Abstract
Sewage surveillance in seven Italian cities between 2005 and 2008, after the introduction of inactivated poliovirus vaccination (IPV) in 2002, showed rare polioviruses, none that were wild-type or circulating vaccine-derived poliovirus (cVDPV), and many other enteroviruses among 1,392 samples analyzed. Two of five polioviruses (PV) detected were Sabin-like PV2 and three PV3, based on enzyme-linked immunosorbent assay (ELISA) and PCR results. Neurovirulence-related mutations were found in the 5′ noncoding region (5′NCR) of all strains and, for a PV2, also in VP1 region 143 (Ile > Thr). Intertypic recombination in the 3D region was detected in a second PV2 (Sabin 2/Sabin 1) and a PV3 (Sabin 3/Sabin 2). The low mutation rate in VP1 for all PVs suggests limited interhuman virus passages, consistent with efficient polio immunization in Italy. Nonetheless, these findings highlight the risk of wild or Sabin poliovirus reintroduction from abroad. Non-polio enteroviruses (NPEVs) were detected, 448 of which were coxsackievirus B (CVB) and 294 of which were echoviruses (Echo). Fifty-six NPEVs failing serological typing were characterized by sequencing the VP1 region (nucleotides [nt] 2628 to 2976). A total of 448 CVB and 294 Echo strains were identified; among those strains, CVB2, CVB5, and Echo 11 predominated. Environmental CVB5 and CVB2 strains from this study showed high sequence identity with GenBank global strains. The high similarity between environmental NPEVs and clinical strains from the same areas of Italy and the same periods indicates that environmental strains reflect the viruses circulating in the population and highlights the potential risk of inefficient wastewater treatments. This study confirmed that sewage surveillance can be more sensitive than acute flaccid paralysis (AFP) surveillance in monitoring silent poliovirus circulation in the population as well as the suitability of molecular approaches to enterovirus typing.
INTRODUCTION
In 1999, a mixed polio vaccination schedule consisting of two doses each of inactivated polio vaccines (IPV) and oral polio vaccines (OPV) was adopted in Italy, passing to a full IPV schedule in August 2002. Polio immunization is mandatory and free of charge, and the nationwide vaccination coverage with 3 IPV doses among children less than 2 years of age is 96.5% (1).
In line with the WHO eradication control requirements (2, 3, 4), a surveillance program of acute flaccid paralysis (AFP) coordinated by the Italian Ministry of Health (MoH) and the Istituto Superiore di Sanità (ISS) has been active since 1997 (5, 6, 7), and both epidemiological and virological data indicate that Italy is free of polio. The last poliomyelitis case due to indigenous wild virus occurred in 1982, and the last imported wild polio case was a poliomyelitis patient from Libya, hosted in Italy for specific medical treatment in 1988 (8). In addition, no vaccine-associated paralytic poliomyelitis (VAPP) case has been recorded since the IPV introduction.
During the last years, the WHO AFP indicators (9) confirmed high performance of the Italian nationwide surveillance but revealed weakness at the regional level, which probably reflects the lower priority given by local public health systems to AFP than to other infectious diseases.
In contrast, due to high levels of immigration across the Mediterranean sea, Italy remains at risk of importing wild poliovirus (PV) from areas of endemicity as well as Sabin and neurovirulent Sabin-derived poliovirus (circulating vaccine-derived poliovirus [cVDPV]) from countries currently using OPV. Because the IPV vaccine does not elicit a consistent mucosal immunity (10), silent transmission of neurovirulent poliovirus might in fact occur through IPV-immunized individuals, favoring possible infection of unvaccinated subjects or children receiving delayed IPV vaccination (11, 12).
In addition to virological investigation of AFP patients, several countries also survey the circulation of wild or Sabin-derived polioviruses by analysis of surface waters and sewage samples (13, 14, 15). As poliovirus is shed from infected subjects both with and without paralysis, environmental poliovirus surveillance is thought to allow sampling the entire population (13, 14, 16, 17, 18, 19). In Italy, screening of inlet sewage was initiated in 2005, targeting densely populated cities with high immigration rates and AFP surveillance indicators below average.
In addition to poliovirus, coxsackievirus (CV), echovirus (Echo), and other enteroviruses (EV) also represent a relevant public health problem in industrialized countries, causing severe diseases that may particularly affect young subjects and that can occur as outbreaks, including meningoencephalitis, myocarditis and pancreatitis, foot and mouth disease, and febrile illness (20, 21, 22, 23, 24).
Because of the different syndromes related to EV, case surveillance is more difficult, and environmental monitoring can have a major role in detecting the circulation of these viruses in the population and in assessing the risk for citizens. Human EVs (HEV) have been recovered in wastewater, river, sea, and recreational waters (25), which may represent efficient vehicles for viral transmission to humans (26). Crop irrigation with treated wastewaters is also common (27, 28, 29), and vegetables or fruits contaminated with viruses contained in sewage are a threat for the consumer. In fact, data from environmental monitoring and the increasing attention paid to water-transmitted diseases have driven technological improvement in wastewater treatment protocols (30, 31).
Data on the distribution of specific EV serotypes in sewage in Italy are currently limited (32, 33, 34).
This study was conducted to investigate the poliovirus strains possibly circulating in Italy between January 2005 and December 2008, in particular, to confirm the absence of imported wild-type PVs after the OPV abrogation in 2002. To address this aim, all PV strains isolated from sewage were characterized in detail by sequence analysis of four genomic regions, together with targeting possible reverted neurovirulent and recombinant strains. In addition to PV, analyses also included the serological and molecular characterization of the HEV serotypes detected in 1,392 sewage samples collected from the wastewater treatment plants (WWTPs) of 7 cities located in different regions. Non-polio enterovirus (NPEV) strains were characterized by WHO seroneutralization testing, nested PCR analysis, and partial genome sequencing.
MATERIALS AND METHODS
Wastewater sampling.
Environmental surveillance was started in 3 cities (Rome, Milan, and Parma) in January 2005 and was extended to Bari, Palermo, and Sassari between April and May. Finally, Naples was included in May 2006. Sewage samples were collected at the WWTP inlet from the collector sewers serving the surveyed urban areas.
Samples from Palermo, Sassari, and Naples were processed and analyzed in the WHO Regional Reference Laboratory (RRL) at the ISS. Samples from Rome, Parma, and Milan were examined at the accredited subnational polio reference laboratories: the Department of Public Health, University of Rome La Sapienza, the Institute of Hygiene, University of Parma, and the Institute of Virology, University of Milan, respectively. Characterization of all poliovirus strains and typing of most of the NPEVs were performed at the ISS.
The number and volume of samples representative for each population were defined according to the WHO guidelines (35). One sample per month was usually taken from WWTPs serving fewer than 100,000 inhabitants and two or more samples per month from WWTPs serving >100,000 inhabitants (Table 1). Occasionally, fewer samples were collected due to logistical problems, whereas extra sewage samples were collected in Palermo, where particular concern was raised by the high flow of migrants from areas of polio endemicity.
TABLE 1.
Sampling per year for each collector and city during environmental surveillance in Italy (2005 to 2008)
| City | Collector sewer locationa | No. of inhabitants in catchment area | No. of samples |
||||
|---|---|---|---|---|---|---|---|
| 2005 | 2006 | 2007 | 2008 | Total | |||
| Milan | MN | 300,000 | 18 | 17 | 23 | 24 | 82 |
| MNE | 300,000 | 18 | 17 | 23 | 24 | 82 | |
| MP | 300,000 | NDb | 17 | 22 | 24 | 63 | |
| Parma | PE | 130,000 | 22 | 24 | 22 | 24 | 92 |
| PO | 160,000 | 22 | 24 | 22 | 24 | 92 | |
| Rome | RN | 490,000 | 18 | 15 | 12 | 0 | 45 |
| RE | 720,000 | 22 | 15 | 12 | 0 | 49 | |
| RS | 1,000,000 | 24 | 17 | 14 | 0 | 55 | |
| Bari | BF | 300,000 | 18 | 24 | 23 | 24 | 89 |
| MB | 300,000 | 18 | 24 | 23 | 24 | 89 | |
| BJ | 300,000 | 18 | 20 | 23 | 24 | 85 | |
| Palermo | AdC | 130,000 | 14 | 24 | 35 | 24 | 97 |
| FV | 70,000 | 14 | 20 | 24 | 24 | 82 | |
| VD | 70,000 | 6 | 10 | 12 | 12 | 40 | |
| JH | 70,000 | 7 | 10 | 11 | 11 | 39 | |
| Naples | NC | 1,000,000 | ND | 34 | 44 | 9 | 87 |
| NT | 700,000 | ND | 30 | 30 | 20 | 80 | |
| NE | 500,000 | ND | 23 | 29 | 20 | 72 | |
| Sassari | SS | 120,000 | 12 | 24 | 24 | 12 | 72 |
| All | 251 | 389 | 428 | 324 | 1392 | ||
MN, Milan Nosedo; MNE, Milan Nosedo Est; MP, Milan Peschiera; PE, Parma Est; PO, Parma Ovest; RN, Rome Nord; RE, Rome Est; RS, Rome Sud; BF, Bari Fesca; MB, Mola di Bari; BJ, Bari Japigia; AdC, Acqua dei Corsari; FV, Fondo Verde; VD, Via Diaz; JH, Jolly Hotel; NC, Naples Cuma; NT, Naples Teduccio; NE, Naples Est; SS, Sassari.
