Abstract
To successfully colonize and eventually kill pine trees, Grosmannia clavigera (Gs cryptic species), the main fungal pathogen associated with the mountain pine beetle (Dendroctonus ponderosae), has developed multiple mechanisms to overcome host tree chemical defenses, of which terpenoids are a major component. In addition to a monoterpene efflux system mediated by a recently discovered ABC transporter, Gs has genes that are highly induced by monoterpenes and that encode enzymes that modify or utilize monoterpenes [especially (+)-limonene]. We showed that pine-inhabiting Ophiostomale fungi are tolerant to monoterpenes, but only a few, including Gs, are known to utilize monoterpenes as a carbon source. Gas chromatography-mass spectrometry (GC-MS) revealed that Gs can modify (+)-limonene through various oxygenation pathways, producing carvone, p-mentha-2,8-dienol, perillyl alcohol, and isopiperitenol. It can also degrade (+)-limonene through the C-1-oxygenated pathway, producing limonene-1,2-diol as the most abundant intermediate. Transcriptome sequencing (RNA-seq) data indicated that Gs may utilize limonene 1,2-diol through beta-oxidation and then valine and tricarboxylic acid (TCA) metabolic pathways. The data also suggested that at least two gene clusters, located in genome contigs 108 and 161, were highly induced by monoterpenes and may be involved in monoterpene degradation processes. Further, gene knockouts indicated that limonene degradation required two distinct Baeyer-Villiger monooxygenases (BVMOs), an epoxide hydrolase and an enoyl coenzyme A (enoyl-CoA) hydratase. Our work provides information on enzyme-mediated limonene utilization or modification and a more comprehensive understanding of the interaction between an economically important fungal pathogen and its host's defense chemicals.
INTRODUCTION
The mountain pine beetle (MPB; Dendroctonus ponderosae Hopkins) has become the most destructive bark beetle in North America. In the past 2 decades, this beetle and its fungal associates have killed over 16 million hectares of pine forest in British Columbia, Canada, and have spread to the east of the Rocky Mountains (1). While lodgepole pine (Pinus contorta) is the predominant host in the MPB epidemic, the MPB can also colonize at least 20 other pine species (2). More recently, this insect-fungal association has expanded its range to Jack pine (Pinus banksiana), a new host at the eastern front of the epidemic in Alberta (3), which has increased the risk that the MPB-fungal association could establish itself in the boreal forest across the Canadian north. Pine trees have both chemical (terpenoids and phenolics) and physical defenses to prevent or limit beetle and fungal attacks. In general, monoterpenoids are fungicidal and repellent to the beetles and contribute to the chemical barrier formed by the tree host (4). Although monoterpenes are energetically costly to synthesize and store (5), these compounds are constitutively produced in the tree phloem, and they are also induced to additional higher levels when a pine tree is wounded or is inoculated with pathogenic fungi (4, 6). Monoterpene compositions vary qualitatively and quantitatively within and between different conifer species (7, 8). While β-phellandrene and β-pinene are the two most abundant monoterpenes in healthy lodgepole pine, limonene is one of the most highly inducible compounds following an MPB attack, and limonene levels are higher in attacked trees than in nonattacked trees (6). In order to successfully colonize the tree phloem, both the beetle and its associated fungi must overcome the tree defense system.
Historically, Grosmannia clavigera, a member of the Ophiostomatales in the Ascomycota, has been shown to be symbiotically associated with both the MPB and its sibling the Jeffrey pine beetle (JPB) (9, 10). While MPB can colonize different pine species, it does not infest Pinus jeffreyi, which is the only known host of the JPB. Because JPB has lower degree of genetic diversity than MPB, the former may have diverged from the latter (11). When inoculated into trees above a certain density, G. clavigera can kill pine trees without the participation of its beetle vector (12, 13). Recent work by Alamouti et al. (14, 15) on epidemic MPB populations in Canada and localized MPB populations in the western and southern United States, as well as on the JPB population in California, have shown that G. clavigera is in fact a genetically heterogeneous species, consisting of at least two distinct cryptic fungal species that are reported as Gs and Gc. While these fungal lineages inhabit distinct ecological niches, they are closely related phylogenetically and may have evolved by adapting to the specific chemistry of their host trees.
Because G. clavigera Gs is one of the most pathogenic fungi carried by MPB in the current epidemic in western North American, diverse genomic resources have been developed for this fungus (16–18). Gs can effectively detoxify monoterpenes and utilize monoterpenes (especially limonene) as a sole carbon source. To decrease the toxicity of monoterpenes that enter Gs cells, the fungus has evolved an active efflux pump system through an ABC transporter (18). A mutant lacking this transporter was more sensitive to monoterpene treatments on malt extract agar (MEA) and was unable to grow on a minimal medium in which monoterpenes were the sole carbon source. However, this ABC transporter mutant was still able to grow and colonize young pine trees and cause tree death. Transcriptome analyses indicate that while the ABC transporter is induced 6 h after treatment with monoterpenes, the genes coding for enzymes that may further mediate monoterpene detoxification were upregulated only after a longer exposure (36 h). DiGuistini et al. (16) proposed that an induced ∼100-kb gene cluster may be involved in detoxifying monoterpenes.
Much is known about the microbial metabolism of limonene and other monoterpenes (19–21), since naturally occurring monoterpenes, including those of pines, as well as their derivatives are widely used in the chemical and biotechnology industry, for example for the production of food flavor compounds, fragrances, and household cleaners (22–24). Bioconversion of monoterpenes can be accomplished with bacteria; more recently, fungi, plants, and algae have been assessed (25–27). Bacteria use at least six pathways to convert limonene. Usually the first step involves oxygenation of monoterpenes, often catalyzed by cytochrome P450s (CYP450s) (21, 28, 29). Depending on the carbon position where oxygenation occurs, the pathways are designated limonene-1,2-oxide (C-1), isopiperitenol (C-3), carveol (C-6), perillyl alcohol (C-7), α-teripenol (C-8), and limonene-8,9-oxide (C-8). Some bacteria (e.g., Pseudonomas spp. and Rhodococcus erythropolis) can utilize limonene as a carbon source through either the perillyl alcohol or limonene-1,2-oxide pathway (27, 30). Some fungi (e.g., Aspergillus cellulosae, Penicillium digitatum, and Fusarium oxysporum) can detoxify limonene through C-1, C-3, C-6, or C-7 oxygenation (31, 32). Further, cometabolic conversion of limonene by enzymes that degrade other compounds is also common in bacteria (25, 33). It has also been suggested that other monoterpenes (e.g., α- and β-pinene) may be detoxified through pathways similar to those for limonene, since limonene was found as an intermediate product in the metabolism of these monoterpenes (27). While much is known about microbial bioconversion of limonene for biotechnological applications, there is little information on limonene utilization by fungi as a carbon source.
Here, we describe mechanisms involved in the tolerance or utilization of monoterpenes by Gs. We describe a set of genes that code for enzymes used by this fungus to modify or metabolize limonene as a carbon source, in parallel to using the ABC-G1 efflux transporter. We propose pathways that may be involved in limonene transformation or degradation by Gs and the potential application of such fungal pathways for bioconversion of limonene.
MATERIALS AND METHODS
Strains and growth conditions.
The fungal isolates used in this study are listed below. Most of them are deposited at the University of Alberta Mycological Herbarium (UAMH); a few were stored in the Breuil Lab at the University of British Columbia (UBC). Grosmannia clavigera strain (Gs [kw1407 or UAMH 11150]) was the first Grosmannia species to have its genome and transcriptome sequenced (16). The G. clavigera strain (Gc) from Pinus ponderosae is from the American Type Culture Collection (ATCC 18086). The other strains from P. jeffreyi (Gc; UAMH 11351) and P. contorta (Gs [UMAH 11154], Leptographium longiclavatum UMAH 4876 and UBC 868AW110622, Ophiostoma montium UAMH 1363 and UBC S5R134E2, and Ceratocystiopsis sp. strains UAMH 10945 and UBC S4118AG, as well as the saprophyte Ophiostoma piceae UMAH 11346 and UAMH 11672), have also been isolated by our group. O. novo-ulmi strains (W2WT and HM75) were a gift from Louis Bernier (University Laval, Québec, Canada). Neurospora crassa strains (4200 and 2489) were obtained from the Fungal Genetics Stock Center in the United States. All knockout mutants were generated from the Gs kw1407 strain and verified by PCR as described previously (34). The mutants have been deposited at UAMH; the deposit numbers are as follows: UAMH 11802, CMQ_6956; UAMH 11803, CMQ_7007; UAMH 11804, CMQ_6887; UAMH 11805, CMQ_7009; UAMH 11806, CMQ_4626; and UAMH 11807, CMQ_1732.
