Abstract
Cytophaga hutchinsonii is an aerobic cellulolytic soil bacterium which was reported to use a novel contact-dependent strategy to degrade cellulose. It was speculated that cellooligosaccharides were transported into the periplasm for further digestion. In this study, we reported that most of the endoglucanase and β-glucosidase activity was distributed on the cell surface of C. hutchinsonii. Cellobiose and part of the cellulose could be hydrolyzed to glucose on the cell surface. However, the cell surface cellulolytic enzymes were not sufficient for cellulose degradation by C. hutchinsonii. An outer membrane protein, CHU_1277, was disrupted by insertional mutation. Although the mutant maintained the same endoglucanase activity and most of the β-glucosidase activity, it failed to digest cellulose, and its cellooligosaccharide utilization ability was significantly reduced, suggesting that CHU_1277 was essential for cellulose degradation and played an important role in cellooligosaccharide utilization. Further study of cellobiose hydrolytic ability of the mutant on the enzymatic level showed that the β-glucosidase activity in the outer membrane of the mutant was not changed. It revealed that CHU_1277 played an important role in assisting cell surface β-glucosidase to exhibit its activity sufficiently. Studies on the outer membrane proteins involved in cellulose and cellooligosaccharide utilization could shed light on the mechanism of cellulose degradation by C. hutchinsonii.
INTRODUCTION
Cellulose is the primary component of plant biomass derived through photosynthetic carbon fixation (1), which is the sustainable source of biofuels and materials available to humans (2). Cellulose is composed of β-1,4-linked glucose chains forming a tightly ordered crystalline structure which is difficult to degrade. Cellulolytic microorganisms often use two well-studied strategies to digest cellulose. Most aerobic fungi secrete extracellular free cellulases, including endoglucanases (EC 3.2.1.4), exoglucanases (cellobiohydrolases) (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21), which act synergistically to digest cellulose (1, 3). Most anaerobic bacteria produce cell surface-anchored multicellulase complexes (cellulosomes) to degrade cellulose (4, 5). However, aerobic Cytophaga hutchinsonii and anaerobic Fibrobacter succinogenes utilize cellulose through a unique mechanism without cellulosomes or free cellulases (6).
C. hutchinsonii is an abundant aerobic cellulolytic Gram-negative soil bacterium (7, 8). Direct contact between C. hutchinsonii cells and insoluble cellulose is necessary for cellulose degradation (9), and most of the cellulase activity appears to be cell associated (8, 10, 11). It was speculated that C. hutchinsonii cells use a contact-dependent digestion mode to utilize cellulose, but details of the mechanism were still unknown. Recently, several genetic manipulation techniques, including transposon mutagenesis and gene disruption with replicative plasmids or suicide vectors, have been developed (12–15) which offer a great opportunity to explore the mysterious mechanism of cellulose degradation by C. hutchinsonii. In a previous study, we found that a cellulose utilization-deficient mutant lost some cellulose binding proteins in the outer membrane. These proteins were thought to play important roles in cellulose degradation (13). Here, one of these cellulose binding proteins, CHU_1277, was inactivated by insertional mutagenesis. The cellulose and cellooligosaccharide utilization abilities of the mutant were studied on both the microbial and enzymatic levels to explore the role of CHU_1277 in cellulose utilization. Based on the findings, a pathway for cellulose degradation on the cell surface of C. hutchinsonii is proposed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Cytophaga hutchinsonii ATCC 33406 was kindly provided by Mark J. McBride and grown in peptone-yeast-glucose (PYG) medium at 30°C (14). Escherichia coli strains were grown in Luria-Bertani medium at 37°C. For testing C. hutchinsonii utilization of different carbohydrate sources, 0.4% (wt/vol) glucose was replaced by 0.2% (wt/vol) cellobiose, 0.2% (wt/vol) cellotriose, 0.2% (wt/vol) cellotetrose, 0.2% (wt/vol) cellopentaose, 0.4% (wt/vol) regenerated amorphous cellulose (RAC), or 0.4% (wt/vol) crystalline cellulose. RAC was prepared as described by Zhang et al. (16). For detection of filter paper utilization, C. hutchinsonii was inoculated into the filter paper, which was preplaced on the top of solid PYG agar without glucose. C. hutchinsonii was grown on PYG solid medium containing 5 g of agar per liter to observe colony spreading. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/ml; erythromycin (Em), 30 μg/ml; chloramphenicol (Cm), 10 μg/ml. The plasmids and primers used are listed in Table 1.
TABLE 1.
