Abstract
From August to September 2008, the Centers for Disease Control and Prevention (CDC) assisted the Alaska Division of Public Health with an outbreak investigation of campylobacteriosis occurring among the residents of Southcentral Alaska. During the investigation, pulsed-field gel electrophoresis (PFGE) of Campylobacter jejuni isolates from human, raw pea, and wild bird fecal samples confirmed the epidemiologic link between illness and the consumption of raw peas contaminated by sandhill cranes for 15 of 43 epidemiologically linked human isolates. However, an association between the remaining epidemiologically linked human infections and the pea and wild bird isolates was not established. To better understand the molecular epidemiology of the outbreak, C. jejuni isolates (n = 130; 59 from humans, 40 from peas, and 31 from wild birds) were further characterized by multilocus sequence typing (MLST). Here we present the molecular evidence to demonstrate the association of many more human C. jejuni infections associated with the outbreak with raw peas and wild bird feces. Among all sequence types (STs) identified, 26 of 39 (67%) were novel and exclusive to the outbreak. Five clusters of overlapping STs (n = 32 isolates; 17 from humans, 2 from peas, and 13 from wild birds) were identified. In particular, cluster E (n = 7 isolates; ST-5049) consisted of isolates from humans, peas, and wild birds. Novel STs clustered closely with isolates typically associated with wild birds and the environment but distinct from lineages commonly seen in human infections. Novel STs and alleles recovered from human outbreak isolates allowed additional infections caused by these rare genotypes to be attributed to the contaminated raw peas.
INTRODUCTION
Campylobacter jejuni is the most commonly implicated pathogen associated with bacterial gastroenteritis worldwide (1). With an estimated 1.3 million cases per year, Campylobacter infection is one of the most common causes of diarrheal illness and hospitalizations in the United States (2).
Although the majority of Campylobacter infections are sporadic, outbreaks do occur. Between 1997 and 2008, there were 262 Campylobacter outbreaks, with a total of 9,136 cases, 159 hospitalizations, and 3 deaths in the United States (3). The great majority of outbreaks are food borne, and dairy products caused the largest number (50%) of illnesses in these outbreaks, followed by produce (10%) and poultry (4%).
Infection with C. jejuni is zoonotic and most commonly associated with food-producing animals (4), including poultry (5), cattle, and sheep (6). C. jejuni is also widely disseminated in the natural environment, which is often contaminated by feces from wildlife, including wild birds and mammals (7–9).
Although fresh produce is less often identified as a source of outbreaks, campylobacters have been isolated from various types of fresh produce, including radishes, cabbage, spinach, bean sprouts, lettuce, parsley, and potatoes (10–13), from samples collected from farms (10), local outdoor markets (10, 13) and supermarkets (11). C. jejuni survives on fresh produce for up to 3 days after contamination at different temperatures, which is long enough for produce to be distributed and eaten (14). In a retrospective case-control study in Wales, the consumption of salad vegetables was concluded to be second only to consumption of chicken as a risk factor for Campylobacter infection (15).
In August 2008, the CDC assisted the Alaska Division of Public Health with an investigation of a Campylobacter outbreak that occurred among the residents of Southcentral Alaska; an epidemiological link was identified between illness and the consumption of raw peas, harvested from a field frequented by wild sandhill cranes (Grus canadensis) (16). Pulsed-field gel electrophoresis (PFGE) testing of C. jejuni isolates from human, pea, and wild bird samples subsequently confirmed the link. There was substantial diversity in PFGE types: 25 different C. jejuni PFGE types were found among the 55 human isolates tested. Four clusters were identified that encompassed 15 human isolates, 2 pea isolates, and 3 fecal isolates from sandhill cranes. It was unclear, however, whether and how the remaining epidemiologically linked infections that were not matched with pea and wild bird samples were associated with the outbreak.
We exploited a rare opportunity to explore the underlying dynamics of disease transmission in a produce-associated Campylobacter outbreak linked to wild birds by characterizing C. jejuni isolates from human, pea, and wild bird samples by using multilocus sequence typing (MLST) (16), with the aim to confirm the source of the outbreak and attribute the remaining epidemiologically linked infections that were not indistinguishable by PFGE from pea and wild bird samples from the outbreak. Studies of well-characterized outbreaks can provide important insights into understanding the routes and vehicles of disease transmission (17, 18).
MATERIALS AND METHODS
C. jejuni isolates.
A total of 59 isolates from humans were included. Forty-three outbreak-associated isolates from stool specimens from epidemiologically linked cases were available from the initial epidemiologic investigation (16). A total of 16 sporadic C. jejuni isolates (including 6 isolates from the initial study) from epidemiologically unrelated illnesses during the same period and in the same area were collected. From the initial study, 40 pea and 31 wild bird C. jejuni isolates derived from 16 environmental samples were also included for analysis (16).
MLST.
C. jejuni isolates were characterized by MLST; the internal sequences of the seven housekeeping loci were amplified and sequenced by use of the primers and reaction conditions described in a previously published scheme (19). BioNumerics v6.1 (Applied Maths) was used for sequence editing and assembly. The Campylobacter MLST database was used for sequence type (ST) and clonal complex assignment (http://pubmlst.org/campylobacter) (20).
Penner serotyping and antimicrobial susceptibility testing.
