Abstract
Quinolones are synthetic antibiotics, and the main cause of resistance to these antimicrobials is mutation of the genes encoding their targets. However, in contrast to the case for other organisms, such mutations have not been found in quinolone-resistant Stenotrophomonas maltophilia isolates, in which overproduction of the SmeDEF efflux pump is a major cause of quinolone resistance. SmeDEF is chromosomally encoded and highly conserved in all studied S. maltophilia strains; it is an ancient element that evolved over millions of years in this species. It thus seems unlikely that its main function would be resistance to quinolones, a family of synthetic antibiotics not present in natural environments until the last few decades. Expression of SmeDEF is tightly controlled by the transcriptional repressor SmeT. Our work shows that plant-produced flavonoids can bind to SmeT, releasing it from smeDEF and smeT operators. Antibiotics extruded by SmeDEF do not impede the binding of SmeT to DNA. The fact that plant-produced flavonoids specifically induce smeDEF expression indicates that they are bona fide effectors regulating expression of this resistance determinant. Expression of efflux pumps is usually downregulated unless their activity is needed. Since smeDEF expression is triggered by plant-produced flavonoids, we reasoned that this efflux pump may have a role in the colonization of plants by S. maltophilia. Our results showed that, indeed, deletion of smeE impairs S. maltophilia colonization of plant roots. Altogether, our results indicate that quinolone resistance is a recent function of SmeDEF and that colonization of plant roots is likely one original function of this efflux pump.
INTRODUCTION
The acquisition of antibiotic resistance by bacterial pathogens is a relevant problem for current medical practice and is also one of the few evolutionary processes amenable to study on a human time scale. Development of resistance can be achieved either by mutation or by acquiring a resistance gene from another microorganism. Since most antibiotics have been isolated from environmental microorganisms (1), it was early suggested that resistance genes originated in natural microbial populations to counteract the inhibitory action of antibiotics present in natural ecosystems (2, 3).
Nevertheless, purely synthetic antimicrobials, such as quinolones, are also in use, and bacteria have developed resistance to these antibacterials. Since there are no quinolone producers in nature, it was supposed that resistance to this family of drugs would be due just to mutations in the genes coding for their targets, the bacterial topoisomerases (4). In the absence of quinolone selective pressure, environmental microorganisms would not require quinolone resistance, and consequently, quinolone resistance genes should not have evolved in nature. Further work showed, however, that this is not true; there are quinolone resistance genes conferring low-level resistance to these drugs via chromosomally encoded multidrug (MDR) efflux pumps (5, 6) or horizontally transferred resistance determinants, such as Qnr (7). However, it is true that despite these findings, mutations in genes encoding bacterial topoisomerases are still the main cause of high-level quinolone resistance in all studied microorganisms. The exception to this rule is Stenotrophomonas maltophilia, a free-living and often plant-associated opportunistic pathogen that is considered a prototype of intrinsically resistant bacteria (8, 9), which carries in its genome several operons encoding multidrug efflux pumps (10, 11). Different works have shown that, in contrast to what happens for other bacterial species, clinical quinolone-resistant S. maltophilia isolates do not display mutations in genes encoding bacterial topoisomerases (12, 13). One characteristic that may explain this situation is that, in contrast to the case in other organisms, in which overproduction of efflux pumps usually renders low-level resistance to quinolones, in most cases below clinical breakpoints, the overproduction of the multidrug efflux pump SmeDEF confers high-level quinolone resistance to S. maltophilia (14, 15). Furthermore, it has been shown that SmeDEF overexpression is frequent among clinical isolates of S. maltophilia and that the level of expression of this efflux pump correlates well with the level of resistance to quinolones (16). Together with the analysis of mutants defective in this efflux pump (17), these results indicate that SmeDEF is an important element in the intrinsic quinolone resistance of S. maltophilia (18) and, upon its overexpression, is a major contributor to acquired resistance to that family of antibiotics by this bacterial species (16, 19).
As stated above, quinolones are synthetic antibiotics which were not present in environmental S. maltophilia habitats until the last few decades. On the other hand, the SmeDEF genes are intrinsic genes present in the genomes of all known S. maltophilia strains, where they have evolved during several million years, rather than being a recent acquisition via horizontal gene transfer. It is difficult to believe that the genes for this element evolved in the genome of S. maltophilia to counteract the activity of a family of antibiotics that did not exist in nature until very recently. Furthermore, although S. maltophilia is a relevant nosocomial pathogen (8), its natural habitats are water and soil, where it can colonize all parts of plants, including the whole endosphere (20). Since efflux pumps of plant-interacting bacteria can be involved in plant colonization/invasion (21–23), we wondered whether SmeDEF may also play this role. Indeed, by using biochemical and functional assays, we found that this efflux pump, which is a major determinant of quinolone resistance in S. maltophilia, is involved in the endophytic colonization of plant roots.
MATERIALS AND METHODS
Bacterial strains and growing conditions.
The bacterial strains used were the wild-type S. maltophilia isolate D457 (15) and its isogenic derivatives MBS411 (D457 ΔsmeE) and D457R (D457 overexpressing SmeDEF) (14, 24). All strains were grown in LB medium at 37°C, unless indicated otherwise.
In silico prediction of the interactions of SmeT with plant exudates.
