Abstract
The genus Listeria is ubiquitous in the environment and includes the globally important food-borne pathogen Listeria monocytogenes. While the genomic diversity of Listeria has been well studied, considerably less is known about the genomic and morphological diversity of Listeria bacteriophages. In this study, we sequenced and analyzed the genomes of 14 Listeria phages isolated mostly from New York dairy farm environments as well as one related Enterococcus faecalis phage to obtain information on genome characteristics and diversity. We also examined 12 of the phages by electron microscopy to characterize their morphology. These Listeria phages, based on gene orthology and morphology, together with previously sequenced Listeria phages could be classified into five orthoclusters, including one novel orthocluster. One orthocluster (orthocluster I) consists of large-genome (∼135-kb) myoviruses belonging to the genus “Twort-like viruses,” three orthoclusters (orthoclusters II to IV) contain small-genome (36- to 43-kb) siphoviruses with icosahedral heads, and the novel orthocluster V contains medium-sized-genome (∼66-kb) siphoviruses with elongated heads. A novel orthocluster (orthocluster VI) of E. faecalis phages, with medium-sized genomes (∼56 kb), was identified, which grouped together and shares morphological features with the novel Listeria phage orthocluster V. This new group of phages (i.e., orthoclusters V and VI) is composed of putative lytic phages that may prove to be useful in phage-based applications for biocontrol, detection, and therapeutic purposes.
INTRODUCTION
Listeria monocytogenes is an important food-borne pathogen responsible for severe infections in both animals and humans (1, 2). Contamination of food and food production facilities by L. monocytogenes continues to be both a major health concern and an economic burden. In the United States alone, Listeria is responsible for an annual average of 1,455 hospitalizations and 255 deaths and estimated economical costs upwards of $2.5 billion (3). To reduce the burden of L. monocytogenes, there is a drive to develop better detection and control strategies; Listeria phages have the potential to help achieve both of these goals. Currently, there are several phage-based control products on the market as well as phage-based detection assays in development (4, 5). Additionally, previous studies have also shown that phages play an important role in the evolution and virulence of many pathogens (6–9); however, we are just beginning to understand how Listeria phages contribute to their hosts' pathogenicity and biology. One prophage, found within the comK gene in many Listeria strains, has recently been shown to excise from the host chromosome without entering the lytic cycle. After phage excision, ComK's function as a transcriptional activator was restored, and the resulting upregulation of late com genes was shown to play a role in host phagosome escape (10). This same mechanism of nonlethal excision and reintegration may also help to shape the evolution of Listeria; a similar comK prophage in one L. monocytogenes strain's chromosome was shown to have undergone repeated rearrangement over the course of 12 years in a food processing plant, while the rest of the chromosome experienced only one SNP (single nucleotide polymorphism) (11).
Listeria phages were previously classified into the following five species, based on observed morphology: 4211, 2671, 2685, H387, and 2389 (12). These morphospecies were validated by the Bacterial Virus Subcommittee of the ICTV (International Committee on Taxonomy of Viruses) (12). Recently reported genomes of Listeria phages range in size from 35.6 to 134.4 kb (13, 14). Molecular and in silico analyses by Dorscht et al. (14) showed that Listeria phages sequenced to date belong to several phylogenetic clades and display a conserved genome organization. Recently, the unique Listeria phage P70 was described by Schmuki et al. (15). P70 has a medium-sized genome (∼67 kb), similar in size to several phages isolated by our group from dairy farms in the state of New York (16). P70 also possesses an unusual elongated capsid not yet seen in other Listeria phages. However, this particular morphology has been observed for Enterococcus faecalis bacteriophage VD13 (17).
In this paper, we present 14 new Listeria phage genomes, one new E. faecalis phage genome, as well as a comparative analysis probing the genomic diversity, morphological diversity, and evolutionary relationships of sequenced phages. To determine relationships between bacteriophages, we performed a cluster analysis based on the presence or absence of orthologous genes; we define a cluster identified from this analysis as an “orthocluster,” a term previously used by Moreno Switt et al. to describe clusters of Enterobacteriaceae-infecting phages from a similar analysis (18). We report genomic and morphological evidence that Listeria phages form at least five conserved orthoclusters, one of which fits into a greater cluster of Listeria and Enterococcus phages that share genomic features and a unique morphology (including phages P70 and VD13).
MATERIALS AND METHODS
Bacterial strains and bacteriophages.
Twelve out of 14 Listeria phages sequenced in this study were isolated from silage samples collected on two dairy farms in New York State between August 2008 and July 2009 (Table 1). Phages were isolated by using three L. monocytogenes host strains: FSL J1-208 (lineage IV, serotype 4a), F2365 (lineage I, serotype 4b), and MACK (lineage II, serotype 1/2a). These 12 Listeria phages exhibited a wide diversity of host ranges, as determined by spot tests against 13 L. monocytogenes isolates representing the nine most common serotypes as well as the four phylogenetic lineages of L. monocytogenes (16). Two Listeria phages, LP-083-2 and LP-030-3, were later isolated as low-level contaminants from LP-083-1 and LP-030-2 cultures, respectively. These contaminants were initially detected by electron microscopy. VD13, an E. faecalis phage isolated from human urogenital secretions and first described in 1975 by Ackermann et al. (17), was obtained from the Felix d'Herelle Reference Center for Bacterial Viruses (Quebec City, Quebec, Canada). Genome sizes of the Listeria phages were previously estimated by pulsed-field gel electrophoresis (PFGE) (16).
TABLE 1.
Bacteriophage general characteristics
| Phage | Isolation host strain | Family | Genome length (bp) | GC content (%) | No. of contigs | No. of predicted genes | No. of tRNAs |
|---|---|---|---|---|---|---|---|
| LP-026 | FSL J1-208 | Siphoviridae | 67,150 | 36.3 | 1 | 115 | 0 |
| LP-030-2 | F2365 | Siphoviridae | 38,275 | 34.8 | 1 | 69 | 0 |
| LP-030-3 | F2365 | Siphoviridae | 41,156 | 36.6 | 1 | 73 | 0 |
| LP-032 | FSL J1-208 | Siphoviridae | 67,040 | 36.3 | 3 | 116 | 0 |
| LP-037 | FSL J1-208 | Siphoviridae | 64,756 | 36.6 | 1 | 114 | 0 |
| LP-048 | MACK | Myoviridae | 133,096 | 36.0 | 1 | 179 | 17 |
| LP-064 | MACK | Myoviridae | 135,279 | 35.9 | 1 | 188 | 17 |
| LP-083-1 | MACK | Siphoviridae | 35,745 | 40.8 | 2 | 57 | 0 |
| LP-083-2 | MACK | Myoviridae | 135,831 | 35.9 | 1 | 180 | 17 |
| LP-101 | MACK | Siphoviridae | 43,767 | 35.5 | 1 | 70 | 0 |
| LP-110 | FSL J1-208 | Siphoviridae | 65,132 | 36.3 | 1 | 114 | 0 |
| LP-114 | FSL J1-208 | Siphoviridae | 66,676 | 36.4 | 1 | 118 | 0 |
| LP-125 | MACK | Myoviridae | 135,281 | 35.9 | 1 | 189 | 17 |
| LP-124 | F2365 | Myoviridae | 135,817 | 35.9 | 1 | 188 | 17 |
| VD13a | ATCC 29200 | Siphoviridae | 55,210 | 40.0 | 1 | 88 | 0 |
Enterococcus phage.