All the strains and mutants were maintained on 1% oxoid malt extract agar (MEA). Fungal growth rates were examined at room temperature (∼22°C). For growth on MEA alone or with monoterpenes, we transferred plugs of actively growing fungal cultures on MEA into the centers of glass petri dishes containing MEA alone or MEA with monoterpene treatments. Colony diameters of three replicates were measured along two perpendicular lines, and the radial growth rates were calculated in millimeters per day. Single monoterpenes or mixtures of monoterpenes {MT; (+)-limonene [LIM], (+)-3-carene, racemic α-pinene, and (−)-β-pinene at a ratio of 5:3:1:1} were applied at 200 μl on two strips of filter paper that were placed inside the lid of the plate. The glass plates were sealed with DuraSeal film (laboratory sealing film, catalog number 89031-573; VWR) and incubated with the lid down in a sealed glass container. For additional details see the work of Wang et al. (18). For growth and utilization of single or mixtures of monoterpenes as a carbon source, a similar procedure was used, with a yeast nitrogen base medium (YNB) that contained no carbon source except the monoterpenes tested (18). The monoterpenes were purchased from Sigma (Oakville, ON, Canada) and were kept in the dark at −20°C.
RNA-seq analysis and qRT-PCR.
We analyzed six transcriptome libraries produced from Gs fungal mycelia grown on (i) MEA (for 2 days), (ii) MEA plus (+)-limonene (for 4 days), (iii) YNB plus mannose (for 5 days), (iv) YNB plus oleic acid (OA) (for 5 days), or (v) YNB plus MT [monoterpene mixture: (+)-limonene, 3-carene, racemic α-pinene, and β-pinene at a ratio of 5:3:1:1] (for 7 days) as described by Lah et al. (35) and Wang et al. (18). For each transcriptome sequencing (RNA-seq) library, samples were collected from three biological replicates and the pooled samples were pair end-sequenced on an Illumina GAIIx (Canada's Michael Smith Genome Science Center, Vancouver, BC, Canada). Sequence filtering, trimming, mapping to the reference genome (16), and RNA-seq analyses were conducted with CLC Genomic Workbench v.4 (CLC Bio) software. Statistical analysis was carried out using Kal's Z-test (36). Classification of gene functions was done using Blast2go. Expression of selected genes was verified by quantitative reverse transcription PCR (qRT-PCR) on the same samples as used for RNA-seq. Data collection and statistical analysis were carried out on the Bio-Rad CFX96 real-time PCR detection system as described by Hesse-Orce et al. (17).
Gs intracellular volatile metabolites: sampling and extraction for GC-MS analyses.
Fungal growth and induction by (+)-limonene were the same as described for the transcriptome collection, except that only 50 μl of analytical-grade (+)-limonene was used for the treatments. On MEA, 50,000 fungal spores were spread on cellophane and incubated for 3 days; then the young mycelia were treated with 50 μl of analytical-grade (+)-limonene for 3 days. After the 3-day incubation period, filter papers with limonene were removed and the plates were left opened in the fume hood for 5 min. Mycelia were then scraped from the film of cellophane and immersed into 1 ml of methyl-tert-butyl ether (MTBE) with 10−5 isobutyl benzene (IBB) as an internal standard. Five to 10 discs of cellophane with spores germinated on MEA were transferred to YNB, and then 50 μl of analytical-grade (+)-limonene was applied on filter paper place in the lid of the petri dish. After 21 days of incubation, mycelia were harvested as described above. Once the samples were transferred into MTBE plus IBB, cells were vortexed for 2 min and incubated overnight on a shaker at 50 rpm at room temperature. Volatile metabolites were then extracted with 0.2 ml of 0.1 M (NH4)2CO3 (pH 8). The top ether layer was removed and placed into a new glass chromatography (GC) vial for analysis. GC-mass spectrometry (GC-MS) analyses were performed on an Agilent 6890A series GC system coupled to an Agilent 5975 Inert XL mass spectrometer under the conditions described by Hall et al. (7). The internal control and different weights of dry mycelia were used to determine the extraction efficiency. For detection of metabolites with alcohol or acid groups, the extracted samples were derivatized with the same amount of N,O-bis(trimethylsilyl)trifluoroacetamide–pentane (1:1) and analyzed by GC-MS. Metabolites were identified by comparison to authentic standards and reference mass spectra from mass spectral libraries (8).
RESULTS
Response of pine ophiostomatoid fungi to monoterpene treatments.
Over the last 10 years, we and others have shown that the microflora associated with the mountain pine beetle (MPB) is more diverse than originally reported (9, 37). MPBs carry multiple Ophiostomatales fungal species, including G. clavigera (which consists of the two cryptic species Gc and Gs) (14, 15), Leptographium longiclavatum, Ophiostoma montium, and Ceratocystiopsis (37, 38). However, little information is available on the relative abilities of these species to tolerate monoterpenes or to utilize them as a single carbon source. On MEA, the above-listed fungal species, isolated from MPB-colonized pine, as well as the saprophyte O. piceae, isolated from pine lumber, tolerated high concentrations (∼1.4 g/liter) of monoterpene mixtures [MT; (+)-limonene, (+)-3-carene, racemic α-pinene, and (−)-β-pinene at a ratio of 5:3:1:1]. For the MPB fungal species, growth rates were either unaffected or reduced by 20 to 40% by monoterpene treatments; in contrast, the elm tree pathogen O. novo-ulmi and the fungal model organism Neurospora crassa were killed by the same treatment (Table 1). We then assessed a minimal medium (YNB) in which monoterpenes were the sole carbon source. Of the Grosmannia species, the epidemic Gs strain isolated from lodgepole pine (P. contorta) and the nonepidemic Gc strain (ATCC 18086) isolated from ponderosa pine (P. ponderosae) grew. Gc isolated from Jeffrey pine (P. jeffreyi) did not grow, indicating that it was unable to utilize the monoterpenes as a carbon source (Table 1). The other MPB-associated fungi mentioned above grew with variable densities, while the saprophyte O. piceae did not grow under this condition. We further compared the degrees of growth of the Gs epidemic strain and the Gc (ATCC 18086) strain on YNB with individual monoterpenes. Of the monoterpenes tested, only limonene supported fungal growth, while 3-carene, α-pinene, and β-pinene either were toxic or did not support growth. Recently, Boone et al. (6) have shown that limonene was the most highly induced monoterpene in lodgepole pine during an MPB mass attack. While both (+)- and (−)-limonene enantiomers supported Gs growth, we report work on (+)-limonene for the metabolic studies described below.
TABLE 1.
