Abstract
Molecular diagnostics for crop diseases can enhance food security by enabling the rapid identification of threatening pathogens and providing critical information for the deployment of disease management strategies. Loop-mediated isothermal amplification (LAMP) is a PCR-based tool that allows the rapid, highly specific amplification of target DNA sequences at a single temperature and is thus ideal for field-level diagnosis of plant diseases. We developed primers highly specific for two globally important rice pathogens, Xanthomonas oryzae pv. oryzae, the causal agent of bacterial blight (BB) disease, and X. oryzae pv. oryzicola, the causal agent of bacterial leaf streak disease (BLS), for use in reliable, sensitive LAMP assays. In addition to pathovar distinction, two assays that differentiate X. oryzae pv. oryzae by African or Asian lineage were developed. Using these LAMP primer sets, the presence of each pathogen was detected from DNA and bacterial cells, as well as leaf and seed samples. Thresholds of detection for all assays were consistently 104 to 105 CFU ml−1, while genomic DNA thresholds were between 1 pg and 10 fg. Use of the unique sequences combined with the LAMP assay provides a sensitive, accurate, rapid, simple, and inexpensive protocol to detect both BB and BLS pathogens.
INTRODUCTION
Severe rice diseases, such as bacterial leaf streak (BLS), caused by Xanthomonas oryzae pv. oryzicola, and bacterial blight (BB), caused by X. oryzae pv. oryzae, are increasing in prevalence in parts of Asia and sub-Saharan Africa and can cause average yield losses of 20 and 50%, respectively (1). Increased incidences of BLS and BB are considered to be the result of the introduction of new susceptible rice varieties, the intensification of cultivation, the absence of adequate phytosanitary controls, and environmental changes, such as rising global temperatures (2, 3). The losses caused by these diseases could jeopardize global food security.
Documenting the extent and distribution of BB and BLS is invaluable to understanding the severity of their threat to rice production. Seed-borne dissemination of X. oryzae pv. oryzicola is a problem in parts of Asia and, presumably, in Africa (4). While clean seed and quarantine programs are prevalent in Asia, these have not yet been developed in Africa. X. oryzae pv. oryzae has been detected in seed, but whether or not this form of transmission is important is still controversial (5–10).
High-quality genome sequences of four strains of X. oryzae pv. oryzae and two strains of X. oryzae pv. oryzicola are publicly available (GenBank accession numbers AYSX00000000 and AYSY00000000, respectively) (11–14). These resources, along with draft genome sequences of another nine X. oryzae strains, provided insights into the genetic diversity among strains within this species, including a unique group of weakly pathogenic X. oryzae strains isolated in the United States (13; V. Verdier, unpublished). In a previous study, we used a comparative genomics approach to develop diagnostic primers that distinguished strains by pathovar (X. oryzae, X. oryzae pv. oryzae, and X. oryzae pv. oryzicola) and differentiated certain groups of strains on the basis of their geographic origin (13, 15). Multilocus sequence analysis and restriction fragment polymorphism analysis have shown that X. oryzae pv. oryzae is composed of two major genetic groups, the Asian and African lineages (16, 17). Pathovar-specific primers have been adopted for identification of X. oryzae pv. oryzae and X. oryzae pv. oryzicola from field-collected leaf samples (4) and from seed samples (International Rice Research Institute Seed Health Unit, personal communication). However, the adoption of these primers for field-level surveys or for routine screens of seed samples by quarantine officials has been limited largely due to the high costs and requirements for sophisticated laboratories to perform the available diagnostic assays.
A recent advance for molecular diagnostics is the adaptation of the loop-mediated isothermal amplification (LAMP) method for the rapid, specific amplification of target DNA sequences at a single temperature (18). Incubation can be accomplished using a simple water bath without the need for expensive equipment (19). LAMP can be more sensitive and less influenced by inhibitors in test samples than PCR, and it can be adapted so that it provides a simple visual discrimination of the test result without requiring electrophoresis or other equipment (20). LAMP assays have been developed for the detection of phytoplasma, viral, bacterial, and fungal plant pathogens as well as the detection of genetically modified crops (21–25, 27, 28). Visual assays in particular are ideally suited for deployment in nonspecialized laboratories with limited equipment and resources or for incorporation into a simple-to-use diagnostic test for use in the field. The increased sensitivity of the LAMP assay coupled with a closed-tube system where no addition of DNA intercalating dye after the reaction is necessary is attractive for regulatory labs. LAMP can be used in epidemiological surveys, to support microbial forensic investigations for quarantine officials.
The intent of the project described here was to develop and evaluate LAMP assays for X. oryzae pathovars to enable surveillance activities in rice fields and testing of traded materials (seeds) in regional quarantine offices. We focused on genomic regions unique for X. oryzae pv. oryzae and X. oryzae pv. oryzicola (15) to develop pathovar-specific LAMP primers that detect and differentiate strains of each pathovar. We show the effectiveness of these assays in detecting these organisms in diverse sample preparations, such as DNA, heat-killed cells, or crude preparations from plant tissue. In addition, we used draft genomic comparisons to develop LAMP assays that distinguish the African and Asian lineages of X. oryzae pv. oryzae.
MATERIALS AND METHODS
Bacterial strains, DNA, and plant samples.
The bacterial strains used in this study are listed in Table 1. Strains of X. oryzae, selected to represent the genetic and geographic diversity of the pathovars, were screened to determine assay specificity. These included 45 strains of X. oryzae pv. oryzae, 40 strains of X. oryzae pv. oryzicola, and 7 strains of a distinct group of X. oryzae isolates recovered in the United States (13, 29). An additional 31 strains representing other plant-pathogenic species and unknown bacteria isolated from rice tissue and seed were tested. Heat-killed cells, genomic DNA, or crude plant exudate were used as the template in the LAMP reactions. Genomic DNA was isolated using either an Easy-DNA kit (Life Technologies, Grand Island, NY) following the manufacturer's recommendations or a DNeasy blood and tissue kit (Qiagen, Inc., Valencia, CA) following the manufacturer's recommendations, except that DNA was eluted in 30 μl of water in the final step. All samples were diluted to 20 ng μl−1 in sterile water. Heat-killed cells were prepared from cultures grown for 24 h on peptone-sucrose agar (PSA) (30) at 28°C, diluted in sterile water to appropriate concentrations, and incubated at 95°C for 10 min. Plant tissue was collected from rice plants at 0, 24, 48, and 72 h postinoculation (hpi) by syringe infiltration with either X. oryzae pv. oryzae PXO99A (a 5-azacytidine-resistant derivative of strain PXO99) or X. oryzae pv. oryzicola BLS256 or MAI10 as previously reported (31). Each inoculum was adjusted to an optical density at 600 nm of 0.2 by dilution in distilled water (about 108 CFU ml−1) prior to inoculation. Tissues were individually ground in a TissueLyser II apparatus (Qiagen, Inc., Valencia, CA) in 1 ml of distilled water and were serially diluted. Diluted ground tissue was sampled from three independent leaves for testing in each appropriate assay, and the experiment was repeated at least twice.
TABLE 1.
