Iberian Peninsula, which was extended for a few years in Spain and spread to Portugal and Morocco indicating that AHSV had overwintered and spread by European Culicoides midges [1], [3].
The serogroup AHSV within the genus Orbivirus of the Reoviridae family consists of nine serotypes (AHSV-1 – AHSV-9). The virus particle contains ten genome segments of double-stranded RNA (dsRNA) encoding seven structural proteins (VP1-VP7). Additionally, at least three non-structural proteins (NS1-NS3) are synthesized in virus infected cells. The virus particle consists of three distinct protein layers, of which the VP2 and VP5 proteins form the outer shell and are the most variable proteins of AHSV. Dominant antigenic sites inducing serotype specific neutralizing antibodies (nAbs) are mainly located on VP2, however, other structural and non-structural proteins – VP3, VP5, VP7, NS1 and NS2 – also induce humoral and cellular immune responses [4], [5], [6], [7], [8], [9].
Since there is no successful treatment for AHS, vaccination is the most important approach to protect horses against AHS. Live-attenuated vaccines (LAVs) obtained by serial passages of AHSV in cell culture are available commercially for most serotypes in South Africa [1]. Although LAVs have been extensively used in South Africa and other African countries, there are still concerns as LAVs cause viremia and could be transmitted by midges. However, the biggest concern of using these vaccines is reassortment between LAVs or with wild type AHSV, which could result in more pathogenic virus variants. Moreover, the recent outbreak of AHSV serotype 9 in Gambia is suspected to be derived from vaccine strains [10]. Currently, LAVs are not licensed in Europe. To overcome safety issues, alternative AHS vaccines are under development including inactivated virus, recombinant VP2, DNA vaccine and vaccinia virus vectors expressing VP2 protein [11], [12], [13], [14], [15], [16], [17], [18], [19].
Outer capsid protein VP2 of orbiviruses determines the serotype and is the main target of nAbs [20], [21], [22], [23]. Vaccination with recombinant VP2 of AHSV serotype 4, 5 or 9 has been reported to induce nAbs and protect horses against homologous AHSV challenge infection [13], [14], [16], [18], [19], [22], [24]. To date, there are no reports regarding the immunogenicity of VP2 proteins of other serotypes of AHSV. In this report, VP2 of all nine AHSV serotypes were produced individually using the baculovirus expression system and their immunogenic activities were investigated by immunization of guinea pigs, singly or in cocktail mixtures. The results demonstrated that recombinant VP2 proteins of all nine AHSV serotypes have the potential to be used as safe subunit vaccines for AHS either individually or in a multi-serotype cocktail.
2. Materials and methods
2.1. Viruses and cells
AHSV reference strains (obtained from ANSES, France) were passaged and amplified in BSR cells, a derivative of the BHK-21 cell line, in Dulbecco's modified Eagle's medium (DMEM) (Sigma) supplemented with 10% fetal bovine serum (Invitrogen). Virus titers were determined by a plaque-forming assay in BSR cells and defined as plaque forming units per ml (pfu/ml) as described [25]. Insect cell lines of Spodoptera frugiperda, Sf9 and Sf21, were cultured at 28 °C in Insect-Xpress (Lonza, Basel, Switzerland) and TC100 medium (Biochrom AG, Berlin, Germany), respectively. TC100 medium was supplemented with 10% fetal bovine serum.
2.2. Preparation of recombinant AHSV VP2 protein
Genome segment 2 of AHSV serotypes 2, 4, 5 and 6, encoding VP2 were obtained from viral dsRNA according to the previously described method [26]. For AHSV serotypes 1, 3, 7, 8 and 9, open reading frames based on amino acid sequences of VP2 proteins (GenBank accession number: CAP04841; U01832; AAN74570; ABI96883, respectively), were designed for optimized expression in insect cells (Gene Art, Regensburg, Germany). VP2 genes were amplified by PCR with specific primers containing BamHI or SmaI site for cloning purposes into the transfer vector pAcYM1 [27].
Recombinant vectors pAcYM1 with VP2 genes were purified and co-transfected into Sf9 cells with linearized baculovirus DNA (strain BAC10:KO1629), using Cellfectin® II Reagent (Invitrogen) according to the manufacturer's instruction. On day six after transfection, 200 μl of the supernatants were transferred to fresh Sf9 cells in 12-wells plates. After the first passage, supernatants were transferred to fresh Sf9 cells every 3–5 days until virus infection was confirmed by light microscopy. The virus titer was measured by standard plaque assay using Sf21 cells.