Penner serotypes were determined by standard methods for heat-stable antigens (21), using a panel of 25 antisera which represent common serotypes in the United States (22). Antimicrobial susceptibility testing was performed at the CDC by using the National Antimicrobial Resistance Monitoring System (NARMS) standard panel of nine antimicrobials (azithromycin, ciprofloxacin, clindamycin, erythromycin, florfenicol, gentamicin, nalidixic acid, telithromycin, and tetracycline) and broth microdilution (Sensititre; Trek Diagnostics), which was performed according to the manufacturer's instructions and interpreted using CLSI criteria when available.
Data analysis.
The genetic relatedness among unique STs from 130 C. jejuni isolates in the study was visualized, along with strains (887 STs; n = 7,044 isolates) from humans, chickens, wild birds, and environmental waters from the Campylobacter PubMLST database (20) as a comparison. A neighbor-joining tree was constructed from a matrix of pairwise differences calculated between the 3,309-bp concatenated nucleotide sequences from the seven MLST housekeeping genes. A consensus tree was derived using MEGA software v5.1 (23), with 1,000 bootstrap replicates. The degrees of linkage disequilibrium within the data set and subsets were measured by calculating the standardized index of association (IAS) (24). If IAS = 0, then alleles at multiple loci are in absolute linkage equilibrium, indicating a freely recombining or panmictic population structure, while if IAS ≠ 0, it suggests the presence of linkage disequilibrium, indicating that recombination has been rare enough or absent to allow the emergence of a clonal population structure. IAS was computed using the START2 package (25). Population differentiation based on estimates of gene flow between C. jejuni subpopulations within the data set was carried out using 3,309-bp concatenated nucleotide sequences from the seven MLST loci, using DnaSP software v5.10.01 (26). Gene flow is measured by a fixation index (FST) value that falls between 0 and 1, where a value of 0 indicates that two populations are genetically indistinguishable and a value of 1 represents completely differentiated populations.
RESULTS
Novel genetic diversity.
Among 130 C. jejuni isolates characterized by MLST, 39 STs and 11 clonal complexes were identified, comprising 26 novel (n = 101; 78% of all isolates) and 13 known (n = 29; 22% of all isolates) STs. Only one of the novel STs (ST-5068) belonged to one of the 11 clonal complexes identified (Table 1). Among the novel STs, 24 of 26 (92%) were composed of newly identified alleles, often observed at multiple MLST loci. ST-5054 was composed entirely of novel alleles at all seven loci. Forty-six novel alleles (31%) were identified among a total of 145 alleles in the data set.
TABLE 1.
Genetic diversity and frequency distribution of 130 C. jejuni isolates from humans, peas, and wild birds from the outbreake,f
| Sequence type | Clonal complexa | MLST allelic profileb |
PFGE profilec |
Penner serotyped | No. of isolates |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human |
Wild bird | Pea | Total | |||||||||||||
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | SmaI | KpnI | Outbreak | Sporadic | ||||||
| Human, pea, and wild bird clusters | ||||||||||||||||
| ST-5061* (cluster A) | UA | 10 | 387‡ | 61 | 423 | 39 | 449‡ | 1 | DBRS 16.1283 | DBRK02.0549 | HS19 | 1 | 1 | 2 | ||
| ST-5059* (cluster B) | UA | 10 | 31 | 25 | 49 | 57 | 7 | 313 | DBRS 16.0127 | DBRK02.0546 | HS8 | 5 | 1 | 1 | 7 | |
| ST-5048* (cluster C) | UA | 10 | 383 | 106 | 419‡ | 57 | 443‡ | 1 | DBRS 16.1256 | DBRK02.0522 | HS18 | 3 | 4 | 7 | ||