Bacterial strains used in this study to validate specificity of each assaya
| Species | Strain | Origin | Host | Source | LAMP result |
|||
|---|---|---|---|---|---|---|---|---|
| X. oryzae pv. oryzicola | X. oryzae pv. oryzae | African lineage X. oryzae pv. oryzae | Asian lineage X. oryzae pv. oryzae | |||||
| X. oryzae pv. oryzae | R-3 | Australia | Oryza sativa | I. Buddenhagen | − | + | − | + |
| X. oryzae pv. oryzae | BAI3 | Burkina Faso | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | BAI4 | Burkina Faso | Oryza glaberrima | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | BAI1 | Burkina Faso | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | BAI2 | Burkina Faso | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | CFBP1948 | Cameroon | O. sativa | J. L. Notteghem | − | + | + | − |
| X. oryzae pv. oryzae | CFBP1952 | Mali | O. sativa | J. L. Notteghem | − | + | + | − |
| X. oryzae pv. oryzae | 94 (35) | China | O. sativa | − | + | − | + | |
| X. oryzae pv. oryzae | A3857 | India | O. sativa | J. E. Leach | − | + | − | + |
| X. oryzae pv. oryzae | IXO16 | Indonesia | O. sativa | − | + | − | + | |
| X. oryzae pv. oryzae | MAFF311018 | Japan | O. sativa | A. Bogdanove | − | + | − | + |
| X. oryzae pv. oryzae | KACC10331 | South Korea | O. sativa | − | + | − | + | |
| X. oryzae pv. oryzae | Xoo197 | South Korea | O. sativa | S. H. Choi | − | + | − | + |
| X. oryzae pv. oryzae | Xoo199 | South Korea | O. sativa | S. H. Choi | − | + | − | + |
| X. oryzae pv. oryzae | MXO90 | Malaysia | O. sativa | − | + | − | + | |
| X. oryzae pv. oryzae | MXO92 | Malaysia | O. sativa | − | + | − | + | |
| X. oryzae pv. oryzae | CFBP1949 | Mali | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | CFBP1951 | Mali | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | MAI1 | Mali | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | MAI9 | Mali | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | R-13 | Myanmar | O. sativa | I. Buddenhagen | − | + | − | + |
| X. oryzae pv. oryzae | NXO537 | Nepal | O. sativa | Tika Adhikari | − | + | − | + |
| X. oryzae pv. oryzae | NXO544 | Nepal | O. sativa | Tika Adhikari | − | + | − | + |
| X. oryzae pv. oryzae | NXO588 | Nepal | O. sativa | Tika Adhikari | − | + | − | + |
| X. oryzae pv. oryzae | NAI2 | Niger | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | NAI5 | Niger | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | NAI6 | Niger | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | NAI7 | Niger | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | NAI8 | Niger | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | NAI9 | Niger | O. sativa | V. Verdier | − | + | + | − |
| X. oryzae pv. oryzae | PXO69 | Philippines | O. sativa | C. M. Vera Cruz | − | + | − | + |
| X. oryzae pv. oryzae | PXO99A | Philippines | O. sativa | A. Bogdanove | − | + | − | + |
| X. oryzae pv. oryzae | PXO86 | Philippines | O. sativa | C. M. Vera Cruz | − | + | − | + |
| X. oryzae pv. oryzae | PXO111 | Philippines | O. sativa | C. M. Vera Cruz | − | + | − | + |
| X. oryzae pv. oryzae | PXO121 | Philippines | O. sativa | C.M. Vera Cruz | − | + | − | + |
| X. oryzae pv. oryzae | PXO130 | Philippines | O. sativa | C. M. Vera Cruz | − | + | − | + |
| X. oryzae pv. oryzae | PXO132 | Philippines | O. sativa | C. M. Vera Cruz | − | + | − | + |
| X. oryzae pv. oryzae | PXO172 | Philippines | O. sativa | C. M. Vera Cruz | − | + | − | + |
| X. oryzae pv. oryzae | PXO183 | Philippines | O. sativa | C. M. Vera Cruz | − | + | − | + |
| X. oryzae pv. oryzae | PXO344 | Philippines | O. sativa | C. M. Vera Cruz | − | + | − | + |
| X. oryzae pv. oryzae | CL-1 | Sri Lanka | O. sativa | I. Buddenhagen | − | + | − | + |
| X. oryzae pv. oryzae | Xoo1 | Thailand | O. sativa | J. E. Leach | − | + | − | + |
| X. oryzae pv. oryzae | Xoo3 | Thailand | O. sativa | J. E. Leach | − | + | − | + |
| X. oryzae pv. oryzae | RS61 | China | O. sativa | J. S. Wang | − | + | − | + |
| X. oryzae pv. oryzicola | RS85 | China | O. sativa | J. S. Wang | + | − | − | x |
| X. oryzae pv. oryzicola | CFBP2286 | Malaysia | O. sativa | V. Verdier | + | − | − | − |
| X. oryzae pv. oryzicola | MAI4 | Mali | O. sativa | V. Verdier | + | − | − | − |
| X. oryzae pv. oryzicola | MAI5 | Mali | O. sativa | V. Verdier | + | − | − | − |
| X. oryzae pv. oryzicola | MAI6 | Mali | O. sativa | V. Verdier | + | − | − | − |
| X. oryzae pv. oryzicola | MAI7 | Mali | O. sativa | V. Verdier | + | − | − | − |
| X. oryzae pv. oryzicola | MAI8 | Mali | O. sativa | V. Verdier | + | − | − | − |
| X. oryzae pv. oryzicola | MAI10 | Mali | O. sativa | V. Verdier | + | − | + | − |
| X. oryzae pv. oryzicola | BLS46 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | x |
| X. oryzae pv. oryzicola | BLS96 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | x |
| X. oryzae pv. oryzicola | BLS98 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | − |
| X. oryzae pv. oryzicola | BLS105 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | − |
| X. oryzae pv. oryzicola | BLS106 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | x |
| X. oryzae pv. oryzicola | BLS111 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | x |
| X. oryzae pv. oryzicola | BLS114 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | − |
| X. oryzae pv. oryzicola | BLS123 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | x |
| X. oryzae pv. oryzicola | BLS125 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | − |
| X. oryzae pv. oryzicola | BLS170 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | x |
| X. oryzae pv. oryzicola | BLS175 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | x |
| X. oryzae pv. oryzicola | BLS179 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | − |
| X. oryzae pv. oryzicola | BLS220 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | x |
| X. oryzae pv. oryzicola | BLS256 | Philippines | O. sativa | A. Bogdanove | + | − | − | x |
| X. oryzae pv. oryzicola | BLS276 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | − |
| X. oryzae pv. oryzicola | BLS281 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | x |
| X. oryzae pv. oryzicola | BLS291 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | x |
| X. oryzae pv. oryzicola | BLS294 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | x |
| X. oryzae pv. oryzicola | BLS333 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | x |
| X. oryzae pv. oryzicola | BLS346 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | − |
| X. oryzae pv. oryzicola | BLS354 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | x |
| X. oryzae pv. oryzicola | BLS356 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | x |
| X. oryzae pv. oryzicola | BLS377 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | − |
| X. oryzae pv. oryzicola | BLS404 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | − |
| X. oryzae pv. oryzicola | BLS413 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | x |
| X. oryzae pv. oryzicola | BLS415 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | x |
| X. oryzae pv. oryzicola | BLS417 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | − |
| X. oryzae pv. oryzicola | BLS420 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | − |
| X. oryzae pv. oryzicola | BLS421 | Philippines | O. sativa | C. M. Vera Cruz | + | − | x | x |
| X. oryzae pv. oryzicola | BLS468 | Philippines | O. sativa | C. M. Vera Cruz | + | − | − | x |
| X. oryzae | X1-8 | USA | O. sativa | C. Gonzalez | − | + | − | − |
| X. oryzae | X8-1A | USA | O. sativa | C. Gonzalez | − | + | − | − |
| X. oryzae | X211-2 | USA | O. sativa | C. Gonzalez | − | + | − | − |
| X. oryzae | X4-2C | USA | O. sativa | C. Gonzalez | − | + | − | − |
| X. oryzae | RU87-17 | USA | O. sativa | C. Gonzalez | − | + | − | − |
| X. oryzae | X11-5A | USA | O. sativa | C. Gonzalez | − | + | − | − |
| X. oryzae | X1-10 | USA | O. sativa | C. Gonzalez | − | + | − | − |
| Xanthomonas sp. | 97 M | Philippines | O. sativa | V. Verdier | − | − | − | − |
| Xanthomonas sp. | M136 | Mali | O. sativa | V. Verdier | − | − | − | − |
| Xanthomonas sp. | SHU36 | Philippines | O. sativa, seed | C. M. Vera Cruz | − | − | − | − |
| Xanthomonas sp. | SHU50 | Philippines | O. sativa, seed | C. M. Vera Cruz | − | − | − | − |
| Xanthomonas sp. | SHU100 | Philippines | O. sativa, seed | C. M. Vera Cruz | − | − | − | − |
| Xanthomonas sp. | SHU147 | Philippines | O. sativa, seed | C. M. Vera Cruz | − | − | − | − |
| Xanthomonas sp. | SHU178 | Philippines | O. sativa, seed | C. M. Vera Cruz | − | − | − | − |
| Xanthomonas sp. | SHU199 | Philippines | O. sativa, seed | C. M. Vera Cruz | − | − | x | x |
| Xanthomonas sp. | SHU202 | Philippines | O. sativa, seed | C. M. Vera Cruz | − | − | − | − |
| Xanthomonas sp. | SHU222 | Philippines | O. sativa, seed | C. M. Vera Cruz | − | − | − | − |
| Xanthomonas sp. | SHU268 | Philippines | O. sativa, seed | C. M. Vera Cruz | − | − | − | − |
| Xanthomonas sp. | SHU303 | Philippines | O. sativa, seed | C. M. Vera Cruz | − | − | x | − |
| Acidovorax avenae pv. avenae | NCPPB3112 | Brazil | Canna indica | NCPPB | − | − | − | x |
| A. avenae | BPJ4821 | Philippines | O. sativa | C. M. Vera Cruz | − | − | x | x |
| A. avenae pv. citrulli | 94-21 | USA | Citrullus lanatus | R. Walcott | − | − | − | − |
| Burkholderia andropogonis | 3549 | USA | Saccharum officinarum | L. E. Claflin | − | − | − | − |
| Burkholderia gladioli | O187 | USA | Allium cepa | H. F. Schwartz | − | − | − | − |
| Curtobacterium flaccumfaciens | B473 | USA | Phaseolous sp. | H. F. Schwartz | − | − | − | − |
| Escherichia coli | DH5α | USA | NA | Life Technologies | − | − | − | − |
| Enterobacter sp. | O121 | USA | Allium cepa | H. F. Schwartz | − | − | − | − |
| Enterobacter sp. | O174 | USA | Allium cepa | H. F. Schwartz | − | − | − | − |
| Pseudomonas marginalis | ATCC 10844 | USA | Lactuca sativa | H. F. Schwartz | − | − | − | − |
| Pseudomonas syringae pv. syringae | M72 | USA | Capsicum annuum | H. F. Schwartz | − | − | − | − |
| P. syringae pv. syringae | M108 | USA | Solanum lycopersicum | H. F. Schwartz | − | − | x | − |
| Pseudomonas viridiflava | ATCC 13223 | USA | Phaseolus coccineus | H. F. Schwartz | − | − | x | − |
| Ralstonia solanacearum | K60 | USA | S. lycopersicum | J. E. Leach | − | − | − | x |
| Xanthomonas axonopodis pv. vesicatoria | 85-10 | USA | Capsicum sp. | A. Bogdanove | − | − | x | − |
| X. axonopodis pv. sojense | 4455 | USA | Glycine max | L. E. Claflin | − | − | − | x |
| Xanthomonas campestris pv. campestris | X1910 | USA | Brassica oleracea | N. Dunlop | − | − | x | − |
| X. campestris pv. carotae | NCPPB1422 | New Zealand | Daucus carota | L. E. Claflin | − | − | − | x |
| Xanthomonas translucens pv. cerealis | NCPPB1836 | USA | Secale cereale | L. E. Claflin | − | − | − | − |
| Unknown | Ven | Venezuela | O. sativa, seed | V. Verdier | − | − | − | − |
+, positive amplification; −, absence of amplification; x, strain not tested; NCPPB, National Collection of Plant Pathogenic Bacteria; NA, not applicable.