2.3. Preparation of soluble VP2 protein
Recombinant baculoviruses expressing AHSV VP2 were used to infect Sf9 cells with a multiplicity of infection (moi) of 5. Infected cells were incubated at 28 °C for 72 h. Then, infected cells were harvested by centrifugation, washed with phosphate buffered saline (PBS) and pelleted by centrifugation. Cell pellets were suspended in 25 mM sodium bicarbonate (NaHCO3, pH 8.39) at 1.0 × 107 cells/ml. Cells were disrupted by dounce homogenization and after centrifugation at 6000 rpm for 3 min, supernatants containing soluble VP2 protein were collected. To examine the amount of VP2 proteins, soluble VP2 were mixed with equal volumes of SDS-PAGE sample buffer (10 mM Tris-HCl, pH 6.8, 2% (w/v) SDS, 2% β-mercaptoethanol, 20% glycerol, 0.05% bromophenol blue). After heating at 95 °C for 1 min, the samples were analyzed by SDS-PAGE with BSA as concentration standard and protein molecular weight standard (Page Ruler, SM0671, Fermentas). Concentrations of all samples were adjusted to 100 μg of VP2 per ml by 25 mM sodium bicarbonate and stored at −80 ° C until use.
2.4. Immunization of soluble VP2 in guinea pigs
All experiments with live animals were performed under the guidelines of the European Community (86/609) and were approved by the Committee on the Ethics of Animal Experiments of the Central Veterinary Institute (Permit numbers: 2011-042 and 2011-170). Adult female guinea pigs were purchased from a registered breeding farm for guinea pigs and were randomly divided into groups of six animals. Nine groups were immunized with VP2 protein from each AHSV serotype, two groups were immunized with cocktails of different combinations of VP2 proteins (one consisting of serotypes 1, 3, 7, 8 and other, serotypes of 2, 4, 5, 6, 9, respectively) and one group was immunized with phosphate buffered saline (PBS). Shortly before immunization, recombinant VP2 proteins or PBS in 1.5 ml were warmed to 37 °C and mixed with an equal volume of Montanide 206VG (Seppic) by vortexing. Each animal received 400 μl containing adjuvanted 50 μg of VP2 protein by subcutaneous injection of 100 μl at four places left and right from the spinal cord. Cocktails contained equal amounts of each VP2; 12.5 μg of each VP2 (1, 3, 7 and 8) or 10 μg of each VP2 (2, 4, 5, 6 and 9). At day 21, all guinea pigs were boosted by the same procedure and with the same amount of proteins. At day 42 of the experiment, animals were sacrificed and sera were collected.
2.5. Detection of neutralizing antibodies (nAbs)
Guinea pig sera collected at the end of the experiment, day 42, were examined for nAbs by plaque reduction based standard neutralizing assay [21]. Briefly, serially 2-fold diluted sera in DMEM were mixed with an equal volume of each AHSV reference strain virus (20–40 pfu/25 μl) and incubated at 37 °C for 60 min in a 5% CO2 incubator. As a control, each virus was mixed with an equal volume of DMEM without any serum. After incubation, 50 μl neutralized viruses were used to infect BSR monolayers in a 12-well plate. After absorption of virus for 1 h at 37 °C, cells were overlaid with DMEM- 1% low-melting agarose gel, followed by incubation at 37 °C for 2–4 days until plaques were visible. The neutralization titers were calculated by the reciprocal value of the maximum dilution, at which the number of plaques showed 50% reduction compared with the serum-free control. The neutralizing tests were performed in duplicate. The average and 95% confidence interval was calculated in each group.
2.6. Immunostaining of AHSV-infected monolayers
Equal volumes of sera from guinea pigs of each group collected at the end of the experiment were pooled and examined for AHSV specific antibodies (Abs) by immunoperoxydase monolayer assay (IPMA). Pooled sera collected prior to immunization (day 0) were used as negative control serum. In brief, BSR monolayers were infected at low multiplicity of infection with each of the reference strains representing all nine AHSV serotypes, respectively. At the beginning of cytopathic effect (CPE), medium was removed and monolayers were washed with PBS, and fixed with methanol/acetone (1:1) according to standard procedures. Monolayers were stained by IPMA with sera diluted 1:500, followed by incubation with conjugated α-guinea pig rabbit serum (DAKO) and stained according to standard procedures [28].
2.7. Phylogenetic tree
Phylogenetic trees of the AHSV VP2 deduced amino acid sequences were constructed using 39 sequences of AHSV VP2 obtained from GenBank by the neighbor-joining method using MEGA 4.1 software.
3. Results
3.1. Expression of recombinant AHSV VP2 of 9 serotypes
Recombinant VP2 proteins of nine AHSV serotypes were expressed in Sf9 cells using the baculovirus expression system with VP2 genes under the control of the polyhedron promoter. Higher expression of VP2 was obtained with codon optimized VP2 genes for serotypes 1, 3, 7, 8 and 9 than with the original VP2 sequences for serotype 2, 4, 5 and 6 (Fig. 1). The differences in VP2 expression were less obvious in Sf21 cells as shown in our previous study [29]. Soluble VP2 protein of each serotype was harvested at 72 h post-infection. Recombinant baculovirus-infected cells were collected by centrifugation and lysed by homogenization in 25 mM sodium bicarbonate. After centrifugation, the clarified supernatant was used for immunization of guinea pigs. Concentrations of VP2 protein were estimated at 100 μg/ml by comparing with bovine serum albumin (BSA) standard on a Coomassie Brilliant Blue stained SDS-PAGE gel.