Docking predictions of the interactions of plant flavonoid exudates (naringenin, phloretin, genistein, apigenin, and quercetin) with SmeT were performed using AutoDock4 software (25), considering the flexibility of the different ligands but not the protein. The docking area was restricted to the protein's binding pocket, and results were analyzed using AutodockTools 1.5.4. Protein-ligand complexes were visualized using PyMol (26).
DNA labeling and EMSA in the presence of plant flavonoid exudates or antibiotics.
The probe for the electrophoretic mobility shift assay (EMSA) was a 158-bp double-stranded DNA fragment containing the SmeT operator site, and 5′-end labeled with [γ-32P]dATP, which was obtained as previously described (27). This probe was incubated with purified SmeT (0.2 μM) in binding buffer [10 mM Tris-HCl, 50 mM KCl, 10 mM MgCl2, 1 mM EDTA, pH 7.2, 50 μg/ml bovine serum albumin, 1 mM dithiothreitol, 5% (vol/vol) glycerol, and 100 μg/ml poly(dI:dC), as a nonspecific competitor DNA] for 20 min at room temperature. Later, increasing concentrations (0, 0.06, 0.12, 0.25, 0.5, 1, and 2 mM) of commercially available pure flavonoids (naringenin, phloretin, coumestrol, genistein, apigenin, and quercetin) or a 3 mM concentration of antibiotics (chloramphenicol, erythromycin, and ciprofloxacin) was added and incubation continued for 15 min more. Retarded complexes were separated in a 6% nondenaturing polyacrylamide gel (37.5:1 acrylamide:bisacrylamide). Electrophoresis was performed at room temperature with 89 mM Tris-borate and 2 mM EDTA buffer for 90 min at 100 V, and the gel was dried before autoradiography.
RNA preparation and real-time RT-PCR.
Fifteen microliters of an S. maltophilia D457 overnight culture was used to inoculate 15 ml of LB medium containing subinhibitory concentrations of plant exudates (15 μg/ml of phloretin and 10 μg/ml of naringenin, quercetin, apigenin, and genistein) until an optical density at 600 nm (OD600) of ≈1.0 was reached; cells were then spun down at 6,000 × g for 10 min at 4°C and immediately frozen on dry ice and stored at −80°C. Total RNA extraction, DNA elimination, RNA integrity verification, DNA absence confirmation, cDNA generation, and real-time PCR were performed as described previously (28). Total RNA was extracted from cell pellets by use of an RNeasy minikit (Qiagen) according to the manufacturer's instructions. To further eliminate any remaining DNA, Turbo DNA-free (Ambion) was used. RNA integrity was verified on a 1% agarose gel, and the absence of DNA was verified by real-time PCR using primers GyrA-RT.fw and GyrA-RT.rv (Table 1). cDNA was obtained from 1 μg RNA by use of a High Capacity cDNA reverse transcription (RT) kit (AB Applied Biosystems). RT-PCR was performed according to the method of Morales et al. (29). Briefly, the first denaturation step, 95°C for 10 min, was followed by 40 temperature cycles (95°C for 15 s, 60°C for 1 min) for amplification and quantification. The GyrA-RT.fw and GyrA-RT.rv primers were used to amplify the housekeeping gene gyrA as described previously (30) (Table 1). Differences in the relative amounts of mRNA for the different genes were determined according to the 2−ΔΔCT method (31). Real-time RT-PCRs were performed using a Power SYBR green kit (Applied Biosystems) as indicated by the manufacturer. In all cases, the mean values for relative mRNA expression obtained in three independent triplicate experiments were considered. Student's t test was used for statistical analysis of the results.
TABLE 1.
Oligonucleotides used in this work
| Oligonucleotide | Sequence | Utilization |
|---|---|---|
| GyrA-RT.fw | CCAGGGTAACTTCGGTTCGA | Internal control gene for RT-PCR |
| GyrA-RT.rv | GCCTCGGTGTATCGCATTG | |
| SmeD-RT.fw | CGGTCAGCATCCTGATGGA | RT-PCR for smeD |
| SmeD-RT.rv | TCAACGCTGACTTCGGAGAACT | |
| SmeY-RT.fw | AGCTGCTGTTCTCCGGTATCA | RT-PCR for smeY |
| SmeY-RT.rv | CACCAGGATGCGCAGGAT | |
| SmeI-RT.fw | ACTGCGATGAACACCGTTACC | RT-PCR for smeI |
| SmeI-RT.rv | CACGTCACCCTGCTTCACTTC | |
| SmeA-RT.fw | ACCCGGTCTATCTGGATGTG | RT-PCR for smeA |
| SmeA-RT.rv | CTCATGGGCATAGGTGCTG | |
| SmeG-RT.fw | AAGAACGTGAAGACCGATGG | RT-PCR for smeG |
| SmeG-RT.rv | CCTTCCTTGACCTTCTGCAC | |
| SmeM-RT.fw | ATTTCCAGGTGCCTGAAGTG | RT-PCR for smeM |
| SmeM-RT.rv | CGCATCAATGGTGCTGAC | |
| SmeO-RT.fw | CAGGAAAGTCCACTGTCGTTC | RT-PCR for smeO |
| SmeO-RT.rv | CACGTCGCCCTTCTTCAC | |
| SmeV-RT.fw | CCAGGGTAACTTCGGTTCGA | RT-PCR for smeV |
| SmeV-RT.rv | GCCTCGGTGTATCGCATTG |
In vivo plant colonization experiments.