Primer design and screening.
The PXO_00080 (conserved hypothetical protein) and Xoryp_010100019045 (putative glycosyltransferase) loci are unique for X. oryzae pv. oryzae and X. oryzae pv. oryzicola, respectively (15). These loci were used to develop LAMP primers that amplify all isolates within each pathovar. In addition, loci that distinguish isolates of X. oryzae pv. oryzae by geographic origin were identified by analyzing the draft genomic sequence of African lineage strains of X. oryzae pv. oryzae (GenBank accession number AYSX00000000) (see Fig. S1 in the supplemental material). The locus specific to Asian populations is PXO_03925 (conserved hypothetical protein, putative lipase). Primers were designed on the basis of all of these unique sequences using either LAMP Designer software (version 1.02; Premier Biosoft, Palo Alto, CA) or PrimerExplorer software (Eiken Chemical Company; https://primerexplorer.jp/e/) and synthesized by Integrated DNA Technologies (Coralville, IA). Four primers (external primers F3 and B3; internal primers FIP and BIP) were designed for each assay. Loop primers were also designed for X. oryzae pv. oryzicola- and X. oryzae pv. oryzae-specific assays. All oligonucleotide sequences are listed in Table 2.
TABLE 2.
LAMP primers for detection of X. oryzae pv. oryzae and X. oryzae pv. oryzicola
| Target | Primer | Sequence (5′–3′) |
|---|---|---|
| X. oryzae pv. oryzae PXO_00080a | F3 | CTTCAAGGCCAAGGACATC |
| B3 | CACGATCTTGCAAGGGAT | |
| FIP | CGGTGCCGGACTGGATTTGCTAGGAATGAGCAATGCA | |
| BIP | GTAGTTGCCGACGGCTACCAGAAGCGTCCTCGTCTAA | |
| LoopF | TTTGAGGTCCCTTTCCACG | |
| LoopB | GTTTGTGCGCCGTCTATC | |
| X. oryzae pv. oryzicola Xoryp_010100019045b | F3 | GGATCACAGTGATCGTGC |
| B3 | CACTTATCGTCCAGTACGC | |
| FIP | CGATGCCGCCTTGATCGAGTTGTACTCCTACGATGAGC | |
| BIP | ACCGAGTCGTTGCAGGTCTCTTGCGAAACACAAGGAA | |
| LoopF | TTGTGACCACGCTGTCATT | |
| LoopB | TCGCCATCTCCAGTCCTAT | |
| African lineage X. oryzae pv. oryzae, hypothetical protein, draft sequencec | F3 | TATTGGGTGCTGCCGATGA |
| B3 | GGCAACCTCACTTCCGTAAG | |
| FIP | ATGTAGCCATCATGCCCGCCTTTTCCCAGATTTGCGAGTCCTT | |
| BIP | GCGCTCTTCGGATGGTAGTGATTTTTGCCATGCTTGTTTTGTGCA | |
| Asian lineage X. oryzae pv. oryzae PXO_03925a | F3 | GGTGGTCAGCGCATCGA |
| B3 | ACTGCTGCTGTTCCAACG | |
| FIP | ATGCTGACGCGCAGCTGCTTTTAGCCCAGATGCCGCAC | |
| BIP | TGACGGCAATGAATACCCCGCTTTTGACCGACTGGCGTGCT |
Designed on the basis of the X. oryzae pv. oryzae PXO99A genome sequence (GenBank accession number PRJNA28127).
Designed on the basis of X. oryzae pv. oryzicola BLS256 genome sequence (GenBank accession number PRJNA54411).
Designed on the basis of X. oryzae pv. oryzae NAI8 (GenBank accession number AYSX00000000) and BAI3 (V. Verdier et al., unpublished data) draft genome sequences.
LAMP.
LAMP reactions (final volume, 12 μl) were performed in a CFX Connect real-time system (Bio-Rad, Hercules, CA) or a Genie II system (Optigene, Sussex, United Kingdom). The reaction mixture contained 7.2 μl Isothermal master mix (Optigene, Sussex, United Kingdom), 32 nM outer primers (F3 and B3), and 0.32 μM inner primers (FIP and BIP). Pathovar-specific assays included 0.16 μM loop primers (LoopF and LoopB). Lastly, 1 μl of template that was either genomic DNA (20 ng μl−1), heat-killed bacterial cells, or serially diluted, ground inoculated tissue (as described above) was added. Assays for African and Asian lineage X. oryzae pv. oryzae isolates did not include loop primers, and the remaining volume was made up with water. The LAMP reaction mixtures in the CFX Connect system were incubated for 60 min at 65°C, followed by melt curve analysis from 65°C to 95°C. Incubations on the Genie II system were for 30 or 60 min at 65°C. All LAMP assays performed for screening purposes were replicated at least twice, and all experiments included no-template controls (water and no template DNA).
Assay specificity and sensitivity.
Assay specificities were established using a pooling strategy to screen large collections of negative controls after the specificity with positive-control strains was initially confirmed. Positive controls were strains used to derive the published genome sequences (strains PXO99A, MAFF311018, and KACC10331) for X. oryzae pv. oryzae and strain BLS256 for X. oryzae pv. oryzicola (11, 12, 14, 32). Nontarget bacterial DNAs were pooled in equimolar concentrations, with the DNA from 10 strains included in each pool. Each negative pool was separately spiked with 1 μl of positive-control genomic DNA to validate detection in a mixed sample. The sensitivity of each assay was determined using serial dilutions of both genomic DNA (10 ng to 1 fg) and heat-killed cells (108 to 101 CFU ml−1). Initial X. oryzae pv. oryzicola assay development included loop primers, but in subsequent testing for specificity and sensitivity, these primers were removed for greater consistency and to reduce the incidences of false-positive results. The volumes in each reaction mixture were replaced with water.
Seed detection.
A lot of clean (known to be free of X. oryzae pv. oryzae and X. oryzae pv. oryzicola) Oryza sativa cv. IR24 seeds was disinfected using 70% ethanol, and then the seeds were rinsed thrice with sterile distilled water and dried in a laminar flow hood. Subsamples of this lot were artificially inoculated by soaking the seeds in bacterial suspensions of X. oryzae pv. oryzae PXO99 and X. oryzae pv. oryzicola BLS256 for 2 h at room temperature for 2 h at 4°C and placed in a laminar flow hood until the seeds were dry. PXO99 is a Philippine strain of X. oryzae pv. oryzae and was used as a control strain in all experiments completed at the International Rice Research Institute; in experiments completed at Colorado State University, PXO99A, a 5-azacytidine-resistant derivative of PXO99 (33), was used as a control strain. The remaining clean seeds were subdivided into 5-g seed lots (approximately 200 seeds). Cell counts were estimated to be 1.1 × 104 CFU seed−1 (PXO99) and 4.6 × 104 CFU seed−1 (BLS256). To test the sensitivity of the pathovar-specific LAMP assays to detect 0.5% contamination of a 5-g seed lot (1 contaminated seed in 200 seeds), a single contaminated seed from the pool of PXO99- or BLS256-contaminated seeds was added to 5 g of clean seed. Thirty samples each of PXO99-contaminated and BLS256-contaminated seed lots were prepared and processed using an protocol for extraction of bacteria from rice seeds by sonication (M. H. R. Nguyen and C. M. Vera Cruz, unpublished). Seed extracts were stored at 0°C. LAMP reactions were carried out as described above with 1-μl aliquots from seed extracts as the template DNA. Each run included one positive DNA control, four nontarget DNA controls, and one no-template control in which 1 μl of water was added to the reaction mix. In experiments screening for X. oryzae pv. oryzae, nontarget controls included X. oryzae pv. oryzicola BLS175, X. oryzae pv. oryzicola BLS256, Acidovorax avenae BPJ4821, and an uncharacterized yellow nonpathogenic seed-associated bacterium named SHU199. X. oryzae pv. oryzae PXO99A genomic DNA was used as a positive control. Experiments using the X. oryzae pv. oryzicola-specific primers included a positive control (X. oryzae pv. oryzicola BLS256) and four nontarget controls (X. oryzae pv. oryzae PXO99, X. oryzae pv. oryzae PXO349, Acidovorax avenae BPJ4821, and SHU199). All DNA controls were normalized to 20 ng μl−1. In analyzing the X. oryzae pv. oryzae-contaminated seed lots, pathovar loop primers (LoopF and LoopB) were used in all tests except those with X. oryzae pv. oryzicola. The reaction mixtures were incubated in the Genie II system (Optigene, Sussex, United Kingdom) at 65°C for 60 min. All LAMP tests were conducted in triplicate. Sensitivity and specificity values were computed using the formulas discussed by Armitage et al. (26).
Visual detection.