Plasmids and primers used in this study
| Plasmid or primer | Description or sequencea | Reference(s) or source |
|---|---|---|
| Plamids | ||
| pLYL03 | ColE1, Bacteroides-Flavobacterium suicide vector, Apr Emr | 12, 27 |
| pLYIN1277 | 800-bp fragment amplified with primers 1277-L and 1277-R inside CHU_1277 cloned into BamHI and XbaI sites of pLYL03; Apr Emr | This study |
| pSKSO8 | pUC ori, Apr Emr, oriC (the replication origin of chromosome of C. hutchinsonii), replicative plasmid of C. hutchinsonii | 14 |
| pCH | pUC ori, Apr Cmr, oriC, vector used for complementation | This study |
| pCH1277 | pCH containing CHU_1277 (promoter of CHU_1277), Apr Cmr | This study |
| pCHP1277 | pCH containing CHU_1277 (promoter of CHU_1284), Apr Cmr | This study |
| Primers | ||
| 1277-L | GCAATTGGATCCAGAAACAGAAGATACACAC | |
| 1277-R | TGTATGTCTAGAATAGCACCACCTTTTAAGC | |
| 1277test-L | TCAATCGCTCTTCGTTCGTC | |
| 1277test-R | GTTTGACTTACCAGCTTCAC | |
| C1277F | AACTGAGCTCTGGGGTTATGGACTTGGAATCG | |
| NdeI-1277F | CGCAGGCATATGTCAACAATGGGAAC | |
| C1277R | GTCCGTCGACTACAGCAAACAAACTATG | |
| 1276RT-1 | TGGGTATCATTCGTGAGGACAG | |
| 1276RT-2 | ATTACGTTGGTAAGGACCTGCA | |
| 1277RT-1 | GGCTTACGTGCATTTGCTAC | |
| 1277RT-2 | TTACGCCACCTCGTGTTGTT | |
| 1278RT-1 | ATGCTGGTAAATGCGGGTTCTA | |
| 1278RT-2 | GAAGGACAGAAATTATCCGTGT | |
| 1279RT-1 | AGCCAGATTGCGGTGGTAGAAG | |
| 1279RT-2 | GTAACCCGGACCCACTCCTGTA |
Restriction sites are underlined.
Gene targeting of CHU_1277.
CHU_1277 was interrupted by insertional inactivation with plasmid pLYL03 as the suicide vector according to the description of Zhu et al. (15). The internal fragment of CHU_1277 was amplified from C. hutchinsonii genomic DNA using the primers 1277-L and 1277-R. The fragment was digested with BamHI and XbaI and cloned into plasmid pLYL03, resulting in plasmid pLYIN1277. The plasmid was transformed into C. hutchinsonii by electroporation and plated on PYG agar containing erythromycin. After incubation at 30°C for 5 to 7 days, the Emr colonies were picked and analyzed by PCR with primers located in the plasmid (1277test-L) and downstream of the TAA stop codon (1277test-R) to determine if homologous recombination insertion had occurred. The PCR-positive colonies with 1.87-kb bands were subcultured for further study.
Detection and localization of CHU_1277.
Outer membrane protein preparation and binding to cellulose were performed as previously described (13), and cellulose binding outer membrane proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The target protein band was sliced and identified by an Ultraflex III matrix-assisted laser desorption ionization–two-stage time of flight mass spectrometer (MALDI-TOF/TOF-MS) (Bruke).
To locate CHU_1277, the buffer-washed proteins were collected by a modification of the procedure described by Jun et al. (17). Briefly, cells cultured in PYG medium were pelleted at 6,000 × g for 15 min. The cells were resuspended in 50 mM piperazine-1,4-bis(2-ethanesulfonic acid) (PIPES) buffer (pH 6.8). The suspension was incubated at 4°C for 5 min with shaking at 150 rpm, followed by centrifugation at 23,000 × g for 20 min to remove the cells. The supernatant containing the buffer-washed proteins was subjected to ultracentrifugation (100,000 × g, 20 min). The sediment was resuspended in PIPES buffer and separated by SDS-PAGE. In this way, the protein band of CHU_1277 was directly detected from the profile of the SDS-PAGE gel.
Reverse transcription-PCR (RT-PCR) analysis.
Total RNA was isolated using a bacterial RNA kit (Omega, Norcross, GA, USA) from 3 ml of exponential-phase cultures of wild-type C. hutchinsonii and a CHU_1277-disrupted mutant grown in PYG medium. Elimination of traces of DNA and synthesis of first-strand cDNA were carried out with a PrimeScript RT reagent kit containing a genomic DNA (gDNA) Eraser from TaKaRa (Dalian, China) according to the manufacturer's instructions. Then the cDNA was used as a template to perform PCR with the primers listed in Table 1 (1276RT-1, 1276RT-2, 1277RT-1, 1277RT-2, 1278RT-1, 1278RT-2, 1279RT-1, and 1279RT-2). RNA without reverse transcription was used as the template in PCR control reactions to determine whether the RNA was free of genomic DNA.
Complementation of a CHU_1277 mutant.
The replicative plasmid pCH used for complementation of genes in C. hutchinsonii was constructed from plasmid pSKSO8 (14), in which the chloramphenicol acetyltransferase gene (cat) was under the control of the ompA promoter from Flavobacterium johnsoniae (18). The map of pCH is shown in Fig. S1 in the supplemental material.
Based on pCH, two plasmids, pCH1277 and pCHP1277, were constructed to complement the CHU_1277 mutant. A fragment spanning CHU_1277, 232 bp upstream of the start codon, according to the genome annotation, and 54 bp downstream of the stop codon, was amplified with primers C1277F and C1277R. The fragment was digested with SacI and SalI and ligated into the corresponding sites of pCH to generate pCH1277. Plasmid pCHP1277 was constructed with CHU_1277 under the control of the constitutive promoter of CHU_1284 described by Xu et al. (14). In detail, a 1.9-kbp fragment spanning CHU_1277 (from 66 bp upstream of the start codon to 54 bp downstream of the stop codon) was amplified with primers NdeI-1277F and C1277R, digested with NdeI and SalI, and ligated into the corresponding sites of pCH. Plasmids pCH1277 and pCHP1277 were electroporated into the CHU_1277 mutant, and transformants were selected by chloramphenicol resistance.
Growth curves, enzymatic assay, and cellulose adhesion test.
Cells of C. hutchinsonii were grown in PYG medium or PY-cellobiose medium, in which 0.2% (wt/vol) cellobiose was used instead of glucose. Incubations were done in 300-ml flasks with shaking (160 rpm) at 30°C, and growth rates were determined by monitoring the optical density at 600 nm of the cultures.