| ST-137 (cluster D) | ST-45 | 4 | 7 | 10 | 4 | 42 | 7 | 1 | DBRS 16.0749 | DBRK02.0524 | HS15 | 5 | 4 | 9 | ||
| ST-5049* (cluster E) | UA | 180 | 172 | 322 | 47 | 39 | 180 | 3 | Unrestricted | DBRK02.0548 | HS29, HS21 | 2 | 2 | 3 | 7 | |
| Pea and wild bird isolates | ||||||||||||||||
| ST-5047* | UA | 180 | 389 | 95 | 47 | 541 | 442 | 3 | 5 | 4 | 9 | |||||
| ST-5057* | UA | 1 | 389 | 325 | 422 | 541 | 180 | 6 | 4 | 5 | 9 | |||||
| ST-5050* | UA | 178 | 7 | 95 | 6 | 354 | 88 | 1 | 6 | 6 | ||||||
| ST-5056* | UA | 178 | 385 | 95 | 129 | 545 | 180 | 48 | 6 | 6 | ||||||
| ST-5052* | UA | 1 | 31 | 323 | 420 | 394 | 89 | 6 | 4 | 1 | 5 | |||||
| ST-5055* | UA | 10 | 280 | 277 | 49 | 394 | 443‡ | 6 | 5 | 5 | ||||||
| ST-5053* | UA | 1 | 2 | 95 | 62 | 472 | 400 | 147 | 1 | 3 | 4 | |||||
| ST-5054* | UA | 285 | 384 | 324 | 421 | 542 | 445 | 312 | 3 | 3 | ||||||
| ST-5073* | UA | 178 | 8 | 327 | 49 | 544 | 28 | 6 | 3 | 3 | ||||||
| ST-5058* | UA | 1 | 2 | 34 | 90 | 543 | 443‡ | 6 | 1 | 1 | 2 | |||||
| ST-5051* | UA | 10 | 385 | 106 | 129 | 39 | 444 | 3 | 1 | 1 | ||||||
| ST-5060* | UA | 10 | 387‡ | 95 | 423 | 39 | 7 | 1 | 1 | 1 | ||||||
| ST-5072* | UA | 290 | 389 | 5 | 418 | 39 | 441 | 6 | 1 | 1 | ||||||
| ST-5074* | UA | 10 | 386 | 61 | 419‡ | 39 | 451 | 1 | 1 | 1 | ||||||
| Human outbreak-associated isolates | ||||||||||||||||
| ST-5062*† | UA | 287 | 6 | 34 | 424 | 394 | 446 | 172 | HS41 | 8 | 9 | |||||
| HS8 | 1 | |||||||||||||||
| ST-468 | UA | 10 | 2 | 50 | 62 | 91 | 73 | 45 | 3 | 3 | ||||||
| ST-5069* | UA | 10 | 6 | 61 | 427 | 548 | 447 | 6 | 3 | 3 | ||||||
| ST-5063* | UA | 10 | 308 | 106 | 47 | 57 | 447 | 1 | 2 | 2 | ||||||
| ST-5065* | UA | 1 | 172 | 5 | 426 | 39 | 231 | 6 | 2 | 2 | ||||||
| ST-5064* | UA | 1 | 82 | 95 | 425 | 546 | 448 | 314 | 1 | 1 | ||||||
| ST-5067* | UA | 286 | 172 | 5 | 426 | 39 | 443‡ | 6 | 1 | 1 | ||||||
| ST-5066* | UA | 10 | 74 | 276 | 47 | 39 | 449‡ | 51 | 2 | 2 | ||||||
| ST-45 | ST-45 | 4 | 7 | 10 | 4 | 1 | 7 | 1 | 2 | 2 | ||||||
| ST-42 | ST-42 | 1 | 2 | 3 | 4 | 5 | 9 | 3 | 1 | 1 | ||||||
| ST-48 | ST-48 | 2 | 4 | 1 | 2 | 7 | 1 | 5 | 1 | 1 | ||||||
| Human sporadic isolates | ||||||||||||||||
| ST-5066* | UA | 10 | 74 | 276 | 47 | 39 | 449‡ | 51 | 1 | 1 | ||||||
| ST-45 | ST-45 | 4 | 7 | 10 | 4 | 1 | 7 | 1 | 3 | 3 | ||||||
| ST-42 | ST-42 | 1 | 2 | 3 | 4 | 5 | 9 | 3 | 1 | 1 | ||||||
| ST-48 | ST-48 | 2 | 4 | 1 | 2 | 7 | 1 | 5 | 1 | 1 | ||||||
| ST-19 | ST-21 | 2 | 1 | 5 | 3 | 2 | 1 | 5 | 1 | 1 | ||||||
| ST-52 | ST-52 | 9 | 25 | 2 | 10 | 22 | 3 | 6 | 1 | 1 | ||||||
| ST-353 | ST-353 | 7 | 17 | 5 | 2 | 10 | 3 | 6 | 1 | 1 | ||||||
| ST-403 | ST-403 | 10 | 27 | 16 | 19 | 10 | 5 | 7 | 1 | 1 | ||||||
| ST-508 | ST-508 | 1 | 6 | 60 | 24 | 12 | 28 | 1 | 1 | 1 | ||||||
| ST-572 | ST-206 | 62 | 4 | 5 | 2 | 2 | 1 | 5 | 1 | 1 | ||||||
| ST-991 | ST-692 | 37 | 52 | 57 | 26 | 107 | 29 | 23 | 1 | 1 | ||||||
| ST-2875 | UA | 2 | 29 | 4 | 48 | 10 | 25 | 57 | 1 | 1 | ||||||
| ST-5068* | ST-42 | 1 | 2 | 3 | 4 | 5 | 9 | 315 | 1 | 1 | ||||||
| Total | 43 | 16 | 31 | 40 | 130 | |||||||||||
UA, unassigned status for a given sequence type.
Newly identified alleles are shown in bold.
PFGE profiles for the restriction enzymes SmaI and KpnI for the five identified clusters (A to E).
Penner serotyping was performed on representatives of human and environmental isolates from clusters A to E and on all ST-5062 isolates, including one from a GBS case.
*, novel STs; †, ST associated with one case of Guillain-Barré syndrome; ‡, overlapping novel alleles found in human and environmental isolates.
The numbers of alleles per locus (% novel) were 14 (28.5), 23 (26), 22 (22.7), 24 (41.6), 22 (31.8), 24 (41.6), and 16 (25) for the aspA, glnA, gltA, glyA, pgm, tkt, and uncA genes, respectively.
Outbreak molecular epidemiology.