The oilseed rape plant (cv. Californium; Kwizda, Austria) was used for in vivo assays. Before inoculation of seeds with bacteria, the surfaces of the seeds were sterilized using a modification of the seed infiltration approach described by Müller and Berg (32). In brief, seeds were treated with a 3% sodium hypochlorite (NaClO) solution for 5 min and subsequently washed three times with sterile distilled water. The numbers of CFU ml−1 of S. maltophilia strains used in the colonization experiments were determined by diluting and plating 100-μl aliquots of overnight cultures of each bacterial strain on LB plates. Colonies were counted after incubation of the plates at 37°C for 24 h. Surface-sterilized seeds were placed in a petri dish with 10 ml of 0.85% NaCl solution containing 106 CFU/ml of each bacterial culture on an orbital shaker at 85 rpm and room temperature for 4 h.
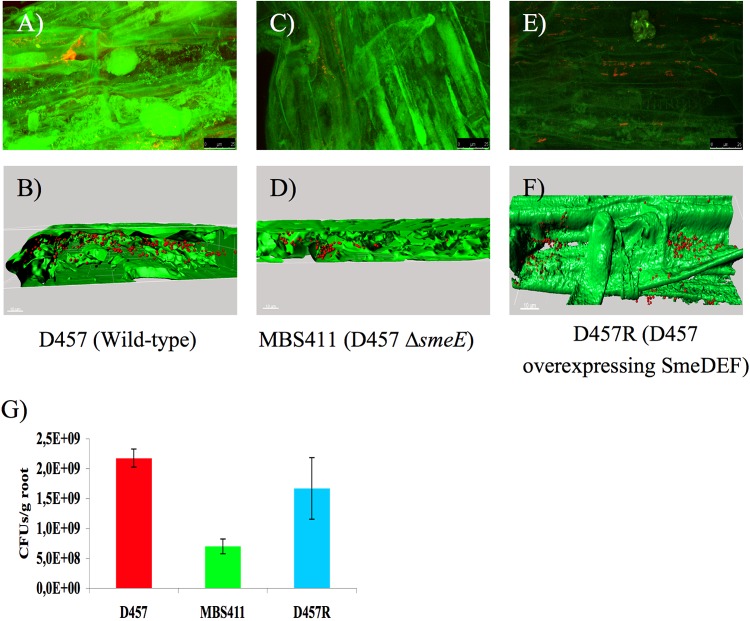
The capability of S. maltophilia strains D457, MBS411, and D57R to colonize plant rhizospheres was checked by use of seed germination pouches (Mega International, MN). Autoclaved pouches were loaded with 5 inoculated seeds each and moisturized with 20 ml of sterile distilled water. Pouches were placed in sterile, covered polypropylene containers to avoid contamination and dehydration and then incubated in a greenhouse at 23 ± 2°C under artificial lighting (16-h light period) for 7 days. Bacteria were reisolated from 7-day-old oilseed rape plants. To achieve this purpose, root sections were sampled into sterile Whirl-Pak bags (Carl Roth, Germany), supplemented with 5 ml of 0.85% NaCl solution, and roughly disintegrated using a mortar and pestle. Serial dilutions of the extract were then plated onto LB plates. After incubation for 24 h at 37°C, colonies were counted and the number of CFU/g root was assessed.
FISH.
Root sections of oilseed rape plants were fixed with 4% paraformaldehyde–phosphate-buffered saline (PBS) (3:1 [vol/vol]). The fixed samples were then stored in PBS–96% ethanol (1:1) at −20°C. In-tube fluorescent in situ hybridization (FISH) was performed as described earlier (33), using commercially available FISH probes purchased from genXpress (Wiener Neudorf, Austria). An equimolar ratio of FISH probes EUC I, EUC II, and EUC III, labeled with the fluorescent dye Cy3, was used to detect the analyzed S. maltophilia strains. The hybridization step was carried out using 35% formamide in a 46°C water bath for 90 min. After hybridization, the samples were washed at 48°C for 15 min. Microscopic images of the samples were captured using a Leica TCS SPE confocal microscope (Leica Microsystems, Wetzlar, Germany). A Leica ACS Apo 63× oil CS objective (numerical aperture [NA] = 1.30) was used to acquire confocal stacks by applying a z-step of 0.4 to 0.8 μm. Three-dimensional (3D) analysis of the confocal laser scanning microscopy (CLSM) stacks was performed with Imaris 7.0 software (Bitplane, Zurich, Switzerland).
RESULTS AND DISCUSSION
Multidrug efflux pumps are relevant elements in the development of antibiotic resistance by bacterial populations (34, 35). However, besides antibiotics, these resistance elements can extrude a wide variety of substrates. Moreover, different efflux pumps from the same bacterium usually present overlapping substrate ranges (22, 36, 37). Given this lack of specificity, inferring function by activity is a difficult task for this family of antibiotic resistance determinants (38).