ND, sampling not done.
Samples (1 liter) of raw sewage were collected during the peak hours of household sewage flow or during 24 h for plants with an automated flow system (Bolzano; Acqua dei Corsari and Fondo Verde, Palermo; Cuma and Napoli Est, Naples).
Sewage and virus concentration.
Sewage was collected in sterile polypropylene bottles and stored at −20°C usually for <3 days, until transportation to the reference laboratories in dry ice or under refrigerated conditions. Samples (500 ml each) were concentrated by the two-phase polyethylene glycol (PEG)-dextran separation method and decontaminated by chloroform extraction, as recommended by the WHO (35). After treatment, 10 ml of sample was obtained, resulting in an approximately 50-fold volume reduction.
Validation of virus concentration from wastewater.
To evaluate the virus detection capability of the WHO concentration procedure, six Palermo raw sewage samples (500 ml each) were autoclaved (121°C for 30 min) to inactivate any virus possibly present. Five samples were spiked with 2 × 105, 2 × 103, 20, 2, and 0.2 50% cell culture infective doses (CCID50) of poliovirus type 1 Sabin, respectively, and a sixth sample served as a negative control (35). Wastewaters were concentrated as described above, and 10 ml was obtained for both spiked and control samples. A 0.5-ml volume of chloroform-extracted sewage concentrate was inoculated onto RD cell monolayers in 50-ml cell culture flasks, in duplicate. Two serial blind passages were performed in the event of a negative first-passage result.
Clinical samples.
Cerebrospinal fluid samples (CSF) were collected from four patients involved in an outbreak of meningitis that occurred in Sassari in December 2005. Stools from a 4-year-old female presenting with Guillain-Barré syndrome reported in Sassari in December 2008 were processed according to guidelines of the polio laboratory manual (36).
Cell cultures and virus isolation.
Virus isolation was performed according to the WHO guidelines for clinical fecal specimens (36), inoculating 0.5 ml of the 10-ml chloroform-extracted sewage concentrate onto two RD and two L20B (selective for polioviruses) cell monolayers in 50-ml cell culture flasks. Two serial blind passages were performed with both cell lines, and samples positive on RD cells were passaged on L20B for specific amplification of poliovirus.
Identification and characterization of viruses.
For rapid virus identification, cytopathic effect (CPE)-positive cell lysates were tested by PCR with primers (PVM1-1 [5′CAA GCA CTT CTG TTT CCC C3′] and PVM1-2 [5′ATT GTC ACC ATA AGC AGC CA3′]) specific for the 5′ noncoding region [5′NCR]) (nucleotide [nt] 179 to 575) (37) common to all enteroviruses.
Samples inducing CPE but with negative enterovirus PCR results were classified as enteric nonenteroviruses (ENEV) and were not investigated further.
Immunostaining of infected cells with monoclonal antibodies (MAbs) specific for each poliovirus serotype, coxsackievirus B1 to B6, echoviruses 4, 6, 9, 11, 30, and 34 (Chemicon International, Temecula, CA), and a pan-enterovirus MAb (Dako, Denmark) was also used for confirmation (38). Virus typing was performed by microneutralization assays using EV serum pools and type-specific poliovirus antisera (RIVM, Bilthoven). Intratypic differentiation of Sabin-like (SL) polioviruses from non-Sabin-like (NSL) polioviruses was performed by enzyme-linked immunosorbent assays (ELISA) using cross-adsorbed antibodies (RIVM) and by reverse transcription-PCR (RT-PCR) with specific primers (CDC) (36). Sequencing of the 5′NCR (nt 179 to 575), VP1 (nt 2482 to 3384 for PV2 and nt 2477 to 3376 for PV3), VP1/2A junction (nt 3241 to 3460), and 3D (nt 6086 to 6376) genomic regions following RT-PCR amplification was also performed for all poliovirus isolates (37, 39). The 5′NCR was also sequenced to confirm the typing of some coxsackievirus and echovirus strains using the primers described above (40, 41).
Sewage EV isolates classified as untypeable by neutralization assays were typed by nucleotide sequencing of the amplicon resulting from a seminested RT-PCR analysis of the VP1 region, including the BC loop (nt 2628 to 2976) encoding a linear antigenic site (42). Sequencing was performed by Macrogen (Korea), using primers AN88 (5′TAC TGG ACC ACC TGG NGG NAY RWA CAT3′) and AN89 (5′CCA GCA CTG ACA GCA GYN GAR AYN GG3′).
The sequences of the VP1 region amplified from HEV isolates were analyzed using the neighbor-joining (NJ) method for clustering. The percentage of identity was determined based on the number of nucleotide substitutions per site. The alignment of sequences was performed using Mega 5.0 software (43). Amino acid sequence alignments were obtained using BioEdit software (44).
Nucleotide sequence accession numbers.
The nucleotide sequences of the 5′NCR, VP1, VP1/2A junction, and 3D regions of polioviruses (Table 2) have been deposited in the GenBank database under accession numbers KJ852629 to KJ852648; those of coxsackieviruses B2 and B5 (Figures 2, 3, and 4) have been deposited under accession numbers KJ867446 to KJ867452 and KJ867453 to KJ867459, respectively.
TABLE 2.
Polioviruses isolated in sewage samples during the surveillance in Italy (2005 to 2008)a
| Cities | Date | Serotype | ITD | Sequence |
|||
|---|---|---|---|---|---|---|---|
| 5′NCR | VP1 | VP1/2A | 3D | ||||
| Parma | June 2005 | PV3 | SL | 472 U > C | |||
| Naples | May 2007 | PV3 | SL | 472 U > C | |||
| 473 C > U | |||||||
| Milan | September 2007 | PV2 | SL | 481 A > G | 2487 T > C | Recombinant S2/S1 | |
| 2736 T > C | |||||||
| Milan | April 2008 | PV2 | SL | 325 U > C | 2909 T> C | 6147 T > C | |
| 481 A > G | 6213 G > A | ||||||
| 6285 C > T | |||||||
| Palermo | June 2008 | PV3 | SL | 472 U > C | 2637 C > T | Recombinant S3/S2 | |
5′NCR, nt 179 to 575; VP1, nt 2482 to 3384 for PV2 and nt 2477 to 3376 for PV3; VP1/2A, nt 3241 to 3460; 3D, nt 6086 to 6376. ITD, intratypic differentiation.
FIG 2.
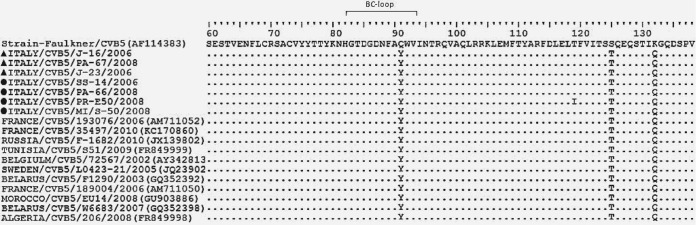
Coxsackievirus B5 multiple amino acid alignments (amino acids [aa] 60 to 138) of the VP1 partial structural protein of sequences of the Faulkner reference strain, seven Italian strains, four untypeable strains (marked by a circle), and three typeable strains (marked by a triangle) and 11 others isolated in different areas in the world and available on GenBank. (Each dot indicates a position where there was no difference in sequence from the reference strain).
FIG 3.

Coxsackievirus B2 multiple amino acid alignments (aa 53 to 121) of the VP1 partial structural protein of sequences of the Ohio-1 reference strain, seven Italian strains, five untypeable strains (marked by a circle), and two typeable strains (marked by a triangle) and 11 others isolated in the different areas of the world and available on GenBank. (Each dot indicates a position where there was no difference in sequence from the reference strain).
FIG 4.