Fig. 1.
Expression of recombinant VP2 proteins of nine AHSV serotypes in Sf9 cells: Proteins were separated on a SDS-PAGE gel and stained by Coomassie Brilliant Blue. VP2 proteins (121–124 kDa) are indicated by arrow heads.
3.2. Immunogenicity of AHSV VP2 proteins in guinea pigs
Guinea pigs were immunized twice with 50 μg of VP2 protein after mixing with an equal volume of Montanide 206VG according to a prime-boost protocol with an interval of 3 weeks. At day 42 of the experiment, sera were collected and tested for the neutralization activities as described in Section 2. Immunization with a single VP2 protein induced serotype specific nAbs (Table 1). Despite the same amount of recombinant VP2 proteins being used in each group and in each guinea pig, serotype specific nAb titers strongly varied between groups, and also between animals within the same group. For example, nAb titers ranged from 37 (95% confidence interval (CI): 27–48) for AHSV-2 to as high as 1365 (95% CI: 942–1788) for AHSV-6.
Table 1.
Titers of neutralizing antibodies (nAbs) raised in guinea pigs.
| Guinea pig groups (n = 6) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 1, 3, 7, 8 | 2, 4, 5, 6, 9 | Control | |
| AHSV 1 | 112 | 11 | ||||||||||
| (48–176) | (2–19) | |||||||||||
| AHSV 2 | 37 | 10 | ||||||||||
| (27–48) | (1–19) | |||||||||||
| AHSV 3 | 341 | 19 | ||||||||||
| (236–447) | (11–28) | |||||||||||
| AHSV 4 | 171 | 12 | ||||||||||
| (118–224) | (9–16) | |||||||||||
| AHSV 5 | 179 | 53 | ||||||||||
| (14–345) | (40–67) | |||||||||||
| AHSV 6 | 1365 | 12 | 117 | |||||||||
| (942–1788) | (3–21) | (58–177) | ||||||||||
| AHSV 7 | 8 | 122 | 20 | |||||||||
| (3–13) | (46–197) | (12–28) | ||||||||||
| AHSV 8 | 3 | 96 | 4 | 2 | ||||||||
| (1 –4) | (10–87) | (2–7) | (0–4) | |||||||||
| AHSV 9 | 24 | 853 | 19 | |||||||||
| (17–31) | (642–1065) | (10–27) | ||||||||||
Single VP2 proteins (1–9) or with cocktails of 4 or 5 different VP2 proteins (1, 3, 7, 8 or 2, 4, 5, 6, 9, respectively) were used to immunize six guinea pigs per group. Titers of nAbs against each of the AHSV serotypes (AHSV-1–AHSV-9) were determined. The average nAb titers were calculated and the 95% confidence interval is indicated between brackets. Titers of nAbs <2 were evaluated as 0.
Two groups of animals injected with the cocktails containing VP2 proteins of serotypes 1, 3, 7 and 8, and of 2, 4, 5, 6 and 9, respectively, had nAb titers to the included AHSV serotypes (Table 1). However, nAb titers against each of the AHSV serotypes were consistently lower than those after immunization with single VP2 proteins. It is possible that this is due to the 4–5 times lower dose per VP2 protein in the cocktails compared to the single VP2 immunization; i.e. 50 μg of VP2 proteins were inoculated for single VP2 vaccination, whereas 10 μg per VP2 in the pentavalent cocktail and 12.5 μg per VP2 in the quadrivalent cocktail was injected. The lower nAb titer by cocktails compared to single VP2 protein varied from 3 to 4 times lower for serotype 5 to ∼45 times lower for serotype 9 after immunization with these VP2 proteins in cocktails (Table 1). Importantly, some cross-neutralization was also detected for a few genetically related serotypes (Fig. 1) [30]. For example, α-AHSV-3 VP2 serum for serotype 7, α-AHSV-5 VP2 serum for serotype 8, and α-AHSV-6 and α-AHSV-9 VP2 serum both showed nAbs titers for the genetically related serotype: i.e. serotype 9, and 6, respectively (Table 1). However, these cross-reactive nAb titers are >40 times lower than the nAb titer against the respective homologous serotypes used for immunization. Further, no significant nAb titers against genetically unrelated serotypes were found. Immunization with VP2 cocktails did not result in significant nAbs titers against genetically unrelated serotypes, and only a very low nAb titer against a related serotype (α-AHSV-5 VP2 serum for serotype 8) could be detected (Table 1).