Fungal monoterpene utilization or tolerancea
| Species | Host | Growth on YNB plus MT | Growth rate (mm/day) |
|
|---|---|---|---|---|
| MEA | MEA plus MT | |||
| G. clavigera (Gs) (UAMH 11150) | Pinus contorta | +++ | 10.8 ± 0.5 | 8.7 ± 0.2 |
| G. clavigera (Gc) (UAMH 1135) | Jeffrey pine | S | 9.0 ± 0.2 | 8.7 ± 0.2 |
| G. clavigera (Gc) (ATCC 18086) | Ponderosa pine | +++ | 10.8 ± 0.4 | 8.8 ± 0.2 |
| G. clavigera (Gs) (UAMH 11154) | P. contorta | +++ | 9.4 ± 0.3 | 7.5 ± 0.2 |
| L. longiclavatum (UAMH 4876) | P. contorta | ++ | 8.6 ± 0.4 | 7.9 ± 0.5 |
| L. longiclavatum (868AW1) | P. contorta | + | 6.0 ± 0.4 | 4.7 ± 0.5 |
| O. montium (UAMH 1363) | P. contorta | ++ | 5.0 ± 0.1 | 4.9 ± 0.1 |
| O. montium (S5R134E2) | P. contorta | ++ | 4.7 ± 0.2 | 4.6 ± 0.1 |
| Ceratocystiopsis sp. (UAMH 10945) | P. contorta | + | 1.4 ± 0.0 | 1.1 ± 0.1 |
| Ceratocystiopsis sp. (S4118AG) | P. contorta | + | 1.3 ± 0.0 | 1.1 ± 0.1 |
| Ophiostoma piceae (UAMH 11346) | P. contorta lumber | S | 3.1 ± 0.1 | 2.4 ± 0.0 |
| O. piceae (UAMH 11672) | P. contorta lumber | S | 3.0 ± 0.1 | 2.4 ± 0.0 |
| O. novo-ulmi (W2WT) | Elm tree | K | 3.7 ± 0.1 | 0 ± 0 |
| O. novo-ulmi (HM75) | Elm tree | K | 4.4 ± 0.2 | 0 ± 0 |
| N. crassa (2489) | Unknown | K | 22.8 ± 0.6 | 0 ± 0 |
| N. crassa (4200) | Unknown | K | 22.5 ± 0.7 | 0 ± 0 |
Fungal isolates from different origins were inoculated on YNB with a mixture of monoterpenes as the carbon source or on MEA with a mixture of monoterpenes as a chemical stress. For each fungal species tested, we assessed two isolates, doing three technical replicates for each isolate. MT, (+)-limonene, (+)-3-carene, racemic α-pinene, and (−)-β-pinene at a ratio of 5:3:1:1. +++, growth similar to that obtained on YNB plus mannose; ++ and +, less growth. S, no growth and the inoculum survived; K, no growth and the inoculum was killed; MEA, malt extract agar; YNB, yeast nitrogen base or minimal medium (see the work of Wang et al. [18]). Values are means ± standard deviations.
Gs genes that were differentially expressed after monoterpene treatment.
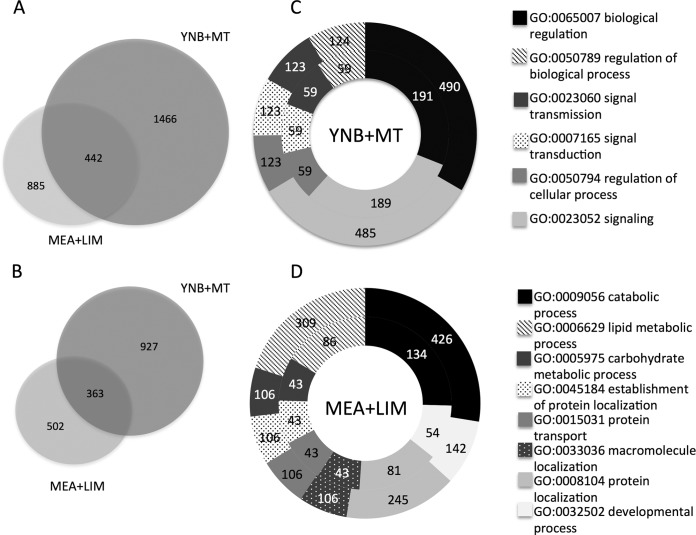
We used RNA-seq data to identify Gs genes that were differentially expressed in fungal mycelia that were grown on (i) MEA with (+)-limonene treatment (MEA plus LIM) for 4 days, compared to MEA alone, and (ii) YNB medium with a monoterpene mixture (YNB plus MT) for 10 days, compared to YNB containing mannose as a carbon source (Fig. 1). While 1,327 genes from mycelia grown on MEA plus LIM and 1,908 genes from mycelia grown on YNB plus MT were significantly upregulated (P value < 0.05), only 442 genes were upregulated with both treatments (Fig. 1A); 1,290 genes were downregulated with MEA plus LIM and 865 for YNB plus MT, with 363 downregulated under both conditions (Fig. 1B).
FIG 1.
Differentially expressed Gs genes on MEA with limonene (MEA plus LIM) versus MEA and on YNB with a monoterpene mixture (YNB plus MT) versus YNB with mannose as a carbon source. (A) Significantly upregulated genes in MEA plus LIM and YNB plus MT (P value < 0.05); (B) significantly downregulated genes in MEA plus LIM and YNB plus MT (P value < 0.05); (C) most important GO terms in YNB plus MT (inner circle) compared with the total GO terms in the Gs genome (outer circle); (D) most important GO terms in MEA plus LIM (inner circle) compared with the total gene ontology (GO) terms in the Gs genome (outer circle).
Using Blast2Go, we assigned putative functions to the differentially expressed genes. For MEA plus LIM, the analysis reported enriched gene categories that were involved in lipid and carbohydrate metabolic processes, transport and localization, and developmental processes (Fig. 1D); for YNB plus MT, the most enriched gene categories were associated with signaling and biological regulation (Fig. 1C). These results appear consistent with macroscopic observations of fungal growth. The fungus grew more rapidly on MEA plus MT than on YNB plus MT, even when exposed to high concentrations of (+)-limonene. On the poorer YNB plus MT medium, on which the fungus had to cope with chemical stresses and the lack of easily assimilable carbon sources, mycelial growth was slow and the mycelia were highly melanized.
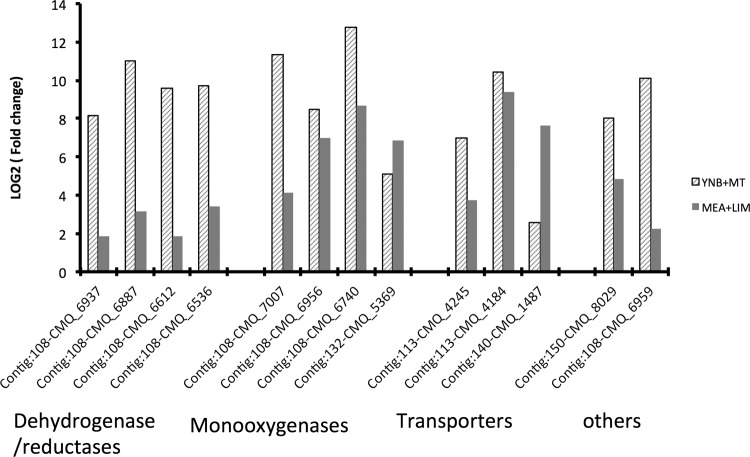
Among the 442 genes that were upregulated under both conditions (Fig. 1A), genes involved in metabolic processes were highly expressed, including those encoding certain oxidoreductases, transferases, and hydrolases. Figure 2 shows 14 genes with fold changes greater than 100 under at least one of the following growth conditions: 4 monooxygenase genes, 4 dehydrogenase/reductase genes, 3 transporter genes (including the monoterpene transporter gene GcABC-G1), 1 acetoacetyl-synthase gene, and 2 genes with unknown function. Of the 14 genes, 8 were located in contig 108 of the reference genome (in an ∼11-kb region at position 942715 to 1043294) and encode 3 monooxygenases, 4 dehydrogenases/reductases, and an acetoacetyl-synthase. Previously, we treated Gs with a more complex mixture of terpenoids than the (+)-limonene or monoterpene mixtures used in this study, and we suggested that an ∼100-kb genomic cluster in contig 108 (GL108: positions 921864 to 104374) may contribute actively to terpene detoxification (16). In the current work, 19 of the 35 genes in this cluster were upregulated in YNB plus MT and 14 were upregulated in MEA plus LIM, with most fold changes larger for YNB plus MT than for MEA plus LIM.
FIG 2.
Upregulated genes with mRNA abundance fold changes greater than 100 under at least one growth condition. Gray bars indicate growth in MEA with limonene (MEA plus LIM) versus MEA, while diagonal bars indicate growth in YNB with a monoterpene mixture (YNB plus MT) versus YNB with mannose as a carbon source.
The genes that were differentially expressed under both growth conditions suggest that Gs may have similar mechanisms to cope with a single monoterpene or a mixture of monoterpenes. It is important to note that MEA provides a richer nutrient environment than YNB; in the former, the fungus can easily metabolize sugars as a carbon source, while in the latter it has to use monoterpenes [i.e., (+)-limonene]. Genes that were upregulated in MEA plus LIM, but not in YNB plus MT, included no obvious enriched functional gene categories (i.e., hydrophobic compound degradation). Genes that were highly upregulated in YNB plus MT (>100×) but downregulated or not significantly upregulated in MEA plus LIM included a four-gene cluster in contig 161 of the reference genome (CMQ_8234, CMQ_8045, CMQ_8299, and CMQ_8094); genes in contig 140 encoding a short-chain dehydrogenase, a cytochrome P450, and a lipase esterase; and a gene in contig 132 encoding a beta-lactamase (Table 2). The 3-hydroxy-3-methylglutaryl coenzyme A (3-hydroxy-3-methylglutaryl-CoA) lyase gene (CMQ_8094) was the gene with the highest level of upregulation when Gs used monoterpene as a carbon source.