Preparation of phage lysates and phage DNA extraction.
Phage lysates were prepared from previously purified stocks, as described previously (16). Lysates were then concentrated by polyethylene glycol precipitation (16). DNA was extracted from the concentrated stocks according to a modified version of the lambda DNA extraction protocol described previously by Sambrook and Russell (19). Modifications to the protocol include an initial addition of DNase I (Promega BioScience, Madison, WI) (final concentration, 5 μg/ml) to the samples followed by an RNase A (Sigma-Aldrich, St. Louis, MO) (30 μg/ml) to the samples followed by an incubation at room temperature for 30 min to remove exogenous nucleic acids from the lysed bacterial host; CaCl2 was added to a final concentration of 2 mM for this step. Nuclease digestions were then stopped by the addition of EDTA (final concentration, 20 mM) and incubation at 65°C for 10 min. Phage capsids were subsequently digested with proteinase K (Roche Applied Science, Penzberg, Germany) (final concentration, 0.2 mg/ml) in the presence of SDS (added to a final concentration of 0.5% [wt/vol]) at 56°C for 1 h. Chloroform-phenol was used to extract DNA, which was then concentrated by ethanol precipitation.
Phage genome sequencing, annotation, and analysis.
Library preparation and DNA sequencing were performed at the Cornell University Life Science Core Laboratory Facilities (Ithaca, NY). LP-032, LP-110, and LP-083-1 genomic DNAs were sequenced by using the Illumina GA II sequencing platform (Illumina Inc., San Diego, CA). Thirty-six-base-pair reads were assembled de novo by using the Velvet algorithm (20). The 13 remaining phages were sequenced on the Illumina HiSeq 2500 platform. One-hundred-base-pair reads were de novo assembled with SPAdes 2.5.1 (21) for the “Twort-like viruses” (described below) and with Velvet for the remaining phages. Contigs with ends that exactly matched the ends of other contigs by at least 20 bp were assembled into larger contigs by using Sequencher v 5.1 (Gene Codes Corp., Ann Arbor, MI). All sequences were submitted to the RAST (available at http://rast.nmpdr.org/) genome annotation service (22) for automatic annotation. Further homology searches of nucleotide sequences and manual annotation of predicted amino acid sequences were performed through NCBI databases (http://www.ncbi.nlm.nih.gov) by using BLAST algorithms (23) as well as through the EMBL-EBI database (http://www.ebi.ac.uk) by using the InterProScan tool (24). Predicted proteins with unknown functions containing conserved domains were annotated “X domain protein” or “X domain-containing protein,” depending on whether the domain made up a majority of the predicted protein sequence or not, respectively (here X represents the name of the conserved domain). Phage genome maps were drawn by using Easyfig v2.1 (25). Average BLAST nucleotide identities were calculated by using jSpecies v1.2.1 (26).
Clustering based on the presence or absence orthologous genes.
Orthologous genes among phage genomes were identified by using OrthoMCL v1.4 (27), using the default settings (BLAST E value cutoff of 1e−5, BLAST identity cutoff of 0, and percent MCL [Markov Cluster] match cutoff of 0). The orthologous gene matrix for the OrthoMCL analysis was converted into a binary matrix (where 1 is presence and 0 is absence). Duplicated genes were collapsed into one entry and scored as being present. A neighbor-joining tree analysis with 1,000 bootstrap replicates was performed with SplitsTree4 (28).
Morphological analysis by transmission electron microscopy.
Twelve sequenced phages were sedimented for 60 min at 25,000 × g in a Beckman J2-21 centrifuge equipped with a JA18.1 fixed-angle rotor, washed in 0.1 M neutral ammonium acetate under the same conditions, deposited onto carbon-coated copper grids, stained with uranyl acetate (2%, pH 4.5) or potassium phosphotungstate (2%, pH 7.0), and examined at 60 kV under a Philips EM300 electron microscope at an instrumental magnification of ×29,700, monitored with T4 phage tails.
Nucleotide sequence accession numbers.
The bacteriophage genome sequences generated in this study have been deposited in GenBank with the following accession numbers: KJ094020 for LP-026, JX120799.2 for LP-030-2, KJ094022 for LP-030-3, KJ094024 to KJ094026 for LP-032, JX126920.2 for LP-037, KJ094033 for LP-048, KJ094029 for LP-064, KJ094027 and KJ094028 for LP-083-1, KJ094030 for LP-083-2, KJ094023 for LP-101, JX126919 for LP-110, KJ094021 for LP-114, KJ094031 for LP-124, JX126918.2 for LP-125, and KJ094032 for VD13. Accession numbers of previously reported phage genomes analyzed in this study are summarized in Table S1 in the supplemental material.
RESULTS
Genomes of newly sequenced Listeria phages can be classified into five distinct groups based on orthologous gene content.
Genome sequencing of 14 Listeria phages and 1 Enterococcus phage, followed by de novo assembly, yielded 13 genome sequences that represented a single contig; phages LP-083-1 and LP-032 represented two and three contigs, respectively. Genome size estimates based on PFGE of LP-081-1 and LP-032 (16) were congruent with the size of the total assembly; thus, despite not being fully closed, most of the genomes of these phages are likely covered by the assembly. Mapping of raw sequencing reads to the assemblies revealed an average coverage across all phage genomes at a very high sequencing depth (71- to 700-fold coverage). Assemblies for five phages, which were subsequently classified as Twort-like viruses (see below for details), showed a single region (of approximately 3 kb) with a high level of homology to the direct terminal repeats previously reported for the Twort-like Listeria phage A511 (13), a member of the Spounavirinae subfamily of tailed phages. Mapping of reads against selected assemblies showed higher read coverage for these regions, further supporting that these sequences represent direct terminal repeats. These repeat sequences are located in a single region of the previously reported Twort-like Listeria phage P100 genome (starting approximately 25 kb from the 3′ end) (29) but were assembled into both ends of the A511 genome. For the phages sequenced here, the terminal repeat sequences were maintained in the location where they assembled (assemblies are available at GenBank; see the accession numbers reported above). Initial RAST annotation (22) of all genomes required considerable subsequent manual refinement to provide high-quality gene function predictions.