3.3. Serotype specificity of guinea pig sera raised against recombinant VP2 proteins
Titers of nAbs raised against different VP2 proteins demonstrated a high level of serotype specificity. Non-neutralizing Abs still could cross-react between serotypes by binding to common epitopes. Therefore, serotype specificity was further studied by immunoperoxydase monolayer assay (IPMA). Pooled sera per group were 500-fold diluted and used in IPMA to immunostain BSR monolayers infected with each of the nine reference AHSV strains. As expected, guinea pig sera raised against single VP2 proteins immunostained monolayers infected with the homologous AHSV serotype (Table 2). Similar to cross-neutralization of genetically related AHSV serotypes, some monolayers infected with genetically related AHSV serotypes were also immunostained. In contrast to the cross-neutralization results (Table 1), AHSV-6 was not recognized by α-AHSV-9 VP2 serum (Table 2). In addition to immunostaining of genetically related AHSV serotypes, some unrelated AHSV reference strains were also recognized in IPMA; e.g. AHSV-8 was recognized not only by α-VP2 sera of AHSV-5 and -8 but also by AHSV-4. AHSV-5 was also recognized by α-VP2 of AHSV-3. In general this immunostaining was weaker than for the respective homologous AHSV serotype (Table 2).
Table 2.
IPMA results with pooled sera raised in guinea pigs.
| Guinea pig groups (n = 6) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 1, 3, 7, 8 | 2, 4, 5, 6, 9 | Control | |
| AHSV 1 | +++ | +++ | ||||||||||
| AHSV 2 | +++ | +++ | ||||||||||
| AHSV 3 | +++ | +++ | ||||||||||
| AHSV 4 | +++ | +++ | ++ | |||||||||
| AHSV 5 | ++ | +++ | +++ | ++ | +++ | |||||||
| AHSV 6 | +++ | +++ | ||||||||||
| AHSV 7 | ++ | +++ | +++ | |||||||||
| AHSV 8 | ++ | +++ | ++ | ++ | ++ | |||||||
| AHSV 9 | ++ | +++ | + | |||||||||
Single VP2 proteins (1–9) or with cocktails of 4 or 5 different VP2 proteins (1, 3, 7, 8 or 2, 4, 5, 6, 9, respectively) were used to immunize six guinea pigs per group. BSR monolayers were infected with each of the AHSV serotypes (AHSV1–AHSV9), and immunostained by binding of guinea pig antibodies followed by binding of conjugated α-guinea pigs rabbit serum. Staining was semi-quantitated by +++, ++, + for strong, medium, and weak staining, respectively.
4. Discussion
VP2 protein of orbiviruses is the major determinant of eliciting nAbs and has been used as recombinant protein-based vaccine in previous studies [17], [21], [22], [23], [31]. Particularly, VP2 of AHSV serotype 4 has been studied extensively by European research groups, as the last European AHS outbreak was caused by this serotype [32]. In this report we studied the immunogenicity of VP2 proteins of all nine AHSV serotypes as a first step in the development of AHS subunit vaccines. This is the first report to show that VP2 of all nine AHSV serotypes induce serotype specific nAbs with slight cross-neutralizing antibodies. The baculovirus expression system was used to produce recombinant VP2 protein of all nine serotypes for induction of nAbs. Further, some VP2 genes were optimized to increase protein expression. Still, quantities of soluble VP2 significantly varied between the different serotypes. Since it is generally known that recombinant VP2 protein of orbivirus is highly insoluble, it is likely that quantities of soluble VP2 proteins vary by differences in expression or solubility [33].
VP2 proteins of each AHSV serotype were produced in insect cells and each induced detectable nAb titers in guinea pigs as an alternative animal model. Previously, purified AHSV VP2 seemed to be less immunogenic in rabbits [21], but as little as 5 μg of VP2 protein in insect cell lysate could protect horses from AHS by induction of nAbs [14]. In this study, guinea pigs were immunized with insect cell lysate containing 50 μg of VP2 to elicit detectable antibodies. Each VP2 elicited serotype specific Abs, but nAb titers varied considerably among different AHSV serotypes, from 37 for AHSV-2 to 1365 for AHSV-6. Further, cross-neutralization antibodies between genetically related serotypes were detected, but most of those cross-neutralizing Abs titers were considerably lower than for the respective serotype. Moreover, some expected cross-reactive nAbs were not detected. This lack of expected cross-reactive nAbs are most likely due to low titer of cross-reactive nAbs compared with serotype specific nAbs. Unfortunately challenge experiments could not be performed in guinea pigs, as horses are the natural host for AHSV. The AHSV infection model using interferon-α knockout mice were recently reported [17]. The use of the small animal model for our future VP2 vaccine study should help to evaluate the vaccine efficacy.