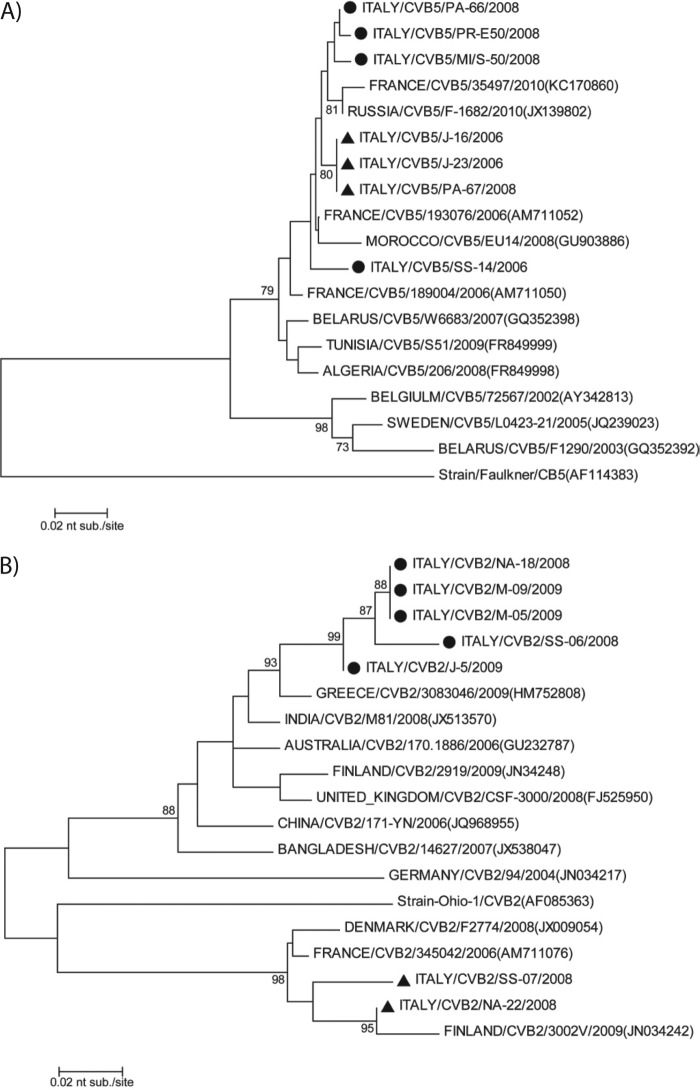
Neighbor-joining phylogenetic tree of the partial nucleotide sequence encoding the VP1 region (nt 2628 to 2976) of coxsackievirus B5 and B2 isolated from environmental surveillance in Italy between 2005 and 2008. (A) Coxsackievirus B5 strains: four serologically untypeable strains (circles), three typeable strains (triangles) identified in this study, and other reference or field strains. (B) Coxsackievirus B2 strains: five serologically untypeable strains (circles), two typeable strains (triangles) identified in this study, and other reference and field strains. GenBank accession numbers are reported. The evolutionary history was analyzed using the neighbor-joining algorithm in the Mega 5 program, with the Kimura-2 parameter. The scale bar indicates the number of nucleotide substitutions per site. Bootstrap values in 1,000 pseudoreplicates >75 are indicated at the branch nodes.
RESULTS
Virus recovery and detection limit.
Sewage samples spiked with 2 × 105 and 2 × 103 CCID50 of Sabin type 1 poliovirus suspension, respectively, produced a CPE upon passage of the first RD cells, while CPE was seen only on the second passage using 20 CCID50 of virus. The sample spiked with 2 CCID50 of Sabin type 1 poliovirus was CPE negative. The resulting detection limit of 20 CCID50/sample is in line with the WHO requirements for a satisfactory environmental surveillance of poliovirus and other enteroviruses (35).
Identification of enteroviruses in sewage samples.
Between January 2005 and December 2008, 1,392 sewage samples were collected from 19 WWTPs in seven cities and analyzed for enteroviruses (Table 1).
Overall, 702 samples gave CPE-positive results in cell cultures, ranging from 41% of 104 samples in 2005 to 56% of 181 samples in 2008. In 680 (96.8%) cases, the enterovirus positivity in cell cultures was confirmed by RT-PCR with 5′NCR-specific primers.
Twenty-two samples that induced CPE in RD cells gave negative results by 5′NCR PCR with primers specific for enteroviruses and were classified as positive for enteric nonenteroviruses (ENEV). These viruses were not investigated further and may possibly have contained norovirus, rotavirus, or other enteric viruses.
Of the 680 enterovirus-positive samples, 612 contained one virus and 68 contained two viruses, yielding a total of 748 enterovirus strains. Seroneutralization tests (RIVM polyclonal sera) identified 5 (0.7%) polioviruses, 415 (55.5%) coxsackievirus B strains, 1 (0.1%) coxsackievirus A strains, 271 (36.2%) echoviruses, and 56 (7.5%) untypeable enteroviruses (NTEVs). After seminested PCR and sequencing, the latter were found to be common enteroviruses, i.e., coxsackievirus B2 (CVB2) (13 samples), CVB3 (9), CVB5 (6), CVB1 (3), CVB4 (2), Echo 11 (8), Echo 6 (5), Echo 13 (4), Echo 7 (3), Echo 19 (1), Echo 25 (1), and Echo 30 (1). Therefore, overall CVB and echovirus totals were 448 and 294, respectively.
Five polioviruses were isolated in four different cities in 2005, 2007, and 2008 (Table 2) and were characterized as three Sabin-like poliovirus 3 and two Sabin-like poliovirus 2 strains by both ELISA (RIVM) and PCR (CDC) tests (36). Their genomes were sequenced and compared with the sequences of PV reference strains (AY184219.1, AY184220.1, and AY184221.1 [accession numbers for PV1, PV2, and PV3, respectively]) (Table 2). Interestingly, all strains contained a 472 (U > C) mutation or a 481 (A > G) mutation, which have been associated with reversion to the neurovirulent phenotype in PV3 and PV2, respectively. Additional mutations were found in the same region, specifically, at nt 473 (C > U) in strain Naples 07 and at nt 325 (U > C) in strain Milan 08. Mutations in the region coding for VP1 (nt 2482 to 3384) were identified only in strain Milan 07, at nt 2487 (T > C) and at nt 2736 (T > C), in strain Milan 08, at nt 2909 (T > C), and in strain Palermo 08, at nt 2637 (C > T). Mutation in VP1 at nt position 2909 of PV2 Milan 08 induced an amino acid substitution (I143T), confirming the fully neurovirulent genotype of this strain. Interestingly, poliovirus isolates PV2 Milan 07 and PV3 Palermo 08 also showed intertypic recombination with other PV serotypes in the region encoding the 3D polymerase (pol) (nt 6086 to 6376) (Table 2): PV2 strain Milan 07 presented a Sabin 1 sequence between nt 6354 to 6109, and PV3 strain Palermo 08 shared nt 6354 to 6109 (typical of Sabin 2). The crossing-over site for isolate Milan 07 was at nt 6230. In PV2 strain Milan 08, three additional mutations were also present in the region encoding the 3D pol, at nt 6147 (T > C), 6213 (G > A), and 6285 (C > T). All mutations present in the VP1, except nt 2909 (T > C) in PV2 strain Milan 08, and in the 3D regions were synonymous, inducing no amino acid change.
Among the 13 different echovirus serotypes isolated, Echo 11 (32.7%) and Echo 6 (37.1%) largely predominated, followed by Echo 4 (16.7%), whereas the proportions of the other serotypes ranged between 0.3% to 3.7% of cases (Table 3). Among coxsackieviruses, serotypes CVB5 and CVB3 were more frequent (both 29.9%), followed by CVB4 (20.0%), CVB2 (15.8%), and CVB1 (4.2%) (Table 3), whereas CVB6 was not isolated in this study. Type A coxsackieviruses were detected very rarely (0.2%); specifically, only serotype CVA24 was detected.
TABLE 3.
Distribution by city of enteroviruses isolated from sewage samples in Italy (2005 to 2008)
| EV serotypea | No. of samples by region and city |
|||||||
|---|---|---|---|---|---|---|---|---|
| North (n = 273) |
Central (n = 113) |
South (n = 362) |
Total | |||||
| Milan | Parma | Rome | Sassari | Naples | Palermo | Bari | ||
| CAV 24 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| CVB 1 | 2 | 1 | 3 | 2 | 1 | 0 | 10 | 19 |
| CVB 2 | 10 | 18 | 7 | 2 | 7 | 8 | 19 | 70 |
| CVB 3 | 10 | 27 | 4 | 4 | 25 | 37 | 27 | 134 |
| CVB 4 | 1 | 30 | 4 | 13 | 30 | 5 | 8 | 90 |
| CVB 5 | 22 | 32 | 27 | 15 | 8 | 20 | 10 | 134 |
| Echo 3 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 4 |
| Echo 4 | 12 | 2 | 0 | 16 | 6 | 2 | 11 | 49 |
| Echo 6 | 67 | 0 | 0 | 1 | 9 | 6 | 13 | 96 |
| Echo 7 | 5 | 0 | 0 | 0 | 0 | 4 | 0 | 9 |
| Echo 9 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 |
| Echo 11 | 15 | 0 | 0 | 13 | 36 | 11 | 34 | 110 |
| Echo 12 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Echo 13 | 8 | 0 | 0 | 0 | 2 | 1 | 0 | 11 |
| Echo 19 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Echo 22 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Echo 25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 |
| Echo 29 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Echo 30 | 1 | 1 | 0 | 2 | 0 | 3 | 1 | 8 |
| PV | 2 | 1 | 0 | 0 | 1 | 0 | 1 | 5 |
| Total | 161 | 112 | 45 | 68 | 127 | 100 | 135 | 748 |
CAV, coxsackievirus A; CVB, coxsackievirus B; Echo, echovirus; PV, poliovirus.