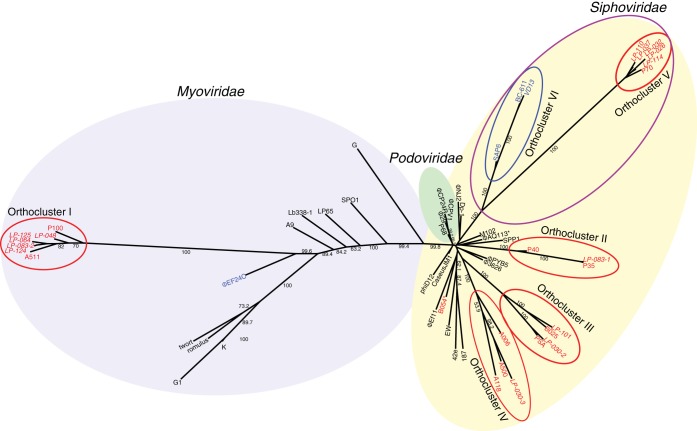
An initial classification of the 15 phages sequenced was performed by a comprehensive cluster analysis, based on the presence or absence of orthologous genes; this analysis also included 11 previously sequenced Listeria phages (for a total of 25 Listeria phages analyzed) as well as an additional 30 firmicute-infecting phages (see Table S1 in the supplemental material for detailed information on all phages). This analysis allowed classification of the 25 Listeria phages into five distinct orthoclusters, designated orthoclusters I to V (see below), which were determined by a cluster analysis based on the presence or absence of orthologous genes; each of these orthoclusters typically contained phages with similar genome sizes, GC contents, and morphologies (Fig. 1). Only a single Listeria phage (B054) (Fig. 1) did not cluster into one of these orthoclusters; this phage was previously described as a myovirus (14, 30) but clustered here with phages classified as belonging to the Siphoviridae. We also identified an orthocluster containing three Enterococcus-infecting Siphoviridae, which was designated orthocluster VI; this orthocluster groups with orthocluster V (bootstrap value, 100). Overall, orthoclustering generally coincided with taxonomical phage families, as determined by morphology; whereas Myoviridae and Siphoviridae formed distinct clusters, the Podoviridae formed a cluster that was closely related to Siphoviridae and included one branch (P68) that did not have high bootstrap support (bootstrap value, 66).
FIG 1.
Neighbor-joining tree based on the presence or absence of orthologous genes (1,000 bootstrap replicates). This tree includes phages sequenced in this study (italicized) as well as other previously sequenced firmicute-infecting phages (see Table S1 in the supplemental material). Listeria phages are labeled in red, and Enterococcus phages are labeled in blue. Red circles indicate Listeria phage orthoclusters, the blue circle indicates Enterococcus phage orthocluster VI, and the purple circle indicates a larger orthocluster containing Listeria and Enterococcus phages.
Listeria phages belonging to the Myoviridae are Twort-like viruses that share high average nucleotide identities and are nearly morphologically identical to previously described phages A511 and P100.
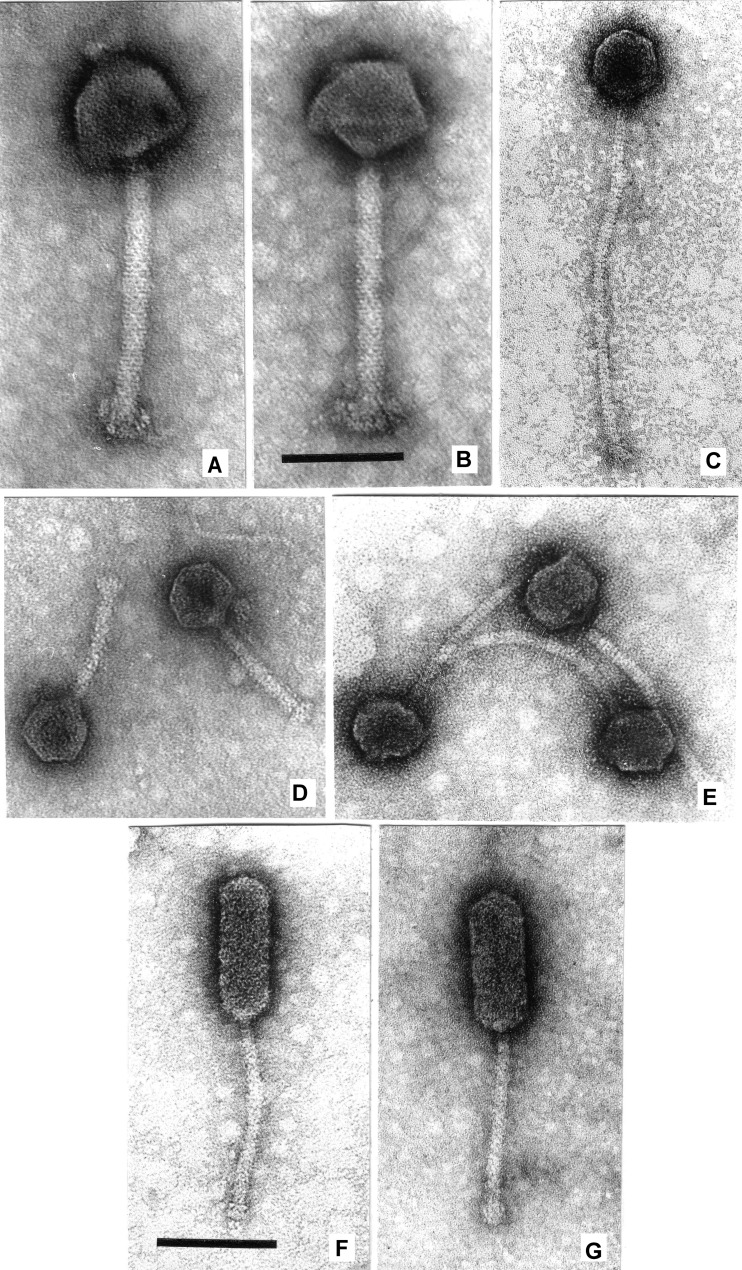
Orthocluster I, which is supported by a bootstrap value of 100, contains phages LP-048, LP-064, LP-083-2, LP-124, and LP-125 (sequenced here) as well as phages A511 and P100. All orthocluster I phages are myoviruses, characterized by long contractile tails, and belong to the genus of Twort-like viruses within the Spounavirinae subfamily (31, 32); the well-studied Listeria phage A511 (30) is part of this group. Bacteriophages classified as Twort-like viruses have been found in other host genera within the Firmicutes, such as Bacillus, Staphylococcus, Enterococcus, and Lactobacillus (31). The genome size for orthocluster I phages ranges from 131 to 138 kb (with an average GC content of 35.9 to 36.0%). Annotation identified 177 to 191 predicted genes as well as 16 to 18 tRNAs in these genomes. The average nucleotide identity of the phages in this group ranges from 91 to 100% across 90 to 100% of their genomes (see Fig. S1 in the supplemental material). Orthocluster I phages encode a putative structural and DNA packaging module of ∼30 kb in length in the center of their genomes. This putative structural module is flanked by a tRNA region (∼5 kb) on its 5′ side and by a putative DNA replication, modification, and recombination module on its 3′ side (∼30 kb). The ends of the genomes for these phages contain large regions (∼30 kb), with almost all genes encoding putative proteins with unknown functions. The heads of the orthocluster I phages examined here (LP-048, LP-083-2, LP-124, and LP-125) show capsomers and, as evidenced by the observation of capsids with pentagonal or hexagonal outlines, are icosahedra (Fig. 2A and B). The heads are separated from the sheaths by a 10-nm-long neck. Extended tails measure 206 by 18 nm (Table 2), show conspicuous transverse striations, and carry a thin base plate with 10-nm-long spikes. Contracted tails measure 90 by 25 nm and display double base plates.