Cells of the mid-exponential phase were gathered through centrifugation (6,000 × g, 10 min) for cellulase activity measurement. For intact cell samples, pelleted cells were washed with Na2HPO4 · KH2PO4 buffer (50 mM, pH 6.8) and resuspended in the same buffer with 0.04% (wt/vol) NaN3. The suspended cells were sonicated to prepare the cell lysates, and the outer membrane proteins were prepared according to a previously described procedure (13). Sodium carboxymethyl cellulose (CMC-Na), p-nitrophenyl beta-d-glucopyranoside (pNPG), and p-nitrophenyl beta-d-cellobioside (pNPC) were all purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as the substrates to measure endoglucanase, β-glucosidase, and exoglucanase activities, respectively, according to previously described methods (13, 19). All of the enzymatic assays were carried out in triplicate, and the protein concentration was quantified as described by Bradford (20).
The relative adhesion rate of C. hutchinsonii cells to Avicel cellulose PH105 (Sigma-Aldrich) was measured by a turbidity-based method as previously described (13, 21).
Cellulose and cellobiose hydrolysis by cells or outer membrane proteins of C. hutchinsonii.
Cells from exponentially growing cultures (200 ml) were harvested (6,000 × g, 10 min), washed with Na2HPO4 · KH2PO4 buffer (50 mM, pH 6.8), and resuspended in the same buffer to a volume of 20 ml. The suspension (5 ml) was mixed with 1% (wt/vol) RAC (5 ml). Glucono-δ-lactone or NaN3 was added to the reaction mixture to a final concentration of 0.2% or 0.04% (wt/vol) separately as needed. The mixture was incubated at 30°C for 6 h, which was followed by centrifugation at 10,000 × g for 10 min. The supernatants containing hydrolytic products were ultrafiltered, lyophilized, and then dissolved with 1 ml of ultrapure water. Hydrolytic products were analyzed by a Dionex Ultra high-performance liquid chromatograph (HPLC) (Sunnyvale, CA, USA) equipped with a Corona ultradetector and XBridge amide column (Waters, Milford, MA, USA). The column was maintained at 50°C and eluted with a mobile phase of acetonitrile-water (65:35, vol/vol) at a flow rate of 0.6 ml/min.
To analyze cellobiose hydrolysis by intact cells or outer membrane proteins, cellobiose was mixed with intact cells or outer membrane proteins and incubated at 30°C. The initial concentration of cellobiose was 0.2% (wt/vol). The protein content of the cells or outer membrane proteins was 0.005% (wt/vol). One milliliter of the mixture was withdrawn at regular time intervals and analyzed by HPLC. The protein concentration was quantified as described by Bradford (20), and all the assays were carried out in triplicate.
RESULTS
Disruption of CHU_1277 in C. hutchinsonii.
The CHU_1277 locus encodes a cellulose binding outer membrane protein, CHU_1277, which consists of 584 amino acids with a molecular mass of 65.3 kDa, according to the genome annotation (13). No signal peptide was detected in CHU_1277 by SignalP, version 4.0 (22), and no transmembrane helices were predicted by TMHMM, version 2.0 (23). However, we examined the DNA sequence and found several potential start codons upstream of the one selected by the automated annotation system. According to two of the potential start codons, CHU_1277 would contain 606 (or 603) amino acids instead of 584 amino acids, and an obvious signal peptide was detected at the amino terminus. The prediction of subcellular localization by PSORTb, version 3.0 (24), also indicated that CHU_1277 was located on the outer membrane (13). This was in accordance with our previous study showing that CHU_1277 is an outer membrane protein. These results implied that the annotation of CHU_1277 was possibly incorrect. The BLAST results for the protein sequence revealed that CHU_1277 showed only weak similarity to a hypothetical membrane protein from Fibrobacter succinogenes (GenBank accession number YP_003249606.1; 17% identity over 584 amino acids), a bacterium speculated to have a cellulolytic mechanism similar to that of C. hutchinsonii (6). The bioinformatic analysis showed that CHU_1277 was a protein with unknown function.
To investigate its physiological role, CHU_1277 was interrupted by insertional inactivation. The plasmid pLYIN1277 was transformed into C. hutchinsonii by electroporation. Cells resistant to erythromycin were subjected to PCR assay with the diagnostic primers 1277test-L and 1277test-R, and an amplicon of 1.87 kbp for the mutant strain which was absent for the wild-type strain verified that homologous recombination had occurred at the CHU_1277 locus (data not shown).
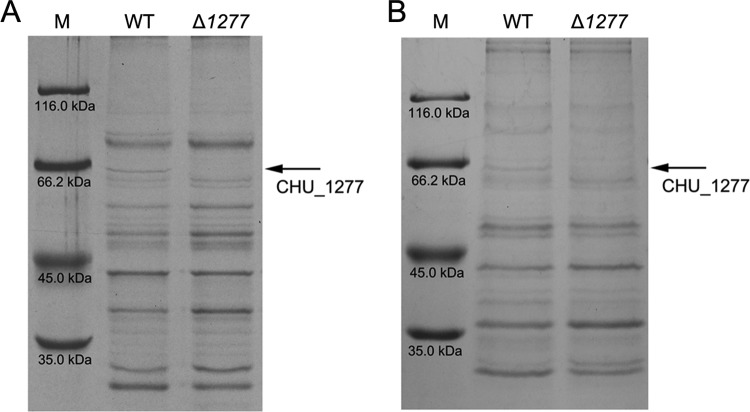
Our previous work showed that CHU_1277 was detected in the cellulose binding outer membrane proteins (13). To determine the disruption of CHU_1277 in the mutant cells, the cellulose binding outer membrane proteins and the buffer-washed proteins of both the wild-type strain and the mutant were prepared and analyzed by SDS-PAGE (Fig. 1). The profiles showed that CHU_1277 was absent from both of the samples of the mutant, indicating that it was disrupted in the mutant cells. CHU_1277 could be washed out from the intact cells by PIPES buffer, indicating that it might be located on the cell surface (Fig. 1B).