TABLE 2.
Upregulated genes showing fold changes of >100 in YNB plus MT but not upregulated in MEA plus LIMa
| Gene | Contig | Description of product | Fold change in: |
|
|---|---|---|---|---|
| MEA plus LIM | YNB plus MT | |||
| CMQ_8234 | 161 | Zinc-binding oxidoreductase | −1.02 | 2,956.39* |
| CMQ_8045 | 161 | C-6 zinc finger domain-containing protein | −1.22 | 339.62* |
| CMQ_8299 | 161 | Acyl-dehydrogenase | −1.55 | 1,482.13* |
| CMQ_8094 | 161 | Hydroxymethylglutaryl-lyase | 5.82 | 18,483.47* |
| CMQ_1732 | 140 | Cytochrome P450 | 2.33 | 430.64* |
| CMQ_371 | 140 | Short-chain dehydrogenase reductase | 1.16 | 1,065.55* |
| CMQ_6642 | 140 | Lipase esterase family protein | 9.6 | 429.18* |
| CMQ_5362 | 132 | Beta-lactamase family protein | −0.09 | 133.84* |
Note that transporters and hypothetical proteins were excluded from the list. Negative values represent downregulation. *, P < 0.05, Kal's Z-test.
Expression of genes potentially involved in fatty acid metabolism.
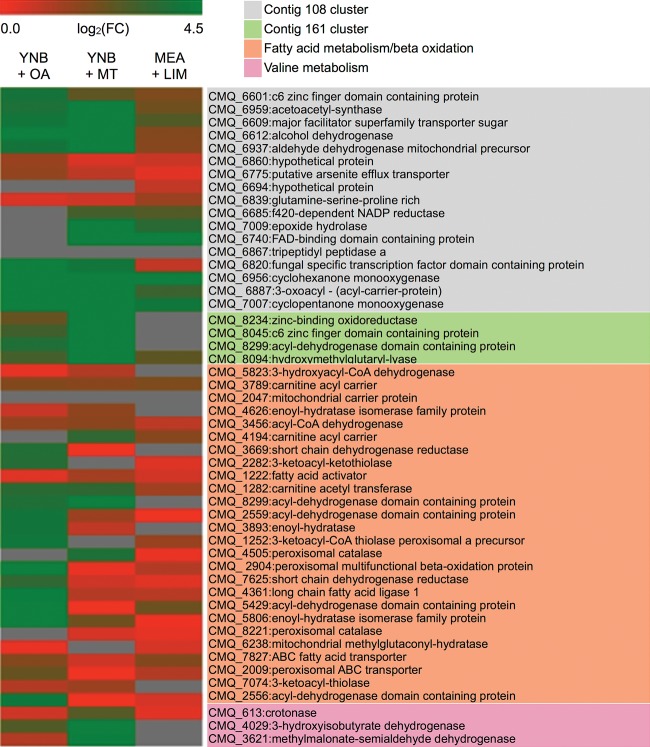
In bacteria, limonene degradation products may be further processed through the beta-oxidation pathway (30). We examined this pathway in Gs. We first used sequences for 24 fatty acid metabolic proteins of Yarrowia lipolytica, Laccaria bicolor, and Magnaporthe grisea to identify the corresponding proteins in Gs (see Table S2 in the supplemental material); then we predicted the subcellular localizations of these Gs proteins with Euk-mPLoc 2.0 (39). We compared the expression of the encoding genes potentially involved in fatty acid degradation when Gs was grown on YNB with oleic acid (OA) or on YNB with monoterpenes (highlighted in light orange in Fig. 3). Gs gene expression profiles indicate that the degradation of OA requires most of the fatty acid metabolic genes, including genes located in peroxisomes and mitochondria, while the degradation of monoterpenes seems to require mainly beta-oxidation genes located in mitochondria (Fig. 3; see also Table S1 in the supplemental material). We noticed that the signature gene for peroxisomal beta-oxidation, the peroxisomal multifunctional protein-encoding gene (CMQ_2904), was upregulated in YNB plus OA but not in YNB plus MT. Three mitochondrial enoyl-CoA hydratases were upregulated in both oleic acid and YNB plus MT (see Table S1). The four-gene cluster in contig 161 (Table 2) (CMQ_8234, CMQ_8045, CMQ_8299, and CMQ_8094) was upregulated only in YNB plus MT; it was not upregulated in either YNB plus OA or MEA plus LIM. Further, four major genes of the valine catabolic pathway were upregulated in YNB plus MT and YNB plus OA, but not in MEA plus MT (highlighted in light purple in Fig. 3). These four genes were the crotonase (CMQ_613), 3-hydroxy-isobutyryl-CoA hydrolase (CMQ_647), 3-hydroxy-isobutyrate dehydrogenase (CMQ_4029), and methylmalonate-semialdehyde dehydrogenase (CMQ_3621) genes.
FIG 3.
Expressed clusters and genes potentially involved in monoterpene degradation. The heatmap was generated by the MultiExperimental Viewer (MeV). Relative abundances of each gene (rows) in each growth condition (columns) are shown as log-transformed fold change (FC) relative to the condition's control. Red versus green shows low versus high FC.
Validation of differentially expressed genes using RT-PCR.
To validate the upregulation of genes potentially related to monoterpene metabolism, we selected genes located in (i) contig 108 (flavin adenine dinucleotide [FAD] binding monooxygenase gene CMQ_6740, Baeyer-Villiger monooxygenase genes CMQ_7007 and CMQ_6956, 3-oxo-carrier protein dehydrogenase reductase gene CMQ_6887, and epoxide hydrolase gene CMQ_7009), (ii) contig 161 (hydroxy-methylgutaryl-CoA lyase gene CMQ_8094 and acyl-dehydrogenase gene CMQ_8299), (iii)) the CYP450 gene (CMQ_1732), and (iv) two genes involved in fatty acid beta-oxidation (enoyl-coA hydratase gene CMQ_4626 and CoA ligase gene CMQ_4361). The results are shown in Fig. 4. Overall, the expression of all of these genes was higher in YNB plus MT than in MEA plus LIM. Except for CYP450 and the two beta-oxidation genes, all of the genes assessed in contigs 108 and 161 showed fold changes greater than 100 in YNB plus MT. The FAD-binding monooxygenase gene, CMQ_6740, had the highest expression in both YNB plus MT versus YNB (3,729×) and MEA plus LIM versus MEA (206×). The fold changes of the two beta-oxidation genes CMQ_4626 and CMQ_4361 were relatively low (<5) under both conditions.
FIG 4.
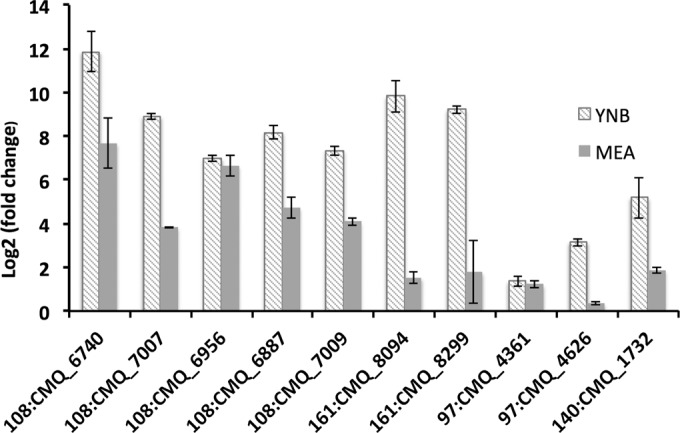
qRT-PCR validates the mRNA abundance of selected genes on YNB plus MT (diagonal bars) and MEA plus LIM (gray bars). Growth and treatment conditions were the same as for transcriptome analyses. mRNA abundance was normalized using the β-tubulin gene, a housekeeping gene; error bars show standard deviations based on three technical replicates.