The enteroviruses identified in the different cities investigated are reported in Table 3. Major differences were shown in the EV serotypes predominating between different cities.
The monthly distribution of enteroviruses isolated during the study period in northern, central, and southern Italy is shown in Fig. 1. With few exceptions, both the CVBs and echoviruses were detected in all months, with a prevalence of CVBs compared to the echoviruses, a trend which did not differ with respect to geographic areas. For CVBs, peaks were observed in the months from May to August and October to November, whereas for the echovirus, peaks appeared between August and September and were more pronounced in the northern and central regions than in the southern regions. Polioviruses were detected only between April and September.
FIG 1.
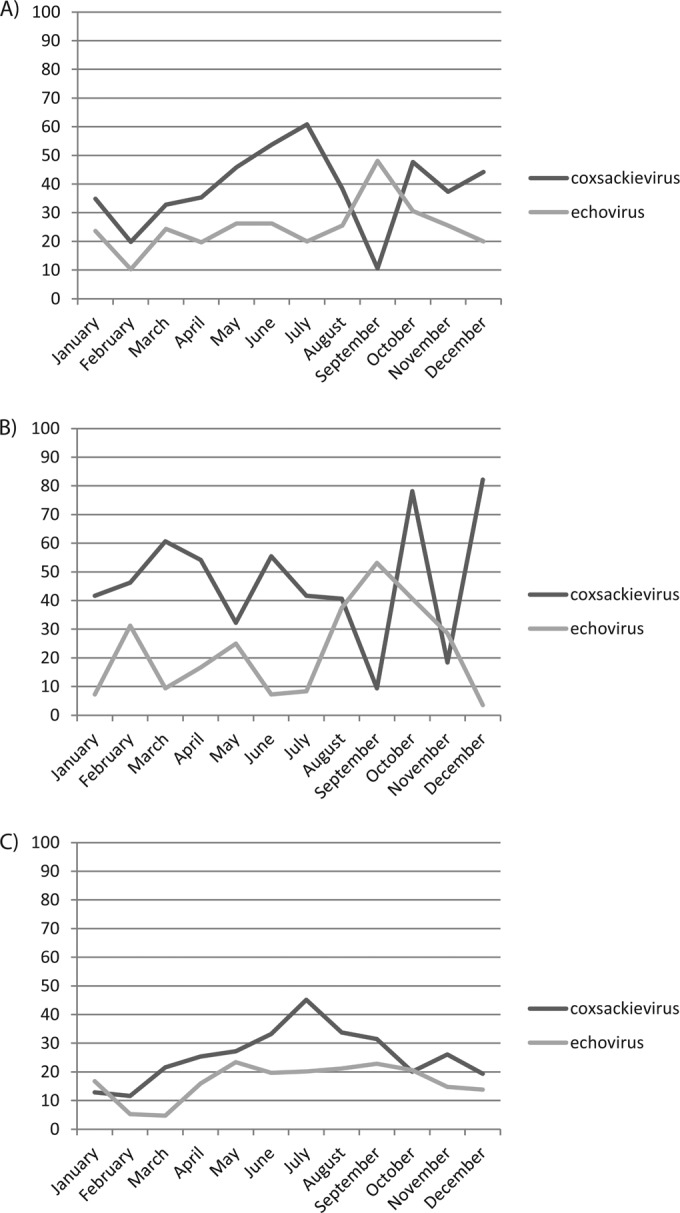
Percentages of geographic and seasonal distribution of enteroviruses isolated in RD cell cultures during monthly routine environmental surveillance in northern (Milan and Parma) (A), central (Rome and Sassari) (B), and southern (Bari, Naples, and Palermo) (C) Italy in 2005 to 2008.
VP1 amino acid sequence analysis of selected CVB5 and CVB2 strains.
The 56 enteroviruses that could not be typed with the RIVM antibodies were subjected to partial sequencing of the VP1 region, including the BC loop, to investigate possible amino acid changes responsible for the lack of neutralization. Of these, four randomly selected CVB5 and five CVB2 strains were compared with other CVB5 and CVB2 strains serotyped in this study and with reference and GenBank strains (Fig. 2 and 3). All test and GenBank CVB5 strains showed identical sequences in this genome region, except for a T119I amino acid change in a single strain. Three further changes (Y91Q, T125S, and Q132K) were shown by all strains compared to the Faulkner reference strain (Fig. 2).
Comparative amino acid sequence analysis of coxsackievirus B2 also showed the same substitution (R54H) for all field strains with respect to the reference Ohio-1 strain. One of the serologically untypeable CVB2 strains presented no further changes, whereas three other strains showed two (K99R and H119E) and another strain three (E117D, G118I, and H119E) additional changes (Fig. 3). In contrast, the two normally serotyped CVB2 strains investigated presented one or two amino acid changes (T84N or T84N and T86A, respectively). The former change was also presented by three GenBank strains of different origins (Fig. 3).
Phylogenetic analysis of CVB5 and CVB2 strains.
Phylogenetic analysis of the VP1 gene sequence of randomly selected CVB5 isolates, classified as typeable or untypeable by serological tests in this study, and 11 CVB5 strains isolated in different years and countries, available in GenBank, was carried out. All Italian strains showed high sequence similarity (99%), grouped with the other global strains within the same cluster, and were distant from the Faulkner reference strain, which presented only 82% identity with field CVB5 isolates (Fig. 4A).
In contrast, the two Italian serotyped CVB2 strains analyzed shared high sequence similarity between themselves (97%) and with the Ohio-1 reference strain, whereas the five strains proving untypeable by serological tests shared lower nucleotide similarity (92%) with the former strains, grouping within a separate cluster. However, all CVB2 strains from this study clustered with other CVB2 strains reported in different countries during the same period (Fig. 4B).
Enteroviruses from clinical cases in Italy.
An Echo 30 strain was isolated from the spinal fluid collected from four patients during an outbreak of meningitis that occurred in Sassari in December 2005 (Table 4). In December 2008, a coxsackievirus B5 strain was isolated in this same city from two stool samples from an AFP case presenting with Guillain-Barré syndrome (Table 4). The 5′NCR (nt 179 to 575) and VP1 (nt 2628 to 2976) sequences of these strains were compared with those of Echo 30 strains isolated from environmental samples in Sassari in October and November 2005 and with that of a CVB5 strain isolated in December 2008. Three Echo 30 strains from meningitis cases showed 100% identity in both the 5′NCR and VP1 with the Echo 30 strain isolated from wastewater few days later. The isolate from the fourth patient showed 100% nucleotide similarity in the 5′NCR and 99.4% in VP1. The CVB5 strains had 100% similarity in both the 5′NCR and VP1 with the CVB5 environmental isolate.
TABLE 4.
Sequence comparison of enteroviruses isolated from clinical and environmental samples in Sassari in the same period and in different years
| Samplea | Date (day[s]/mo/yr) | Sample source | Disease | Virus | % 5′NCR identityb | % VP1 identityc |
|---|---|---|---|---|---|---|
| SS/105 | 22/12/2005 | Liquor | Meningitis | Echo 30 | 100 | 100 |
| SS/205 | 22/12/2005 | Liquor | Meningitis | Echo 30 | 100 | 100 |
| SS/305 | 22/12/2005 | Liquor | Meningitis | Echo 30 | 100 | 100 |
| SS/405 | 22/12/2005 | Liquor | Meningitis | Echo 30 | 100 | 99.4 |
| SS-05-09 | 30/10/2005 | Wastewater | Echo 30 | |||
| SS-05-11 | 30/11/2005 | Wastewater | Echo 30 | |||
| SS/505 | 13–14/12/2008 | Stool | AFP (GBS) | CVB 5 | 100 | 100 |
| SS-08-23 | 15/12/2008 | Wastewater | CVB 5 |
Patients: SS/105, SS/205, SS/305, SS/405, and SS/505; SS, Sassari.
Nucleotide identity between nucleotides 179 and 575 of 5′NCR regions in human case samples and environmental samples.
Nucleotide identity between nucleotides 2628 and 2976 of VP1 regions in human case samples and environmental samples.
DISCUSSION
In recent years, several studies investigating viral pollution in surface waters have been carried out on both river and coastal waters of Italy (45, 46, 47, 48, 49, 50, 51). In contrast, only a few recent investigations have been performed using detailed serotyping of enterovirus strains in Italian wastewaters (32, 33, 34, 52) compared to a higher number of similar studies reported from other countries (53, 54, 55, 56, 57, 58).
This study was undertaken as a supplement to the AFP surveillance in Italy to monitor the possible presence of wild-type (WPV) or vaccine-derived (cVDPV) poliovirus or other enterovirus strains in wastewaters and to obtain further evidence in support of the maintenance of the polio-free status of Italy.
For this purpose, a network of laboratories using standardized methods for sample collection and virus isolation was established.
No wild polioviruses were isolated from the environmental samples examined during the study period, supporting the epidemiological data deriving from AFP surveillance (6, 7, 59).