FIG 1.
SDS-PAGE of the wild-type (WT) strain of C. hutchinsonii and the CHU_1277-disrupted mutant (Δ1277). (A) SDS-PAGE of outer membrane proteins binding to cellulose; (B) SDS-PAGE of buffer-washed proteins. Lane M, molecular mass marker.
Phenotypic characteristics of the mutant.
Many phenotypic characteristics, including growth, cellulose degradation, colony spreading, and adhesion to cellulose of the mutant cells, were studied to investigate the physiological role of CHU_1277.
The growth properties of the CHU_1277 mutant and the wild-type strain in liquid medium with different carbohydrates as the sole carbon source are shown in Table 2. The results indicate that the mutant grew well in glucose medium but could not utilize RAC and crystalline cellulose. The mutant grew slowly and poorly in cellooligosaccharide medium (from cellobiose to cellopentaose). The growth curves of the mutant in glucose and cellobiose liquid medium were further determined and compared with those of the wild type. In glucose medium, the mutant had a growth rate similar to that of the wild-type strain, and its final cell density was almost the same as that of the wild-type strain (Fig. 2A). When the glucose-grown cells were inoculated into cellobiose culture, the mutant showed a reduced growth rate in the exponential phase, and its lag phase was about 45 h longer than that of the wild-type strain.
TABLE 2.
Growth of the wild type of C. hutchinsonii and the CHU_1277-disrupted mutant on different carbohydrates
| Strain | Growth on the indicated carbohydratea |
||||||
|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | RAC | MCC | |
| Wild type | + + | + + | + + | + + | + + | + + | + + |
| CHU_1277 mutant | + + | + | + | + | + | − | − |
Growth is indicated as follows: + +, approximately that of the wild type; +, less than that of the wild type; and −, same as that of the negative control. Carbohydrates are the following: G1, glucose; G2, cellobiose; G3, cellotriose; G4, cellotetrose; G5, cellopentaose; RAC, regenerated amorphous cellulose; and MCC, microcrystalline cellulose.
FIG 2.
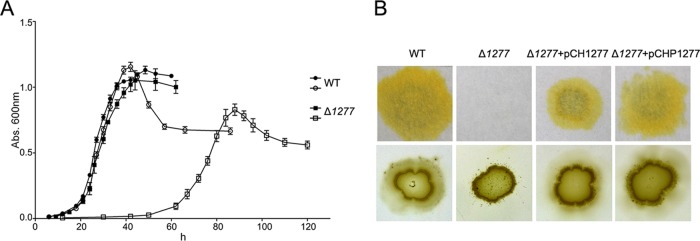
Phenotypic characteristics of the wild-type (WT) strain of C. hutchinsonii and the CHU_1277-disrupted mutant (Δ1277). (A) Growth curves of the cells in glucose and cellobiose medium. The closed and open symbols stand for growth of the cells in glucose and cellobiose, respectively. (B) Filter paper degradation and colony spreading of the bacterial cells on soft agar. Δ1277+pCH1277, the CHU_1277 mutant complemented with pCH1277; Δ1277+pCHP1277, the CHU_1277 mutant complemented with pCHP1277. Abs, absorbance.
A filter paper plate assay could directly reflect the utilization of the filter paper by the C. hutchinsonii cells. On the filter paper plate, the wild-type strain could completely digest filter paper after 7 days, while the CHU_1277-disrupted mutant showed no sign of growth, even after 15 days (Fig. 2B). This suggested that the mutant cells had lost the ability to utilize filter paper, which was in accordance with the result that the mutant cells showed no growth in RAC and crystalline cellulose medium. In our previous study, the wild-type C. hutchinsonii cells exhibited colony spreading on the surface of a PYG plate with 0.5% agar (13). As shown in Fig. 2B, the wild-type cells spread out evenly as a thin layer, while the mutant cells showed an unusual colony-spreading pattern, with satellite colonies spread out from the periphery of the mutant inoculant. It seemed that the mutant had lost the associated moving ability of colony-forming cells but still retained the moving ability of individual cells on the soft-agar surface (Fig. 2B).
The relative adhesion rate of the mutant cells to cellulose was about 93%, which was similar to that of the wild-type strain although CHU_1277 is a cellulose binding protein. This indicated that more proteins were involved in cellulose binding with cells.
RT-PCR analysis and complementation of the CHU_1277 mutant.
In the genome of C. hutchinsonii, CHU_1277 was arranged in the same direction as its surrounding genes (Fig. 3A). To investigate whether the transcription of the surrounding genes was affected by the insertion in CHU_1277, RT-PCR was performed as described in Materials and Methods. The amplicons corresponding to CHU_1276, CHU_1277, CHU_1278, and CHU_1279 were all present in the wild-type strain of C. hutchinsonii while the CHU_1277 amplicon was not obtained in the CHU_1277-disrupted mutant (Fig. 3B). The presence of amplicons corresponding to CHU_1276, CHU_1278, and CHU_1279 in the CHU_1277-disrupted mutant indicated that the insertion in CHU_1277 did not affect the transcription of the surrounding genes.