FIG 2.
Electron micrographs. (A) Listeria phage LP-083-2 with an extended tail and hexagonal head. The presence of capsomers is hinted at by small asperities on the capsid. (B) LP-083-2 with a pentagonal head. (C) Phage LP-030-3. (D) Two particles of phage LP-083-1. (E) Three particles of phage LP-030-2. (F) Listeria phage LP-032. (G) Enterococcus phage VD13. Phages were stained with uranyl acetate and visualized at a final magnification of ×297,000. Panels A to E and panels F and G, respectively, are shown at the same scale; bars indicate 100 nm.
TABLE 2.
Main dimensions of Listeria phages and Enterococcus phage VD13
| Family | Phage(s) | Genus or species | Head diam (nm) | Tail diam (nm) | Stain(s)a | No. of particles measured |
|---|---|---|---|---|---|---|
| Myoviridae | LP-048, LP-083-2, LP-124, LP-125 | Twort | 86 | 206 × 18 | UA | 14 |
| Siphoviridae | LP-030-3 | 2671 | 55 | 297 × 8 | UA, PT | 12 |
| LP-030-2 | 2389 | 53 | 160 × 7–10 | UA, PT | 12 | |
| LP-083-1 | P35 | 57 | 100 × 8 | UA | 5 | |
| LP-026, LP-032, LP-037, LP-110 | 123 × 44b | 162 × 7–8 | UA, PT | 20 | ||
| VD13 | VD13 | 113 × 43b | 145 × 8 | UA | 10 |
PT, phospotungstate; UA, uranyl acetate.
Dimensions represent length by diameter for phages with an elongated capsid morphology.
Siphoviridae Listeria phages with small genomes (<42 kb) form three separate groups with distinct genomic and morphological characteristics.
Three orthoclusters identified here (orthoclusters II, III, and IV) (Fig. 1) represent small-genome Siphoviridae of the B1 morphotype; all phages in these three orthoclusters analyzed in this study have genome sizes of between 36 kb and 43 kb (Table 3). The B1 morphotype is very common and comprises phages with long noncontractile tails and isometric heads (33).
TABLE 3.
Genomic characteristics and morphology of Listeria and Enterococcus phage orthoclusters
| Orthocluster | Phages | Host | Genome size (kb) | No. of predicted genes | GC content (%) | % nucleotide identity across % of genome | Morphology |
|---|---|---|---|---|---|---|---|
| I | A511, LP-048, LP-064, LP-083-2, LP-124, LP-125, P100 | Listeria | 131–138 | 177–191 | 35.9–36.0 | 91–100 across 90–100 | Myoviridae |
| II | LP-083-1, P35, P40 | Listeria | 36 | 56–62 | 39.3–40.8 | 61–99 across 15–96 | Siphoviridae (B1) |
| III | B025, LP-030-2, LP-101, PSA | Listeria | 36–43 | 59–70 | 34.8–35.5 | 85–91 across 22–83 | Siphoviridae (B1) |
| IV | A006, A118, A500, LP-030-3 | Listeria | 38–41 | 62–73 | 35.5–36.6 | 84–99 across 2–69 | Siphoviridae (B1) |
| V | LP-026, LP-032, LP-037, LP-110, LP-114, P70 | Listeria | 65–67 | 114–119 | 33.3–33.6 | 95–100 across 81–98 | Siphoviridae (B3) |
| VI | BC-611, SAP6, VD13 | Enterococcus | 54–59 | 43–88a | 40.0–40.4 | 87–94 across 61–77 | Siphoviridae (B3) |
Phages in this group were annotated by using different methodologies.
Orthocluster II, which is supported by a bootstrap value of 100, contains phage LP-083-1 (sequenced here) as well as two previously reported Listeria phages (P35 and P40) (Fig. 1). The phages in this orthocluster have genome lengths of approximately 36 kb (with an average GC content of 39.3 to 40.8%) and remarkably short tails of only about 100 by 8 nm. Annotation identified 56 to 62 predicted genes in their genomes. The average nucleotide identity of the phages in this group ranges from 61 to 99% across 15 to 96% of their genomes. Despite LP-083-1 being one of two phages with genome assemblies resulting in more than one contig, LP-083-1 and P35 are highly similar (99% average nucleotide identity over ≥94% of their genomes) and show a high level of synteny (see Fig. S2A in the supplemental material). Consistent with a previously reported comparison of phages P35 and P40 (14), we identified a putative structural and DNA packaging module at the 5′ end of LP-083-1, including a group of three adjacent genes clearly annotated as encoding (i) a large terminase subunit, (ii) a portal protein, and (iii) a capsid morphogenesis domain-containing protein (see Fig. S2A in the supplemental material). Other than these genes, genes in this region could not be annotated as encoding specific functions (thus representing “hypothetical proteins”). Subsequent regions at the 3′ end of this module contain genes encoding lysis functions (e.g., a holin), followed by genes annotated as encoding DNA replication functions, followed by genes encoding putative regulatory functions, including a Cro/C1-type helix-turn-helix domain protein and a MazG-like pyrophosphohydrolase domain protein. We did not identify genes encoding integrase functions in LP-083-1, suggesting that this phage is a putative obligate lytic phage, consistent with previous reports for P35 and P40 (14, 34). LP-083-1 is characterized by an unusually short tail that terminates in a bushel of short spikes (Fig. 2D); this morphology is nearly identical to that reported previously for P35 and P40 (14).