The isolation of 5 Sabin-like polioviruses demonstrated that environmental surveillance is an effective supplemental support to the AFP surveillance to verify the absence of wild or cVDPV neurovirulent polioviruses in circulation in the country. In fact, since the introduction of IPV vaccination in 2002, no polioviruses have been isolated from the AFP cases notified in Italy, whereas environmental surveillance was able to detect the presence of Sabin-like polioviruses, although rarely. The low degree of mutation found in the genomes of the 5 Sabin-like polioviruses isolated suggests that these viruses have undergone only limited circulation in the population, which implies the effectiveness of polio immunization and a high level of protection against polio in Italy.
Since the use of vaccination with poliovirus Sabin vaccine in Italy was dismissed in 2002, the poliovirus Sabin-like strains found in the environment have most likely been excreted by children immunized with OPV abroad or by contacts of vaccinees and should thus be considered imported viruses. Italy has in fact experienced a high rate of immigration during the last 10 years. Nevertheless, the presence of Sabin-like revertant poliovirus strains with mutations associated with neurovirulence in the environment may represent a risk of disease for both unvaccinated subjects and immunodeficient patients. In Italy, some delay in the administration of the first dose of IPV has been reported in some regions, although rarely. Moreover, although vaccination is mandatory in Italy, some parents refuse vaccination for their children because of prejudice or religious problems even if this implies losing access to public education. Poliovirus might therefore replicate unnoticed in immunodeficient subjects and might be transmitted to contact persons if they are unvaccinated.
The presence of intertypic recombination in the 3D region of the poliovirus isolated in Milan and Palermo is interesting, even though similar recombinations were previously detected in poliovirus isolated in several other countries (41, 60, 61). Increased transmissibility over that of Sabin parental strains has been suggested to be the consequence of a recombinant event (62). The two PV2 isolates from Milan most likely represented separate events since the April 08 isolate lacks the recombination in the polymerase 3D that was found for the earlier September 07 isolate.
Most of the polioviruses were isolated between April and June, months when more tourists are present in Italy and immigration increases. The lack of isolation of poliovirus type 1 Sabin-like strains might be related to the reduced replication of this serotype in the human gut (63, 64) or might suggest lower resistance in the environment. In fact, PV1 was isolated from sewage with lower frequency than other PV serotypes following introduction of IPV vaccine in New Zealand (65).
In contrast to polioviruses, non-polio enteroviruses were detected during all seasons in our study. This findings are, however, unusual for countries with a temperate climate, where the majority of enterovirus infections usually occur in summer and autumn, and resemble the situation in tropical countries (66, 67, 68). The large presence of non-polio enteroviruses (coxsackievirus type B and echoviruses) in sewage demonstrates that these viruses have a large diffusion in the population and that they might result in severe diseases (meningitis, encephalitis, myocarditis, etc.). The massive environmental dissemination of these pathogens to receiving water bodies (seas, rivers, etc.) and the very low infectious dose of most enteric viruses emphasizes the risk to public health, considering that legal requirements for microbiological monitoring of wastewaters in Italy and other European countries address only bacterial parameters (69, 70, 71, 72, 73). According to the WHO, the high rate of NPEV in the environment can be considered a proof that the environmental samples were processed and analyzed appropriately and that the cold chain used during specimen transportation was effective in preserving virus infectivity.
The frequencies of detection of different NPEV species differed in the various geographical areas and differed between years, but overall, coxsackievirus B strains, in particular, CVB5, CVB3, and CVB4, were isolated more often than echovirus strains. These findings are in line with data reported from enterovirus surveillances in Europe and with some studies on sewage surveillance (56, 74, 75). CVB5 is an emerging serotype that has been correlated with some outbreaks of meningitis and other diseases in the last 10 years (76, 77, 78), but no clinical cases have been reported in Italy or (probably) investigated by detailed serotyping. The rare isolation of coxsackievirus A in our study might indicate low circulation of these strains in the Italian population, although it might also be related to the lower susceptibility of RD cells to infection with CVA.
The most prevalent echoviruses were serotype 11 and 6, in particular, in southern and northern cities, respectively. These echoviruses are associated with childhood diseases, specifically, meningitis, respiratory, and gastrointestinal illnesses for Echo 11, febrile illness for Echo 4, and outbreaks of respiratory diseases for Echo 6 (79, 80). Echovirus 30, which was the virus most commonly isolated during meningitis outbreaks in several countries (81, 82, 83, 84, 85), was detected with low frequency in the present study.
However, it should mentioned that the use of RD cells in this study may have led to underestimation of some HEVs serotypes, since not all HEVs can be cultivated in these cells. The use of multiple cell lines would optimize isolation of some other enteroviruses (86), but that benefit is counterbalanced by the increased workload and expenses due to reagents and consumables compared to those of the rapid and cheaper molecular techniques.
It should also be taken into account that there are still a number of HEV serotypes that fail to replicate on many cell cultures (87), including CVA of HEV-C, whose detection is very important for the possibility of recombination with PV (88).
Sequence analysis of the serologically untypeable enteroviruses that could be identified only by molecular methods revealed scattered mutations in their genomes, which has been previously reported to occur during replication in humans. Some of these mutations might have affected binding of polyclonal antibodies and strain identification by neutralization assays. In fact, some mutations involved amino acid substitutions in VP1, which is one of the outer capsid proteins containing the BC loop. However, no differences were found in the amino acid sequence in comparisons between CVB5 strains that were typeable or not typeable by neutralization tests in the region sequenced, while two VP1 mutations were identified between typeable or some untypeable CVB2 strains. An additional substitution was present at amino acid 84 within the BC loop in other CVB2 strains. However, our findings do not allow us to confirm a possible role of these mutations in resistance to antibody neutralization, as was previously suggested by others (89). The BC loop is the only known linear antigenic site in poliovirus; the other sites are conformational, and a more detailed study would therefore involve the sequencing of all capsid proteins and conformational analysis.
Mutations that possibly affect strain recognition by antibodies may endanger enterovirus typing, and new antisera or monoclonal antibodies may be needed to update serological reagent panels from time to time. However, our data confirm that isolates failing serological typing may be successfully investigated by molecular techniques.
Phylogenetic analyses of the Italian CVB5 and CVB2 strains suggest that both were highly similar to contemporary strains isolated in other parts of the world available in GenBank while being very different from the reference strains Faulkner and Ohio. The Italian CVB2 strains that were untypeable by antibody assays were more distantly related to the reference strain than the typeable strains, and the distance from the tight cluster of isolates identified most likely suggests a separate introduction and evolution of CVB2.
Combining conventional cell culture isolation methods with RT-PCR allowed confirmation of the presence of enterovirus genome in cells showing CPE in a few additional hours and is more effective than direct molecular detection, particularly in cases of low viral concentrations and/or RT-PCR inhibitors in environmental samples. It should also be considered that maintaining adequate supplies of HEV typing antibody pools for the Global Polio Laboratory network will be difficult in the future.
Overall, the results of our study indicate that extension of wastewater screening to other cities of Italy could be helpful both to supplement AFP surveillance, particularly in areas with larger immigration, and to investigate possible risks of other human diseases and epidemic outbreaks correlated to non-polio enteroviruses, such as enterovirus meningitis, possibly linked to waterborne and food-borne transmission routes.
In conclusion, although AFP surveillance remains the gold standard for poliovirus surveillance, supplemental activities can be useful to implement a satisfactory overall surveillance performance. From our study, sewage surveillance appears to be more sensitive than AFP surveillance in monitoring the silent circulation of poliovirus in the population in countries such as Italy where polio clinical cases have been absent for a long time and could be considered more generally for the posteradication era.
Finally, the systematic collection of environmental samples for investigation of poliovirus circulation may be used conveniently for monitoring other enteric agents such as adenovirus, rotavirus, norovirus, and hepatitis virus.
ACKNOWLEDGMENTS
We are grateful to the following members of the Environmental Surveillance Study Group for their valuable contribution: A. Calvani and V. Martini (Department of Public Health and Infectious Diseases, University of Rome La Sapienza); E. Centurione, L. Bubba, and M. Gambino (Department of Biomedical Sciences for Health, University of Milan); D. Ceriani (ARPA Lombardy); M. Labianca (Department DIMO, Hygiene Unit, University of Bari); C. Cesari (Department of Biomedical, Biotechnological and Translational Sciences, University of Parma); G. Ferrari (AMPS, Parma); P. L. Ciappeddu (Department of Biomedical Sciences, University of Sassari); and G. Gullo and M. G. Cucchiara (AMAP S.p.A., Palermo).
This work was partially supported by grants from the Italian Ministry of Health, from CCM (AFP surveillance in Italy; research of poliovirus and other enteroviruses in immunodeficient patients and in the environment) (2005 to 2009), and from the WHO(TSA EU/06086241/237/22 and TSA EU/08/60850) (laboratory support for AFP surveillance in Italy, Albania, Bosnia, Serbia, Kosovo, Bulgaria, and Greece) (2007 to 2008).