FIG 3.
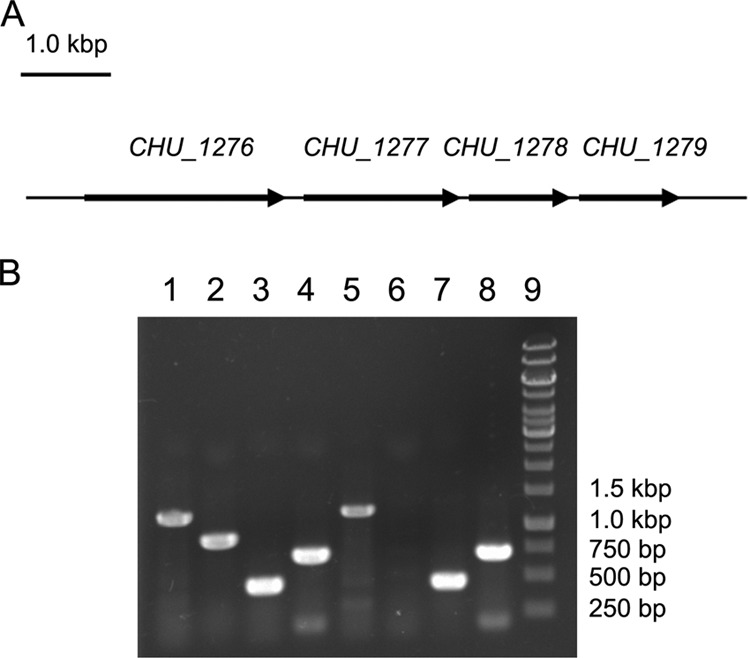
RT-PCR analysis of the transcription of CHU_1277 and the surrounding genes. (A) Illustration of CHU_1277 and the surrounding genes; (B) RT-PCR analysis of the transcription of CHU_1277 and the surrounding genes in the wild-type (WT) strain of C. hutchinsonii and the CHU_1277-disrupted mutant (Δ1277). Lanes 1 to 4, PCRs performed with WT cDNA; lanes 5 to 8, PCRs performed with cDNA of the CHU_1277 mutant; lane 9, marker. The following primers were used for PCRs: 1276RT-1 and 1276RT-2 for lanes 1 and 5; 1277RT-1 and 1277RT-2 for lanes 2 and 6; 1278RT-1 and 1278RT-2 for lanes 3 and 7; 1279RT-1 and 1279RT-2 for lanes 4 and 8.
To verify that the defects in cellulose degradation and colony spreading of the mutant were caused by the disruption of CHU_1277, complementation of the mutant was carried out with plasmids pCH1277 (with its own promoter region) and pCHP1277 (with the constitutive promoter of CHU_1284) as described in Materials and Methods. As shown in Fig. 2B, both of the complemented strains could restore the ability to grow on filter paper and spread on soft-agar surfaces. These results proved that the defects in cellulose degradation and colony spreading of the mutant were caused by the inactivation of CHU_1277.
Cellulase activity determination.
In order to investigate why the mutant cells could not digest cellulose, cellulase activities of the wild-type and the mutant cells were analyzed. Since the mutant could not grow in cellulose medium, cells cultured on PYG medium were collected for cellulase activity assays. As shown in Fig. 4, both endoglucanase activities and β-glucosidase activities could be detected, while no exoglucanase activities were detected. The result was consistent with genomic analysis confirming that the genome of C. hutchinsonii encodes many putative endoglucanases and β-glucosidases but has no homologs of exoglucanases.
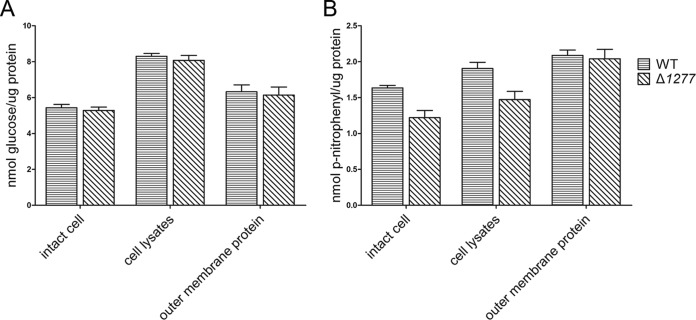
FIG 4.
Endoglucanase and β-glucosidase activities of the wild-type (WT) strain of C. hutchinsonii and the CHU_1277-disrupted mutant (Δ1277). (A) Endoglucanase activity of the intact cells, cell lysates, and outer membrane proteins of the wild-type strain and the CHU_1277 mutant. Endoglucanase activity was determined using CMC-Na as the substrate, and the reducing end concentration was measured using the dinitrosalicylic acid procedure. (B) β-Glucosidase activity of the intact cells, cell lysates, and outer membrane proteins of the wild-type strain and the CHU_1277 mutant. β-Glucosidase activity was determined using pNPG as the substrate, and the released p-nitrophenol was determined by absorption at 410 nm. Error bars indicate standard errors.
Compared with the enzyme activities in cell lysates, which represented total enzyme activities of the cells, the intact cell (enzyme activities on the cell surface) possessed approximately 65% of endoglucanase activity and about 86% of the β-glucosidase activity. (Fig. 4A and B). This indicated that the majority of the cellulase activities were on the cell surface.
