Abstract
In this issue of Blood, Parrilla Castellar et al present a large cohort of anaplastic lymphoma kinase (ALK)-negative anaplastic large cell lymphoma (ALCL) in which they try to understand the diagnostic and the prognostic role of DUSP22 and TP63 in ALK-negative ALCL.1
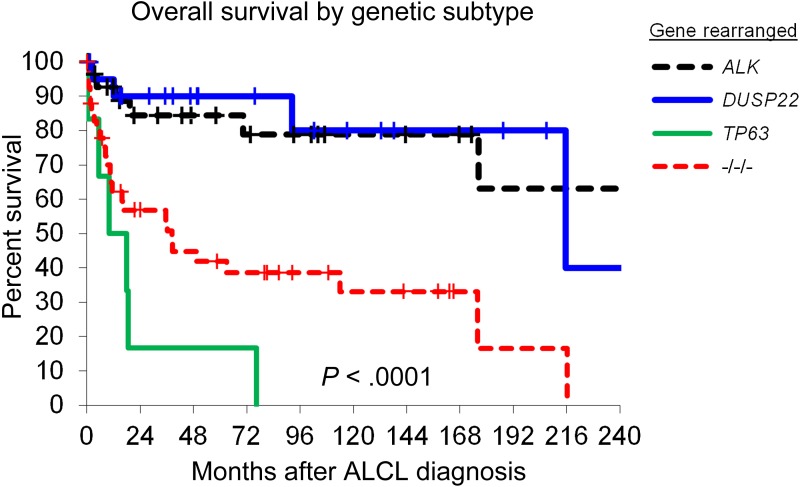
Overall survival in patients with ALCL, stratified by rearrangements of ALK, DUSP22, and TP63. −/−/−, triple-negative cases lacking all 3 rearrangements. See Figure 1B in the article by Parilla Castellar et al that begins on page 1473.
ALCL is a mature aggressive T-cell lymphoma. The definition of this disease has changed over the last decades. It was initially defined by the histomorphology of the lymphoma cells (anaplastic); the development of the CD30 antibody and its application in immunohistochemistry revealed that the morphological spectrum of the disease is much broader than anticipated.2 The discovery of ALK translocations and ALK expression in a subset of ALCL provided a very specific lymphoma marker. With the detection of ALK by immunohistochemistry, ALK-positive ALCLs, despite their morphological variability, became a precisely defined lymphoma entity.3 In fact lymphomas lacking virtually all histological criteria of ALCL like the lymphohistiocytic or small cell variant can be unambigiously identified as ALK-positive ALCL by detection of ALK expression, leading to a nomenclature that in some instances might appear contradictory (eg, anaplastic large cell lymphoma, small cell variant).
A large group of lymphomas morphologically resembling typical (common type) ALCL lack ALK rearrangements and expression. This group of ALK-negative ALCLs thus far consists of 2 subtypes. First is the primary cutaneous ALCL, which is defined by a combination of both histopathology (morphology and immunohistochemistry) and clinical presentation as a tumor primarily arising in the skin without clinical relevant dissemination. Second is ALK-negative ALCL, which is a primarily nodal lymphoma. The current principles of distinguishing ALK-negative ALCL from the rather large and heterogeneous group of peripheral T-cell lymphomas not otherwise specified (PTCL) is based primarily on histopathological features: strong and abundant expression of CD30, anaplastic lymphoma cells, hallmark cells and cohesively appearing (sheet-like), or intrasinusoidal growth. However, any of the features, including the expression of CD30, might also occur in PTCL. Thus, ALK-negative ALCL presents a challenge in lymphoma classification, and the current World Health Organization classification includes ALK-negative ALCL only as a provisional entity.4 In fact, there has been controversy of whether ALK-negative ALCL should be considered ALCL or a variant of PTCL. Over the last few years, molecular analysis of PTCL and ALK-negative ALCL identified new genetic lesions that seemed to occur primarily in ALK-negative ALCL and included chromosomal rearrangements of DUSP225 and TP63.6 These genetic markers have been suggested to better define and diagnose ALK-negative ALCL. However, several questions remained unsolved. Because there is a morphological gray zone between ALK-negative ALCL and PTCL, how “ALCL-like” are the lymphomas with chromosomal rearrangements of these genes? Are these lymphomas “real” ALCL or rather an indication of the molecular heterogeneity of PTCL? What is the clinical relevance of these translocations?