Most of the human outbreak-associated isolates (31 of 43 [72%]) and pea and wild bird isolates (67 of 71 [94%]) were represented by novel STs, while most of the comparable sporadic human isolates (13 of 16 [81%]) were represented by known STs. From a total of 39 STs identified in our data set, 20 STs were seen only among human isolates, and 14 STs were seen only among pea and wild bird isolates (Table 1). However, five clusters with five different STs were identified that contained both human outbreak-associated isolates and pea and wild bird isolates (clusters A to E) (Table 1). The clustering of isolates showed strong agreement between MLST, PFGE, and Penner serotyping results. Clusters A to D corresponded to the four combined SmaI-KpnI PFGE patterns identified in human isolates that were indistinguishable from pea and wild bird isolates in the initial investigation (16). Cluster E isolates contained isolates from all three sample types, including two human outbreak-associated isolates, three isolates from peas, and two isolates from wild bird feces. The two human outbreak-associated isolates from cluster E were unrestricted by PFGE using SmaI but were grouped by an indistinguishable KpnI-restriction PFGE pattern in the initial investigation (16).
ST-5062 did not belong to a cluster but was the most common subtype found among human outbreak-associated isolates from nine patients. The SmaI-KpnI PFGE restriction patterns and Penner serotype (HS:41) were also indistinguishable among eight of these nine patients. One of these eight patients developed Guillain-Barré syndrome (GBS). The isolate from the ninth patient had a different SmaI-KpnI PFGE restriction pattern and Penner serotype (HS:8).
Apart from one human isolate from cluster D, found to be resistant to ciprofloxacin and nalidixic acid, all isolates within the five clusters were pan-susceptible to the nine antimicrobials tested.
Population structure.
To provide additional molecular evidence for the link between wild birds and human illness during the outbreak, we explored the underlying C. jejuni population structure between the human outbreak-associated isolates and pea and wild bird isolates that did not cluster during the outbreak.
The presence of novel alleles was a distinct feature among the five clusters of human outbreak-associated isolates and pea and wild bird isolates identified. However, novel alleles were also widely distributed among human outbreak-associated isolates and pea and wild bird isolates that did not cluster during the outbreak. In particular, four novel alleles (glnA-387, glyA-419, tkt-443, and tkt-449), from clusters A, B, C, and E, were also found among human outbreak-associated isolates and pea and wild bird isolates that did not cluster (Table 1).
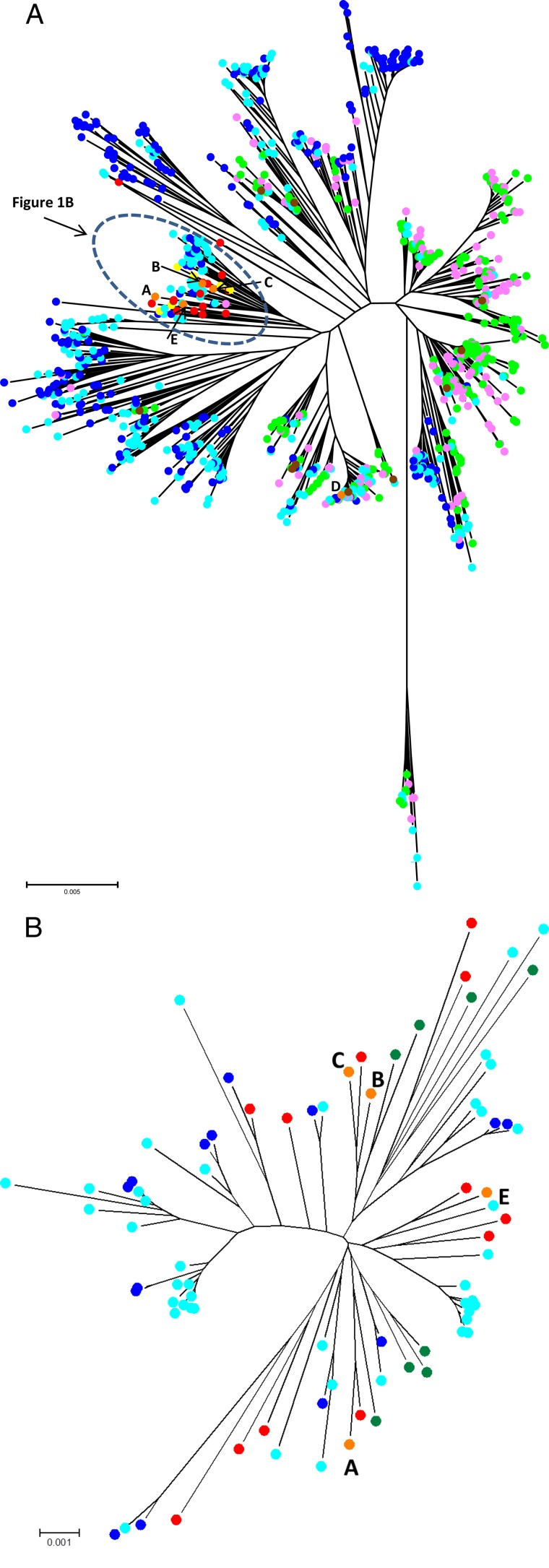
We visualized the C. jejuni population structure of our data set (all 39 STs) in conjunction with isolates (n = 7,044) from the PubMLST database (887 STs) that originated from humans (green), chickens (pink), wild birds (dark blue), and environmental waters (light blue) by using a neighbor-joining tree (Fig. 1A). A host association of C. jejuni clades was apparent, as distinct clades with predominantly “human and chicken isolates” and “wild bird and environmental water isolates” were evident. C. jejuni STs from our human outbreak-associated isolates and pea and wild bird isolates collected during the outbreak were genetically closely related (yellow, human outbreak-associated isolates; red, pea and wild bird isolates; orange, clusters A, B, C, and E) (Fig. 1B). Additionally, these isolates clustered within clades associated with the “wild bird and environmental water isolates” in the PubMLST database. Cluster D isolates (ST-137, ST-45 complex; orange) were grouped among members of a genetically diverse complex frequently associated with avian sources, including chickens and wild birds (8, 27). In contrast, sporadic human C. jejuni isolates from Alaska (brown) were well dispersed among clades associated with “human and chicken isolates.”