A visual LAMP detection protocol for the detection and identification of the X. oryzae pathovars was adapted to reduce cost and the requirement for sophisticated equipment. Assays were performed in conventional thermal cyclers or a water bath at 65°C for 60 min. The 25-μl reaction mix contained 2.5 μl 10× isothermal amplification buffer (New England BioLabs, Ipswich, MA), 1.4 mM deoxynucleoside triphosphates, 6 mM additional MgSO4 for a final MgSO4 concentration of 8 mM (New England BioLabs, Ipswich, MA), 0.8 M betaine (Sigma-Aldrich, St. Louis, MO), 4 U Bst DNA polymerase large fragment or Bst DNA polymerase 2.0 (New England BioLabs, Ipswich, MA), 0.32 μM primers FIP and BIP, 32 nM primers F3 and B3, and 0.16 μM primers LoopF and LoopB (loop primers in pathovar-specific assays only) with 1 μl of 20 ng μl−1 DNA, heat-killed cells, or plant extract. Mineral oil (EMD Millipore, Darmstadt, Germany) was added on top of the reaction mixture (20 μl) to minimize the introduction of aerosolized product into the work spaces. Amplification was terminated by heat inactivation at 80°C for 3 min. After incubation, the tubes were individually opened in a separate lab and 0.5 to 1 μl of Quant-IT Pico green reagent (Invitrogen, Carlsbad, CA) was added. The reaction mixtures were incubated at room temperature for 5 min and then observed under normal and UV light either for a change of color from orange to green or for fluorescence.
RESULTS
Primer design and specificity and sensitivity of LAMP assays.
At least five different primer sets were predicted for each unique sequence and were used to develop LAMP assays specific for each of X. oryzae pv. oryzae and X. oryzae pv. oryzicola (designated pathovar-specific primers) and for geographically distinct lineages of X. oryzae pv. oryzae (African versus Asian; designated geography-specific primers). After initial screening with control DNAs (X. oryzae pv. oryzae PXO99A and BAI3, X. oryzae pv. oryzicola BLS256), the primer sets listed in Table 2 were used for testing. The ratios of primer concentrations were based on previous reports and consultation with colleagues (2, 4, 6; A. Bühlmann, personal communication) and did not require optimization.
LAMP assays were tested for specificity and efficiency with a panel comprising 44 X. oryzae pv. oryzae strains, 38 X. oryzae pv. oryzicola strains, 7 X. oryzae strains, 11 Xanthomonas species strains (the species were unknown, but they were determined not to be X. oryzae by multiplex PCR [15] and multiple-sequence alignment [B. Cottyn et al., unpublished data]), and 19 strains representing eight different bacterial genera. The assays were performed using the Isothermal master mix (Optigene, Sussex, United Kingdom) in a CFX Connect real-time system (Bio-Rad, Hercules, CA) or a Genie II system (Optigene, Sussex, United Kingdom) (Table 1).
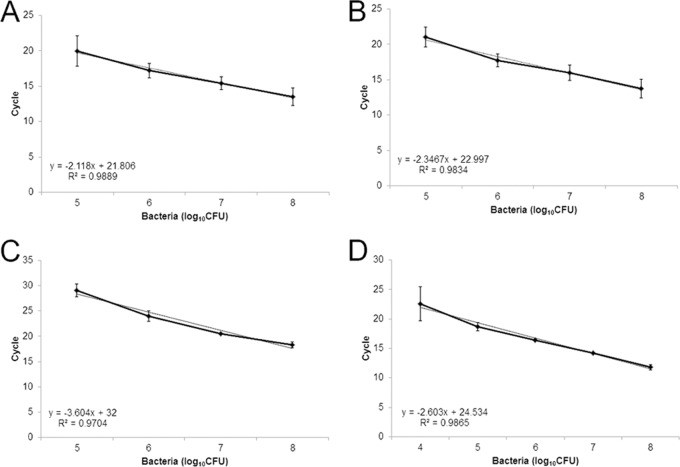
Genomic DNA, diluted to concentrations ranging from 10 ng to 1 fg, was used to establish the sensitivity of each assay. A no-template control (water) was included in each experimental replication. The thresholds of detection were 10 pg for the X. oryzae pv. oryzae-specific assay (Fig. 1A), 1 fg for the X. oryzicola pv. oryzicola-specific assay (Fig. 1B), 1 ng for the African lineage X. oryzae pv. oryzae-specific assay (Fig. 1C), and 1 pg for the Asian lineage X. oryzae pv. oryzae-specific assay (Fig. 1D). As few as 105 CFU ml−1 bacterial cells were detected using both pathovar-specific assays and the African lineage X. oryzae pv. oryzae assay (Fig. 2A to C). The Asian lineage X. oryzae pv. oryzae-specific assay detected as few as 104 CFU/ml, though there was more variation in the technical replicates at the lower concentrations (Fig. 2D). There were no false-positive results for any assay with the no-template control, confirming the specificity of the target region and the primers. Loop primers designed to be specific for X. oryzae pv. oryzicola contributed to the amplification of specific targets; however, false-positive results were identified more often when they were included during initial validations. To address this concern in subsequent testing, loop primers were removed and the volume was replaced with water. This change in protocol reduced the sensitivity and therefore the incidence of false-positive results for this assay by increasing the time to positivity. So, while these primers are reported in Table 2, we recommend conducting this assay without them. Overall, the adapted primers were specific and sensitive in the LAMP assay. In addition, using a comparative genomics approach with draft genome sequences, we identified loci that differentiated X. oryzae pv. oryzae lineages on the basis of geographic origin.
FIG 1.
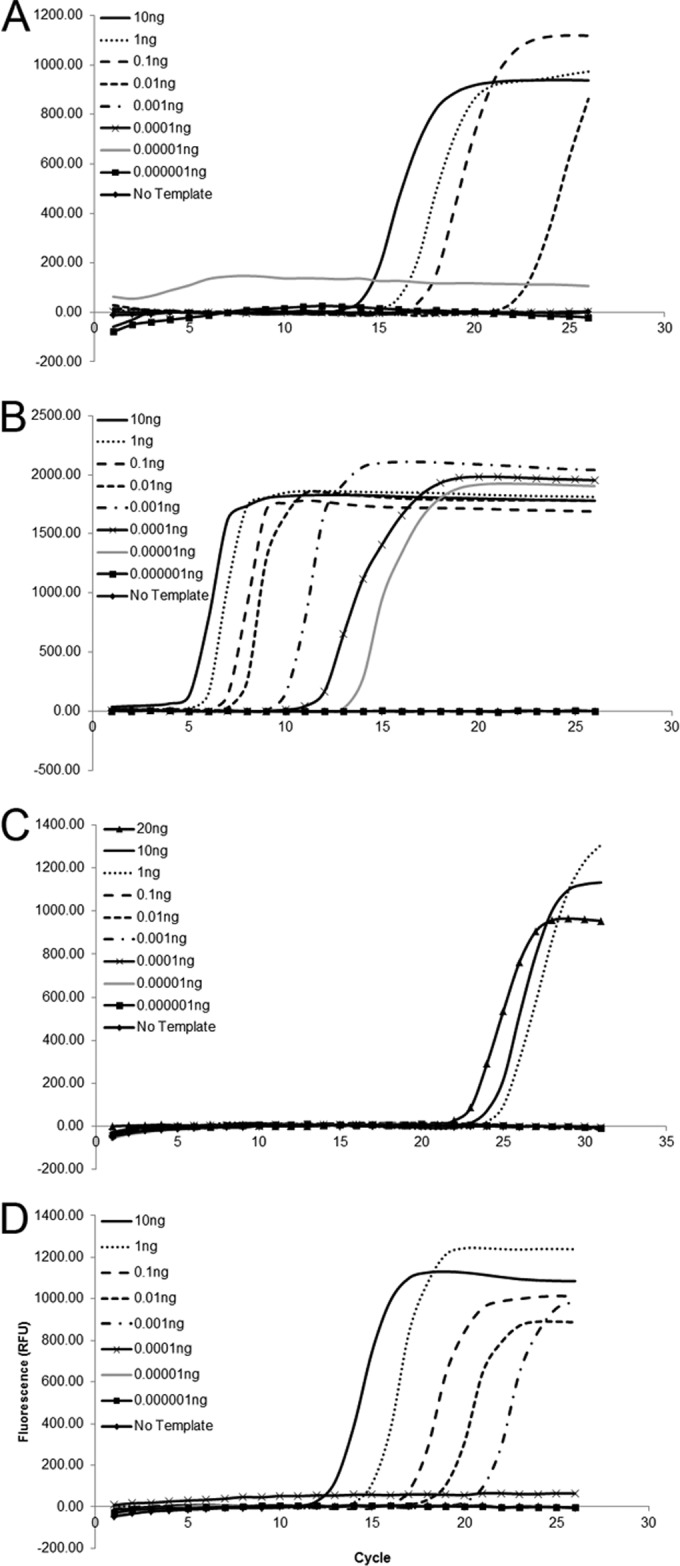
Sensitivities of X. oryzae pv. oryzae-specific (A), X. oryzae pv. oryzicola-specific (B), African lineage X. oryzae pv. oryzae-specific (C), and Asian lineage X. oryzae pv. oryzae-specific (D) LAMP assays. Each diluted DNA was tested at least three times.
FIG 2.
Standard curves showing the sensitivity of detection by LAMP using dilutions of heat-killed cells. (A) X. oryzae pv. oryzae-specific primers with X. oryzae pv. oryzae PXO99A heat-killed cells; (B) X. oryzae pv. oryzicola-specific primers with X. oryzae pv. oryzicola BLS256 heat-killed cells; (C) African lineage X. oryzae pv. oryzae-specific primers using X. oryzae pv. oryzae BAI3 heat-killed cells; (D) Asian lineage X. oryzae pv. oryzae-specific primers using X. oryzae pv. oryzae PXO99A heat-killed cells. Each dilution was tested three times. Bars represent the standard deviation of the mean, and the associated R2 values obtained after linear regression analysis are provided.