Orthocluster III, which is also supported by a bootstrap value of 100, comprises phages LP-030-2 and LP-101 (both sequenced here) as well as the previously described temperate Listeria phages PSA and B025 (Fig. 1). The genome size for phages in this orthocluster ranges from 36 to 43 kb (with an average GC content of 34.8 to 35.5%); annotation identified 59 to 70 predicted genes in these genomes. The average nucleotide identity of the phages in this group ranges from 85 to 91% across 22 to 83% of their genomes. While all phages in this orthocluster show a similar genome organization (see Fig. S2B in the supplemental material), genome alignments and cluster analysis clearly show that these phages represent two subgroups (supported by a bootstrap value of 100). LP-030-2 and PSA show high levels of synteny and genome conservation, whereas LP-101 and B025 are more divergent (see Fig. S2B in the supplemental material). Genomes of all phages in orthocluster III contain, at their 5′ end, a putative structural and DNA packaging module that starts with genes encoding the small and large terminase subunits. Among the 16 to 18 genes in these putative structural and DNA packaging modules, 8 to 10 were annotated as encoding specific functions (see Fig. S2B in the supplemental material). Whereas there was little to no nucleotide identity between the putative structural modules of these two subgroups, the 3′ region of all four orthocluster II phage genomes showed considerable nucleotide similarity; this region encodes putative DNA replication and regulatory function proteins. All phages in this orthocluster encode an integrase, suggesting a temperate life-style. Both B025 and PSA have previously been reported to be temperate phages (30, 35). LP-030-2 was morphologically identified as belonging to morphospecies 2389 of the Siphoviridae (Fig. 2E), which is characterized by relatively flexible tails that sometimes show pointed tips (12).
Orthocluster IV, which is equally supported by a bootstrap value of 100, contains phage LP-030-3 (sequenced here) as well as the previously described temperate Listeria phages A118, A500, and A006. The genome size for phages in this orthocluster ranges from 38 to 41 kb (with average GC contents of 34.8 to 35.5%); annotation identified 62 to 73 predicted genes in these genomes. The average nucleotide identity of the phages in this group ranges from 84 to 99% across 2 to 69% of their genomes (see Fig. S2C in the supplemental material); A006 was the most divergent phage found within this group. Together, all four phages in orthocluster IV share 11 orthologous genes. Consistent with previously reported comparisons of A118, A500, and A006 (14), we identified a putative structural and DNA packaging module at the 5′ end of LP-030-3. Among the 17 putative genes in this putative structural and DNA packaging module, 12 were annotated as encoding specific functions (see Fig. S2C in the supplemental material). The nucleotide identity between LP-030-3 and A118 was greatest within a region immediately at the 3′ end of the integrase (the presence of which supports LP-030-3 as a putative temperate phage). This region (∼7 kb) encodes four proteins with putative regulatory functions (a C1-like repressor, a Cro-like repressor, an antirepressor, and a winged helix-turn-helix DNA-binding domain protein), four conserved domain-containing proteins (Ig-like domain, cobalt ABC transporter domain, zinc ribbon domain, and DUF955), as well as nine hypothetical proteins with no predicted functions. There was also considerable nucleotide identity between the putative structural and DNA packaging modules of A118 and LP-030-3. Morphologically, LP-030-3 was found to belong to morphospecies 2671 of the Siphoviridae (Fig. 2C), which is characterized by very long, rigid tails (36, 37) with transverse striations and spikes that sometimes appear as a six-pointed star (12).
Five Listeria phages belonging to the Siphoviridae family closely resemble P70 by nucleotide sequence and more distantly resemble Enterococcus phage VD13 genomically, yet they all share a nearly identical unique morphology.
Two orthoclusters identified here (orthoclusters V and VI of Listeria and Enterococcus phages, respectively) form a larger orthocluster, with a bootstrap support value of 100 (Fig. 1). All examined phages within this orthocluster are Siphoviridae of the rare B3 morphotype (33), with genome sizes of between 54 kb and 67 kb (Table 3). The B3 morphotype represents phages possessing long noncontractile tails and prolate heads with a length-to-width ratio of 2.7 to 5.5 (12). The tails of LP-026, LP-032, LP-037, LP-110, and VD13 have no collars and terminate in a bulb of indistinct fibers or spikes (Fig. 2F and G). While phages in orthocluster V do not show homology at the nucleotide level with those in orthocluster VI, there is considerable amino acid similarity and conserved gene synteny across parts of the phage chromosomes (see Fig. S3 in the supplemental material). Additionally, nine orthologous genes are found in all orthocluster V and VI phages but not in any of the other phages included in the ortholog analysis. These orthologous genes are putatively involved in structural (e.g., capsid morphogenesis protein and tail protein) and DNA replication (e.g., DNA primase and crossover junction endodeoxyribonuclease) functions.
Orthocluster V, which, like the others, is supported by a bootstrap value of 100, contains Listeria-infecting bacteriophages LP-026, LP-032, LP-037, LP-110, and LP-114 as well as previously described Listeria phage P70 (15). The phages in this orthocluster have genome lengths ranging from 65 to 67 kb (with an average GC content of 33.3 to 33.6%). Annotation identified 114 to 119 predicted genes in these genomes. The average nucleotide identity of the phages in this group ranges from 95 to 100% across 81 to 98% of their genomes. Despite being one of two phages with genome assemblies resulting in more than one contig, LP-032 is highly similar to LP-026 (100% nucleotide identity across ≥97% of their genomes), and both phages show high levels of gene synteny (see Fig. S3 in the supplemental material). Consistent with previous observations of P70, phages in the group were found to encode lysis functions (holin and endolysin) in a genome region between the small terminase subunit (5′ end of the genomes) (see Fig. S3 in the supplemental material) and a putative structural and DNA packaging module. Among the 17 genes in the putative structural and DNA packaging module, 10 were annotated as encoding specific functions, all of which are related to structure or DNA packaging. The phages also possess a putative DNA replication, modification, and recombination module located 3′ of the structural module; this module contains 8 to 9 genes that encode specific DNA-related functions (e.g., DNA primase, DNA polymerase, and crossover junction endodeoxyribonuclease). Interestingly, each orthocluster V phage contains 4 to 5 HNH homing endonucleases. Four of these endonucleases are found in all six phages; however, P70 and LP-114 both contain a fifth HNH homing endonuclease that presumably exists as an intron within the phage-encoded DNA polymerase. LP-114's DNA polymerase is interrupted at nucleotide position 1480 (of the 2,265-bp gene) by the putative intron; both the 5′ and 3′ fragments of the polymerase show homology at the nucleotide level (>96% identical) to the other orthocluster V DNA polymerases.