Endoglucanase activities of the intact cell, cell lysates, and outer membrane proteins of the wild-type cells and the mutant were almost the same, indicating that endoglucanases were unaffected by the disruption of CHU_1277 (Fig. 4A). The β-glucosidase activity of outer membrane proteins of the mutant was also the same as that of the wild-type strain, while the activities on the cell surface and in cell lysates of the mutant were only 74% and 77% of the wild-type strain. This result seems to suggest that the quantity of β-glucosidases of the mutant cells did not change, but their apparent activity was reduced by the disruption of CHU_1277 (Fig. 4B).
Cellulose hydrolysis.
Since significant cellulolytic enzyme activities were detected on the cell surface and in outer membrane proteins of C. hutchinsonii, the cellulose hydrolysates of the intact cells and outer membrane proteins were further analyzed. Cellulose hydrolysate of the intact cells was detected only for the wild-type because the mutant could not live on cellulose.
After incubation of C. hutchinsonii cells with RAC for 6 h, the supernatant was concentrated and analyzed by HPLC. Results showed that there was a small amount of cellobiose and little glucose in the extracellular mixture (Fig. 5B). When 0.2% (wt/vol) glucono-δ-lactone was added to the reaction mixture, which should repress β-glucosidase activity, the amounts of cellobiose, cellotriose, and cellotetrose were significantly increased, while the concentration of glucose did not change (Fig. 5B). These cellooligosaccharide products should mainly come from the degradation of cellulose by cell surface endoglucanase since the β-glucosidase activity was suppressed. With the addition of 0.04% (wt/vol) NaN3, which should inhibit ATP production through the respiratory chain reaction, the ATP-driven transport and assimilation of the cell were inhibited. The extracellular cellooligosaccharide products were further degraded to glucose by the cell surface β-glucosidases. So no cellotriose or cellotetrose was detected under this condition, and cellobiose decreased to a small amount while the accumulation of glucose significantly increased (Fig. 5B).
FIG 5.
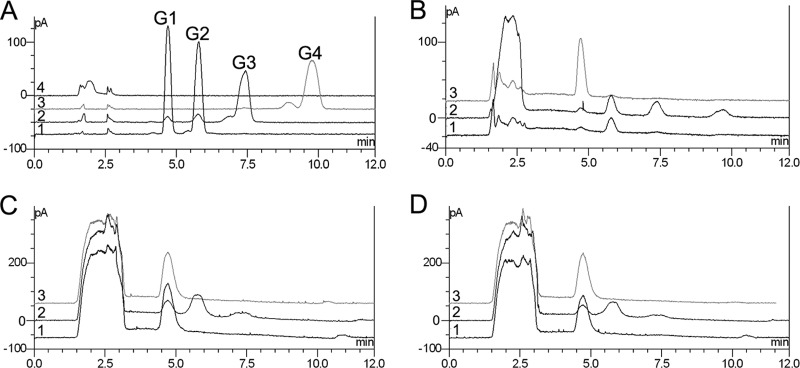
HPLC analyses of hydrolytic products of RAC by intact cells and outer membrane proteins of C. hutchinsonii. (A) Cellooligosaccharide standards. Line 1, glucose and cellobiose; line 2, cellotriose; line 3, cellotetrose; line 4, RAC; G1, glucose; G2, cellobiose; G3, cellotriose; G4, cellotetrose. (B to D) RAC hydrolysates of the intact cells of the wild type (B), the outer membrane proteins of the wild type (C), and the outer membrane proteins of the CHU_1277-disrupted mutant (D) were analyzed. Line 1, RAC was incubated with the intact cells; line 2, 0.2% glucono-δ-lactone was added; line 3, 0.04% NaN3 was added.
Outer membrane proteins extracted from the wild-type cells were also proven to have the ability to hydrolyze cellulose to glucose by HPLC. And no other cellooligosaccharides were detected. When β-glucosidase activity was repressed by the addition of glucono-δ-lactone, more cellobiose and cellotriose were present in the hydrolytic products, and the amount of glucose was reduced. This result was similar to the result of RAC hydrolysis by intact cells. The addition of NaN3 did not change the composition or the quantity of the reaction products, suggesting that NaN3 had no direct effect on enzymatic activity (Fig. 5C). There was no difference between the wild-type strain and the mutant with respect to cellulose hydrolytic activity by the outer membrane proteins (Fig. 5D), implying that cellulolytic enzymes in the outer membrane of the mutant cells were unaffected.
Cellobiose hydrolysis.
To compare the cellobiose hydrolytic abilities of the wild-type strain and the mutant, cellobiose hydrolysates of the intact cells and the outer membrane proteins were analyzed by HPLC. As shown in Fig. 6A, cellobiose was quickly hydrolyzed to glucose in the wild-type cells, while hydrolysis by the mutant cells was very slow. After a 4.5-h incubation, only 35% of cellobiose remained in the reaction mixture of the wild-type strain while 75% remained in the mutant mixture. This revealed that the actual cellobiose hydrolytic ability of the mutant was significantly reduced. However, the cellobiose hydrolytic ability of mutant outer membrane proteins was almost the same as that of the wild-type proteins (Fig. 6B). Cellobiose was completely hydrolyzed into glucose after a 14.5-h incubation. The result that the quantity and activity of β-glucosidase in extracted outer membrane proteins of the mutant cells were not changed was in accordance with the β-glucosidase activity assay.
FIG 6.