Cross-reactive Abs to genetically related AHSV serotypes were shown by IPMA with lower Ab titers than serotype specific reactions, except for AHSV-5 and AHSV-8, in which α-AHSV-5 VP2 serum reacted strongly to both AHSV-5 and AHSV-8, and vice versa. Interestingly, no cross neutralization Abs between AHSV-5 and AHSV-8 were detected. It would be thought that more antibodies to non-neutralizing than to neutralizing domains of AHSV-5 and AHSV-8 VP2 were elicited. These variations in the feasibility of eliciting non-neutralizing Abs and nAbs between serotypes could contribute the considerable differences in the nAb titers. Although the crystal structure of AHSV VP2 has not been solved, neutralizing domains on the secondary structure containing amino acid 199–689 of VP2 were demonstrated [34]. To avoid eliciting non-neutralizing Abs, expression and immunization of only neutralization domain of VP2 may help to induce nAbs more efficiently. In contrast to AHSV-5 and -8, VP2 of AHSV serotype 9 induced nAbs against serotype 6 (nAb titer of 12 with 95% CI: 3–21) which was not detectable by IPMA, suggesting that the non-nAb is not necessarily higher than nAb. This phenomenon is probably due to the structural similarity and dissimilarity between VP2s of relevant serotypes.
Here, we have also studied two cocktails of four or five VP2 proteins. The results suggested a dose-dependent immune response, since all serotype specific nAb titers were lower after immunization with cocktails of VP2 proteins (10/12.5 μg of each VP2 per animal) than those with individual VP2 immunization (50 μg of VP2 per animal). However, this reduction was not linearly related to the amount of injected VP2. The reduction of 4–5 fold VP2 protein in cocktails resulted in 4 to 40 fold reduced nAb titers compared to single VP2 immunization; e.g. for serotype 5, 179 by single and 53 by cocktail VP2 (±30% difference), and for serotype 9, 853 by single and 19 by cocktail VP2 (±2% difference). This might suggest a negative interference between some of the VP2 proteins in cocktails to induce nAbs. The lower serotype specific nAb titer after immunization with cocktails of VP2 proteins could also be due to the simultaneous presentation of various serotype specific epitopes to the immune system or due to the immunodominance of certain serotype specific epitopes. Thus, formulation of VP2 cocktails to protect horses against all included serotypes is also complicated by differences in immunogenicity and possible interference between VP2 proteins to induce humoral immune responses.
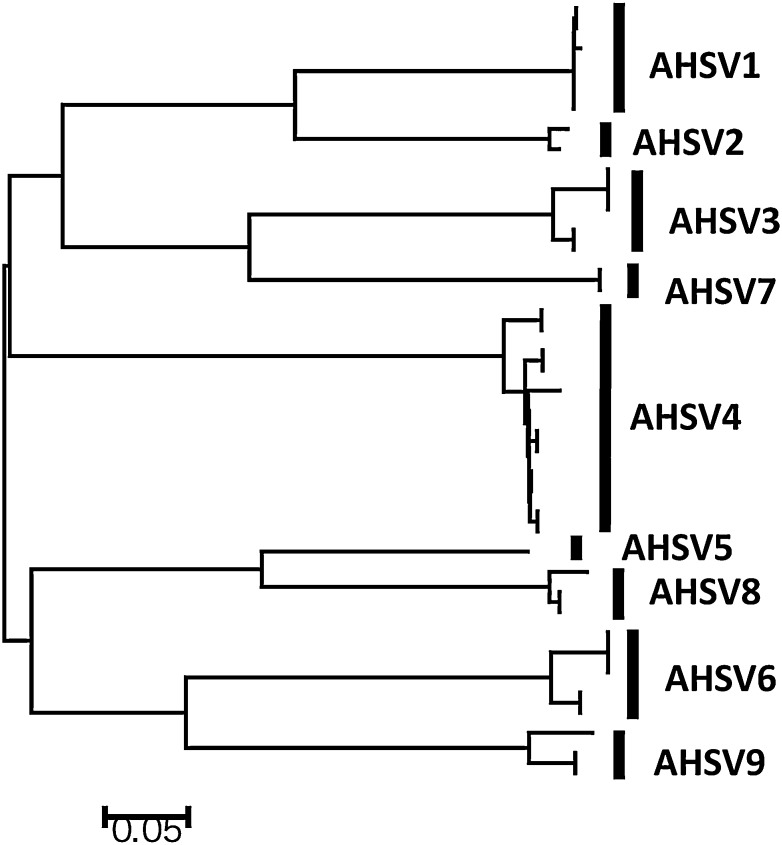
We intended to induce humoral responses against closely related and possibly more serotypes than included in the cocktail by a prime-boost approach [35]. However, genetically related VP2 proteins 3 and 7, or 5 and 8, (Fig. 2) in each of the cocktails did not increase nAbs titers against their related serotypes. No nAbs were detected against unrelated serotypes (Table 1). Further, nAb titers against each VP2 protein differed strongly after immunization with a cocktail or with single VP2 protein. Non-neutralizing Abs were raised by cocktails of VP2 proteins; i.e. Abs against serotype 4, 5 and 9 by the cocktail of 1, 3, 7, 8, and Abs against serotype 8 by the cocktail of 2, 4, 5, 6, 9 (Table 2). Perhaps, AHSV serotypes have common epitopes on VP2 but these differ in avidity or affinity for these Abs. As a result, binding to epitopes occurs and will immunostain AHSV infected monolayers but this binding will not neutralize AHSV.