Knockout of genes potentially involved in limonene modification or utilization.
We used Agrobacterium-mediated gene knockout in Gs to further investigate the roles of some highly expressed genes in (+)-limonene modification or utilization. First, we generated a mutant by deleting a three-gene cluster in contig 108 (CMQ_6956, CMQ_6887, and CMQ_7007). While this mutant grew well on MEA, it was unable to utilize (+)-limonene as a carbon source. Then, by deleting each of these genes separately, as well as three other genes that were highly induced by limonene (CMQ_4626, CMQ_1732, and CMQ_7009), we generated six additional mutants (Table 3). We confirmed the deletion of each gene by sequencing PCR products that included the full-length gene and the 5′ and 3′ flanking regions (34). The six mutants grew similarly to the parental strain (wild type [WT]) on MEA, MEA plus LIM, and YNB plus mannose. However, when the mutants were inoculated on YNB and provided with (+)-limonene as the only carbon source, four of the mutants were unable to grow (Table 3). The two Baeyer-Villiger monooxygenase (BVMO) mutants were killed by (+)-limonene treatment, while the enoyl-CoA hydratase mutant (i.e., with a change in the second gene in the mitochondrial beta-oxidation cycle) and the 3-oxoacyl-(acyl carrier protein) mutant, which did not grow with (+)-limonene, were able to grow when transferred onto MEA (survived). The epoxide hydrolase and CYP450 mutants were still able to utilize limonene as a carbon source, and while the mycelial density of the epoxide hydrolase mutant was much lower than that of the wild type, we noted no significant growth difference between the WT and the CYP450 mutant (Table 3). On YNB plus OA (i.e., with OA as the sole carbon source), the wild type and all the mutants displayed similar degrees of growth, except for the enoyl-CoA hydratase mutant, whose mycelia were of low density and less melanized.
TABLE 3.
Gs mutant phenotypes on YNB with limonene as a single carbon source

Two distinct Baeyer-Villiger monooxygenases contribute to limonene utilization by Gs.
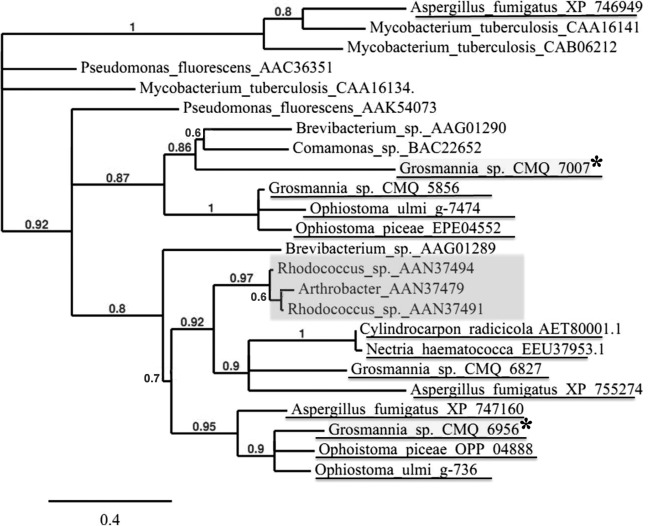
We showed that genes for two FAD-binding monooxygenases (CMQ_7007 and CMQ_6956), both of which were in contig108 (positions 1043294 and 1039559), were strictly required for the utilization of limonene by Gs. These two monooxygenases have the characteristics of Baeyer-Villiger monooxygenases, which catalyze the conversion of ketones to either ester or lactone (40). While CMQ_7007 contains a typical BVMO signature (FXGXXXHXXXWP/D), CMQ_6956 has an atypical one (FXGXXXHXXX), and these two proteins shared only 32% identity. We selected sequences of 24 functionally characterized BVMOs from different microorganisms and generated a maximum likelihood phylogenetic tree (Fig. 5). CMQ_7007 and CMQ_6956 were placed into two distinct clades. CMQ_7007 seems more closely related to bacterial cyclopentanone monooxygenase (CPMO), while CMQ_6956 was grouped into a clade of typical fungal monooxygenases and located in the same clade as other typical cyclohexanone monooxygenases (CHMO). CMQ_7007 was more highly induced in YNB plus MT (fold change, 2,571.45) than in MEA plus LIM (fold change, 356), while CMQ_6956 was more highly induced in MEA plus LIM (fold change, 128) than in YNB plus MT (fold change, 17.57). CMQ_6956, but not CMQ_7007, had a homologue (OP_04888) in the non-limonene-utilizing fungus O. piceae. These results suggest that CMQ_6956 may specifically target monoterpenes or terpene-related compounds in earlier stages of limonene degradation, while CMQ_7007 may have broader specificity and act on degraded compounds later in the pathway (i.e., linear terpenes or fatty acids).
FIG 5.
Maximum likelihood phylogenetic tree of Baeyer-Villiger monooxygenases from 12 species. Amino acid sequences were retrieved from GenBank and aligned using multiple-sequence comparisons by log expectation (MUSCLE); poorly aligned positions and divergent regions were removed using Gblocks, and the maximum likelihood tree was generated by PhyLM (www.phylogeny.fr). The two Gs BVMOS are highlighted by asterisks. The typical bacterial cyclohexanone monooxygenase group is highlighted in light gray.
Identifying limonene bioconversion metabolites using GC-MS.
To obtain additional information on metabolites produced by Gs in the presence of (+)-limonene, we used GC-MS to characterize some intracellular volatile compounds in fungal mycelia grown on either MEA (3 days) or YNB (21 days), with and without (+)-limonene. On MEA plus LIM, we found at least four groups of limonene derivatives formed presumably by oxygenation or hydroxylation of limonene at different carbon positions: carveol/carvone/dihydrocarvone, p-mentha-2,8-dienol, perillyl alcohol, and isopiperitenol (Table 4). Carvone was the most abundant compound under this condition. On YNB plus LIM, the fungus produced a less complex metabolic profile but still showed a peak of carvone and p-mentha-2,8-dienol. However, limonene-1,2-diol, which was almost nondetectable in MEA plus LIM, was the major peak under this condition.
TABLE 4.
Limonene intermediates detected by GC-MS for Gs grown on either MEA or YNB with (+)-limonene
| Intermediate | Retention time (min) | Medium(a) |
|---|---|---|
| trans-p-Mentha-2,8-dineol | 22.143 | MEA |
| Dihydrocarvone | 22.548 | MEA and YNB |
| cis-p-Mentha-2,8-dineol | 22.91 | MEA |
| cis-Carveol | 23.012 | MEA |
| Carvone | 23.3 | MEAa and YNB |
| trans-Carveol | 23.356 | MEA and YNB |
| Isopiperitenone | 23.80 | MEA |
| Fatty acid compound | 23.921 | MEA and YNB |
| p-Mentha-1,8-dine-9-ol | 24.35 | MEA |
| Perillyl alcohol | 24.396 | MEA |
| Limonene-1,2-diol | 25.326 | MEA and YNBa |
| 1,4-Hydroxy-2-oxolimoneneb | 7.2 | YNB |
Highest peak.
Detected from derivatized sample.
We also assessed whether intracellular metabolites were present in the Gs mutants (knockout mutants in monooxygenases, epoxide hydrolase, and CYP450) or in the Gc strain isolated from Jeffrey pine, and the O. piceae pine saprophyte, neither of which can use limonene as a carbon source. When grown on MEA plus LIM, all of the Gs mutants, Gc from Jeffrey pine, and O. piceae produced carvone and p-mentha-2,8-dienol; however, we did not detect limonene-1,2-diol in either Gc from Jeffrey pine or O. piceae. The epoxide hydrolase mutant produces a level of limonene-1,2-diol similar to that produced by Gs (wild type) on YNB plus LIM after 21 days of incubation. We also derivatized metabolite extracts using BSTFA-pentane (1:1) and examined the fatty acid and alcohol profiles. On the MEA plus LIM samples, we could not observe obvious peaks different from those of the control, while on YNB plus LIM, we observed 1-hydroxy-2-oxolimonene (Table 4), which has been reported for bacteria when limonene was used through the C-1 oxygenation pathway (limonene 1,2-oxide and limonene-1,2-diol) (30). In our toxicity assays, limonene-1,2-diol was one of the least toxic intermediates; it showed almost no inhibition on fungal growth, while other intermediates, such as carvone, dihydrocarvone, carveol, perillyl alcohol, and limonene epoxide, were toxic and able to kill the fungus when applied at the same concentration as limonene.