FIG 1.
(A) The genetic relatedness and diversity of 39 unique STs from 130 C. jejuni isolates in the study were visualized, along with 887 STs (n = 7,044 isolates) from various sources, available in the PubMLST database, for comparison. Isolates from the Alaska outbreak included isolates from peas and wild birds (red), humans (yellow), and overlapping clusters A to E (orange). Isolates from the MLST database included isolate collections from humans (green), chickens (pink), environmental waters (light blue), and wild birds (dark blue). (B) Expanded view of the genetic relatedness of clusters A, B, C, and E with closely related isolates from the circled portion of panel A.
We also assessed the genetic similarity between the human outbreak-associated isolates and pea and wild bird isolates and their genetic divergence relative to the sporadic human isolates. The current outbreak data set was divided into subsets of novel STs (representing outbreak, pea, and wild bird isolates) and known STs (representing sporadic human isolates). Differences in the degree of clonality between these subsets were detected by calculating the standardized index of association (IAS) as a measure of linkage disequilibrium. The IAS values (P < 0.001) for the subsets using representative STs were 0.0526 for the subset of novel STs and 0.3057 for the subset of known STs. The difference in IAS values indicated that there were distinct C. jejuni subpopulations with differing degrees of clonality. The IAS value of 0.0526 for the novel ST subset denotes a subpopulation with a higher rate of recombination and a less clonal population structure than those of the known ST subset, which had a higher IAS value. The gene flow between the novel and known ST subsets appears to be limited, as the pairwise FST value of 0.192 indicates considerable population differentiation.
DISCUSSION
Given the association with a wild bird source and an unusual transmission vehicle, the 2008 Alaska Campylobacter raw pea outbreak offered a unique opportunity to extend our current understanding of the epidemiology of C. jejuni infections.
In this investigation, we sought to examine the association between the epidemiologically linked human C. jejuni infections and pea and wild bird samples that could not be linked using PFGE data from the initial study (16).
We explored genetic differences between C. jejuni isolates from pea and wild bird samples and those from human infections from the outbreak. Among the human isolates characterized in this study, most human outbreak-associated isolates belonged to novel STs and were therefore highly suggestive of unique infective sources. Similarly, all but one ST (ST-137) identified from pea and wild bird samples were novel. In contrast, the majority of sporadic human isolates belonged to known STs and clonal complexes that are globally widespread among human infections (http://pubmlst.org/campylobacter/). Despite the high genetic divergence normally shown among environmental C. jejuni isolates, we were still able to observe five clusters of overlapping STs among the genetically similar human outbreak-associated isolates and pea and wild bird isolates. In addition, the occurrence of ST-5049 in all three sample sources (cluster E) provides a definitive link that illustrates the transmission route of C. jejuni during the outbreak.
We attributed additional human cases from the outbreak that are likely to be linked to wild bird sources by exploring the underlying C. jejuni population structure. Based on the possession of identical novel alleles, we linked additional possible human outbreak-associated isolates without identical sequence type matches to those from the peas and wild birds. In our data set, approximately one-third of alleles and 67% of STs were novel and could not be grouped into clonal complexes. In particular, ST-5054, with three wild bird isolates, was composed entirely of novel alleles at seven MLST loci. This finding was similar to that of a study of New Zealand river waters, where 55% of C. jejuni isolates consisted of novel STs, and ST-2381 was composed of six new alleles (28). In several other studies, the occurrence of isolates with novel alleles and STs unassigned to clonal complexes has similarly emerged as a dominant characteristic of genetically divergent C. jejuni isolates found primarily among wildlife and environmental sources (28–30). By clearly demonstrating the dominance of novel C. jejuni strains from wild birds and the peas they contaminated (and fed on), our study lends further support to the hypothesis of the existence of environmental/wildlife-associated C. jejuni strains (30). A distinct distribution of genotypes between wildlife and farmed animals could therefore signify independent disease transmission routes to humans from these sources (29). However, the ecology of C. jejuni among environmental sources is complex, so the precise transmission pathways leading to human infection from these sources have remained difficult to determine. Nonetheless, the absence of the C. jejuni ST-21 complex among human outbreak-associated isolates and pea and wild bird isolates in the current study, a lineage which is thought to be ubiquitous and adapted to a wide range of sources (8), is consistent with this notion.
From the radial neighbor-joining tree constructed, we have shown that human outbreak-associated isolates that did not group in one of the five identified clusters (clusters A to E) with pea and wild bird isolates were genetically closely related and were located within clades associated with wild bird and environmental isolates in the PubMLST database. Similarly, four of the five clusters (clusters A, B, C, and E) identified in the study were also located within the same wild bird/environmental clade. The fact that cluster D was not placed in these clades is not an unusual observation. ST-137 isolates from this cluster belong to the ST-45 complex, which has frequently been associated with human infections and avian sources (8).