The expression of efflux pumps is usually tightly downregulated by specific transcriptional regulators encoded by genes located just upstream of the operons containing the structural genes for the efflux pumps (38, 39). The silencing of the expression of efflux pumps that is observed under regular laboratory growing conditions is likely because their overexpression challenges bacterial physiology (28, 40–42). Consequently, these elements are expressed at high levels only when bacteria face the proper conditions, in the presence of functional effectors capable of binding their transcriptional regulators (43). We reasoned that identifying these effectors might give insight into the original functional roles of multidrug efflux pumps, such as SmeDEF. This determinant contributes to intrinsic and acquired resistance to quinolones in S. maltophilia. Expression of SmeDEF is regulated by the local repressor SmeT (27). Mutations in this transcriptional factor cause overexpression of SmeDEF and resistance to different antibiotics, including quinolones (19, 24). However, acquisition of resistance through this mechanism compromises S. maltophilia's ecological behavior, since SmeDEF overexpression causes a severe fitness cost to S. maltophilia as measured in pairwise competition tests (40).
Analysis of the SmeT structure shows that it presents an effector binding pocket capable of accommodating different effectors, e.g., triclosan (43). Binding of these effectors causes the release of SmeT from its operator and consequently triggers smeDEF expression (27, 43). Given that S. maltophilia is a root endophyte, we wondered whether plant exudates could be SmeT effectors. As the first approach to address this question, we studied the ability of SmeT to accommodate plant-produced flavonoids in its binding pocket by using a modeling approach (Fig. 1). As shown in Fig. 1, plant-produced compounds may indeed dock into SmeT's binding pocket, which makes them potential effectors of the SmeDEF repressor. To validate these predictions, we analyzed the effects that antibiotics, which are substrates of SmeDEF, and the studied plant-produced compounds may have on the capability of SmeT to bind its operator. As shown in Fig. 2, none of the tested antibiotics inhibited the binding of SmeT to its DNA operator, whereas the addition of plant-synthesized compounds produced the release of SmeT from its operator, even at concentrations lower than those tested for the antibiotics. These results confirmed those of the docking predictions and indicate that plant flavonoid exudates are good effectors of SmeT, whereas antibiotics are not.
FIG 1.
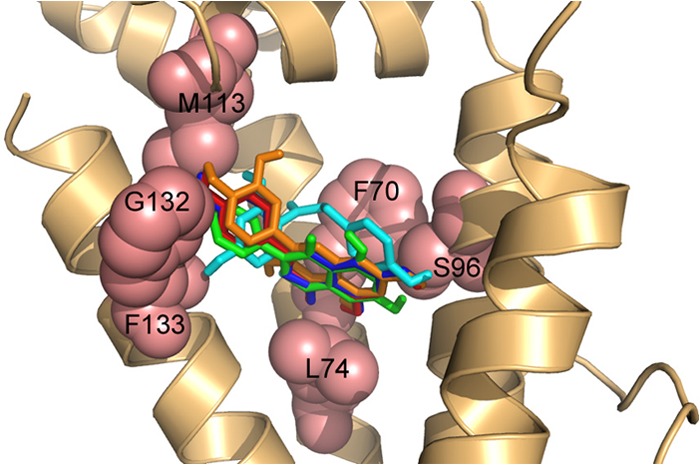
Docking of plant flavonoid exudates to SmeT. The cartoon representation shows the best binding conformations for the analyzed ligands in the pocket of the protein. Binding energies for the five ligands were similar (in the range of −5.7 to −5.0 kcal/mol), and the effector-protein complex was stabilized mainly by hydrophobic interactions. Relevant residues for binding (F70, S96, M113, G132, F133, and L74) are shown as salmon-colored spheres. The positioning of four ligands (apigenin [red], genistein [green], naringenin [blue], and quercetin [orange]) partially overlaps, whereas only one of the aromatic rings of phloretin (cyan) shows an orientation similar to those of the other ligands.
FIG 2.
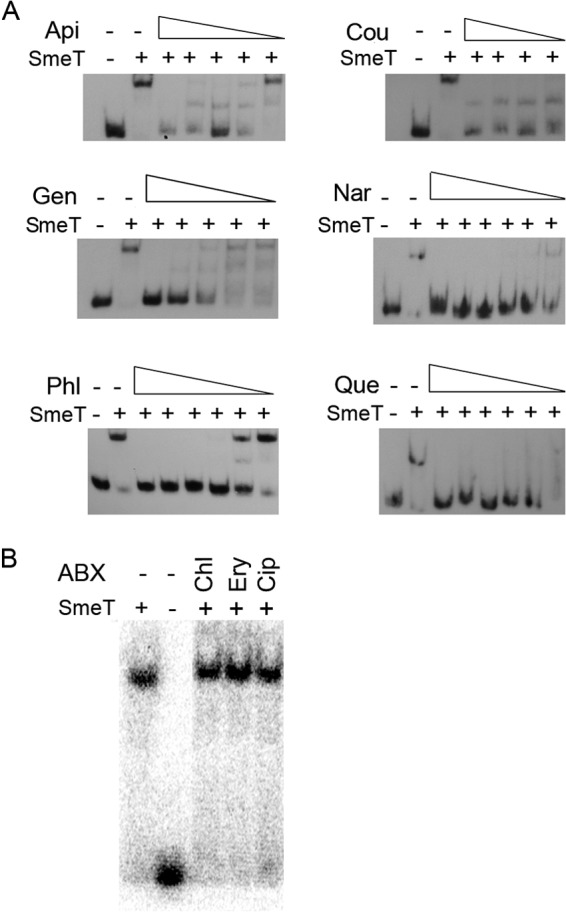
Effects of antibiotics and plant flavonoid exudates on the ability of SmeT to bind its operator DNA. A γ-32P-labeled 158-bp double-stranded DNA fragment containing the SmeT operator site (lanes 1) was incubated with SmeT (0.2 μM) for 20 min at room temperature (lanes 2). Subsequently, increasing concentrations of plant exudates, to a maximum of 2 mM (A), or antibiotics at a fixed concentration of 3 mM (B) were added, and the mixtures were incubated at room temperature for 15 more minutes. Retarded complexes were separated in a 6% nondenaturing polyacrylamide gel. As shown in panel A, all the tested flavonoids could break the SmeT-DNA complex, whereas antibiotics, which are substrates of SmeDEF, could not break this complex (B). Api, apigenin; Cou, coumestrol; Gen, genistein; Nar, naringenin; Phl, phloretin; Que, quercetin; ABX, antibiotics; Chl, chloramphenicol; Ery, erythromycin; Cip, ciprofloxacin.