DISCUSSION
Pines that are attacked by insects or microorganisms release preformed and induced terpenoids, a major class of conifer chemical defense compounds (4). Many pine bark beetles and microbes are sensitive to terpenoids and can be inhibited or killed by these chemicals, while others, like the mountain pine beetle (MPB) and its associated fungi, are able to colonize their host trees because they have evolved mechanisms to overcome the toxicity of terpenoids, more specifically, monoterpenes. In this work, we showed that the two G. clavigera cryptic species (Gs and Gc) that are associated with MPB and the Jeffrey pine beetle, along with other pine ophiostomatoid fungi (e.g., Leptographium longiclavatum, O. montium, Ceratocystiopsis, and the saprophyte O. piceae), which are initially inhibited by high levels of monoterpenes, can resume growth after lag phases of various lengths (15, 18). While most of the MPB-associated fungi can use monoterpenes (i.e., limonene) as a carbon source, one of the Grosmannia cryptic species (Gc from Jeffrey pine) and the saprophyte were unable to.
Successful plant and animal pathogens, such as Nectria haematococca and other fungi, use multiple mechanisms for detoxifying plant antimicrobial compounds (41), and our results indicate that this is also the case for Gs. In previous work, we showed that a fungal ABC efflux transporter was a major mechanism by which Gs copes with the diffusion of monoterpenes into its cells in artificial media or when inoculated into young pine trees. This efflux pump excretes and maintains low levels of monoterpenes inside the fungal cells (18). This ABC transporter is present in pine beetle-associated fungi and highly induced in both monoterpene-utilizing (Gs) and nonutilizing species (e.g., Gc isolates from Jeffrey pine and O. piceae) (15, 18, 42). Here, we describe a second mechanism that allows this fungus to deal with monoterpenoid toxicity: modification or degradation of monoterpenes, in particular, (+)-limonene.
While it is well established that bacteria can use limonene as a carbon source, through either carvone or limonene-1,2-diol pathways, less information is available for fungi (27, 30). Recently, Bicas et al. (24) showed that Penicillium digitatum can grow on limonene as a single carbon source and produces α-terpineol as a major metabolic product (24). In our work, on the basis of combining information from fungal growth, gene deletion mutants, GC-MS, and transcriptome profiling, we propose a (+)-limonene degradation pathway for Gs (see Table S3 in the supplemental material) in which (+)-limonene is oxygenated at the C-1 carbon position, followed by the hydrolysis of the resulting epoxide, as has been proposed for the bacterium Rhodococcus erythropolis (30). Initial steps in bacterial limonene degradation involve four types of enzymes: a luciferase-like monooxygenase, a cofactor-independent epoxide hydrolase, a dichlorophenol-indophenol-dependent (DCPIP) dehydrogenase, and a Baeyer-Villiger monooxygenase (BVMO). While in Gs, we found an upregulated DCPIP dehydrogenase gene and a BVMO gene, no Gs orthologues were found for the first two bacterial enzymes, which suggested that the bacterial proteins for these steps are different from those used by Gs. It is possible that an FAD-binding monooxygenase (CMQ_6740) may be responsible for the epoxidation step, since the gene encoding this enzyme was highly expressed when the fungus was grown on (+)-limonene. Further, Alamouti et al. (15) showed that this gene is silent due to the insertion of a stop codon in the Gc cryptic species, which is unable to use limonene as a carbon source. However, it is also possible that this initial step may be carried out by other monooxygenases, like cytochrome P450, as has been suggested for bacteria (29). While we observed a highly induced CYP450 gene when the fungus was grown on limonene, mutating this gene did not prevent the utilization of limonene. Whether other CYP450s are responsible for the oxygenation of limonene in the degradation pathway needs to be further explored. In contrast to R. erythropolis, which possesses a unique epoxide hydrolase that is not a classic α-β hydrolase (43), Gs has such a classical enzyme (CMQ_7009), and this was significantly upregulated when (+)-limonene was the only substrate. While knocking out this gene in Gs did not totally abolish (+)-limonene utilization, as the mutant was still able to grow, mycelium density was lighter than in the wild type. It is likely that limonene epoxide conversion to limonene-1,2-diol may be mediated by this epoxide hydrolase, along with other α-β hydrolases.
Among other enzymes proposed to be involved in the degradation pathway were two BVMOs. BVMOs are flavoprotein monooxygenases that catalyze the insertion of one atom of oxygen into a linear or cyclic ketone, yielding the corresponding esters or lactones. In many bacteria, a BVMO along with a lactone hydrolase is necessary for ring opening during limonene utilization (44, 45). The roles of BVMOs in the degradation of limonene by fungi have not been reported. Not only were our two Gs BVMO mutants unable to grow on (+)-limonene, but also the inocula were killed by (+)-limonene. The two BVMO genes are colocated in the genome and coexpressed during (+)-limonene utilization. Further, phylogenetic analysis placed these two proteins in different clades, so we hypothesize that the two enzymes likely target different substrates: one (CMQ_6956) may catalyze the oxidation before the ring opening of the monoterpene skeleton, while the other (CMQ_7007) may be involved in breaking down the C10 ketone to a C8 compound (see Table S3 in the supplemental material). A similar oxidation function has been reported for the bacterial BVMO characterized in R. erythropolis, which has broad substrate specificities and can convert 1-hydroxy-2-oxolimonene, dihydrocarvone, and menthone to their corresponding lactones (45). For Gs, both BVMO enzymes are required for limonene utilization, but the corresponding genes need to be cloned and expressed, and the substrate specificity of their enzymes needs to be confirmed.
We can also make suggestions about how putative intermediates in limonene conversion by Gs can be gradually broken down after ring opening of the terpene skeleton. In bacteria (R. erythropolis and Pseudomonas species), it has been proposed that the intermediates might be channeled into the beta-oxidation pathway for further processing and utilization (27). In Gs, our data indicated that only one atypical mitochondrial beta-oxidation reaction occurred; the fungus may then use enzymes involved the valine metabolic pathway before the metabolites are channeled into the tricarboxylic acid (TCA) cycle. Several enoyl coenzyme A (enoyl-CoA) hydratases and acyl-CoA dehydrogenases contributed to oleic acid utilization; we have shown that one of each of these enzymes (CMQ_4626 and CMQ_8299) participates in (+)-limonene utilization (see Table S1 in the supplemental material). In addition to the gene cluster in contig 108 (FunGene accession no. GL629729), which is the major gene group contributing to monoterpene degradation, the fungus also needed a small gene cluster in contig 161 (FunGene accession no. GL629788; positions 1005268 to 1016769) for limonene utilization.
Finally, Gs may also modified limonene through additional enzymes and pathways. When either limonene-utilizing or nonutilizing fungi were grown on MEA plus LIM, we identified traces of limonene-1,2-diol, the main metabolite produced on minimal medium, and larger amounts of carvone or p-mentha-2, 8-dienol. While multiple fungal limonene-converting pathways have been reported, typically only one of the conversion products was present in larger amounts. For example, Penicillium digitatum DSM 62480 produces α-terpineol, carveol, and p-mentha-2,8-dienol from limonene, but only α-terpineol is found in high concentrations (46). Similarly, Cladosporium species that convert (+)-limonene into limonene-1,2-diol can also produce small amounts of dihydrocarvone (47). While Gs can structurally modify (+)-limonene at four different carbon positions, the most abundant metabolite on MEA plus LIM was carvone. Further, while nonutilizers, like O. piceae, modify limonene into carvone or p-mentha-2,8-dienol, at this point it is unclear whether these oxygenated metabolites are further channeled into other degradation pathways or are excreted by the fungal cells. Duetz et al. (20) suggested that the diversity of monoterpene modification products could be explained by nonspecific activity of monooxygenases or CYP450s rather than by the action of multiple enzymes that have different regiospecificities. For example, Aspergillus cellulose is known for its low CYP450 regiospecificity and produces a mixture of five types of oxygenated limonene: isopiperitenol, limonene-1,2-diol, carveol, perillyl alcohol, and α-terpineol (48). While we were not able to find CYP450s that were induced by limonene and were homologous to those of Gs and O. piceae, we noted that homologous FAD-binding monooxygenasegenes were expressed in both Gs (CMQ_7140) and O. piceae (OPP_08337). Whether this monooxygenase is responsible for modifying limonene to carvone (a major intermediate) and p-mentha-2,8-dienol (a minor metabolite) needs to be determined. Interestingly, we found that the homologous BVMO genes in Gs (CMQ_6956) and in O. piceae (OPP_04888) were both induced by limonene. These BVMOs may be involved in the oxidation of dihydrocarvone, a downstream product of carvone. We hypothesize that this metabolite cannot be further degraded by O. piceae because this saprophyte lacks some of the enzymes (e.g., those encoded by genes on Gs contigs 108 and 161) required for the degradation of limonene.