Footnotes
Published ahead of print 9 May 2014
REFERENCES
- 1.Patti AM, Martini V, Calvani A, Vulcano A, Zotti C, Sudano L, Gasparini R, Barbi M, Brusaferro S, Carraro V, Simeoni J, Poli A, Tanzi ML, Bonanni P, Battaglia MA, Carducci AL, D'Errico M, Iorio AM, Vallone S, Grasso GM, Triassi M. 2008. La sorveglianza della poliomielite in Italia: stato immunitario della popolazione di età 0–14 anni. Ann. Ig. 20(Suppl 3):15–22 [Google Scholar]
- 2.de Gourville E, Duintjer Tebbens RJ, Sangrujee N, Pallansch MA, Thompson KM. 2006. Global surveillance and the value of information: the case of the global polio laboratory network. Risk Anal. 26:1557–1569. 10.1111/j.1539-6924.2006.00845.x [DOI] [PubMed] [Google Scholar]
- 3.Pallansch MA, Sandhu HS. 2006. The eradication of polio—progress and challenges. N. Engl. J. Med. 355:2508–2511. 10.1056/NEJMp068200 [DOI] [PubMed] [Google Scholar]
- 4.Aylward B, Yamada T. 2011. The polio endgame. N. Engl. J. Med. 364:2273–2275. 10.1056/NEJMp1104329 [DOI] [PubMed] [Google Scholar]
- 5.Fiore L, Novello F, Grandolfo ME. 1996. Eradicazione della poliomielite e sorveglianza delle paralisi flaccide in Italia, p 43 In Rapporti ISTISAN 96/22 Istituto Superiore di Sanità, Rome, Italy [Google Scholar]
- 6.Fiore L, Novello F, Simeoni P, Amato C, Vellucci L, De Stefano D, Grandolfo ME, Luzzi I. 1999. Surveillance of acute flaccid paralysis in Italy: 1996–1997. AFP Study Group. Acute flaccid paralysis. Eur. J. Epidemiol. 15:757–763 [DOI] [PubMed] [Google Scholar]
- 7.Ministry of Health Italy. 2007. Attività per l'eradicazione globale della poliomielite. Stato della sorveglianza della paralisi flaccida acuta e del contenimento di laboratorio dei Poliovirus selvaggi in Italia nell'anno 2007. 0001451-P-13/01/2009 Ministry of Health Italy, Rome, Italy [Google Scholar]
- 8.Novello F, Lombardi F, Amato C, Santoro R, Fiore L, Grandolfo ME, Pasquini P. 1987. Paralytic poliomyelitis in Italy (1981–85). Eur. J. Epidemiol. 3:54–60. 10.1007/BF00145073 [DOI] [PubMed] [Google Scholar]
- 9.WHO. 2003. Recommended standards for surveillance of selected vaccine preventable diseases: vaccines and biologicals, p 31–34 WHO/V&B/03.01. WHO, Geneva, Switzerland [Google Scholar]
- 10.Hird TR, Grassly NC. 2012. Systematic review of mucosal immunity induced by oral and inactivated poliovirus vaccines against virus shedding following oral poliovirus challenge. PLoS Pathog. 8:e1002599. 10.1371/journal.ppat.1002599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander JP, Ehresmann K, Seward J, Wax G, Harriman K, Fuller S, Cebelinski EA, Chen Q, Jorba J, Kew OM, Pallansch MA, Oberste MS, Schleiss M, Davis JP, Warshawsky B, Squires S, Hull HF; Vaccine-Derived Poliovirus Investigations Group. 2009. Transmission of imported vaccine-derived poliovirus in an undervaccinated community in Minnesota. J. Infect. Dis. 199:391–397. 10.1086/596052 [DOI] [PubMed] [Google Scholar]
- 12.Anis E, Kopel E, Singer SR, Kaliner E, Moerman L, Moran-Gilad J, Sofer D, Manor Y, Shulman LM, Mendelson E, Gdalevich M, Lev B, Gamzu R, Grotto I. 2013. Insidious reintroduction of wild poliovirus into Israel, 2013. Euro Surveill. 18:pii=20586 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20586 [DOI] [PubMed] [Google Scholar]
- 13.Roivainen M, Blomqvist S, Al-Hello H, Paananen A, Delpeyroux F, Kuusi M, Hovi T. 2010. Highly divergent neurovirulent vaccine-derived polioviruses of all three serotypes are recurrently detected in Finnish sewage. Euro Surveill. 15:pii=19566 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19566 [PubMed] [Google Scholar]
- 14.Zurbriggen S, Tobler K, Abril C, Diedrich S, Ackermann M, Pallansch MA, Metzler A. 2008. Isolation of Sabin-like polioviruses from wastewater in a country using inactivated polio vaccine. Appl. Environ. Microbiol. 74:5608–5614. 10.1128/AEM.02764-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dedepsidis E, Kyriakopoulou Z, Pliaka V, Kottaridi C, Bolanaki E, Levidiotou-Stefanou S, Komiotis D, Markoulatos P. 2007. Retrospective characterization of a vaccine-derived poliovirus type 1 isolate from sewage in Greece. Appl. Environ. Microbiol. 73:6697–6704. 10.1128/AEM.00535-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hovi T, Blomqvist S, Nasr E, Burns CC, Sarjakoski T, Ahmed N, Savolainen C, Roivainen M, Stenvik M, Laine P, Barakat I, Wahdan MH, Kamel FA, Asghar H, Pallansch MA, Kew OM, Gary HE, Jr, deGourville EM, El Bassioni L. 2005. Environmental surveillance of wild poliovirus circulation in Egypt—balancing between detection sensitivity and workload. J. Virol. Methods 126:127–134. 10.1016/j.jviromet.2005.02.002 [DOI] [PubMed] [Google Scholar]
- 17.El Bassioni L, Barakat I, Nasr E, de Gourville EM, Hovi T, Blomqvist S, Burns C, Stenvik M, Gary H, Kew OM, Pallansch MA, Wahdan MH. 2003. Prolonged detection of indigenous wild polioviruses in sewage from communities in Egypt. Am. J. Epidemiol. 158:807–815. 10.1093/aje/kwg202 [DOI] [PubMed] [Google Scholar]
- 18.Chowdhary R, Dhole TN. 2008. Interrupting wild poliovirus transmission using oral poliovirus vaccine: environmental surveillance in high-risks area of India. J. Med. Virol. 80:1477–1488. 10.1002/jmv.21230 [DOI] [PubMed] [Google Scholar]
- 19.Shulman LM, Manor Y, Sofer D, Handsher R, Swartz T, Delpeyroux F, Mendelson E. 2006. Neurovirulent vaccine-derived polioviruses in sewage from highly immune populations. PLoS One 1:e69. 10.1371/journal.pone.0000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tapparel C, Siegrist F, Petty TJ, Kaiser L. 2013. Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. 14:282–293. 10.1016/j.meegid.2012.10.016 [DOI] [PubMed] [Google Scholar]
- 21.Palacios G, Oberste MS. 2005. Enteroviruses as agents of emerging infectious diseases. J. Neurovirol. 11:424–433. 10.1080/13550280591002531 [DOI] [PubMed] [Google Scholar]
- 22.Yajima T. 2011. Viral myocarditis: potential defense mechanisms within the cardiomyocyte against virus infection. Future Microbiol. 6:551–566. 10.2217/fmb.11.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhoades RE, Tabor-Godwin JM, Tsueng G, Feuer R. 2011. Enterovirus infections of the central nervous system. Virology 411:288–305. 10.1016/j.virol.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hober D, Sane F, Jaïdane H, Riedweg K, Goffard A, Desailloud R. 2012. Immunology in the clinic review series; focus on type 1 diabetes and viruses: role of antibodies enhancing the infection with Coxsackievirus-B in the pathogenesis of type 1 diabetes. Clin. Exp. Immunol. 168:47–51. 10.1111/j.1365-2249.2011.04559.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pianetti A, Baffone W, Citterio B, Casaroli A, Bruscolini F, Salvaggio L. 2000. Presence of enteroviruses and reoviruses in the waters of the Italian coast of the Adriatic Sea. Epidemiol. Infect. 125:455–462. 10.1017/S0950268899004604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajtar B, Majek M, Polański Ł, Polz-Dacewicz M. 2008. Enteroviruses in water environment—a potential threat to public health. Ann. Agric. Environ. Med. 15:199–203 [PubMed] [Google Scholar]
- 27.Steele M, Odumeru J. 2004. Irrigation water as source of foodborne pathogens on fruit and vegetables. J. Food Prot. 67:2839–2849 [DOI] [PubMed] [Google Scholar]
- 28.Muñoz I, Tomàs N, Mas J, García-Reyes JF, Molina-Díaz A, Fernández-Alba AR. 2010. Potential chemical and microbiological risks on human health from urban wastewater reuse in agriculture. Case study of wastewater effluents in Spain. J. Environ. Sci. Health B 45:300–309. 10.1080/03601231003704648 [DOI] [PubMed] [Google Scholar]