To ascertain whether the capability of binding to SmeT correlates with smeDEF induction, expression of smeD was measured in the presence and absence of plant exudates. As shown in Fig. 3, plant exudates triggered smeD expression (in all cases, P < 0.05), which supports the hypothesis that these compounds are bona fide effectors of SmeT.
FIG 3.
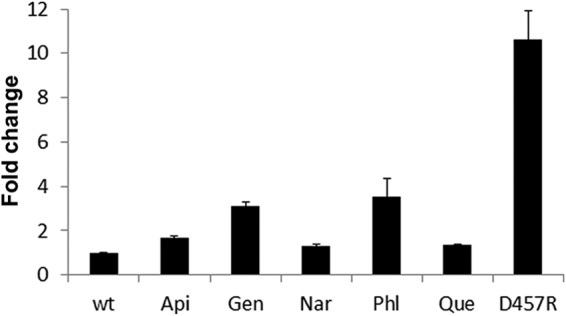
The presence of plant exudates triggers smeD expression. The amounts of smeD mRNA in the presence of plant exudates were measured by real-time RT-PCR, and the fold changes estimated with respect to the value determined for the wild-type strain (wt) grown in the absence of exudates are shown. As shown, mRNA levels were increased in the presence of apigenin (Api), genistein (Gen), naringenin (Nar), phloretin (Phl), and quercetin (Que). D457R is a mutant strain in which expression of smeDEF is fully derepressed due to a mutation that inactivates SmeT.
To analyze if the observed induction was specific for SmeDEF, the effect of phloretin, the flavonoid triggering the highest induction levels, on the expression of the previously described S. maltophilia tripartite efflux pumps (10) was analyzed by measuring the expression of the first gene of each of the operons, namely, smeA, smeD, smeV, smeY, smeG, smeM, smeO, and smeI. As shown in Fig. 4, phloretin preferentially induced smeD (P = 0.027), and a minor effect was also observed for smeO (P = 0.048), and maybe for smeG (not statistically significant; P = 0.16), while the expression of the other efflux pumps did not change in the presence of the flavonoid. The D457R mutant lacks a functional SmeT repressor, and this causes smeDEF overexpression. Since phloretin did not further increase the expression of any of the tested efflux pumps in this mutant (Fig. 4), most likely the induction observed in the wild-type strain is attributable to SmeT, a feature in agreement with the EMSA results displayed in Fig. 2. Although smeG and smeO are not regulated by SmeT, the regulators of both operons in which these genes are present belong to the TetR family of transcriptional repressors. The induction by phloretin of the expression of smeO (and maybe smeG) suggests that this flavonoid may bind other TetR-type regulators besides SmeT. In line with this reasoning, previous work showed that phloretin binds TtgR, a TetR regulator homologous to SmeT (44). Since phloretin induces smeDEF expression, it might be possible that it is extruded by SmeDEF, which would make the D457R mutant overproducing this efflux pump more resistant and a mutant lacking the efflux pump more susceptible to the presence of high concentrations of the flavonoid. To analyze this possibility, the wild-type strain and mutants lacking smeE or presenting constitutive high-level SmeDEF expression were grown in the absence or presence of phloretin (300 μg/ml). Consistent with previously published information (40), SmeDEF overexpression conferred a fitness cost to S. maltophilia in the absence of phloretin (Fig. 5A). However, in the presence of phloretin, the mutant overexpressing SmeDEF presented normal growth, whereas the wild-type strain was impaired and the mutant lacking smeE did not grow. These results strongly suggest that phloretin is a SmeDEF substrate.
FIG 4.
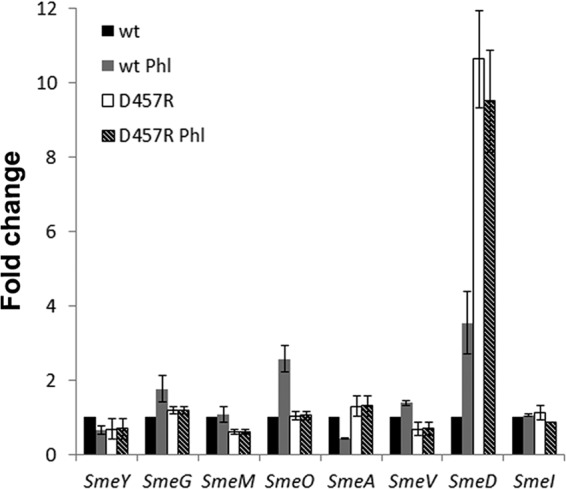
Effects of phloretin on the expression of S. maltophilia operons encoding multidrug efflux pumps. The mRNA levels of the first genes of the operons encoding MDR determinants in the chromosome of S. maltophilia (10), i.e., smeA, smeD, smeV, smeY, smeG, smeM, smeO, and smeI, were analyzed by real-time RT-PCR with the wild-type D457 strain (black) and the D457R strain (white), a mutant strain in which expression of smeDEF is fully derepressed due to a mutation that inactivates SmeT. The expression in the presence of phloretin was measured by real-time RT-PCR with D457 (gray) and D457R (stripes). The fold changes were estimated with respect to the value determined for the wild-type strain grown in the absence of the flavonoid.