Detection of X. oryzae pv. oryzae and X. oryzae pv. oryzicola from seed and crude plant extracts.
Artificially inoculated seed lots were used to assess the capacity of the pathovar-specific LAMP assays to detect bacteria at a 0.5% contamination level in 5-g seed samples (Table 3). Using the X. oryzae pv. oryzicola pathovar-specific LAMP primers, BLS256 DNA was detected in 30 out of 30 samples, giving a sensitivity of 100%. None of the 30 seed samples contaminated with X. oryzae pv. oryzae PXO99 was amplified with the X. oryzae pv. oryzicola-specific LAMP primers (0% false-positive detection). The sensitivity of the X. oryzae pv. oryzae-specific LAMP primers was 93.3%, with target strain PXO99 being detected in 28 out of 30 contaminated seed lots. Five of 30 seed samples contaminated with X. oryzae pv. oryzicola BLS256 were found to be positive by use of the X. oryzae pv. oryzae-specific LAMP primers (specificity, 83.3%); i.e., the nontarget organism was detected in at least two of three technical replications. The trials were done with little or no optimization needed, but due to the high sensitivity and robustness of the primers designed for use in the X. oryzae pv. oryzicola-specific LAMP assay, the loop primers were excluded from the reaction mix to prevent random false-positive results. Representative amplification curves for each pathovar-specific assay and appropriate controls are shown in Fig. 3A and B for X. oryzae pv. oryzicola and X. oryzae pv. oryzae, respectively. Pairwise inoculations were used to demonstrate consistent specificity for detection of the presence of either organism. As described above, a single contaminated seed of either X. oryzae pv. oryzae PXO99 or X. oryzae pv. oryzicola was added to 5 g of clean seed. The X. oryzae pv. oryzae PXO99-contaminated lots were tested with the X. oryzae pv. oryzicola-specific assay, and conversely, the X. oryzae pv. oryzicola BLS256-contaminated seed lots were tested with the X. oryzae pv. oryzae-specific assay. Nontarget DNAs from Acidovorax avenae BPJ4821 or Xanthomonas sp. strain SHU199 were included in each experiment as negative controls alongside no-template controls and did not amplify with either assay (Table 1).
TABLE 3.
LAMP detection of X. oryzae pv. oryzae and X. oryzae pv. oryzicola using pathovar-specific assays in seed lots (200 seeds) inoculated with one seed carrying 104 CFU of X. oryzae pv. oryzae PXO99 or X. oryzae pv. oryzicola BLS256c
| Sample no. | No. of samples with amplification from seed/total no. of samples tested by: |
|||
|---|---|---|---|---|
|
X. oryzae pv. oryzicola-specific LAMP assay |
X. oryzae pv. oryzae-specific LAMP assay |
|||
| X. oryzae pv. oryzicola BLS256 | X. oryzae pv. oryzae PXO99 | X. oryzae pv. oryzae PXO99 | X. oryzae pv. oryzicola BLS256 | |
| 1 | 3/3 | 0/3 | 2/3 | 2/3 |
| 2 | 3/3 | 0/3 | 3/3 | 0/3 |
| 3 | 3/3 | 0/3 | 3/3 | 0/3 |
| 4 | 3/3 | 0/3 | 3/3 | 0/3 |
| 5 | 3/3 | 0/3 | 3/3 | 2/3 |
| 6 | 3/3 | 0/3 | 2/3 | 0/3 |
| 7 | 3/3 | 0/3 | 1/3 | 1/3 |
| 8 | 3/3 | 0/3 | 3/3 | 0/3 |
| 9 | 3/3 | 0/3 | 0/3 | 1/3 |
| 10 | 3/3 | 0/3 | 2/3 | 0/3 |
| 11 | 3/3 | 0/3 | 3/3 | 0/3 |
| 12 | 3/3 | 0/3 | 3/3 | 2/3 |
| 13 | 3/3 | 0/3 | 3/3 | 0/3 |
| 14 | 3/3 | 0/3 | 3/3 | 3/3 |
| 15 | 3/3 | 0/3 | 3/3 | 0/3 |
| 16 | 3/3 | 0/3 | 3/3 | 0/3 |
| 17 | 3/3 | 0/3 | 3/3 | 0/3 |
| 18 | 3/3 | 0/3 | 3/3 | 0/3 |
| 19 | 3/3 | 0/3 | 3/3 | 0/3 |
| 20 | 3/3 | 0/3 | 3/3 | 0/3 |
| 21 | 3/3 | 0/3 | 3/3 | 0/3 |
| 22 | 3/3 | 1/3 | 2/3 | 0/3 |
| 23 | 3/3 | 0/3 | 2/3 | 0/3 |
| 24 | 3/3 | 0/3 | 3/3 | 3/3 |
| 25 | 3/3 | 0/3 | 3/3 | 0/3 |
| 26 | 3/3 | 1/3 | 3/3 | 0/3 |
| 27 | 3/3 | 0/3 | 2/3 | 1/3 |
| 28 | 3/3 | 1/3 | 3/3 | 0/3 |
| 29 | 3/3 | 0/3 | 2/3 | 0/3 |
| 30 | 3/3 | 0/3 | 3/3 | 0/3 |
| BPJ4821a | 0/9 | 0/9 | 0/9 | 0/9 |
| SHU199a | 0/9 | 0/9 | 0/9 | 0/9 |
| Water (NTC) | 0/9 | 1/9 | 0/9 | 0/9 |
| Total no. of positive samplesb | 30 | 0 | 28 | 5 |
| Total no. of negative samplesb | 0 | 30 | 2 | 25 |
Genomic DNAs from bacteria isolated from seeds (strains BPJ4829 and SHU199) were included as negative controls and are referenced in Table 1.
The total numbers of positive and negative samples refer to inoculated seed preparations; negative and no-template controls were not included in this sum or for sensitivity and specificity calculations.
The level of contamination was 0.5%. Each sample was analyzed in triplicate against each primer set, where the presence of an amplification curve at least two out of three runs was interpreted as a positive result. NTC, no-template control. The sensitivity and specificity of the X. oryzae pv. oryzicola-specific LAMP assay were both 100%. The sensitivity and specificity of the X. oryzae pv. oryzae-specific LAMP assay were 93.33% and 83.33, respectively.
FIG 3.
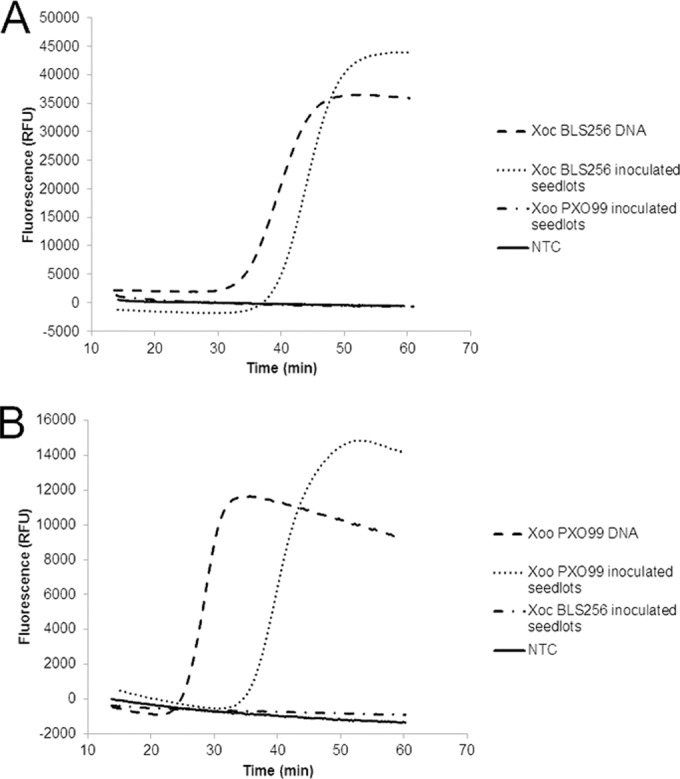
LAMP detection of X. oryzae pv. oryzicola (A) and X. oryzae pv. oryzae (B) using pathovar-specific assays and seed extracts from 5-g seed lots with 0.5% contamination (1 seed carrying 104 CFU out of the 200 seeds) and from DNA extracts (20 ng μl−1 in TE [Tris-EDTA] buffer). Mean fluorescence data for seed extracts were obtained from the amplification curves of 10 seed lots per isolate against each primer set. X. oryzae pv. oryzae (Xoo) PXO99 DNA and X. oryzae pv. oryzicola (Xoc) BLS256 DNA were tested, and no-template controls (NTC) consisting of water and nontarget DNAs served as negative controls and failed to amplify in any run with either primer set in at least three independent runs. Data were collected on a Genie II system (Optigene, Sussex, United Kingdom) and were normalized to the background fluorescence.