Fig. 2.
Phylogenetic analysis of AHSV VP2 protein: VP2 sequences from a total of 39 isolates of 9 serotypes (obtained from GenBank) were used to deduce amino acid residues in order to generate the phylogenetic tree by the neighbor-joining method using MEGA 4.1 software.
Currently used cocktails of live-attenuated vaccines (LAVs) induce a broader protection. Even LAV for serotype 5 and 9 are not included, and protection against AHSV-5 and -9 are achieved by serotype-related LAVs for serotype 8 and 6, respectively [36]. However, when using cocktails of LAVs it was also suggested that there are substantial differences in cross-reactivity between serotypes; e.g. cross-reactivity between AHSV-5 and -8 seems to be stronger than between AHSV-6 and -9 [37]. Importantly, undesirable events such as reversion to virulence and reassortment between LAVs or with field virus are highly likely. Furthermore, LAVs induce an immune response against all viral proteins and are therefore not ‘DIVA’ (differentiating infected from vaccinated animals) vaccines. In contrast, VP2 subunit vaccine induces Abs solely against VP2, and horses vaccinated with VP2 subunit vaccines should therefore be seronegative for VP7 antibodies. An AHSV infection results rapidly in seroconversion for VP7 antibody and VP7 is the target for several commercially available tests to detect AHSV infections. DIVA testing by these commercially available tests will be very supportive in combination with vaccination with VP2 subunit vaccine. Thus, rapid control of AHS outbreaks as well as confirming the virus-free status of animals for international movements irrespective of the vaccination status can be achieved with the current available and extensively validated VP7 ELISA.
In summary, we demonstrated that multi-serotype VP2 subunit vaccines for AHS are potentially feasible, as shown here by immunization of guinea pigs as an alternative animal model. The guinea pig model can be initially used for immunogenicity studies in order to reduce experiments in horses. The considerable difference in immunogenicity between VP2 proteins in guinea pigs has to be taken into account and should be investigated further prior to the formulation of single as well as cocktail VP2 subunit vaccines for African horse sickness.
Acknowledgements
This study was funded by the EU (FP7, ORBIVAC, number 245266) and Biotechnical and Biological Sciences Research Council (Grant number BB/K015168/1). We are grateful to the animal caretakers of the Central Veterinary Institute of Wageningen University for their assistance and handling of experiments with guinea pigs.
References
- 1.Mellor P.S., Hamblin C. African horse sickness. Vet Res. 2004;35(July–August (4)):445–466. doi: 10.1051/vetres:2004021. [DOI] [PubMed] [Google Scholar]
- 2.Bouayoune H., Touti J., el Hasnaoui H., Baylis M., Mellor P.S. The Culicoides vectors of African horse sickness virus in Morocco: distribution and epidemiological implications. Arch Virol Suppl. 1998;14:113–125. doi: 10.1007/978-3-7091-6823-3_12. [DOI] [PubMed] [Google Scholar]
- 3.Capela R., Purse B.V., Pena I., Wittman E.J., Margarita Y., Capela M. Spatial distribution of Culicoides species in Portugal in relation to the transmission of African horse sickness and bluetongue viruses. Med Vet Entomol. 2003;17(June (2)):165–177. doi: 10.1046/j.1365-2915.2003.00419.x. [DOI] [PubMed] [Google Scholar]
- 4.Huismans H., van der Walt N.T., Cloete M., Erasmus B.J. Isolation of a capsid protein of bluetongue virus that induces a protective immune response in sheep. Virology. 1987;157(March (1)):172–179. doi: 10.1016/0042-6822(87)90326-6. [DOI] [PubMed] [Google Scholar]
- 5.Roy P., Urakawa T., Van Dijk A.A., Erasmus B.J. Recombinant virus vaccine for bluetongue disease in sheep. J Virol. 1990;64(May (5)):1998–2003. doi: 10.1128/jvi.64.5.1998-2003.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wade-Evans A.M., Pullen L., Hamblin C., O‘Hara R., Burroughs J.N., Mertens P.P. African horsesickness virus VP7 sub-unit vaccine protects mice against a lethal, heterologous serotype challenge. J Gen Virol. 1997;78(July (Pt 7)):1611–1616. doi: 10.1099/0022-1317-78-7-1611. [DOI] [PubMed] [Google Scholar]