FIG 5.
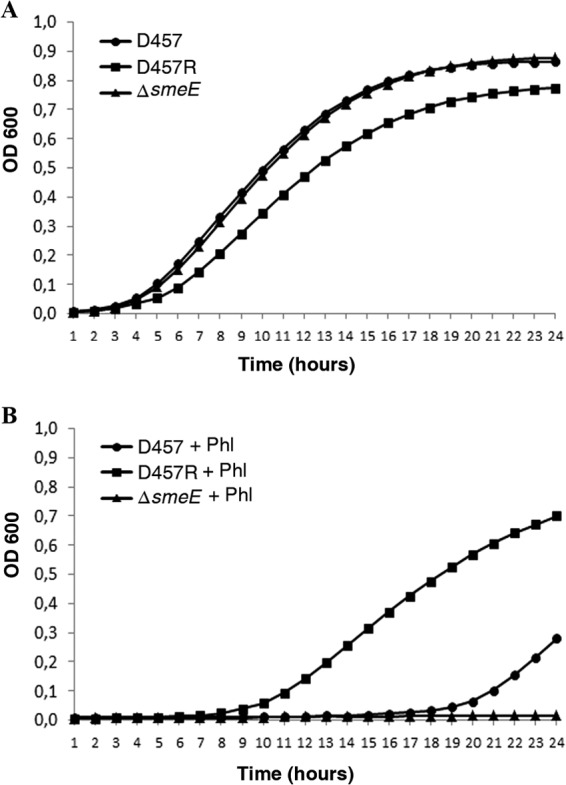
Overexpression of SmeDEF reduces the susceptibility of S. maltophilia to phloretin. The figure shows the growth of the wild-type strain and mutants either overproducing or lacking smeE. (A) Growth in the absence of phloretin. (B) Growth in the presence of phloretin at 300 μg/ml. As shown, the susceptibility to phloretin correlates with the SmeDEF expression level.
As mentioned earlier, MDR efflux pumps are expressed at their highest levels only when needed. Bacteria can determine this need by sensing their environment and identifying the proper effectors. Since the effectors triggering SmeDEF overexpression are plant exudates, not antibiotics, it is conceivable that one original function of this efflux pump was the interaction with plant roots. To study this possibility, we measured the capability for endophytic colonization of an S. maltophilia mutant lacking smeE in comparison with its wild-type parental counterpart. The D457R mutant, which overexpresses SmeDEF, was also used as a control. As shown in Fig. 6, the mutant lacking smeE was impaired in colonizing plant roots (P = 0.0000000046), whereas the differences observed between the D457R mutant and the wild-type strain were not significant (P = 0.082). This indicates that SmeDEF is required for achieving a normal colonization level. The fact that the D457R mutant does not present enhanced colonization capabilities can be explained by three different (not mutually exclusive) hypotheses. (i) It has been shown that SmeDEF overexpression compromises in vitro S. maltophilia growth (Fig. 5A) (40); the potential benefits on colonization will be compensated by such impairment. (ii) When S. maltophilia grows on plants, the presence of flavonoids induces smeDEF in the wild-type strain, to levels similar to those observed for D457R. (iii) The colonization of the plant requires a given threshold of SmeDEF expression, and increasing the level does not enhance colonization. Altogether, our results support the hypothesis that one original function for which SmeDEF has been selected in S. maltophilia is colonization of plants, not resistance against the action of antibiotics such as quinolones.
FIG 6.
Colonization of oilseed rape roots by S. maltophilia. Colonization was visualized by FISH. (A) Wild-type strain D457. (C) smeE-defective mutant strain MBS411. (E) D457R mutant that overexpresses SmeDEF. 3D analysis of the CLSM stacks of D457 (B), MBS411 (D), and D457R (F) was performed with Imaris 7.0 software. (G) Quantification of the numbers of cells colonizing the roots. Results shown are mean values for three independent replicates.
Our results indicate that antibiotic resistance genes, which are causing problems at hospitals, have not necessarily always evolved in the field to counteract the production of antibiotics by surrounding competitors (45, 46). Rather, enzymes or efflux pumps with substrates similar to an antibiotic can render resistance when the antimicrobial is present. However, this should not downplay their role as significant antibiotic resistance elements but solely points out that, on occasion, they evolved for other purposes in nature. It has been stated that the amounts of antibiotics in natural ecosystems are likely very low for a general role as inhibitors (47–51). However, the wide use of antibiotics by human beings has changed this situation, and there are different ecosystems, including individuals under medical treatment, where antibiotics constitute a highly relevant selective pressure (48).
ACKNOWLEDGMENTS
Work in our laboratory is supported by grant BIO2011-25255 from the Spanish Ministry of Economy and Competitivity, grant S2010/BMD2414 (PROMPT) from CAM, the Spanish Network for Research on Infectious Diseases (grant REIPI RD12/0015) via the Instituto de Salud Carlos III, and grant HEALTH-F3-2011-282004 (EVOTAR) from the European Union. G.G.-L. has been the recipient of an FPI fellowship.