By assigning novel STs to represent wildlife and environmental isolates and known STs to represent common lineages seen in human infections, we observed genetically distinct C. jejuni subpopulations with different rates of recombination and limited genetic mixing. The difference between IAS values obtained for the subsets suggests a C. jejuni population structure that consists of a subpopulation of highly successful epidemic clones (human infection/food isolates) superimposed on the background of a genetically divergent subpopulation that has a higher rate of recombination and a less clonal structure (wildlife/environmental isolates). This “epidemic” population structure was first proposed for Neisseria meningitidis and may also apply to C. jejuni (31). Indeed, given the higher rate of recombination and weaker ancestral signal, it is not a surprising observation that rare and novel STs have continually been found among environmental C. jejuni isolates (28, 30, 32–34) that remain unassigned to clonal complexes. Based on the pairwise FST value calculated (0.192), we have also shown evidence for limited gene flow between the known and novel ST subsets. This can be compared to values of 0.026 (35) and 0.019 (36) between human and poultry isolates, 0.199 between cattle and wild birds, 0.009 between wild birds and farmland environment (29), and 0.932 between C. jejuni and C. coli (37).
From a public health perspective, we have shown that despite considerable genetic divergence from lineages commonly seen in human infections, C. jejuni from wild birds in this study had the capacity to cause gastroenteritis, GBS, and a large-scale outbreak under favorable circumstances. However, several possible explanations can be speculated for C. jejuni strains that did not overlap between human outbreak-associated isolates and those found in peas and wild birds. First, certain C. jejuni strains from this outbreak may have poor pathogenicity against humans, given the genetic divergence from common strains circulating in human infections. Second, immunity among humans who have had prolonged exposure to strains found in peas and wild birds may not lead to illness after the consumption of contaminated peas. Third, C. jejuni strains genetically identical to those found in humans were simply not captured against the background of a highly diverse genetic pool in the wild bird population, due to the sample size. Lastly, the survival of C. jejuni strains on peas may have been short-lived, limiting the infectious dose required to cause an infection.
It is worth noting that only one of eight patients who shared the same ST (ST-5062), SmaI-KpnI PFGE profile, and Penner serotype (HS:41) developed GBS. This observation indicates that the development of GBS after Campylobacter infection could vary depending on host factors, even when individuals are infected with seemingly genetically identical strains. Therefore, a correlation between MLST genotypes and the onset of GBS could be difficult to determine, though several studies have found significant associations (38, 39). Nonetheless, our result has added further support to the described association between infection with HS:41 and the development of GBS, even from a wild bird source. Furthermore, patients with specific HLA alleles have been shown to be more likely to be predisposed to GBS (40).
From the antimicrobial susceptibility testing performed, only one isolate belonging to a known ST (ST-137) was found to be resistant, showing resistance to fluoroquinolones. Among isolates with novel STs, none showed resistance to any of the nine antimicrobials. This suggests that C. jejuni in wild birds may not have been exposed to antimicrobials that are often used in animals reared for food production. Indeed, the incidence of resistance to antimicrobials among wild bird C. jejuni isolates in Sweden was previously shown to be low (41).
We explored the unique opportunity to investigate the rare occurrence of a Campylobacter outbreak linked to produce and wild birds. Although poultry has been identified as the major source of campylobacteriosis from source attribution studies, the source of a considerable proportion of illnesses remains unidentified (42, 43). Our study has provided new insights into the disease transmission dynamics from the environmental reservoir, highlighting the significance of C. jejuni from produce and/or environmental sources and its potential to cause human disease under favorable circumstances. This study was made possible through timely sample collection and close collaboration between epidemiologists, sanitarians, and microbiologists who worked on the investigation. Additional studies involving isolation and characterization of C. jejuni from the environment would be helpful for building a more complete picture for source attribution efforts.
ACKNOWLEDGMENTS
This publication made use of the Campylobacter jejuni Multi Locus Sequence Typing website (http://pubmlst.org/campylobacter/), developed by Keith Jolley and Man-Suen Chan and sited at the University of Oxford (20). The development of this site was funded by the Wellcome Trust.
We thank the Alaska Division of Public Health's Infectious Disease Program and State Public Health Laboratory and the Alaska Department of Environmental Conservation staff, who assisted with the investigation, as well as Katherine Ross and Shellie Smith for their assistance and Lynne Lucher for her valuable advice and guidance.