- 29.Viau E, Bibby K, Paez-Rubio T, Peccia J. 2011. Toward a consensus view on the infectious risks associated with land application of sewage sludge. Environ. Sci. Technol. 45:5459–5469. 10.1021/es200566f [DOI] [PubMed] [Google Scholar]
- 30.Simmons FJ, Xagoraraki I. 2011. Release of infectious human enteric viruses by full-scale wastewater utilities. Water Res. 45:3590–3598. 10.1016/j.watres.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 31.Francy DS, Stelzer EA, Bushon RN, Brady AM, Williston AG, Riddell KR, Borchardt MA, Spencer SK, Gellner TM. 2012. Comparative effectiveness of membrane bioreactors, conventional secondary treatment, and chlorine and UV disinfection to remove microorganisms from municipal wastewaters. Water Res. 46:4164–4178. 10.1016/j.watres.2012.04.044 [DOI] [PubMed] [Google Scholar]
- 32.Petrinca AR, Donia D, Pierangeli A, Gabrieli R, Degener AM, Bonanni E, Diaco L, Cecchini G, Anastasi P, Divizia M. 2009. Presence and environmental circulation of enteric viruses in three different wastewater treatment plants. J. Appl. Microbiol. 106:1608–1617. 10.1111/j.1365-2672.2008.04128.x [DOI] [PubMed] [Google Scholar]
- 33.Pellegrinelli L, Binda S, Chiaramonte I, Primache V, Fiore L, Battistone A, Fiore S, Gambino M, Bubba L, Barbi M. 2013. Detection and distribution of culturable human enteroviruses through environmental surveillance in Milan, Italy. J. Appl. Microbiol. 115:1231–1239. 10.1111/jam.12321 [DOI] [PubMed] [Google Scholar]
- 34.Cesari C, Colucci ME, Veronesi L, Giordano R, Paganuzzi F, Affanni P, Bracchi MT, Capobianco E, Ferrari G, Tanzi ML. 2010. Detection of enteroviruses from urban sewage in Parma. Acta Biomed. 81:40–46 [PubMed] [Google Scholar]
- 35.WHO. 2003. Guidelines for environmental surveillance of poliovirus circulation. WHO/V&B/03.03. WHO, Geneva, Switzerland [Google Scholar]
- 36.WHO. 2004. Polio laboratory manual. WHO/IVB/04.10. WHO, Geneva, Switzerland [Google Scholar]
- 37.Kilpatrick DR, Iber JC, Chen Q, Ching K, Yang SJ, De L, Mandelbaum MD, Emery B, Campagnoli R, Burns CC, Kew O. 2011. Poliovirus serotype-specific VP1 sequencing primers. J. Virol. Methods 174:128–130. 10.1016/j.jviromet.2011.03.020 [DOI] [PubMed] [Google Scholar]
- 38.Di Lonardo A, Buttinelli G, Amato C, Novello F, Ridolfi B, Fiore L. 2002. Rapid methods for identification of poliovirus isolates and determination of polio neutralizing antibody titers in human sera. J. Virol. Methods 101:189–196. 10.1016/S0166-0934(01)00437-2 [DOI] [PubMed] [Google Scholar]
- 39.Kew OM, Mulders MN, Lipskaya GY, da Silva EE, Pallansch MA. 1995. Molecular epidemiology of polioviruses. Semin. Virol. 6:401–414. 10.1016/S1044-5773(05)80017-4 [DOI] [Google Scholar]
- 40.Zoll GJ, Melchers WJG, Kopecka H, Jambroes G, Van der Poel HJA, Galama JMD. 1992. General primer-mediated polymerase chain reaction for detection of enteroviruses: application for diagnostic routine and persistent infections. J. Clin. Microbiol. 30:160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guillot S, Caro V, Cuervo N, Korotkova E, Combiescu M, Persu A, Aubert-Combiescu A, Delpeyroux F, Crainic R. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434–8443. 10.1128/JVI.74.18.8434-8443.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nix WA, Oberste MS, Pallansch MA. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 44:2698–2704. 10.1128/JCM.00542-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 45.Pianetti A, Baffone W, Citterio B, Casaroli A, Bruscolini F, Salvaggio L. 2000. Presence of enteroviruses and reoviruses in the waters of the Italian coast of the Adriatic Sea. Epidemiol. Infect. 125:455–462. 10.1017/S0950268899004604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danovaro R, Armeni M, Corinaldesi C, Mei ML. 2003. Viruses and marine pollution. Mar. Pollut. Bull. 46:301–304. 10.1016/S0025-326X(02)00461-7 [DOI] [PubMed] [Google Scholar]
- 47.De Donno A, Liaci D, Bagordo F, Guido M, Carducci A. 2005. Evaluation of viral and bacterial contamination of coastal seawater. Ig. Sanità Pubbl. 61:585–600 (In Italian.) [PubMed] [Google Scholar]
- 48.Masciopinto C, La Mantia R, Carducci A, Casini B, Calvario A, Jatta E. 2007. Unsafe tap water in households supplied from groundwater in the Salento region of southern Italy. J. Water Health 5:129–148. 10.2166/wh.2006.054 [DOI] [PubMed] [Google Scholar]
- 49.Muscillo M, La Rosa G, Marianelli C, Zaniratti S, Capobianchi MR, Cantiani L, Carducci A. 2001. A new RT-PCR method for the identification of reoviruses in seawater samples. Water Res. 35:548–556. 10.1016/S0043-1354(00)00282-7 [DOI] [PubMed] [Google Scholar]
- 50.Verani M, Casini B, Battistini R, Pizzi F, Rovini E, Carducci A. 2006. One-year monthly monitoring of Torque teno virus (TTV) in river water in Italy. Water Sci. Technol. 54:191–195 [DOI] [PubMed] [Google Scholar]
- 51.Donia D, Dell'Amico MC, Petrinca AR, Martinucci I, Mazzei M, Tolari F, Divizia M. 2012. Presence of hepatitis E RNA in mussels used as bio-monitors of viral marine pollution. J. Virol. Methods 186:198–202. 10.1016/j.jviromet.2012.06.007 [DOI] [PubMed] [Google Scholar]
- 52.Battistone A, Buttinelli G, Bonomo P, Fiore S, Amato C, Mercurio P, Cicala A, Simeoni J, Foppa A, Triassi M, Pennino F, Fiore L. 26 November 2013. Detection of enteroviruses in influent and effluent flow samples from wastewater treatment plants in Italy. Food Environ. Virol. 10.1007/s12560-013-9132-2 [DOI] [PubMed] [Google Scholar]
- 53.Zheng H, Lu J, Zhang Y, Yoshida H, Guo X, Liu L, Li H, Zeng H, Fang L, Mo Y, Yi L, Chosa T, Xu W, Ke C. 2013. Prevalence of nonpolio enteroviruses in the sewage of Guangzhou City, China, from 2009 to 2012. Appl. Environ. Microbiol. 79:7679–7683. 10.1128/AEM.02058-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klement C, Kissova R, Lengyelova V, Stipalova D, Sobotova Z, Galama JM, Bopegamage S. 2013. Human enterovirus surveillance in the Slovak Republic from 2001 to 2011. Epidemiol. Infect. 141:2658–2662. 10.1017/S0950268813000563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shukla D, Kumar A, Srivastava S, Idris MZ, Dhole TN. 2013. Environmental surveillance of enterovirus in Northern India using an integrated shell vial culture with a semi-nested RT PCR and partial sequencing of the VP1 gene. J. Med. Virol. 85:505–511. 10.1002/jmv.23441 [DOI] [PubMed] [Google Scholar]
- 56.Costán-Longares A, Mocé-Llivina L, Avellón A, Jofre J, Lucena F. 2008. Occurrence and distribution of culturable enteroviruses in wastewater and surface waters of north-eastern Spain. J. Appl. Microbiol. 105:1945–1955. 10.1111/j.1365-2672.2008.03954.x [DOI] [PubMed] [Google Scholar]
- 57.Wieczorek M, Kuryk Ł, Witek A, Diuwe A, Litwińska B. 2013. The detection of enteroviruses in sewage using Caco-2 cells. Pol. J. Microbiol. 62:97–100 [PubMed] [Google Scholar]
- 58.Kargar M, Sadeghipour S, Nategh R. 2009. Environmental surveillance of non-polio enteroviruses in Iran. Virol. J. 6:149. 10.1186/1743-422X-6-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruggeri FM, Fiore L. 2012. Vaccine preventable viral diseases and risks associated with waterborne transmission. Ann. Ist. Super. Sanita 48:460–472. 10.4415/ANN_12_04_12 [DOI] [PubMed] [Google Scholar]
- 60.Savolainen-Kopra C, Samoilovich E, Kahelin H, Hiekka AK, Hovi T, Roivainen M. 2009. Comparison of poliovirus recombinants: accumulation of point mutations provides further advantages. J. Gen. Virol. 90:1859–1868. 10.1099/vir.0.010942-0 [DOI] [PubMed] [Google Scholar]