Footnotes
Published ahead of print 16 May 2014
REFERENCES
- 1.Waksman SA, Woodruff HB. 1940. The soil as a source of microorganisms antagonistic to disease-producing bacteria. J. Bacteriol. 40:581–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benveniste R, Davies J. 1973. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. U. S. A. 70:2276–2280. 10.1073/pnas.70.8.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies JE. 1997. Origins, acquisition and dissemination of antibiotic resistance determinants. Ciba Found. Symp. 207:15–27 [PubMed] [Google Scholar]
- 4.Hernandez A, Sanchez MB, Martinez JL. 2011. Quinolone resistance: much more than predicted. Front. Microbiol. 2:22. 10.3389/fmicb.2011.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen SP, McMurry LM, Hooper DC, Wolfson JS, Levy SB. 1989. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob. Agents Chemother. 33:1318–1325. 10.1128/AAC.33.8.1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper DC. 1999. Mechanisms of fluoroquinolone resistance. Drug Resist. Updat. 2:38–55. 10.1054/drup.1998.0068 [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Martinez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. 10.1016/S0140-6736(97)07322-4 [DOI] [PubMed] [Google Scholar]
- 8.Brooke JS. 2012. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 25:2–41. 10.1128/CMR.00019-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez MB, Hernandez A, Martinez JL. 2009. Stenotrophomonas maltophilia drug resistance. Future Microbiol. 4:655–660. 10.2217/fmb.09.45 [DOI] [PubMed] [Google Scholar]
- 10.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9:R74. 10.1186/gb-2008-9-4-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rocco F, De Gregorio E, Colonna B, Di Nocera PP. 2009. Stenotrophomonas maltophilia genomes: a start-up comparison. Int. J. Med. Microbiol. 299:535–546. 10.1016/j.ijmm.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 12.Ribera A, Domenech-Sanchez A, Ruiz J, Benedi VJ, Jimenez de Anta MT, Vila J. 2002. Mutations in gyrA and parC QRDRs are not relevant for quinolone resistance in epidemiological unrelated Stenotrophomonas maltophilia clinical isolates. Microb. Drug Resist. 8:245–251. 10.1089/10766290260469499 [DOI] [PubMed] [Google Scholar]
- 13.Valdezate S, Vindel A, Saez-Nieto JA, Baquero F, Canton R. 2005. Preservation of topoisomerase genetic sequences during in vivo and in vitro development of high-level resistance to ciprofloxacin in isogenic Stenotrophomonas maltophilia strains. J. Antimicrob. Chemother. 56:220–223. 10.1093/jac/dki182 [DOI] [PubMed] [Google Scholar]
- 14.Alonso A, Martinez JL. 2000. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 44:3079–3086. 10.1128/AAC.44.11.3079-3086.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alonso A, Martinez JL. 1997. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso A, Martinez JL. 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:1879–1881. 10.1128/AAC.45.6.1879-1881.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Leon G, Sanchez MB, Martinez JL. 2012. The inactivation of intrinsic antibiotic resistance determinants widens the mutant selection window for quinolones in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 56:6397–6399. 10.1128/AAC.01558-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Li XZ, Poole K. 2001. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:3497–3503. 10.1128/AAC.45.12.3497-3503.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez P, Alonso A, Martinez JL. 2004. Regulatory regions of smeDEF in Stenotrophomonas maltophilia strains expressing different amounts of the multidrug efflux pump SmeDEF. Antimicrob. Agents Chemother. 48:2274–2276. 10.1128/AAC.48.6.2274-2276.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu B, Liu H, Tian WX, Fan XY, Li B, Zhou XP, Jin GL, Xie GL. 2012. Genome sequence of Stenotrophomonas maltophilia RR-10, isolated as an endophyte from rice root. J. Bacteriol. 194:1280–1281. 10.1128/JB.06702-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burse A, Weingart H, Ullrich MS. 2004. NorM, an Erwinia amylovora multidrug efflux pump involved in in vitro competition with other epiphytic bacteria. Appl. Environ. Microbiol. 70:693–703. 10.1128/AEM.70.2.693-703.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez JL, Sanchez MB, Martinez-Solano L, Hernandez A, Garmendia L, Fajardo A, Alvarez-Ortega C. 2009. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33:430–449. 10.1111/j.1574-6976.2008.00157.x [DOI] [PubMed] [Google Scholar]
- 23.Palumbo JD, Kado CI, Phillips DA. 1998. An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J. Bacteriol. 180:3107–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez P, Alonso A, Martinez JL. 2002. Cloning and characterization of SmeT, a repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. Antimicrob. Agents Chemother. 46:3386–3393. 10.1128/AAC.46.11.3386-3393.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30:2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLano WL. 2002. The PyMOL molecular graphics system. http://www.pymol.org