Amplifications were delayed when seeds were tested compared to the time of amplification when pure DNA was used as the template in specificity tests. The fluorescence values for the detection of amplification in the X. oryzae pv. oryzae-specific assay (in relative fluorescence units [RFU]) were also higher than those in the specificity and sensitivity assays. Seed detection experiments were conducted with a Genie II system (Optigene, Sussex, United Kingdom), while sensitivity tests were completed on a CFX Connect real-time system (Bio-Rad, Hercules, CA). We attribute the variation in these results to the two platforms used for detecting amplification as well as the nature of the samples. Seed extracts may contain contaminants that slow the amplification reaction, as was described in a watermelon system detecting cucumber mottle mosaic virus (34), and possibly influence fluorescence detection capabilities. However, data were evaluated for the presence or absence of an exponential amplification compared to the result for the negative controls, and for these tests, the amplifications were specific for primer/DNA combinations and were consistent with all previous results.
Crude extracts from inoculated rice leaf tissue also served as viable templates for all of the reported LAMP assays (Table 4). Representative data from the X. oryzae pv. oryzicola-specific LAMP assay are illustrated in Fig. 4, and the threshold detected correlates with the threshold detected when heat-killed cells were used (starting concentration, 107 CFU ml−1).
TABLE 4.
LAMP detection of X. oryzae pv. oyrzae and X. oryzae pv. oryzicola in leaf tissue inoculated by syringe infiltration at 72 hpic
| Strain or samplea | No. of samples positive/total no. of samples amplified with primer set specific for: |
|||
|---|---|---|---|---|
| Pathovar-specific X. oryzae pv. oryzae | Pathovar-specific X. oryzae pv. oryzicola | African lineage of X. oryzae pv. oryzae | Asian lineage of X. oryzae pv. oryzae | |
| PXO99A | 3/3 | 0/3 | 0/3 | 3/3 |
| MAI1 | 3/3 | 0/3 | 3/3 | 0/3 |
| BLS256 | 0/3 | 3/3 | 0/3 | 0/3 |
| Uninoculated leaf | 0/3 | 0/3 | 0/3 | 0/3 |
| No-template controlb | — | — | — | — |
The strains are referenced in Table 1.
No-template controls contained water and were included in each reaction. —, the absence of any amplification signal.
Three independently inoculated leaves were analyzed at least twice.
FIG 4.
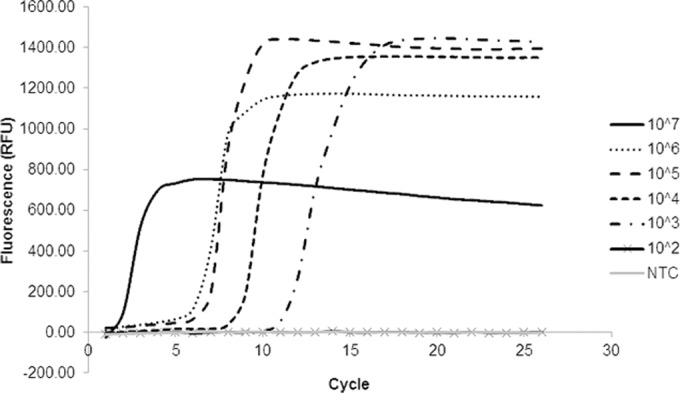
Sensitivity of X. oryzae pv. oryzicola-specific primers in a LAMP assay using ground tissue inoculated with X. oryzae pv. oryzicola MAI10, sampled at 48 hpi, and then serially diluted.
Interestingly, although viable bacteria could not be recovered, X. oryzae pv. oryzae strain PXO99A was correctly detected by both the X. oryzae pv. oryzae-specific and Asian lineage X. oryzae pv. oryzae-specific assays in leaf samples that had been inoculated 23 years earlier and stored at room temperature. These samples did not amplify with the X. oryzae pv. oryzicola-specific or African lineage X. oryzae pv. oryzae-specific primers, confirming that the assays are robust and can detect target bacteria in diverse sample preparations (data not shown).
Visual detection of LAMP products.
A visual detection protocol was adapted and tested for all LAMP primers. Chemistries including hydroxynaphthol blue, Gel Red (Biotium, In., Hayward, CA), and ethidium bromide (data not shown) did not perform as reliably or clearly as the SYBR stain Quant-IT Pico green reagent (Life Technologies, Grand Island, NY) added postincubation. SYBR green was able to detect DNA directly in heat-killed cells to the same threshold as the Isothermal master mix using a thermal cycler. The results of SYBR green-stained reactions are shown in Fig. 5 for the X. oryzae pv. oryzae-specific assay. A water bath was successfully used for incubation, and crude inoculated plant extract was amplified by each specific primer set designed (data not shown).
FIG 5.
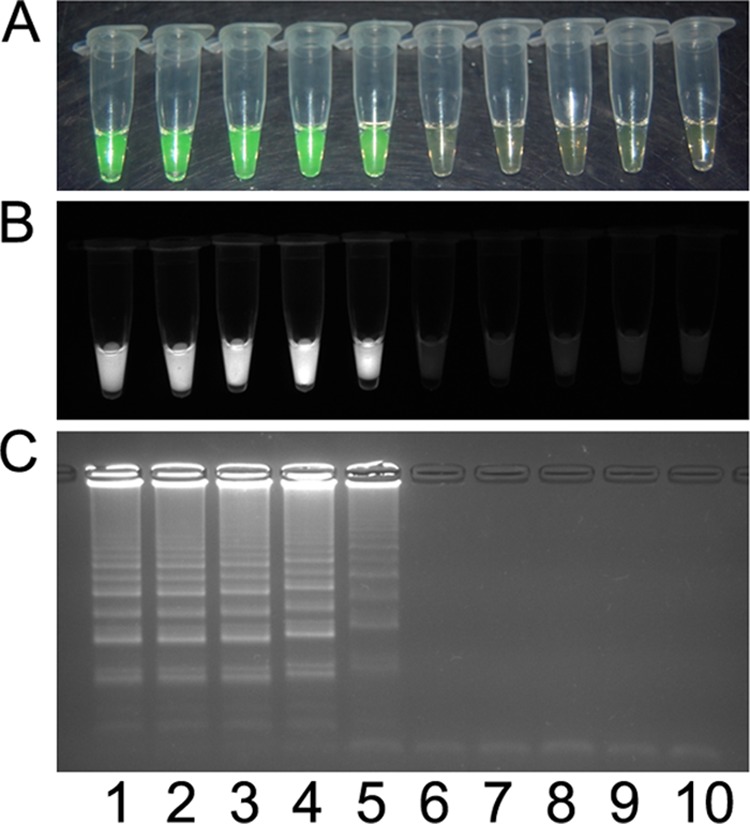
Visual detection of X. oryzae pv. oryzae PXO99A heat-killed cells using the pathovar-specific LAMP assay. The assay was used to test a dilution series consisting of 108 CFU ml−1 (lane 1), 107 CFU ml−1 (lane 2), 106 CFU ml−1 (lane 3), 105 CFU ml−1 (lane 4), 104 CFU ml−1 (lane 5), 103 CFU ml−1 (lane 6), 102 CFU ml−1 (lane 7), and 101 CFU ml−1 (lane 8); X. oryzae pv. oryzicola BLS256 (lane 9); and a no-template control (lane 10). Products were detected by using 1 μl Quant-IT Pico green reagent (Life Technologies, Grand Island, NY) under visual light, where a positive result changes from orange to green (A), or ultraviolet light, where a positive result fluoresces (B), or by 1.5% agarose gel electrophoresis, where a positive result is a laddered product (C).
DISCUSSION
Adaptation of previously designed specific conventional PCR primers to LAMP resulted in a reliable, sensitive, specific, and robust test to detect and differentiate X. oryzae pv. oryzae and X. oryzae pv. oryzicola. The primers and LAMP assays were validated on a wide diversity of bacterial strains, including a large collection of both X. oryzae pathovars as well as other Xanthomonas species and other genera of bacteria, to demonstrate primer specificity and assay reliability. The assay with the X. oryzae pv. oryzicola-specific primers was the most sensitive. The Asian lineage X. oryzae pv. oryzae-specific primers, X. oryzae pv. oryzae-specific primers, and African lineage X. oryzae pv. oryzae-specific primers were slightly less sensitive but still detected 1 ng of genomic DNA. Differences in assay sensitivity are not likely due to the copy number of the target, because, where sequences are known, all loci are present in single copies. Therefore, we predict that the inherent efficiency of annealing for each primer set causes this variation. Regardless, the sensitivity thresholds among the four assays developed were consistent with those previously reported for other plant-pathogenic bacteria, ranging from 10 fg to 0.01 ng genomic DNA and 103 to 104 CFU ml−1 (22, 35–37, 42), and correlate with the equivalent range of 103 to 106 genome copies based on the 5.2-Mbp X. oryzae pv. oryzae PXO99A genome and the 4.8-Mbp X. oryzae pv. oryzicola genome (11, 12).
We previously used comparative genomic approaches with whole-genome (15) and draft genome (13) sequences for the design of pathovar-specific primers. In this study, we mirrored this approach and took advantage of a comparison of draft genomes (our unpublished data) that identified sequences differentiating geographically distinct populations of X. oryzae pv. oryzae, i.e., populations from Africa or Asia. The ability to differentiate the geographic origin of a strain could facilitate epidemiological and surveillance studies, along with the monitoring of imported seed at quarantine stations. The specificity of all primers after adaptation to LAMP-based assays was confirmed, and no cross-reactivity to other xanthomonads or bacterial genera was detected (Table 1). Furthermore, no false-positive amplification occurred in water (no-template) controls. These results confirm the utility of the use of the draft sequence for the development of unique primers and the streamlined adaptation of conventional PCR to LAMP. Because of their specificity and sensitivity, the primers and assays will be useful as forensic tools in quarantine offices, in epidemiological studies, and for seed certification.