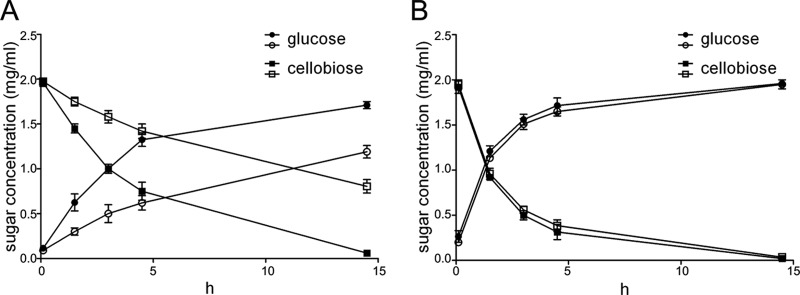
HPLC analyses of cellobiose hydrolysate by the intact cells and outer membrane proteins of the wild-type (WT) strain of C. hutchinsonii and the CHU_1277-disrupted mutant (Δ1277). (A) Cellobiose hydrolysate of the intact cells of the wild-type strain and the CHU_1277 mutant; (B) cellobiose hydrolysate of the outer membrane proteins of the wild-type strain and the CHU_1277 mutant. The closed and open symbols represent cellobiose hydrolysis of the wild-type strain and the CHU_1277 mutant, respectively.
DISCUSSION
Direct contact between the cells and insoluble cellulose was thought to be necessary for cellulose degradation by C. hutchinsonii (9), and most of the cellulase activity appears to be cell associated (8, 10, 11). A possible cellulose utilization model was proposed according to which individual cellulose molecules are transported into the periplasmic space and degraded by the endoglucanases there (25). In this study, we found that most of the endoglucanase and β-glucosidase activities were detected on the cell surface rather than in the periplasmic space as speculated before. Moreover, part of the cellulose could be hydrolyzed to glucose on the cell surface (Fig. 5). The extracted outer membrane proteins containing free endoglucanases and β-glucosidases also had the ability to hydrolyze cellulose to glucose in vitro. We propose that on the cell surface of C. hutchinsonii, cellulose could be partly depolymerized to cellooligosaccharides by cell surface endoglucanases and that the cellooligosaccharides could be further hydrolyzed to glucose by cell surface glucosidases. Some recent studies on cell-bound endoglucanases support this view. Zhu et al. reported that an outer membrane endoglucanase, ChCel5A, from C. hutchinsonii could hydrolyze microcrystalline cellulose to cellobiose and cellotriose (15). Another cell surface endoglucanase, CHU_2103, was shown to be a processive endoglucanase (26). However, whether cell surface degradation was the main mechanism of cellulose utilization by C. hutchinsonii was still unknown.
When cellulose was degraded by the intact cells, small amounts of glucose and cellobiose could be detected, as shown in Fig. 5B. NaN3 could block the ATP-driven transport and assimilation. Therefore, with the addition of NaN3, the amount of extracellular cellulose hydrolysate increased significantly. Only a small amount of reducing sugar could be detected in the dynamic process without NaN3, suggesting that the cellulose degradation and ATP required for assimilation of the hydrolysate might be tightly coupled. That might be one of the reasons why no reducing sugar was detected extracellularly in many previous studies (7, 10, 11).
Cellobiose is one of the few substrates that can be used by C. hutchinsonii as the sole carbon and energy source (8), which is also the main intermediate product of cellulose degradation. In the study of cellobiose hydrolysis by the living cells of C. hutchinsonii, cellobiose was hydrolyzed to glucose mainly during the lag phase, and glucose acted as the main carbon source to support further growth of the bacterium (see Fig. S2 in the supplemental material). Quantitative analysis showed that 2 mg/ml of cellobiose was completely hydrolyzed and that 1.7 mg/ml glucose was accumulated extracellularly, which represented about 85% of the theoretical yield (Fig. 6). All of these results indicated that degradation of cellobiose into glucose by cell surface β-glucosidases was the main mechanism of cellobiose utilization by C. hutchinsonii. This is significantly different from previous studies that indicated that most aerobic fungi secrete free β-glucosidases, while most anaerobic bacteria use cytoplasmic β-glucosidases to degrade cellobiose intracellularly (1). The isolation and further study of the cell surface β-glucosidases are now being undertaken in our lab.
CHU_1277, coding an outer membrane cellulose binding protein, was inactivated by insertional mutagenesis. The CHU_1277-disrupted mutant cells were deficient in cellulose utilization and grew slowly and poorly in different cellooligosaccharide media. The apparent β-glucosidase activity and the cellobiose hydrolytic ability of the intact mutant cells were dramatically decreased. However, the outer membrane proteins extracted from the mutant cells have β-glucosidase activity and cellobiose hydrolytic activity similar to those of the wild-type strain in vitro (Fig. 5 and 6). These results implied that the quantity and activity of the β-glucosidase in the outer membrane of the mutant cells were unaffected. Since the cellulose adhesion rate of mutant cells was not significantly changed, we speculated that the direct or indirect interaction between β-glucosidase and CHU_1277 is necessary for the cell surface enzyme to achieve sufficient activity. This revealed that in addition to cellulolytic enzymes, certain outer membrane proteins were also necessary for effective cellulose utilization by C. hutchinsonii. Further study of how CHU_1277 affects cellulose and cellooligosaccharide utilization will provide more insights into the novel mechanism of cellulose degradation by C. hutchinsonii .
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (2011CB707402) and the National Natural Science Foundation of China (31170051 and 31371262).
We sincerely thank Mark J. McBride (University of Wisconsin—Milwaukee) for providing C. hutchinsonii ATCC 33406. We thank Edward C. Mignot, Shandong University, for linguistic advice.
Footnotes
Published ahead of print 16 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00687-14.