Orthocluster V phages are morphologically nearly identical to Enterococcus phage VD13 (Fig. 2F and G). The Listeria phages and VD13 have the same aspect and approximately the same dimensions (Table 2). The VD13 genome closely resembles those of two preliminarily described Enterococcus phages (38, 39). Together, with a bootstrap support value of 100, these three Enterococcus phages form orthocluster VI (Fig. 1). The phages in this orthocluster have genome sizes ranging from 54 to 59 kb (with average GC contents of 40.0 to 40.4%). The average nucleotide identity of the phages in this group ranges from 87 to 94% across 61 to 77% of their genomes. The putative structural module observed in orthocluster V phages is also present in orthocluster VI phages. Between one and five HNH homing endonucleases are found within orthocluster VI phages, one of which (located directly downstream of DNA polymerase) is found in VD13, BC-611, and all orthocluster V phages.
DISCUSSION
In this study, we sequenced and analyzed 14 Listeria phages and 1 Enterococcus phage to obtain information on genetic diversity, genome organization, gene functions, morphological characteristics of these phages, and their relatedness to previously sequenced firmicute-infecting phages. It appears that (i) Listeria phages can be grouped into clusters of lytic phages with limited genomic variation as well as into more diverse clusters of temperate phages, (ii) the classic morphology-based classification scheme is in concordance with genomic-based cluster analysis, and (iii) lytic Listeria and Enterococcus phages that share orthologous gene contents and morphologies represent a novel phage group that includes two orthoclusters. As publically available bacterial genomes now far outnumber available bacteriophage genomes (40), the contribution of these Listeria phage genomes expands our understanding of Listeria and Listeria phage diversity.
Listeria phages can be grouped into clusters of lytic phages with limited genomic variation as well as into more diverse groups of temperate phages.
Comparative genomics revealed five conserved orthoclusters of Listeria phages and one additional orthocluster of Enterococcus phages, which is closely related to Listeria phage orthocluster V. With the high level of similarity between previously reported complete phage genomes (13–15) and genome sequences reported here (including those with multiple contigs [LP-032 and LP-083-1]), it is extremely unlikely that assembly gaps resulted in a failure to identify genes affecting the orthoclustering of phages sequenced in this study. Two orthoclusters (orthoclusters III and IV) contain phages previously characterized as temperate phages, e.g., A118 and PSA (41, 42); as expected, genomes of these phages encode an integrase, which enables the phage chromosome to integrate into the host chromosome through site-specific recombination. Three Listeria phage orthoclusters (orthoclusters I, II, and V) contain phages previously reported to be obligate lytic phages, e.g., P100 and P35 (14, 29). The three putative lytic phage orthoclusters appear to be more highly conserved at the nucleotide level and exhibit greater gene synteny than the two putative temperate phage orthoclusters (orthoclusters III and IV). This finding is consistent with the reported relationships of Enterobacteriaceae-infecting phages, which were shown previously to form well-defined orthoclusters of lytic phages, while the temperate phages showed weaker relationships to one another (18). This could potentially be due to an increased likelihood of temperate phages undergoing recombination, a mechanism that is known to contribute to the diversification of phage populations (43–45) and is responsible for phages being described as “mosaic in nature” (46). Consistent with other studies that showed extensive mosaicism in temperate phages (47–49), our data set supports a model where recombination and mosaicism occur at a greater frequency in temperate phages than in lytic phages. This model is, however, challenged by the relatively diverse lytic ΦKZ-related phages of Pseudomonas, for which there is strong evidence for considerable recombination within the group (50); hence, future large-scale comparative genomic studies should further test this hypothesis across phage-host systems.
Our data set reported here also provides some initial insight into the global distribution of different firmicute-infecting phage groups. For example, phages isolated in North America (sequenced in this study) clustered with phages isolated in Europe (A511, P100, and P70) (15, 29, 30) and in East Asia (SAP6 and BC-611) (38, 39). Three orthoclusters (orthoclusters I, V, and VI, all lytic) each contained phages isolated on different continents. This finding of globally distributed related phages is consistent with previous studies of marine viruses (51), mycobacteriophages (52), and Enterobacteriaceae-infecting phages (18). The phages within each of the three putative obligate lytic orthoclusters are closely related and share significant gene synteny with one another, suggesting that they are not subject to frequent recombination with unrelated phages. This may have implications for phage-based applications, as phages from orthoclusters that show relative genomic stability may be less likely to aid in the horizontal transfer of resistance or virulence genes, a serious potential consequence of introducing phages into the environment (53).
The classic morphology-based classification scheme is in concordance with genomic-based cluster analysis.
Combined analysis of clustering based on orthologous gene content and associated morphological characterization data for the firmicute-infecting phages showed that the Myoviridae, Siphoviridae, and Podoviridae generally formed distinct clusters. The Myoviridae formed a single large cluster (with a few exceptions, as discussed below). Similarly, the few Podoviridae included in the genomic analysis in Fig. 1 formed a single cluster (although one podovirus, phiP68, clustered with a bootstrap support value of only 67). While the Siphoviridae did form a single large cluster, this cluster included a branch comprised solely of Podoviridae. Similarly to our findings, ortholog-based clustering of mycobacteriophages has also shown that phages representing Myoviridae and Siphoviridae clustered by morphological family (52). Despite the overall convergence of morphological family assignment and orthoclustering-based analyses, we also observed some potentially noteworthy exceptions. Specifically, two phages that were morphologically classified as Myoviridae clustered with Siphoviridae (i.e., B054 clusters with ΦEf11, and ΦAQ113 clusters with SPP1). Because of these inconsistencies, phages B054 and ΦAQ113 should be reexamined.
We also observed that orthoclusters within a family (e.g., within the Siphoviridae family) show distinct morphological features. For example, orthoclusters II, III, and IV have icosahedral heads, while orthoclusters V and VI have elongated heads. Orthocluster II phages also have considerably shorter tails than those of phages of the other Siphoviridae orthoclusters. Orthocluster V Listeria phages had a slightly longer head and tail than those of VD13 (orthocluster VI); these differences could be due to the use of different electron microscopes and calibration procedures but may alternatively represent a true difference, such as head elongation in orthocluster V phages by the addition of rows of capsomers. This was reported previously for Salmonella phage 7-11, which has elongated capsids and produces 11 head size classes, with each differing by about 11 nm and ranging from very long heads to isometric capsids (54). Morphological analyses also showed differences among phage isolates that grouped into the same orthocluster. For example, B025 has a considerably longer tail (252 nm) than those of other orthocluster III phages, PSA and LP-030-2 (180 nm and 160 nm, respectively). Consistent with this observation, B025 encodes a longer tape measure protein (∼1,600 amino acids) than those of PSA and LP-030-2 (∼1,000 amino acids); the link between tape measure protein length and tail length is well established in the literature (55–57). While these differences were observed within orthocluster III, the differences were further supported by the presence of well-defined subclusters within this orthocluster, which further indicate the heterologous nature of orthocluster III. Our data set thus not only shows that genomic analysis allows basic morphological group classification of firmicute-infecting phages but also shows how genomic data may allow prediction of specific morphological features. As large data sets of phage genomes become available, one can thus imagine an increased ability to predict phage morphology as long as high-quality electron microscopy data are available for appropriate reference phages.