- 7.Jones L.D., Williams T., Bishop D., Roy P. Baculovirus-expressed nonstructural protein NS2 of bluetongue virus induces a cytotoxic T-cell response in mice which affords partial protection. Clin Diagn Lab Immunol. 1997;4(May (3)):297–301. doi: 10.1128/cdli.4.3.297-301.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrew M., Whiteley P., Janardhana V., Lobato Z., Gould A., Coupar B. Antigen specificity of the ovine cytotoxic T lymphocyte response to bluetongue virus. Vet Immunol Immunopathol. 1995;47(August (3–4)):311–322. doi: 10.1016/0165-2427(94)05410-t. [DOI] [PubMed] [Google Scholar]
- 9.Janardhana V., Andrew M.E., Lobato Z.I., Coupar B.E. The ovine cytotoxic T lymphocyte responses to bluetongue virus. Res Vet Sci. 1999;67(December (3)):213–221. doi: 10.1053/rvsc.1999.0306. [DOI] [PubMed] [Google Scholar]
- 10.Oura C.A., Ivens P.A., Bachanek-Bankowska K., Bin-Tarif A., Jallow D.B., Sailleau C. African horse sickness in the Gambia: circulation of a live-attenuated vaccine-derived strain. Epidemiol Infect. 2012;140(March (3)):462–465. doi: 10.1017/S095026881100080X. [DOI] [PubMed] [Google Scholar]
- 11.Ozawa Y., Bahrami S. African horse-sickness killed-virus tissue culture vaccine. Can J Comp Med Vet Sci. 1966;30(November (11)):311–314. [PMC free article] [PubMed] [Google Scholar]
- 12.House J.A., Lombard M., Dubourget P., House C., Mebus C.A. Further studies on the efficacy of an inactivated African horse sickness serotype 4 vaccine. Vaccine. 1994;12(February (2)):142–144. doi: 10.1016/0264-410x(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 13.Romito M., Du Plessis D.H., Viljoen G.J. Immune responses in a horse inoculated with the VP2 gene of African horsesickness virus. Onderstepoort J Vet Res. 1999;66(June (2)):139–144. [PubMed] [Google Scholar]
- 14.Roy P., Bishop D.H., Howard S., Aitchison H., Erasmus B. Recombinant baculovirus-synthesized African horsesickness virus (AHSV) outer-capsid protein VP2 provides protection against virulent AHSV challenge. J Gen Virol. 1996;77(September (Pt 9)):2053–2057. doi: 10.1099/0022-1317-77-9-2053. [DOI] [PubMed] [Google Scholar]
- 15.Guthrie A.J., Quan M., Lourens C.W., Audonnet J.C., Minke J.M., Yao J. Protective immunization of horses with a recombinant canarypox virus vectored vaccine co-expressing genes encoding the outer capsid proteins of African horse sickness virus. Vaccine. 2009;27(July (33)):4434–4438. doi: 10.1016/j.vaccine.2009.05.044. [DOI] [PubMed] [Google Scholar]
- 16.Chiam R., Sharp E., Maan S., Rao S., Mertens P., Blacklaws B. Induction of antibody responses to African horse sickness virus (AHSV) in ponies after vaccination with recombinant modified vaccinia Ankara (MVA) PloS one. 2009;4(6):e5997. doi: 10.1371/journal.pone.0005997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castillo-Olivares J., Calvo-Pinilla E., Casanova I., Bachanek-Bankowska K., Chiam R., Maan S. A modified Vaccinia Ankara Virus (MVA) vaccine expressing African Horse Sickness Virus (AHSV) VP2 protects against AHSV challenge in an IFNAR −/− Mouse Model. PloS one. 2011;6(1):e16503. doi: 10.1371/journal.pone.0016503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scanlen M., Paweska J.T., Verschoor J.A., van Dijk A.A. The protective efficacy of a recombinant VP2-based African horsesickness subunit vaccine candidate is determined by adjuvant. Vaccine. 2002;20(January (7–8)):1079–1088. doi: 10.1016/s0264-410x(01)00445-5. [DOI] [PubMed] [Google Scholar]
- 19.Alberca B., Bachanek-Bankowska K., Cabana M., Calvo-Pinilla E., Viaplana E., Frost L. Vaccination of horses with a recombinant modified vaccinia Ankara virus (MVA) expressing African horse sickness (AHS) virus major capsid protein VP2 provides complete clinical protection against challenge. Vaccine. 2014;32(June (29)):3670–3674. doi: 10.1016/j.vaccine.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burrage T.G., Trevejo R., Stone-Marschat M., Laegreid W.W. Neutralizing epitopes of African horsesickness virus serotype 4 are located on VP2. Virology. 1993;196(October (2)):799–803. doi: 10.1006/viro.1993.1537. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Torrecuadrada J.L., Iwata H., Venteo A., Casal I., Roy P. Expression and characterization of the two outer capsid proteins of African horsesickness virus: the role of VP2 in virus neutralization. Virology. 1994;202(July (1)):348–359. doi: 10.1006/viro.1994.1351. [DOI] [PubMed] [Google Scholar]
- 22.Stone-Marschat M.A., Moss S.R., Burrage T.G., Barber M.L., Roy P., Laegreid W.W. Immunization with VP2 is sufficient for protection against lethal challenge with African horsesickness virus Type 4. Virology. 1996;220(June (1)):219–222. doi: 10.1006/viro.1996.0304. [DOI] [PubMed] [Google Scholar]