Footnotes
Published ahead of print 16 May 2014
REFERENCES
- 1.World Health Organization. 2012. The global view of campylobacteriosis: report of an expert consultation. World Health Organization, Utrecht, The Netherlands: http://apps.who.int/iris/bitstream/10665/80751/1/9789241564601_eng.pdf [Google Scholar]
- 2.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.p11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor EV, Herman K, Ailes EC, Fitzgerald C, Yoder J, Mahon BE, Tauxe RV. 2013. Common source outbreaks of Campylobacter infection in the United States, 1997–2008. Epidemiol. Infect. 141:987–996. 10.1017/S0950268812001744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphrey T, O'Brien S, Madsen M. 2007. Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117:237–257. 10.1016/j.ijfoodmicro.2007.01.006 [DOI] [PubMed] [Google Scholar]
- 5.Corry JEL, Atabay HI. 2001. Poultry as a source of Campylobacter and related organisms. J. Appl. Microbiol. 90:96S–114S. 10.1046/j.1365-2672.2001.01358.x [DOI] [PubMed] [Google Scholar]
- 6.Stanley K, Jones K. 2003. Cattle and sheep farms as reservoirs of Campylobacter. J. Appl. Microbiol. 94:104S–113S. 10.1046/j.1365-2672.94.s1.12.x [DOI] [PubMed] [Google Scholar]
- 7.Broman T, Waldenström J, Dahlgren D, Carlsson I, Eliasson I, Olsen B. 2004. Diversities and similarities in PFGE profiles of Campylobacter jejuni isolated from migrating birds and humans. J. Appl. Microbiol. 96:834–843. 10.1111/j.1365-2672.2004.02232.x [DOI] [PubMed] [Google Scholar]
- 8.Colles FM, Jones K, Harding RM, Maiden MCJ. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409–7413. 10.1128/AEM.69.12.7409-7413.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen L, Nielsen EM, Engberg J, On SLW, Dietz HH. 2001. Comparison of genotypes and serotypes of Campylobacter jejuni isolated from Danish wild mammals and birds and from broiler flocks and humans. Appl. Environ. Microbiol. 67:3115–3121. 10.1128/AEM.67.7.3115-3121.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai LC, Ghazali FM, Bakar FA, Lee HY, Suhaimi LRA, Talib SA, Nakaguchi Y, Nishibuchi M, Radu S. 2009. Occurrence of thermophilic Campylobacter spp. contamination on vegetable farms in Malaysia. J. Microbiol. Biotechnol. 19:1415–1420. 10.4014/jmb.0901.0002 [DOI] [PubMed] [Google Scholar]
- 11.Chai LC, Robin T, Ragavan UM, Gunsalam JW, Bakar FA, Ghazali FM, Radu S, Kumar MP. 2007. Thermophilic Campylobacter spp. in salad vegetables in Malaysia. Int. J. Food Microbiol. 117:106–111. 10.1016/j.ijfoodmicro.2007.02.014 [DOI] [PubMed] [Google Scholar]
- 12.Kumar A, Agarwal RK, Bhilegaonkar KN, Shome BR, Bachhil VN. 2001. Occurrence of Campylobacter jejuni in vegetables. Int. J. Food Microbiol. 67:153–155. 10.1016/S0168-1605(01)00433-0 [DOI] [PubMed] [Google Scholar]
- 13.Park CE, Sanders GW. 1992. Occurrence of thermotolerant campylobacters in fresh vegetables sold at farmers' outdoor markets and supermarkets. Can. J. Microbiol. 38:313–316. 10.1139/m92-052 [DOI] [PubMed] [Google Scholar]
- 14.Kärenlampi R, Hänninen ML. 2004. Survival of Campylobacter jejuni on various fresh produce. Int. J. Food Microbiol. 97:187–195. 10.1016/j.ijfoodmicro.2004.04.019 [DOI] [PubMed] [Google Scholar]
- 15.Evans MR, Ribeiro CD, Salmon RL. 2003. Hazards of healthy living: bottled water and salad vegetables as risk factors for Campylobacter infection. Emerg. Infect. Dis. 9:1219–1225. 10.3201/eid0910.020823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner TJ, Fitzgerald C, Xavier C, Klein R, Pruckler J, Stroika S, McLaughlin JB. 2011. Outbreak of campylobacteriosis associated with consumption of raw peas. Clin. Infect. Dis. 53:26–32. 10.1093/cid/cir249 [DOI] [PubMed] [Google Scholar]
- 17.Clark CG, Bryden L, Cuff WR, Johnson PL, Jamieson F, Ciebin B, Wang GH. 2005. Use of the Oxford multilocus sequence typing protocol and sequencing of the flagellin short variable region to characterize isolates from a large outbreak of waterborne Campylobacter sp. strains in Walkerton, Ontario, Canada. J. Clin. Microbiol. 43:2080–2091. 10.1128/JCM.43.5.2080-2091.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanninen ML, Haajanen H, Pummi T, Wermundsen K, Katila ML, Sarkkinen H, Miettinen I, Rautelin H. 2003. Detection and typing of Campylobacter jejuni and Campylobacter coli and analysis of indicator organisms in three waterborne outbreaks in Finland. Appl. Environ. Microbiol. 69:1391–1396. 10.1128/AEM.69.3.1391-1396.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dingle KE, Colles FM, Wareing DRA, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJL, Urwin R, Maiden MCJ. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14–23. 10.1128/JCM.39.1.14-23.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolley KA, Chan MS, Maiden MCJ. 2004. mlstdbNet-distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. 10.1186/1471-2105-5-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penner JL, Hennessy JN. 1980. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J. Clin. Microbiol. 12:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patton CM, Nicholson MA, Ostroff SM, Ries AA, Wachsmuth IK, Tauxe RV. 1993. Common somatic O and heat-labile serotypes among Campylobacter strains from sporadic infections in the United States. J. Clin. Microbiol. 31:1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haubold B, Hudson RR. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847–848. 10.1093/bioinformatics/16.9.847 [DOI] [PubMed] [Google Scholar]