In a multicenter approach 73 ALK-negative ALCLs and 32 ALK-positive ALCLs were analyzed for morphology, immunophenotype, and chromosomal rearrangements of DUSP22 and TP63. The authors of the study conducted the histopathological review in a 2-step process, in which the pathologists evaluated the diagnosis first based on morphology and CD30 staining alone and a second step that considered the immunophenotype including ALK expression. During the review, the pathologists were blinded for the result of the genetic analysis (even for ALK status), as well as for the results of the other pathologists. All 3 genetic rearrangements (ALK, DUSP22, and TP63) were mutually exclusive, and a subgroup of lymphomas lacked all of them. Interestingly, DUSP22-rearranged lymphomas fulfilled the morphological criteria of ALCL to a very high degree as reflected by a high level of agreement among pathologists. TP63-rearranged and lymphomas lacking both rearrangements had less agreement in the blinded review. Taking the occasionally subjective nature of morphology assessment into consideration, the agreement among the 3 pathologists might be used to measure to which extent the lymphoma morphologically resembles ALCL. The data indicate that all genetic subtypes, but especially DUSP22 lymphomas, are ALCL according to our current definition. DUSP22-rearranged lymphomas might indeed present a prototype ALCL using purely morphological criteria, although other diagnostic features of ALCL such as expression of epithelial membrane antigen and cytotoxic granules are less frequent in this subtype. Nevertheless, the study supports the concept of the World Health Organization classification to consider ALK-negative ALCL as a separate entity distinct from PTCL. However, the selection criteria to include cases in the current study based on ALCL morphology might have influenced the result. PTCL was not included to confirm the absence of these genetic lesions in T-cell lymphomas other than ALK-negative ALCL.
In addition to the diagnostic value, the study also provided insight into the clinical relevance of genetic rearrangements in ALK-negative ALCL. The outcomes for ALCLs carrying rearrangements of DUSP22 and ALK-positive ALCLs were similarly good, whereas TP63-rearranged lymphomas and lymphomas without any of the 3 genetic aberrations did much worse (see figure). Thus, molecular heterogeneity of ALK-negative ALCL might influence clinical decisions when managing these patients. It is noteworthy to mention that DUSP22 rearrangements have been detected in cutaneous CD30-positive lymphoproliferative disease including lymphomatoid papulosis and cutaneous ALCL. However, all lymphomas analyzed in the study by Parrilla Castellar et al were reported to be nodal lymphomas, although clinical details, eg, regarding the presentation and skin involvement, were not given. Nevertheless, future studies will help in understanding the biological implications of this genetic similarity. Furthermore, functional studies will be needed to understand why very diverse genetic aberrations, eg, of ALK and DUSP22, occur in lymphomas that are morphologically and clinically so similar.
Footnotes
Conflict-of-interest disclosure: The author declares no competing financial interests.
REFERENCES
- 1.Parilla Castellar ER, Jaffe ES, Said JW, et al. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood. 2014;124(9):1473–1480. doi: 10.1182/blood-2014-04-571091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pileri S, Falini B, Delsol G, et al. Lymphohistiocytic T-cell lymphoma (anaplastic large cell lymphoma CD30+/Ki-1 + with a high content of reactive histiocytes). Histopathology. 1990;16(4):383–391. doi: 10.1111/j.1365-2559.1990.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 3.Pulford K, Lamant L, Morris SW, et al. Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)-ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood. 1997;89(4):1394–1404. [PubMed] [Google Scholar]
- 4.Swerdlow SH, Campo E, Harris N, et al. Lyon, France: IARC; 2008. WHO Classification of Tumors of the Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 5.Feldman AL, Dogan A, Smith DI, et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood. 2011;117(3):915–919. doi: 10.1182/blood-2010-08-303305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasmatzis G, Johnson SH, Knudson RA, et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood. 2012;120(11):2280–2289. doi: 10.1182/blood-2012-03-419937. [DOI] [PMC free article] [PubMed] [Google Scholar]