REFERENCES
- 1.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506–577. 10.1128/MMBR.66.3.506-577.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynd LR, Wyman CE, Gerngross TU. 1999. Biocommodity engineering. Biotechnol. Prog. 15:777–793. 10.1021/bp990109e [DOI] [PubMed] [Google Scholar]
- 3.Zhang YH, Lynd LR. 2004. Toward an aggregated understanding of enzymatic hydrolysis of cellulose: noncomplexed cellulase systems. Biotechnol. Bioeng. 88:797–824. 10.1002/bit.20282 [DOI] [PubMed] [Google Scholar]
- 4.Bayer EA, Belaich JP, Shoham Y, Lamed R. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521–554. 10.1146/annurev.micro.57.030502.091022 [DOI] [PubMed] [Google Scholar]
- 5.Beguin P, Lemaire M. 1996. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit. Rev. Biochem. Mol. Biol. 31:201–236. 10.3109/10409239609106584 [DOI] [PubMed] [Google Scholar]
- 6.Wilson DB. 2008. Three microbial strategies for plant cell wall degradation. Ann. N. Y. Acad. Sci. 1125:289–297. 10.1196/annals.1419.026 [DOI] [PubMed] [Google Scholar]
- 7.Walker E, Warren FL. 1938. Decomposition of cellulose by Cytophaga. I. Biochem. J. 32:31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie G, Bruce DC, Challacombe JF, Chertkov O, Detter JC, Gilna P, Han CS, Lucas S, Misra M, Myers GL, Richardson P, Tapia R, Thayer N, Thompson LS, Brettin TS, Henrissat B, Wilson DB, McBride MJ. 2007. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl. Environ. Microbiol. 73:3536–3546. 10.1128/AEM.00225-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larkin JM. 1989. Nonphotosynthetic, nonfruiting gliding bacteria, p 2010–2138 In Staley JT, Bryant MP, Pfennig N, Holt JG. (ed), Bergey's manual of systematic bacteriology, vol 3 Williams & Wilkins, Baltimore, MD [Google Scholar]
- 10.Chang WT, Thayer DW. 1977. The cellulase system of a Cytophaga species. Can. J. Microbiol. 23:1285–1292. 10.1139/m77-192 [DOI] [PubMed] [Google Scholar]
- 11.Stanier RY. 1942. The Cytophaga group: a contribution to the biology of myxobacteria. Bacteriol. Rev. 6:143–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride MJ, Baker SA. 1996. Development of techniques to genetically manipulate members of the genera Cytophaga, Flavobacterium, Flexibacter, and Sporocytophaga. Appl. Environ. Microbiol. 62:3017–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji X, Xu Y, Zhang C, Chen N, Lu X. 2012. A new locus affects cell motility, cellulose binding, and degradation by Cytophaga hutchinsonii. Appl. Microbiol. Biotechnol. 96:161–170. 10.1007/s00253-012-4051-y [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, Ji X, Chen N, Li P, Liu W, Lu X. 2012. Development of replicative oriC plasmids and their versatile use in genetic manipulation of Cytophaga hutchinsonii. Appl. Microbiol. Biotechnol. 93:697–705. 10.1007/s00253-011-3572-0 [DOI] [PubMed] [Google Scholar]
- 15.Zhu Y, Zhou H, Bi Y, Zhang W, Chen G, Liu W. 2013. Characterization of a family 5 glycoside hydrolase isolated from the outer membrane of cellulolytic Cytophaga hutchinsonii. Appl. Microbiol. Biotechnol. 97:3925–3937. 10.1007/s00253-012-4259-x [DOI] [PubMed] [Google Scholar]
- 16.Zhang YH, Cui J, Lynd LR, Kuang LR. 2006. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 7:644–648. 10.1021/bm050799c [DOI] [PubMed] [Google Scholar]
- 17.Jun HS, Qi M, Gong J, Egbosimba EE, Forsberg CW. 2007. Outer membrane proteins of Fibrobacter succinogenes with potential roles in adhesion to cellulose and in cellulose digestion. J. Bacteriol. 189:6806–6815. 10.1128/JB.00560-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S, Bagdasarian M, Kaufman MG, Bates AK, Walker ED. 2007. Mutational analysis of the ompA promoter from Flavobacterium johnsoniae. J. Bacteriol. 189:5108–5118. 10.1128/JB.00401-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang YHP, Hong J, Ye X. 2009. Cellulase assays. Methods Mol Biol. 581:213–231. 10.1007/978-1-60761-214-8_14 [DOI] [PubMed] [Google Scholar]
- 20.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 21.Gong J, Forsberg CW. 1989. Factors affecting adhesion of Fibrobacter succinogenes subsp. succinogenes S85 and adherence-defective mutants to cellulose. Appl. Environ. Microbiol. 55:3039–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 23.Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175–182 [PubMed] [Google Scholar]
- 24.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson DB. 2009. Evidence for a novel mechanism of microbial cellulose degradation. Cellulose 16:723–727. 10.1007/s10570-009-9326-9 [DOI] [Google Scholar]
- 26.Zhang C, Wang Y, Li Z, Zhou X, Zhang W, Zhao Y, Lu X. 2014. Characterization of a multi-function processive endoglucanase CHU_2103 from Cytophaga hutchinsonii. Appl. Microbiol. Biotechnol. 10.1007/s00253-014-5640-8 [DOI] [PubMed] [Google Scholar]
- 27.Li LY, Shoemaker NB, Salyers AA. 1995. Location and characteristics of the transfer region of a Bacteroides conjugative transposon and regulation of transfer genes. J. Bacteriol. 177:4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.