- 25.Jolley KA, Feil EJ, Chan MS, Maiden MCJ. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231. 10.1093/bioinformatics/17.12.1230 [DOI] [PubMed] [Google Scholar]
- 26.Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497. 10.1093/bioinformatics/btg359 [DOI] [PubMed] [Google Scholar]
- 27.Manning G, Dowson CG, Bagnall MC, Ahmed IH, West M, Newell DG. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370–6379. 10.1128/AEM.69.11.6370-6379.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter PE, McTavish SM, Brooks HJL, Campbell D, Collins-Emerson JM, Midwinter AC, French NP. 2009. Novel clonal complexes with an unknown animal reservoir dominate Campylobacter jejuni isolates from river water in New Zealand. Appl. Environ. Microbiol. 75:6038–6046. 10.1128/AEM.01039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwan PSL, Barrigas M, Bolton FJ, French NP, Gowland P, Kemp R, Leatherbarrow H, Upton M, Fox AJ. 2008. Molecular epidemiology of Campylobacter jejuni populations in dairy cattle, wildlife, and the environment in a farmland area. Appl. Environ. Microbiol. 74:5130–5138. 10.1128/AEM.02198-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.French N, Barrigas M, Brown P, Ribiero P, Williams N, Leatherbarrow H, Birtles R, Bolton E, Fearnhead P, Fox A. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7:1116–1126. 10.1111/j.1462-2920.2005.00782.x [DOI] [PubMed] [Google Scholar]
- 31.Maynard Smith J, Smith NH, Orourke M, Spratt BG. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. U. S. A. 90:4384–4388. 10.1073/pnas.90.10.4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes LA, Bennett M, Coffey P, Elliott J, Jones TR, Jones RC, Lahuerta-Marin A, Leatherbarrow AH, McNiffe K, Norman D, Williams NJ, Chantrey J. 2009. Molecular epidemiology and characterization of Campylobacter spp. isolated from wild bird populations in northern England. Appl. Environ. Microbiol. 75:3007–3015. 10.1128/AEM.02458-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McTavish SM, Pope CE, Nicol C, Campbell D, French N, Carter PE. 2009. Multilocus sequence typing of Campylobacter jejuni, and the correlation between clonal complex and pulsed-field gel electrophoresis macrorestriction profile. FEMS Microbiol. Lett. 298:149–156. 10.1111/j.1574-6968.2009.01712.x [DOI] [PubMed] [Google Scholar]
- 34.Levesque S, Frost E, Arbeit RD, Michaud S. 2008. Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk, and environmental water in Quebec, Canada. J. Clin. Microbiol. 46:3404–3411. 10.1128/JCM.00042-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Haan CPA, Kivisto R, Hakkinen M, Rautelin H, Hanninen ML. 2010. Decreasing trend of overlapping multilocus sequence types between human and chicken Campylobacter jejuni isolates over a decade in Finland. Appl. Environ. Microbiol. 76:5228–5236. 10.1128/AEM.00581-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ragimbeau C, Schneider F, Losch S, Even J, Mossong J. 2008. Multilocus sequence typing, pulsed-field gel electrophoresis, and fla short variable region typing of clonal complexes of Campylobacter jejuni strains of human, bovine, and poultry origins in Luxembourg. Appl. Environ. Microbiol. 74:7715–7722. 10.1128/AEM.00865-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dingle KE, Colles FM, Falush D, Maiden MCJ. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340–347. 10.1128/JCM.43.1.340-347.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen LN, Sheppard SK, McCarthy ND, Maiden MCJ, Ingmer H, Krogfelt KA. 2010. MLST clustering of Campylobacter jejuni isolates from patients with gastroenteritis, reactive arthritis and Guillain-Barré syndrome. J. Appl. Microbiol. 108:591–599. 10.1111/j.1365-2672.2009.04444.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Islam Z, van Belkum A, Wagenaar JA, Cody AJ, de Boer AG, Tabor H, Jacobs BC, Talukder KA, Endtz HP. 2009. Comparative genotyping of Campylobacter jejuni strains from patients with Guillain-Barré syndrome in Bangladesh. PLoS One 4:e7257. 10.1371/journal.pone.0007257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fekih-Mrissa N, Klai S, Zaouali J, Gritli N, Mrissa R. 2014. Association of HLA-DR/DQ polymorphism with Guillain-Barré syndrome in Tunisian patients. Clin. Neurol. Neurosurg. 121:19–22. 10.1016/j.clineuro.2014.03.014 [DOI] [PubMed] [Google Scholar]
- 41.Waldenstrom J, Mevius D, Veldman K, Broman T, Hasselquist D, Olsen B. 2005. Antimicrobial resistance profiles of Campylobacter jejuni isolates from wild birds in Sweden. Appl. Environ. Microbiol. 71:2438–2441. 10.1128/AEM.71.5.2438-2441.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson DJ, Gabriel E, Leatherbarrow AJ, Cheesbrough J, Gee S, Bolton E, Fox A, Fearnhead P, Hart CA, Diggle PJ. 2008. Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203. 10.1371/journal.pgen.1000203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheppard SK, Dallas JF, Strachan NJC, MacRae M, McCarthy ND, Wilson DJ, Gormley FJ, Falush D, Ogden ID, Maiden MCJ, Forbes KJ. 2009. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48:1072–1078. 10.1086/597402 [DOI] [PMC free article] [PubMed] [Google Scholar]