Overall, Gs and additional MPB associates have developed multiple mechanisms to deal with pine monoterpenes. The first line of defense for Gs to deal with monoterpenes is an ABC transporter that excretes and maintains low levels of monoterpenes in fungal cells (18). The remaining monoterpenes [i.e., (+)-limonene] in the fungal cells can be further detoxified through enzymatic modification or utilization. Gs required two Baeyer-Villiger monooxygenases to break down limonene before using this carbon source mainly through the mitochondrial β-oxidation pathway. It is likely that another FAD-binding monooxygenase, truncated in Gc, that does not use limonene is also a key enzyme in Gs. Additional work is required to confirm the substrate of the different enzymes for which we have created mutants and for other enzymes that were highly induced in the presence of monoterpenes. We anticipate that other conifer inhabiting-ophiostomatoid fungi have evolved similar pathways to cope with tree defense chemicals.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karen Reid for excellent project and laboratory management in the laboratory of J.B. and in support of the Tria project, and we thank the undergraduate students who have worked in the laboratory of C.B. on this project. We thank G. Robertson for help with the graphs and suggestions on the text, Massoumi Alamouti for help with the phylogenetic tree, and Sajeet Harridas for software training.
This work was funded by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC; grant to J.B. and C.B.) and by Genome Canada, Genome BC, and Genome Alberta (grant to J.B. and C.B.) in support of the Tria project (www.thetriaproject.ca).
Footnotes
Published ahead of print 16 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00670-14.
REFERENCES
- 1.Cullingham CI, Cooke JEK, Dang S, Davis CS, Cooke BJ, Coltman DW. 2011. Mountain pine beetle host-range expansion threatens the boreal forest RID G-5906-2011. Mol. Ecol. 20:2157–2171. 10.1111/j.1365-294X.2011.05086.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Safranyik L, Carroll AL, Regniere J, Langor DW, Riel WG, Shore TL, Peter B, Cooke BJ, Nealis VG, Taylor SW. 2010. Potential for range expansion of mountain pine beetle into the boreal forest of North America. Can. Entomol. 142:415–442. 10.4039/n08-CPA01 [DOI] [Google Scholar]
- 3.de la Giroday HC, Carroll AL, Aukema BH. 2012. Breach of the northern Rocky Mountain geoclimatic barrier: initiation of range expansion by the mountain pine beetle. J. Biogeogr. 39:1112–1123. 10.1111/j.1365-2699.2011.02673.x [DOI] [Google Scholar]
- 4.Bohlmann J. 2012. Pine terpenoid defences in the mountain pine beetle epidemic and in other conifer pest interactions: specialized enemies are eating holes into a diverse, dynamic and durable defence system. Tree Physiol. 32:943–945. 10.1093/treephys/tps065 [DOI] [PubMed] [Google Scholar]
- 5.Keeling CI, Bohlmann J. 2006. Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol. 170:657–675. 10.1111/j.1469-8137.2006.01716.x [DOI] [PubMed] [Google Scholar]
- 6.Boone CK, Aukema BH, Bohlmann J, Carroll AL, Raffa KF. 2011. Efficacy of tree defense physiology varies with bark beetle population density: a basis for positive feedback in eruptive species. Can. J. For. Res. 41:1174–1188. 10.1139/X11-041 [DOI] [Google Scholar]
- 7.Hall DE, Robert JA, Keeling CI, Domanski D, Quesada AL, Jancsik S, Kuzyk MA, Hamberger B, Borchers CH, Bohlmann J. 2011. An integrated genomic, proteomic and biochemical analysis of (+)-3-carene biosynthesis in Sitka spruce (Picea sitchensis) genotypes that are resistant or susceptible to white pine weevil. Plant J. 65:936–948. 10.1111/j.1365-313X.2010.04478.x [DOI] [PubMed] [Google Scholar]
- 8.Hall DE, Yuen MMS, Jancsik S, Quesada AL, Dullat HK, Li M, Henderson H, Arango-Velez A, Liao NY, Docking RT, Chan SK, Cooke JEK, Breuil C, Jones SJM, Keeling CI, Bohlmann J. 2013. Transcriptome resources and functional characterization of monoterpene synthases for two host species of the mountain pine beetle, lodgepole pine (Pinus contorta) and jack pine (Pinus banksiana). BMC Plant Biol. 13:80. 10.1186/1471-2229-13-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson-Jeffrey RC, Davidson RW. 1968. Three new Europhium species with Verticicladiella imperfect states on blue-stained pine. Can. J. Bot. 46:1523–1527. 10.1139/b68-210 [DOI] [Google Scholar]
- 10.Six DL, Paine TD. 1997. Ophiostoma clavigerum is the mycangial fungus of the Jeffrey pine beetle, Dendroctonus jeffreyi. Mycologia 89:858–866. 10.2307/3761106 [DOI] [Google Scholar]
- 11.Kelley ST, Farrell BD, Mitton JB. 2000. Effects of specialization on genetic differentiation in sister species of bark beetles. Heredity 84:218–227. 10.1046/j.1365-2540.2000.00662.x [DOI] [PubMed] [Google Scholar]
- 12.Yamaoka Y, Hiratsuka Y, Maruyama PJ. 1995. The ability of Ophiostoma clavigerum to kill mature lodgepole pine trees. Eur. J. Pathol. 25:401–404. 10.1111/j.1439-0329.1995.tb01355.x [DOI] [Google Scholar]
- 13.Lee S, Kim J-J, Breuil C. 2006. Pathogenicity of Leptographium longiclavatum associated with Dendroctonus ponderosae and Pinus contorta. Can. J. For. Res. 36:2864–2872. 10.1139/x06-194 [DOI] [Google Scholar]
- 14.Alamouti SM, Wang V, DiGuistini S, Six DL, Bohlmann J, Hamelin RC, Feau N, Breuil C. 2011. Gene genealogies reveal cryptic species and host preferences for the pine fungal pathogen Grosmannia clavigera. Mol. Ecol. 20:2581–2602. 10.1111/j.1365-294X.2011.05109.x [DOI] [PubMed] [Google Scholar]
- 15.Alamouti S, Haridas S, Feau N, Robertson G, Bohlmann J, Breuil C. 2014. Comparative genomics of the pine pathogens and the beetle symbionts in the genus Grosmannia. Mol. Biol. Evol. 10.1093/molbev/msu102 [DOI] [PubMed] [Google Scholar]
- 16.DiGuistini S, Wang Y, Liao NY, Taylor G, Tanguay P, Feau N, Henrissat B, Chan SK, Hesse-Orce U, Alamouti SM, Tsui CKM, Docking RT, Levasseur A, Haridas S, Robertson G, Birol I, Holt RA, Marra MA, Hamelin RC, Hirst M, Jones SJM, Bohlmann J, Breuil C. 2011. Genome and transcriptome analyses of the mountain pine beetle-fungal symbiont Grosmannia clavigera, a lodgepole pine pathogen. Proc. Natl. Acad. Sci. U. S. A. 108:2504–2509. 10.1073/pnas.1011289108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hesse-Orce U, DiGuistini S, Keeling CI, Wang Y, Li M, Henderson H, Docking TR, Liao NY, Robertson G, Holt RA, Jones SJM, Bohlmann J, Breuil C. 2010. Gene discovery for the bark beetle-vectored fungal tree pathogen Grosmannia clavigera. BMC Genomics 11:536. 10.1186/1471-2164-11-536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Lim L, DiGuistini S, Robertson G, Bohlmann J, Breuil C. 2013. A specialized ABC efflux transporter GcABC-G1 confers monoterpene resistance to Grosmannia clavigera, a bark beetle-associated fungal pathogen of pine trees. New Phytol. 197:886–898. 10.1111/nph.12063 [DOI] [PubMed] [Google Scholar]