Whole-genome sequencing and electron microscopy reveal a novel group of Listeria and Enterococcus phages.
In this study, we found an orthocluster (orthocluster V, as determined by the shared presence or absence of orthologous genes) of Listeria phages that clustered with an orthocluster (orthocluster VI) of Enterococcus phages. This greater orthocluster consists of putative lytic phages that are completely unlike the better-defined lytic firmicute-infecting Spounavirinae. These two phage orthoclusters (orthoclusters V and VI) share a unique morphology (B3), characterized by very long heads. However, VD13 produces malformations such as mottled heads and polyheads and particles with giant heads (17), neither of which are observed in Listeria phages. The six orthocluster V Listeria phages have medium-sized genomes (∼65 kb) and share a high level of sequence similarity to each other but share only a few genes with other Listeria phages (e.g., DNA polymerase and HNH homing endonuclease). They do, however, show homology at the amino acid level and share many orthologous genes with the three orthocluster VI E. faecalis phages; together, these two orthoclusters form a greater orthocluster and also show a conserved gene synteny. These two orthoclusters (orthoclusters V and VI) were found to share 9 orthologs not found in any of the other 46 phages included in this analysis. These unique orthologs encoded putative proteins with structural and DNA replication functions, further supporting this greater cluster as a biologically relevant group of phages. Orthoclusters V and VI are closely related, regardless of their host genera, much like all the members of the Twort-like Myoviridae and “phi29-like” Podoviridae.
Importantly, members of orthoclusters V and VI have been reported to be lytic phages (15, 17, 38, 39). They may thus prove useful for phage-based applications, e.g., for the control and detection of L. monocytogenes and as potential therapeutics for E. faecalis. Whereas most of these phages have shown narrow host ranges, orthocluster V phage LP-037 was found to lyse a majority of L. monocytogenes strains that were classified as “persistent” (repeatedly isolated over time) in a smoked fish processing facility (58). The availability of novel lytic phages also provides new opportunities for development of phage cocktails that contain several phages with distinct genomic contents (e.g., containing not only Twort-like viruses).
Supplementary Material
ACKNOWLEDGMENTS
We thank Roger Hendrix for granting us permission to use the unpublished genome of Bacillus phage G (GenBank accession no. JN638751.1). We also thank Andrew Kropinski for permission to use the unpublished genome sequence of Bacillus phage phi29 (accession no. NC_011048.1).
This research was supported in part by Cornell University Agricultural Experiment Station federal formula funds, project no. NYC-143445, received from the National Institute of Food and Agriculture (NIFA), U.S. Department of Agriculture.
Any opinions, findings, conclusions, or recommendations expressed in the publication are those of the authors and do not necessarily reflect the view of the National Institute of Food and Agriculture (NIFA) or the U.S. Department of Agriculture.
M.W. serves as a scientific advisor for and has a financial interest in Sample6, a company that is producing bacteriophage-based diagnostics for food-borne pathogens.
Footnotes
Published ahead of print 16 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00720-14.
REFERENCES
- 1.Hedberg C. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:840–842. 10.3201/eid0506.990624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farber JM, Peterkin PI. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batz MB, Hoffmann S, Morris JG. 2012. Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J. Food Prot. 75:1278–1291. 10.4315/0362-028X.JFP-11-418 [DOI] [PubMed] [Google Scholar]
- 4.Sulakvelidze A. 2013. Using lytic bacteriophages to eliminate or significantly reduce contamination of food by foodborne bacterial pathogens. J. Sci. Food Agric. 93:3137–3146. 10.1002/jsfa.6222 [DOI] [PubMed] [Google Scholar]
- 5.Schmelcher M, Loessner MJ. 2014. Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 4:e28137. 10.4161/bact.28137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udden SMN, Zahid MSH, Biswas K, Ahmad QS, Cravioto A, Nair GB, Mekalanos JJ, Faruque SM. 2008. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc. Natl. Acad. Sci. U. S. A. 105:11951–11956. 10.1073/pnas.0805560105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM. 2010. Satellite phage TLCφ enables toxigenic conversion by CTX phage through dif site alteration. Nature 467:982–985. 10.1038/nature09469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laanto E, Bamford JK, Laakso J, Sundberg L-R. 2012. Phage-driven loss of virulence in a fish pathogenic bacterium. PLoS One 7:e53157. 10.1371/journal.pone.0053157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd EF. 2012. Bacteriophage-encoded bacterial virulence factors and phage-pathogenicity island interactions. Adv. Virus Res. 82:91–118. 10.1016/B978-0-12-394621-8.00014-5 [DOI] [PubMed] [Google Scholar]
- 10.Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. 2012. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150:792–802. 10.1016/j.cell.2012.06.036 [DOI] [PubMed] [Google Scholar]
- 11.Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, Galagan JE, Birren BW, Ivy RA, Sun Q, Graves LM, Swaminathan B, Wiedmann M. 2008. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9:539. 10.1186/1471-2164-9-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ackermann H-W, DuBow MS. 1987. Viruses of prokaryotes. CRC Press, Boca Raton, FL [Google Scholar]
- 13.Klumpp J, Dorscht J, Lurz R, Bielmann R, Wieland M, Zimmer M, Calendar R, Loessner MJ. 2008. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of Gram-positive bacteria. J. Bacteriol. 190:5753–5765. 10.1128/JB.00461-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorscht J, Klumpp J, Bielmann R, Schmelcher M, Born Y, Zimmer M, Calendar R, Loessner MJ. 2009. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J. Bacteriol. 191:7206–7215. 10.1128/JB.01041-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmuki MM, Erne D, Loessner MJ, Klumpp J. 2012. Bacteriophage P70: unique morphology and unrelatedness to other Listeria bacteriophages. J. Virol. 86:13099–13102. 10.1128/JVI.02350-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vongkamjan K, Switt AM, den Bakker HC, Fortes ED, Wiedmann M. 2012. Silage collected from dairy farms harbors an abundance of listeriaphages with considerable host range and genome size diversity. Appl. Environ. Microbiol. 78:8666–8675. 10.1128/AEM.01859-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackermann H-W, Caprioli T, Kasatiya SS. 1975. A large new Streptococcus bacteriophage. Can. J. Microbiol. 21:571–574. 10.1139/m75-080 [DOI] [PubMed] [Google Scholar]