Using the available public databases, such as GenBank (http://www.ncbi.nlm.nih.gov/GenBank/), the Pathosystems Resource Integration Center (PATRIC; http://patricbrc.org/), and the Comprehensive Phytopathogen Genomics Resource (CPGR; http://cpgr.plantbiology.msu.edu), we searched for the relationships of loci used for primer development with annotated genes in existing curated genomes. The X. oryzae pv. oryzicola-specific locus (Xoryp_010100019045) was the only gene with an available annotation. It encodes a predicted glucosyltransferase with sequence similarity to rhamnose-glucose polysaccharide assembly protein F50 (RgpF) of Streptococcus mutans and shows high similarity to a recently reported X. oryzae pv. oryzicola virulence factor, wxocB (38). This locus is flanked by a methyltransferase on one side and a cluster of ABC O-antigen lipopolysaccharide (LPS) transporters on the other side, consistent with the clustering of LPS gene islands (39).
PXO_00080, which is unique to all X. oryzae pv. oryzae strains and absent in X. oryzae pv. oryzicola, encodes an uncharacterized protein. The African lineage X. oryzae pv. oryzae locus was identified in the draft sequence of NAI8 (our unpublished data), a strain originally from Niger (16), and bears no similarity to any publicly available sequence. Its presence in the draft genome sequences of MAI1 and BAI3 was confirmed (our unpublished data). The Asian lineage X. oryzae pv. oryzae locus is also annotated as a hypothetical protein, but it has similarity to the lipase family of proteins. X. oryzae pv. oryzae PXO_00080 loci have sequence similarity to plasmids found in two related species of Xanthomonas, including Xanthomonas arboricola pv. pruni CFBP 5530 plasmid pXap41 (GenBank accession number FR875157) and X. axonopodis pv. citri 306 plasmid pXAC33 (GenBank accession number AE008924). For example, four mismatches occur between the PXO_00080 locus and the plasmid sequences in question, and two of our primers include those mismatches. While the presence of mismatches does not prove that the primer would not amplify the nontarget sequences and we did not test the specific strains containing these plasmids, our use of a highly diverse and comprehensive panel of negative-control strains for testing primers increases our confidence that the assays are specific for X. oryzae pv. oryzae. Furthermore, the likelihood that X. arboricola pv. pruni and X. axonopodis pv. citri are present on rice tissue or seed is unlikely.
Out of 90 assays (30 independently inoculated seed lots tested in triplicate), only five false-positive results were generated using X. oryzae pv. oryzae-specific primers. This 83.3% error rate is consistent with that for LAMP assays detecting bacteria reported previously (22). These data further emphasize the importance of using reliable positive controls and replicated experiments to validate all results. For critical samples, further testing to confirm the results using a multiplex PCR or individual PCRs targeting different unique loci, as reported by Lang et al. (2010) (15), is recommended. False-positive results were detected more often when we used the loop primers designed for the X. oryzae pv. oryzicola-specific assay. While in some assays loop primers play an important role in enhancing sensitivity, their removal can reduce the rates of false-positive results, presumably by reducing the sensitivity of the assay, resulting in a more reliable and robust assay.
An advantage of LAMP over conventional PCR is that sophisticated equipment is not required for the LAMP protocols. We detected X. oryzae pv. oryzicola and X. oryzae pv. oryzae directly from leaf tissue ground in water (data not shown) using the SYBR green detection system, demonstrating the feasibility of using the assay on field samples. Because the reaction is isothermal, a water bath or thermos with hot water could be used for reactions, simplifying the process for use in the field. We estimate that the current cost for a single 12-μl reaction mixture is $1. The availability of improved DNA polymerases was a key factor in improving the efficiency and accuracy of the LAMP technology (40). New formulations of the enzyme Bst 2.0, including one requiring a warm start (Bst 2.0 Warm Start; New England BioLabs, Ipswich, MA), have improved the efficiency and multiplexing capabilities of custom-developed reaction mixes. Because of the sensitivity of the LAMP assays, extreme care must be taken to avoid aerosolization of the amplified products. This is particularly important when using postreaction detection methods. We added mineral oil to the top of each reaction mixture to avoid contamination when tubes are opened to add the detecting dye. Liang et al. (41) recently developed a technology to separate the DNA intercalating dye from the samples during incubation using a temperature-sensitive wax. We did not test this approach, but it could be very useful for field-based LAMP assays.
In agriculture, the ability to work with crude extracts from plants and minimal equipment enhances the value of the LAMP technology for rapid detection for etiological or epidemiological studies or for regulatory purposes. In this study, we demonstrate the utility of the assay with leaf and seed extracts. Most recently, Randhawa et al. (25) reported the detection of genetically modified organisms using a LAMP based on promoter sequences. Researchers will be able to quickly determine the sources of contaminated material or outbreaks in the field using LAMP. In future work, surveys conducted using the LAMP assays that have been reported will provide a reliable picture of the presence of these diseases across a region or country.
Supplementary Material
ACKNOWLEDGMENTS
This research was partially supported by the International Livestock Research Institute (ILRI) and Biosciences eastern and central Africa (BecA) as part of a subcontract of funding from the Swedish Ministry for Foreign Affairs through the Swedish International Development Agency. J. M. Lang, P. Langlois, L. R. Triplett, and J. E. Leach were supported by the Colorado State Agricultural Experiment Station. V. Verdier (IRD) was supported by a Marie Curie IOF fellowship (EU grant PIOF-GA-2009-235457) at Colorado State University (Department of Bioagricultural Sciences and Plant Management).
We kindly thank Andreas Bühlmann, Alma Perez-Mendez, and Lawrence Goodridge for advising us on primer design and for informative advice and discussions. We recognize Steve Milligton, Optigene, who provided reagents, equipment, and generous technical support. Additionally, we thank Ana Bossa, Loïc Deblais, and Hongxia Liu for providing inoculated rice tissue.
Footnotes
Published ahead of print 16 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00274-14.
REFERENCES
- 1.Ou SH. 1985. Rice disease, 2nd ed. Association Applied Biology, Surrey, United Kingdom [Google Scholar]
- 2.Verdier V, Vera Cruz C, Leach JE. 2012. Controlling rice bacterial blight in Africa: needs and prospects. J. Biotechnol. 159:320–328. 10.1016/j.jbiotec.2011.09.020 [DOI] [PubMed] [Google Scholar]
- 3.Garrett K, Dendy S, Frank E, Rouse M, Travers S. 2006. Climate change effects on plant disease: genomes to ecosystems. Annu. Rev. Phytopathol. 44:489–509. 10.1146/annurev.phyto.44.070505.143420 [DOI] [PubMed] [Google Scholar]
- 4.Wonni I, Cottyn B, Detemmerman L, Dao S, Ouedraogo L, Sarra S, Tekete C, Poussier S, Corral R, Triplett L, Koita O, Koebnik R, Leach J, Szurek B, Maes M, Verdier V. 2014. Analysis of Xanthomonas oryzae pv. oryzicola population in Mali and Burkina Faso reveals a high level of genetic and pathogenic diversity. Phytopathology 104:520–531. 10.1094/PHYTO-07-13-0213-R [DOI] [PubMed] [Google Scholar]
- 5.Sakthivel N, Mortensen C, Mathur S. 2001. Detection of Xanthomonas oryzae pv. oryzae in artificially inoculated and naturally infected rice seeds and plants by molecular techniques. Appl. Microbiol. Biotechnol. 56:435–441. 10.1007/s002530100641 [DOI] [PubMed] [Google Scholar]
- 6.Kauffman HE, Reddy APK. 1975. Seed transmission studies of Xanthomonas oryzae in rice. Phytopathology 65:663–666. 10.1094/Phyto-65-663 [DOI] [Google Scholar]
- 7.Mew TW, Misra JK. 1994. A manual of rice seed health testing. International Rice Research Institute, Los Banos, Philippines [Google Scholar]
- 8.Devadath S, Thri Murty VS. 1984. Role of seed in survival and transmission of Xanthomonas campestris pv. oryzae causing bacterial blight of rice. J. Phytopathol. 110:15–19. 10.1111/j.1439-0434.1984.tb00735.x [DOI] [Google Scholar]
- 9.Gnanamanickam SS, Shigaki T, Medalla ES, Mew TW, Alvarez AM. 1994. Problems in detection of Xanthomonas oryzae pv. oryzae in rice seed and potential for improvement using monoclonal antibodies. Plant Dis. 78:173–178. 10.1094/PD-78-0173 [DOI] [Google Scholar]