- 27.Hernandez A, Mate MJ, Sanchez-Diaz PC, Romero A, Rojo F, Martinez JL. 2009. Structural and functional analysis of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. J. Biol. Chem. 284:14428–14438. 10.1074/jbc.M809221200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olivares J, Alvarez-Ortega C, Linares JF, Rojo F, Kohler T, Martinez JL. 2012. Overproduction of the multidrug efflux pump MexEF-OprN does not impair Pseudomonas aeruginosa fitness in competition tests, but produces specific changes in bacterial regulatory networks. Environ. Microbiol. 14:1968–1981. 10.1111/j.1462-2920.2012.02727.x [DOI] [PubMed] [Google Scholar]
- 29.Morales G, Ugidos A, Rojo F. 2006. Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ. Microbiol. 8:1764–1774. 10.1111/j.1462-2920.2006.01061.x [DOI] [PubMed] [Google Scholar]
- 30.Gould VC, Avison MB. 2006. SmeDEF-mediated antimicrobial drug resistance in Stenotrophomonas maltophilia clinical isolates having defined phylogenetic relationships. J. Antimicrob. Chemother. 57:1070–1076. 10.1093/jac/dkl106 [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 32.Müller H, Berg G. 2008. Impact of formulation procedures on the effect of the biocontrol agent Serratia plymuthica HRO-C48 on Verticillium wilt in oilseed rape. BioControl 53:905–916. 10.1007/s10526-007-9111-3 [DOI] [Google Scholar]
- 33.Cardinale M, Vieira de Castro J, Jr, Muller H, Berg G, Grube M. 2008. In situ analysis of the bacterial community associated with the reindeer lichen Cladonia arbuscula reveals predominance of Alphaproteobacteria. FEMS Microbiol. Ecol. 66:63–71. 10.1111/j.1574-6941.2008.00546.x [DOI] [PubMed] [Google Scholar]
- 34.Li XZ, Nikaido H. 2009. Efflux-mediated drug resistance in bacteria: an update. Drugs 69:1555–1623. 10.2165/11317030-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toone EJ, Nikaido H. 2011. Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 77:1–60. 10.1002/9780470920541.ch1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piddock LJ. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4:629–636. 10.1038/nrmicro1464 [DOI] [PubMed] [Google Scholar]
- 37.Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE., Jr 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976–980. 10.1126/science.1083137 [DOI] [PubMed] [Google Scholar]
- 38.Grkovic S, Brown MH, Skurray RA. 2001. Transcriptional regulation of multidrug efflux pumps in bacteria. Semin. Cell Dev. Biol. 12:225–237. 10.1006/scdb.2000.0248 [DOI] [PubMed] [Google Scholar]
- 39.Grkovic S, Brown MH, Skurray RA. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671–701. 10.1128/MMBR.66.4.671-701.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alonso A, Morales G, Escalante R, Campanario E, Sastre L, Martinez JL. 2004. Overexpression of the multidrug efflux pump SmeDEF impairs Stenotrophomonas maltophilia physiology. J. Antimicrob. Chemother. 53:432–434. 10.1093/jac/dkh074 [DOI] [PubMed] [Google Scholar]
- 41.Linares JF, Lopez JA, Camafeita E, Albar JP, Rojo F, Martinez JL. 2005. Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1384–1391. 10.1128/JB.187.4.1384-1391.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez P, Linares JF, Ruiz-Diez B, Campanario E, Navas A, Baquero F, Martinez JL. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50:657–664. 10.1093/jac/dkf185 [DOI] [PubMed] [Google Scholar]
- 43.Hernandez A, Ruiz FM, Romero A, Martinez JL. 2011. The binding of triclosan to SmeT, the repressor of the multidrug efflux pump SmeDEF, induces antibiotic resistance in Stenotrophomonas maltophilia. PLoS Pathog. 7:e1002103. 10.1371/journal.ppat.1002103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alguel Y, Meng C, Teran W, Krell T, Ramos JL, Gallegos MT, Zhang X. 2007. Crystal structures of multidrug binding protein TtgR in complex with antibiotics and plant antimicrobials. J. Mol. Biol. 369:829–840. 10.1016/j.jmb.2007.03.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez JL. 2008. Antibiotics and antibiotic resistance genes in natural environments. Science 321:365–367. 10.1126/science.1159483 [DOI] [PubMed] [Google Scholar]
- 46.Martinez JL, Fajardo A, Garmendia L, Hernandez A, Linares JF, Martinez-Solano L, Sanchez MB. 2009. A global view of antibiotic resistance. FEMS Microbiol. Rev. 33:44–65. 10.1111/j.1574-6976.2008.00142.x [DOI] [PubMed] [Google Scholar]
- 47.Davies J. 2006. Are antibiotics naturally antibiotics? J. Ind. Microbiol. Biotechnol. 33:496–499. 10.1007/s10295-006-0112-5 [DOI] [PubMed] [Google Scholar]
- 48.Fajardo A, Linares JF, Martinez JL. 2009. Towards an ecological approach to antibiotics and antibiotic resistance genes. Clin. Microbiol. Infect. 15(Suppl 1):S14–S16. 10.1111/j.1469-0691.2008.02688.x [DOI] [PubMed] [Google Scholar]
- 49.Linares JF, Gustafsson I, Baquero F, Martinez JL. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. U. S. A. 103:19484–19489. 10.1073/pnas.0608949103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yim G, Wang HH, Davies J. 2007. Antibiotics as signalling molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362:1195–1200. 10.1098/rstb.2007.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yim G, Wang HH, Davies J. 2006. The truth about antibiotics. Int. J. Med. Microbiol. 296:163–170. 10.1016/j.ijmm.2006.01.039 [DOI] [PubMed] [Google Scholar]