- 61.Avellón A, Cabrerizo M, de Miguel T, Pérez-Breña P, Tenorio A, Pérez JL, de Aragón MV, Trallero G. 2008. Paralysis case and contact spread of recombinant vaccine-derived poliovirus, Spain. Emerg. Infect. Dis. 14:1807–1809. 10.3201/eid1411.080517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haddad-Boubaker S, Ould-Mohamed-Abdallah MV, Ben-Yahia A, Triki H. 2010. Genetic recombination in vaccine poliovirus: comparative study in strains excreted in course of vaccination by oral poliovirus vaccine and circulating strains. Pathol. Biol. (Paris) 58:420–425 (In French.) 10.1016/j.patbio.2009.01.004 [DOI] [PubMed] [Google Scholar]
- 63.Benyesh-Melnick M, Melnick JL, Rawls WE, Wimberly I, Oro JB, Ben-Porath E, Rennick V. 1967. Studies of the immunogenicity, communicability, and genetic stability of oral polio vaccine administered in the winter. Am. J. Epidemiol. 86:112–136 [DOI] [PubMed] [Google Scholar]
- 64.Alexander JP, Gary HE, Pallansch MA. 1997. Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J. Infect. Dis. 175(Suppl 1):S176–S182. 10.1093/infdis/175.1.176 [DOI] [PubMed] [Google Scholar]
- 65.Huang QS, Greening G, Baker MG, Grimwood K, Hewitt J, Hulston D, van Duin L, Fitzsimons A, Garrett N, Graham D, Lennon D, Shimizu H, Miyamura T, Pallansch MA. 2005. Persistence of oral polio vaccine virus after its removal from the immunisation schedule in New Zealand. Lancet 366:394–396. 10.1016/S0140-6736(05)66386-6 [DOI] [PubMed] [Google Scholar]
- 66.Khetsuriani N, Lamonte-Fowlkes A, Oberst S, Pallansch MA, Centers for Disease Control and Prevention 2006. Enterovirus surveillance—United States, 1970–2005. MMWR Surveill. Summ. 55:1–20 [PubMed] [Google Scholar]
- 67.van der Sanden SM, Koopmans MP, van der Avoort HG. 2013. Detection of human enteroviruses and parechoviruses as part of the national enterovirus surveillance in the Netherlands, 1996–2011. Eur. J. Clin. Microbiol. Infect. Dis. 32:1525–1531. 10.1007/s10096-013-1906-9 [DOI] [PubMed] [Google Scholar]
- 68.Fox JP. 1964. Epidemiological aspects of coxsackie and echo virus infections in tropical areas. Am. J. Public Health Nations Health 54:1134–1142. 10.2105/AJPH.54.7.1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parlamento Italiano. 1999. Pubblicato nella Gazzetta Ufficiale n. 246 del 20 ottobre 2000—Supplemento Ordinario n. 172. http://www.camera.it/parlam/leggi/deleghe/99152dl.htm Parlamento Italiano, Rome, Italy [Google Scholar]
- 70.Hot D, Legeay O, Jacques J, Gantzer C, Caudrelier Y, Guyard K, Lange M, Andréoletti L. 2003. Detection of somatic phages, infectious enteroviruses and enterovirus genomes as indicators of human enteric viral pollution in surface water. Water Res. 37:4703–4710. 10.1016/S0043-1354(03)00439-1 [DOI] [PubMed] [Google Scholar]
- 71.Bosch A. 1995. The survival of enteric viruses in the water environment. Microbiologia 11:393–396 [PubMed] [Google Scholar]
- 72.Bosch A. 1998. Human entroviruses in water environment: a minireview. Int. Microbiol. 1:191–196 [PubMed] [Google Scholar]
- 73.Rzezutka A, Cook N. 2004. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 28:441–453. 10.1016/j.femsre.2004.02.001 [DOI] [PubMed] [Google Scholar]
- 74.Antona D, Lévêque N, Chomel JJ, Dubrou S, Lévy-Bruhl D, Lina B. 2007. Surveillance of enterovirus in France, 2000–2004. Eur. J. Clin. Microbiol. Infect. Dis. 26:403–412. 10.1007/s10096-007-0306-4 [DOI] [PubMed] [Google Scholar]
- 75.Klemola P, Kaijalainen S, Ylipaasto P, Roivainen M. 2008. Diabetogenic effects of the most prevalent enteroviruses in Finnish sewage. Ann. N. Y. Acad. Sci. 1150:210–212. 10.1196/annals.1447.012 [DOI] [PubMed] [Google Scholar]
- 76.Groneck P, Jahn P, Schuler-Lüttmann S, Beyrer K. 2011. Neonatal enterovirus meningitis: transmission via parents during rooming-in and current epidemiology in Germany. Z. Geburtshilfe Neonatol. 215:1–5 (In German.). 10.1055/s-0030-1255024 [DOI] [PubMed] [Google Scholar]
- 77.Ramelli GP, Simonetti GD, Gorgievski-Hrisoho M, Aebi C, Bianchetti MG. 2004. Outbreak of coxsackie B5 virus meningitis in a Scout camp. Pediatr. Infect. Dis. J. 23:86–87 [DOI] [PubMed] [Google Scholar]
- 78.Rezig D, Fares W, Seghier M, Yahia AB, Touzi H, Triki H. 2011. Update on molecular characterization of coxsackievirus B5 strains. J. Med. Virol. 83:1247–1254. 10.1002/jmv.22084 [DOI] [PubMed] [Google Scholar]
- 79.Nairn C, Clements GB. 1999. A study of enterovirus isolations in Glasgow from 1977 to 1997. J. Med. Virol. 58:304–312 [PubMed] [Google Scholar]
- 80.Trallero G, Avellon A, Otero A, De Miguel T, Pérez C, Rabella N, Rubio G, Echevarria JE, Cabrerizo M. 2010. Enterovirus in Spain over the decade 1998–2007: virological and epidemiological studies. J. Clin. Virol. 47:170–176. 10.1016/j.jcv.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 81.Lukashev AN, Ivanova OE, Eremeeva TP, Gmyl LV. 2008. Analysis of echovirus 30 isolates from Russia and new independent states revealing frequent recombination and reemergence of ancient lineages. J. Clin. Microbiol. 46:665–670. 10.1128/JCM.02386-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cabrerizo M, Echevarria JE, González I, de Miguel T. 2008. Molecular epidemiological study of HEV-B enteroviruses involved in the increase in meningitis cases occurred in Spain during 2006. J. Med. Virol. 80:1018–1024. 10.1002/jmv.21197 [DOI] [PubMed] [Google Scholar]
- 83.Lévêquea N, Jacquesa J, Renoisa F, Antona D, Abelyd M, Chomele JJ, Andréolettia L. 2010. Phylogenetic analysis of echovirus 30 isolated during the 2005 outbreak in France reveals existence of multiple lineages and suggests frequent recombination events. J. Clin. Virol. 48:137–141. 10.1016/j.jcv.2010.03.011 [DOI] [PubMed] [Google Scholar]
- 84.Milia MG, Cerutti F, Gregori G, Burdino E, Allice T, Ruggiero T, Proia M, De Rosa G, Enrico E, Lipani F, Di Perri G, Ghisetti V. 2013. Recent outbreak of aseptic meningitis in Italy due to echovirus 30 and phylogenetic relationship with other European circulating strains. J. Clin. Virol. 58:579–583. 10.1016/j.jcv.2013.08.023 [DOI] [PubMed] [Google Scholar]
- 85.Savolainen-Kopra C, Paananen A, Blomqvist S, Klemola P, Simonen ML, Lappalainen M, Vuorinen T, Kuusi M, Lemey P, Roivainen M. 2011. A large Finnish echovirus 30 outbreak was preceded by silent circulation of the same genotype. Virus Genes 42:28–36. 10.1007/s11262-010-0536-x [DOI] [PubMed] [Google Scholar]
- 86.Prim N, Rodríguez G, Margall N, Del Cuerpo M, Trallero G, Rabella N. 2013. Combining cell lines to optimize isolation of human enterovirus from clinical specimens: report of 25 years of experience. J. Med. Virol. 85:116–120. 10.1002/jmv.23426 [DOI] [PubMed] [Google Scholar]
- 87.Rodríguez RA, Gundy PM, Gerba CP. 2008. Comparison of BGM and PLC/PRC/5 cell lines for total culturable viral assay of treated sewage. Appl. Environ. Microbiol. 74:2583–2587. 10.1128/AEM.00626-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Combelas N, Holmblat B, Joffret ML, Colbère-Garapin F, Delpeyroux F. 2011. Recombination between poliovirus and coxsackie A viruses of species C: a model of viral genetic plasticity and emergence. Viruses 3:1460–1484. 10.3390/v3081460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Norder H, Bjerregaard L, Magnius L, Lina B, Aymard M, Chomel JJ. 2003. Sequencing of “untypeable” enteroviruses reveals new types, EV-77 and EV-78, within human enterovirus type B and substitutions in the BC loop of the VP1 protein for known types. J. Gen. Virol. 84:827–836. 10.1099/vir.0.18647-0 [DOI] [PubMed] [Google Scholar]