- 10.Xie GL, Mew TW. 1998. A leaf inoculation method for detection of Xanthomonas oryzae pv. oryzicola from rice seed. Plant Dis. 82:1007–1011. 10.1094/PDIS.1998.82.9.1007 [DOI] [PubMed] [Google Scholar]
- 11.Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D, Madupu R, Puiu D, Radune D, Shumway M, Trapnell C, Aparna G, Jha G, Pandey A, Patil PB, Ishihara H, Meyer DF, Szurek B, Verdier V, Koebnik R, Dow JM, Ryan RP, Hirata H, Tsuyumu S, Won Lee S, Seo Y-S, Sriariyanum M, Ronald PC, Sonti RV, Van Sluys M-A, Leach JE, White FF, Bogdanove AJ. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9:204. 10.1186/1471-2164-9-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogdanove AJ, Koebnik R, Lu H, Furutani A, Angiuoli SV, Patil PB, Van Sluys MA, Ryan RP, Meyer DF, Han SW, Aparna G, Rajaram M, Delcher AL, Phillippy AM, Puiu D, Schatz MC, Shumway M, Sommer DD, Trapnell C, Benahmed F, Dimitrov G, Madupu R, Radune D, Sullivan S, Jha G, Ishihara H, Lee SW, Pandey A, Sharma V, Sriariyanun M, Szurek B, Vera-Cruz CM, Dorman KS, Ronald PC, Verdier V, Dow JM, Sonti RV, Tsuge S, Brendel VP, Rabinowicz PD, Leach JE, White FF, Salzberg SL. 2011. Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 193:5450–5464. 10.1128/JB.05262-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Triplett LR, Hamilton JP, Buell CR, Tisserat NA, Verdier V, Zink F, Leach JE. 2011. Genomic analysis of Xanthomonas oryzae isolates from rice grown in the United States reveals substantial divergence from known X. oryzae pathovars. Appl. Environ. Microbiol. 77:3930–3937. 10.1128/AEM.00028-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee B-M, Park Y-J, Park D-S, Kang H-W, Kim J-G, Song E-S, Park I-C, Yoon U-H, Hahn J-H, Koo B-S, Lee G-B, Kim H, Park H-S, Yoon K-O, Kim J-H, Jung C, Koh N-H, Seo J-S, Go S-J. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33:577–586. 10.1093/nar/gki206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang JM, Hamilton JP, Diaz MGQ, Van Sluys MA, Burgos MRG, Vera Cruz CM, Buell CR, Tisserat NA, Leach JE. 2010. Genomics-based diagnostic marker development for Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola. Plant Dis. 94:311–319. 10.1094/PDIS-94-3-0311 [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez C, Szurek B, Manceau C, Mathieu T, Sere Y, Verdier V. 2007. Molecular and pathotypic characterization of new Xanthomonas oryzae strains from West Africa. Mol. Plant Microbe Interact. 20:534–546. 10.1094/MPMI-20-5-0534 [DOI] [PubMed] [Google Scholar]
- 17.Hajri A, Brin C, Zhao S, David P, Feng J, Koebnik R, Szurek B, Verdier V, Boureau T, Poussier S. 2012. Multilocus sequence analysis and type III effector repertoire mining provide new insights into the evolutionary history and virulence of Xanthomonas oryzae. Mol. Plant Pathol. 13:288–302. 10.1111/j.1364-3703.2011.00745.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomlinson J. 2013. In-field diagnostics using loop-mediated isothermal amplification, p 291–300 In Dickinson M, Hodgetts J. (ed), Phytoplasma. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 20.Kaneko H, Kawana T, Fukushima E, Suzutani T. 2007. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 70:499–501. 10.1016/j.jbbm.2006.08.008 [DOI] [PubMed] [Google Scholar]
- 21.Bühlmann A, Pothier JF, Tomlinson JA, Frey JE, Boonham N, Smits THM, Duffy B. 2013. Genomics-informed design of loop-mediated isothermal amplification for detection of phytopathogenic Xanthomonas arboricola pv. pruni at the intraspecific level. Plant Pathol. 62:475–484. 10.1111/j.1365-3059.2012.02654.x [DOI] [Google Scholar]
- 22.Bühlmann A, Pothier JF, Rezzonico F, Smits THM, Andreou M, Boonham N, Duffy B, Frey JE. 2013. Erwinia amylovora loop-mediated isothermal amplification (LAMP) assay for rapid pathogen detection and on-site diagnosis of fire blight. J. Microbiol. Methods 92:332–339. 10.1016/j.mimet.2012.12.017 [DOI] [PubMed] [Google Scholar]
- 23.Villari C, Tomlinson JA, Battisti A, Boonham N, Capretti P, Faccoli M. 2013. Use of loop-mediated isothermal amplification for detection of Ophiostoma clavatum, the primary blue stain fungus associated with Ips acuminatus. Appl. Environ. Microbiol. 79:2527–2533. 10.1128/AEM.03612-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Q-W, Yu C, Zhang S-Y, Yang C-Y, Miriam K, Zhang W-N, Dou D-L, Tao X-R. 2012. One-step detection of bean pod mottle virus in soybean seeds by the reverse-transcription loop-mediated isothermal amplification. Virol. J. 9:187. 10.1186/1743-422X-9-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randhawa GJ, Singh M, Morisset D, Sood P, Zel J. 2013. Loop-mediated isothermal amplification: rapid visual and real-time methods for detection of genetically modified crops. J. Agric. Food Chem. 61:11338–11346. 10.1021/jf4030085 [DOI] [PubMed] [Google Scholar]
- 26.Armitage P, Berry G, Matthews J. 2002. Statistical methods in medical research, 4th ed. Blackwell, Oxford, United Kingdom [Google Scholar]
- 27.Yasuhara-Bell J, Kubota R, Jenkins DM, Alvarez AM. 2013. Loop-mediated amplification of the Clavibacter michiganensis subsp. michiganensis micA gene is highly specific. Phytopathology 103:1220–1226. 10.1094/PHYTO-03-13-0078-R [DOI] [PubMed] [Google Scholar]
- 28.Ash GJ, Lang JM, Triplett LR, Stodart B, Verdier V, Vera Cruz C, Rott PC, Leach JE. 2014. Development of a genomics-based LAMP (loop-mediated isothermal amplification) assay for detection of Pseudomonas fuscovaginae from rice. Plant Dis. 98:909–915. 10.1094/PDIS-09-13-0957-RE [DOI] [PubMed] [Google Scholar]
- 29.Jones RK, Barnes LW, Gonzalez CF, Leach JE, Alvarez AM, Benedict AA. 1989. Identification of low virulence strains of Xanthomonas campestris pv. oryzae from rice in the United States. Phytopathology 79:984–990. 10.1094/Phyto-79-984 [DOI] [Google Scholar]
- 30.Karganilla A, Paris-Natural M, Ou SH. 1973. A comparative study of culture media for Xanthomonas oryzae. Philipp. Agric. 57:141–152 [Google Scholar]
- 31.Verdier V, Triplett LR, Hummel AW, Corral R, Cernadas RA, Schmidt CL, Bogdanove AJ, Leach JE. 2012. Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol. 196:1197–1207. 10.1111/j.1469-8137.2012.04367.x [DOI] [PubMed] [Google Scholar]
- 32.Ochiai H, Inoue V, Takeya M, Sasaki A, Kaku H. 2005. Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn. Agric. Res. Q. 39:275–287. 10.6090/jarq.39.275 [DOI] [Google Scholar]
- 33.Mew TW, Vera CMC, Medalla ES. 1992. Changes in race frequency of Xanthomonas oryzae pv. oryzae in response to rice cultivars planted in the Philippines. Plant Dis. 76:1029. 10.1094/PD-76-1029 [DOI] [Google Scholar]
- 34.Li J, Wei Q, Liu Y, Tan X, Zhang W, Wu J, Charimbu MK, Hu B, Cheng Z, Yu C, Tao X. 2013. One-step reverse transcription loop-mediated isothermal amplification for the rapid detection of cucumber green mottle mosaic virus. J. Virol. Methods 193:583–588. 10.1016/j.jviromet.2013.07.059 [DOI] [PubMed] [Google Scholar]
- 35.Li X, Nie J, Ward L, Madani M, Hsiang T, Zhao Y, De Boer SH. 2009. Comparative genomics-guided loop-mediated isothermal amplification for characterization of Pseudomonas syringae pv. phaseolicola. J. Appl. Microbiol. 107:717–726. 10.1111/j.1365-2672.2009.04262.x [DOI] [PubMed] [Google Scholar]
- 36.Rigano LA, Marano MR, Castagnaro AP, Do Amaral AM, Vojnov AA. 2010. Rapid and sensitive detection of citrus bacterial canker by loop-mediated isothermal amplification combined with simple visual evaluation methods. BMC Microbiol. 10:176. 10.1186/1471-2180-10-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubota R, Vine BG, Alvarez AM, Jenkins DM. 2008. Detection of Ralstonia solanacearum by loop-mediated isothermal amplification. Phytopathology 98:1045–1051. 10.1094/PHYTO-98-9-1045 [DOI] [PubMed] [Google Scholar]
- 38.Yamashita Y, Tsukioka Y, Tomihisa K, Nakano Y, Koga T. 1998. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J. Bacteriol. 180:5803–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Vinogradov EV, Bogdanove AJ. 2013. Requirement of the lipopolysaccharide O-chain biosynthesis gene wxocB for type III secretion and virulence of Xanthomonas oryzae pv. oryzicola. J. Bacteriol. 195:1959–1969. 10.1128/JB.02299-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori Y, Kanda H, Notomi T. 2013. Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J. Infect. Chemother. 19:404–411. 10.1007/s10156-013-0590-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang C, Cheng S, Chu Y, Wu H, Zou B, Huang H, Xi T, Zhou G. 2013. A closed-tube detection of loop-mediated isothermal amplification (LAMP) products using a wax-sealed fluorescent intercalator. J. Nanosci. Nanotechnol. 13:3999–4005. 10.1166/jnn.2013.6497 [DOI] [PubMed] [Google Scholar]
- 42.Gosch C, Gottsberger RA, Stich K, Fischer TC. 2012. Blue EaLAMP—a specific and sensitive method for visual detection of genomic Erwinia amylovora DNA. Eur. J. Plant Pathol. 134:835–845. 10.1007/s10658-012-0059-5 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.