- 19.de Carvalho C, da Fonseca M. 2006. Biotransformation of terpenes. Biotechnol. Adv. 24:134–142. 10.1016/j.biotechadv.2005.08.004 [DOI] [PubMed] [Google Scholar]
- 20.Duetz W, Bouwmeester H, van Beilen J, Witholt B. 2003. Biotransformation of limonene by bacteria, fungi, yeasts, and plants. Appl. Microbiol. Biotechnol. 61:269–277. 10.1007/s00253-003-1221-y [DOI] [PubMed] [Google Scholar]
- 21.Li H, Lan W, Cai C, Zhou Y, Lin Y. 2011. Biotransformation of limonene by microorganisms. Prog. Chem. 23:2318–2325 [Google Scholar]
- 22.Bohlmann J, Keeling CI. 2008. Terpenoid biomaterials. Plant J. 54:656–669. 10.1111/j.1365-313X.2008.03449.x [DOI] [PubMed] [Google Scholar]
- 23.Schwab W, Davidovich-Rikanati R, Lewinsohn E. 2008. Biosynthesis of plant-derived flavor compounds. Plant J. 54:712–732. 10.1111/j.1365-313X.2008.03446.x [DOI] [PubMed] [Google Scholar]
- 24.Bicas JL, Dionisio AP, Pastore GM. 2009. Bio-oxidation of terpenes: an approach for the flavor industry. Chem. Rev. 109:4518–4531. 10.1021/cr800190y [DOI] [PubMed] [Google Scholar]
- 25.Rasoul-Amini S, Fotooh-Abadi E, Ghasemi Y. 2011. Biotransformation of monoterpenes by immobilized microalgae. J. Appl. Phycol. 23:975–981. 10.1007/s10811-010-9625-4 [DOI] [Google Scholar]
- 26.Prieto SGA, Perea JA, Ortiz VLCC. 2011. Microbial biotransformation of (r)-(+)-limonene by Penicillium digitatum DSM 62840 for producing (r)-(+)-terpineol. Vitae Rev. Fac. Quim. Farm. 18:163–172 [Google Scholar]
- 27.Bicas JL, Fontanille P, Pastore GM, Larroche C. 2008. Characterization of monoterpene biotransformation in two pseudomonads. J. Appl. Microbiol. 105:1991–2001. 10.1111/j.1365-2672.2008.03923.x [DOI] [PubMed] [Google Scholar]
- 28.Marostica Junior MR, Pastore GM. 2009. Limonene and its oxyfunctionalized compounds: biotransformation by microorganisms and their role as functional bioactive compounds. Food Sci. Biotechnol. 18:833–841 [Google Scholar]
- 29.Schewe H, Mirata MA, Holtmann D, Schrader J. 2011. Biooxidation of monoterpenes with bacterial monooxygenases. Process Biochem. 46:1885–1899. 10.1016/j.procbio.2011.06.010 [DOI] [Google Scholar]
- 30.van der Werf MJ, Swarts HJ, de Bont JAM. 1999. Rhodococcus erythropolis DCL14 contains a novel degradation pathway for limonene. Appl. Environ. Microbiol. 65:2092–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan Q, Day DF. 1998. Organic co-solvent effects on the bioconversion of (R)-(+)-limonene to (R)-(+)-alpha-terpineol. Process Biochem. 33:755–761. 10.1016/S0032-9592(98)00046-6 [DOI] [Google Scholar]
- 32.Tan Q, Day DF. 1998. Bioconversion of limonene to alpha-terpineol by immobilized Penicillium digitatum. Appl. Microbiol. Biotechnol. 49:96–101. 10.1007/s002530051143 [DOI] [Google Scholar]
- 33.van der Werf MJ. 2000. Purification and characterization of a Baeyer-Villiger mono-oxygenase from Rhodococcus erythropolis DCL14 involved in three different monocyclic monoterpene degradation pathways. Biochem. J. 347:693–701. 10.1042/0264-6021:3470693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, DiGuistini S, Wang TCT, Bohlmann J, Breuil C. 2010. Agrobacterium-meditated gene disruption using split-marker in Grosmannia clavigera, a mountain pine beetle associated pathogen. Curr. Genet. 56:297–307. 10.1007/s00294-010-0294-2 [DOI] [PubMed] [Google Scholar]
- 35.Lah L, Haridas S, Bohlmann J, Breuil C. 2012. The cytochromes P450 of Grosmannia clavigera: genome organization, phylogeny, and expression in response to pine host chemicals. Fungal Genet. Biol. 10.1016/j.fgb.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 36.Kal A, van Zonneveld A, Benes V, van den Berg M, Koerkamp M, Albermann K, Strack N, Ruijter J, Richter A, Dujon B, Ansorge W, Tabak H. 1999. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol. Biol. Cell 10:1859–1872. 10.1091/mbc.10.6.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S, Kim JJ, Breuil C. 2006. Diversity of fungi associated with the mountain pine beetle, Dendroctonus ponderosae and infested lodgepole pines in British Columbia. Fungal Divers. 22:91–105 [Google Scholar]
- 38.Plattner A, Kim J, Reid J, Hausner G, Lim YW, Yamaoka Y, Breuil C. 2009. Resolving taxonomic and phylogenetic incongruence within species Ceratocystiopsis minuta. Mycologia 101:878–887. 10.3852/08-132 [DOI] [PubMed] [Google Scholar]
- 39.Chou K, Shen H. 2010. A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLoS One 5:e9931. 10.1371/journal.pone.0009931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leisch H, Morley K, Lau PCK. 2011. Baeyer-Villiger monooxygenases: more than just green chemistry. Chem. Rev. 111:4165–4222. 10.1021/cr1003437 [DOI] [PubMed] [Google Scholar]
- 41.Coleman JJ, White GJ, Rodriguez-Carres M, VanEtten HD. 2011. An ABC transporter and a cytochrome P450 of Nectria haematococca MPVI are virulence factors on pea and are the major tolerance mechanisms to the phytoalexin pisatin. Mol. Plant Microbe Interact. 24:368–376. 10.1094/MPMI-09-10-0198 [DOI] [PubMed] [Google Scholar]
- 42.Haridas S, Wang Y, Lim L, Alamouti SM, Jackman S, Docking R, Robertson G, Birol I, Bohlmann J, Breuil C. 2013. The genome and transcriptome of the pine saprophyte Ophiostoma piceae, and a comparison with the bark beetle-associated pine pathogen Grosmannia clavigera. BMC Genomics 14:373. 10.1186/1471-2164-14-373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbirato F, Verdoes JC, de Bont JAM, van der Werf MJ. 1998. The Rhodococcus erythropolis DCL14 limonene-1,2-epoxide hydrolase gene encodes an enzyme belonging to a novel class of epoxide hydrolases. FEBS Lett. 438:293–296. 10.1016/S0014-5793(98)01322-2 [DOI] [PubMed] [Google Scholar]
- 44.van der Vlugt-Bergmans CJB, van der Werf MJ. 2001. Genetic and biochemical characterization of a novel monoterpene epsilon-lactone hydrolase from Rhodococcus erythropolis DCL14. Appl. Environ. Microbiol. 67:733–741. 10.1128/AEM.67.2.733-741.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Werf MJ, Boot AM. 2000. Metabolism of carveol and dihydrocarveol in Rhodococcus erythropolis DCL14. Microbiology 146:1129–1141 [DOI] [PubMed] [Google Scholar]
- 46.Demyttenaere J, Van Belleghem K, De Kimpe N. 2001. Biotransformation of (R)-(+)- and (S)-(−)-limonene by fungi and the use of solid phase microextraction for screening. Phytochemistry 57:199–208. 10.1016/S0031-9422(00)00499-4 [DOI] [PubMed] [Google Scholar]
- 47.Mukherjee BB, Kraidman G, Hill I. 1973. Synthesis of glycols by microbial transformation of some monocyclic terpenes. Appl. Microbiol. 25:447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noma Y, Yamasaki S, Asakawa Y. 1992. Biotransformation of limonene and related compounds by Aspergillus cellulosae. Phytochemistry 31:2725–2727. 10.1016/0031-9422(92)83619-A [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.