- 23.Urakawa T., French T.J., Adachi Y., Fukusho A., LeBlois H., Flamand M. Synthesis of recombinant baculoviruses expressing the outer capsid protein VP2 of five BTV serotypes and the induction of neutralizing antibodies to homologous and heterologous BTV serotypes. Virus Res. 1994;31(February (2)):149–161. doi: 10.1016/0168-1702(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 24.du Plessis M., Cloete M., Aitchison H., Van Dijk A.A. Protein aggregation complicates the development of baculovirus-expressed African horsesickness virus serotype 5 VP2 subunit vaccines. Onderstepoort J Vet Res. 1998;65(December (4)):321–329. [PubMed] [Google Scholar]
- 25.Boyce M., Roy P. Recovery of infectious bluetongue virus from RNA. J Virol. 2007;81(March (5)):2179–2186. doi: 10.1128/JVI.01819-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maan S., Rao S., Maan N.S., Anthony S.J., Attoui H., Samuel A.R. Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J Virol Methods. 2007;143(August (2)):132–139. doi: 10.1016/j.jviromet.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 27.Matsuura Y., Possee R.D., Overton H.A., Bishop D.H. Baculovirus expression vectors: the requirements for high level expression of proteins, including glycoproteins. J Gen Virol. 1987;68(May (Pt 5)):1233–1250. doi: 10.1099/0022-1317-68-5-1233. [DOI] [PubMed] [Google Scholar]
- 28.Wensvoort G., Terpstra C., Boonstra J., Bloemraad M., Van Zaane D. Production of monoclonal antibodies against swine fever virus and their use in laboratory diagnosis. Vet Microbiol. 1986;12(July (2)):101–108. doi: 10.1016/0378-1135(86)90072-6. [DOI] [PubMed] [Google Scholar]
- 29.Kanai Y., Athmaram T.N., Stewart M., Roy P. Multiple large foreign protein expression by a single recombinant baculovirus: a system for production of multivalent vaccines. Protein Expr Purif. 2013;91(September (1)):77–84. doi: 10.1016/j.pep.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Coetzer J.A.W., Guthrie A.J. In: Infectious diseases of livestock. 2nd ed. Coetzer J.A.W., Tustin R.C., editors. Oxford University Press; Cape Town, Southern Africa: 2004. African horse sickness; pp. 1231–1246. [Google Scholar]
- 31.Stewart M., Dovas C.I., Chatzinasiou E., Athmaran T.N., Papanastassopoulou M., Papadopoulos O. Protective efficacy of Bluetongue virus-like and subvirus-like particles in sheep: presence of the serotype-specific VP2, independent of its geographic lineage, is essential for protection. Vaccine. 2012;30(March (12)):2131–2139. doi: 10.1016/j.vaccine.2012.01.042. [DOI] [PubMed] [Google Scholar]
- 32.Mellor P.S. African horse sickness: transmission and epidemiology. Vet Microbiol. 1986;12(July (2)):101–108. [PubMed] [Google Scholar]
- 33.Capocefalo A., Franceschi V., Mertens P.P., Castillo-Olivares J., Cavirani S., Di Lonardo E. Expression and secretion of Bluetongue virus serotype 8 (BTV-8)VP2 outer capsid protein by mammalian cells. J Virol Methods. 2010;169(November (2)):420–424. doi: 10.1016/j.jviromet.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Torrecuadrada J.L., Langeveld J.P., Meloen R.H., Casal J.I. Definition of neutralizing sites on African horse sickness virus serotype 4 VP2 at the level of peptides. J Gen Virol. 2001;82(October (Pt 10)):2415–2424. doi: 10.1099/0022-1317-82-10-2415. [DOI] [PubMed] [Google Scholar]
- 35.Potgieter A.C., Cloete M., Pretorius P.J., van Dijk A.A. A first full outer capsid protein sequence data-set in the Orbivirus genus (family Reoviridae): cloning, sequencing, expression and analysis of a complete set of full-length outer capsid VP2 genes of the nine African horsesickness virus serotypes. J Gen Virol. 2003;84(May (Pt 5)):1317–1326. doi: 10.1099/vir.0.18919-0. [DOI] [PubMed] [Google Scholar]
- 36.von Teichman B.F., Smit T.K. Evaluation of the pathogenicity of African Horsesickness (AHS) isolates in vaccinated animals. Vaccine. 2008;26(September (39)):5014–5021. doi: 10.1016/j.vaccine.2008.07.037. [DOI] [PubMed] [Google Scholar]
- 37.von Teichman B.F., Dungu B., Smit T.K. In vivo cross-protection to African horse sickness Serotypes 5 and 9 after vaccination with Serotypes 8 and 6. Vaccine. 2010;28(September (39)):6505–6517. doi: 10.1016/j.vaccine.2010.06.105. [DOI] [PubMed] [Google Scholar]