- 18.Moreno Switt AI, Orsi RH, den Bakker HC, Vongkamjan K, Altier C, Wiedmann M. 2013. Genomic characterization provides new insight into Salmonella phage diversity. BMC Genomics 14:481. 10.1186/1471-2164-14-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20.Zerbino DR. 2002. Using the Velvet de novo assembler for short-read sequencing technologies. John Wiley & Sons, Inc, Hoboken, NJ: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19:455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 24.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120. 10.1093/nar/gki442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106:19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 29.Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. 2005. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 43:301–312. 10.1016/j.yrtph.2005.08.005 [DOI] [PubMed] [Google Scholar]
- 30.Zink R, Loessner MJ. 1992. Classification of virulent and temperate bacteriophages of Listeria spp. on the basis of morphology and protein analysis. Appl. Environ. Microbiol. 58:296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klumpp J, Lavigne R, Loessner MJ, Ackermann H-W. 2010. The SPO1-related bacteriophages. Arch. Virol. 155:1547–1561. 10.1007/s00705-010-0783-0 [DOI] [PubMed] [Google Scholar]
- 32.Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann H-W, Kropinski AM. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9:224. 10.1186/1471-2180-9-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ackermann HW, Eisenstark A. 1974. The present state of phage taxonomy. Intervirology 3:201–219. 10.1159/000149758 [DOI] [PubMed] [Google Scholar]
- 34.Hodgson DA. 2000. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 35:312–323. 10.1046/j.1365-2958.2000.01643.x [DOI] [PubMed] [Google Scholar]
- 35.Loessner MJ, Estela LA, Zink R, Scherer S. 1994. Taxonomical classification of 20 newly isolated Listeria bacteriophages by electron microscopy and protein analysis. Intervirology 37:31–35 [DOI] [PubMed] [Google Scholar]
- 36.Ackermann H-W, Audurier A, Rocourt J. 1981. Morphologie de bactériophages de Listeria monocytogenes. Ann. Virol. 132:371–382 [Google Scholar]
- 37.Ortel S, Ackermann H-W. 1985. Morphologie von neuen Listeria-Phagen. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 260:423–427. 10.1016/S0176-6724(85)80062-6 [DOI] [PubMed] [Google Scholar]
- 38.Lee YD, Park JH. 2012. Complete genome sequence of enterococcal bacteriophage SAP6. J. Virol. 86:5402–5403. 10.1128/JVI.00321-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horiuchi T, Sakka M, Hayashi A, Shimada T, Kimura T, Sakka K. 2012. Complete genome sequence of bacteriophage BC-611 specifically infecting Enterococcus faecalis strain NP-10011. J. Virol. 86:9538–9539. 10.1128/JVI.01424-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bibby K. 2014. Improved bacteriophage genome data is necessary for integrating viral and bacterial ecology. Microb. Ecol. 67:242–244. 10.1007/s00248-013-0325-x [DOI] [PubMed] [Google Scholar]
- 41.Loessner MJ, Inman RB, Lauer P, Calendar R. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35:324–340. 10.1046/j.1365-2958.2000.01720.x [DOI] [PubMed] [Google Scholar]
- 42.Zimmer M, Sattelberger E, Inman RB, Calendar R, Loessner MJ. 2003. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed + 1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 50:303–317. 10.1046/j.1365-2958.2003.03684.x [DOI] [PubMed] [Google Scholar]
- 43.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Hendrix RW, Hatfull GF. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–182. 10.1016/S0092-8674(03)00233-2 [DOI] [PubMed] [Google Scholar]
- 44.Mmolawa PT, Schmieger H, Heuzenroeder MW. 2003. Bacteriophage ST64B, a genetic mosaic of genes from diverse sources isolated from Salmonella enterica serovar Typhimurium DT 64. J. Bacteriol. 185:6481–6485. 10.1128/JB.185.21.6481-6485.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casjens SR, Thuman-Commike PA. 2011. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 411:393–415. 10.1016/j.virol.2010.12.046 [DOI] [PubMed] [Google Scholar]
- 46.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U. S. A. 96:2192–2197. 10.1073/pnas.96.5.2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Botstein D, Herskowitz I. 1974. Properties of hybrids between Salmonella phage P22 and coliphage lambda. Nature 251:584–589. 10.1038/251584a0 [DOI] [PubMed] [Google Scholar]
- 48.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. 10.1006/jmbi.2000.3729 [DOI] [PubMed] [Google Scholar]
- 49.Tang F, Bossers A, Harders F, Lu C, Smith H. 2013. Comparative genomic analysis of twelve Streptococcus suis (pro)phages. Genomics 101:336–344. 10.1016/j.ygeno.2013.04.005 [DOI] [PubMed] [Google Scholar]
- 50.Cornelissen A, Hardies SC, Shaburova OV, Krylov VN, Mattheus W, Kropinski AM, Lavigne R. 2012. Complete genome sequence of the giant virus OBP and comparative genome analysis of the diverse ΦKZ-related phages. J. Virol. 86:1844–1852. 10.1128/JVI.06330-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. 10.1371/journal.pbio.0040368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko C-C, Weber RJ, Patel MC, Germane KL, Edgar RH, Hoyte NN, Bowman CA, Tantoco AT, Paladin EC, Myers MS, Smith AL, Grace MS, Pham TT, O'Brien MB, Vogelsberger AM, Hryckowian AJ, Wynalek JL, Donis-Keller H, Bogel MW, Peebles CL, Cresawn SG, Hendrix RW. 2010. Comparative genomic analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J. Mol. Biol. 397:119–143. 10.1016/j.jmb.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meaden S, Koskella B. 2013. Exploring the risks of phage application in the environment. Front. Microbiol. 4:358. 10.3389/fmicb.2013.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moazamie N, Ackermann H-W, Murthy MR. 1979. Characterization of two Salmonella Newport bacteriophages. Can. J. Microbiol. 25:1063–1072. 10.1139/m79-163 [DOI] [PubMed] [Google Scholar]
- 55.Katsura I, Hendrix RW. 1984. Length determination in bacteriophage lambda tails. Cell 39:691–698. 10.1016/0092-8674(84)90476-8 [DOI] [PubMed] [Google Scholar]
- 56.Katsura I. 1987. Determination of bacteriophage λ tail length by a protein ruler. Nature 327:73–75. 10.1038/327073a0 [DOI] [PubMed] [Google Scholar]
- 57.Abuladze NK, Gingery M, Tsai J, Eiserling FA. 1994. Tail length determination in bacteriophage T4. Virology 199:301–310. 10.1006/viro.1994.1128 [DOI] [PubMed] [Google Scholar]
- 58.Vongkamjan K, Roof S, Stasiewicz MJ, Wiedmann M. 2013. Persistent Listeria monocytogenes subtypes isolated from a smoked fish processing facility included both phage susceptible and resistant isolates. Food Microbiol. 35:38–48. 10.1016/j.fm.2013.02.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.