Abstract
Next-generation sequencing (NGS) opens up exciting possibilities for improving our knowledge of environmental microbial diversity, allowing rapid and cost-effective identification of both cultivated and uncultivated microorganisms. However, library preparation, sequencing, and analysis of the results can provide inaccurate representations of the studied community compositions. Therefore, all these steps need to be taken into account carefully. Here we evaluated the effects of DNA extraction methods, targeted 16S rRNA hypervariable regions, and sample origins on the diverse microbes detected by 454 pyrosequencing in marine cold seep and hydrothermal vent sediments. To assign the reads with enough taxonomic precision, we built a database with about 2,500 sequences from Archaea and Bacteria from deep-sea marine sediments, affiliated according to reference publications in the field. Thanks to statistical and diversity analyses as well as inference of operational taxonomic unit (OTU) networks, we show that (i) while DNA extraction methods do not seem to affect the results for some samples, they can lead to dramatic changes for others; and (ii) the choice of amplification and sequencing primers also considerably affects the microbial community detected in the samples. Thereby, very different proportions of pyrosequencing reads were obtained for some microbial lineages, such as the archaeal ANME-1, ANME-2c, and MBG-D and deltaproteobacterial subgroups. This work clearly indicates that the results from sequencing-based analyses, such as pyrosequencing, should be interpreted very carefully. Therefore, the combination of NGS with complementary approaches, such as fluorescence in situ hybridization (FISH)/catalyzed reporter deposition (CARD)-FISH or quantitative PCR (Q-PCR), would be desirable to gain a more comprehensive picture of environmental microbial communities.
INTRODUCTION
Elucidating how biodiversity is distributed and determining the underlying mechanisms that drive these patterns are central questions in ecology. However, the majority of microorganisms that occur in natural environments cannot be cultured or isolated using traditional cultivation techniques (1–3), which limits our knowledge of environmental microbial diversity. Culture-independent studies, such as molecular characterization, have provided new insights into our understanding of microbial community structures, especially for populations inhabiting extreme environments, such as marine cold seeps and hydrothermal vents, whose growth conditions may be difficult to mimic in the laboratory (4). Today, high-throughput sequencing allows the characterization of microbial communities with unprecedented levels of coverage (5–9). While molecular analyses provide a less biased picture of environmental microbial communities than culture-based methods, each step of the analysis needs to be taken into account. Indeed, the DNA extraction method, PCR amplification, or data analysis can all lead to distortions of the compositions of analyzed samples (10–18).
Extraction is often pointed out as a key step in obtaining DNAs from all microorganisms present in one studied environment (10, 11, 13, 14, 19–22). However, DNA extraction from sediment samples may be difficult. The presence of complex organic matter and enzymatic inhibitors, such as humic substances, can interfere with enzymatic reactions (23–25). Additionally, some microorganism cells might be resistant to lysis (e.g., resting cells, spores, and cysts), especially in extreme environments (26). Indeed, microbial cell envelopes vary in structure between and within microbial lineages, and some cells are more easily disrupted than others (13, 27, 28). Thus, optimization of DNA extraction methods is often necessary to access a correct representation of whole microbial communities.
The DNA amplification step required for most sequencing methods is also critical. Sequencing of the 16S rRNA gene has been used widely to assess the phylogenetic diversity in environmental samples (4, 29–31). This gene is composed of alternating conserved and hypervariable regions (32). The presence of conserved regions enables PCR amplifications using “universal” primers supposed to bind even to DNAs of unknown organisms (32). DNA primers can be designed to be complementary to two distant conserved regions. PCR can then be done by using forward and reverse primers leading to a PCR fragment including both conserved and variable regions. Amplified products are then used to evaluate microbial diversity and to study taxonomic affiliations and evolutionary relationships between organisms (32). However, 16S rRNA gene hypervariable regions exhibit different degrees of variability according to the studied phylogenetic groups; therefore, no unique hypervariable region allows discrimination between all known microbial lineages (33–35). Thus, working on partial 16S rRNA gene sequences may bias analyses of diversity and species richness recovery (33, 35, 36). Compared to other high-throughput sequencing technologies, pyrosequencing (Roche 454) is an attractive method for 16S rRNA gene sequencing, as relatively long reads are obtained (14, 37), though it does not allow access to the full length of the gene (i.e., about 1,550 bp). Therefore, in pyrosequencing studies, the choice of primers (14) and sequenced regions may significantly affect the estimates of operational taxonomic unit (OTU) richness and microbial diversity analysis (15, 33, 38).
In this study, we analyzed the effects of two major steps of microbial diversity characterization using the 16S rRNA gene. Three physicochemically contrasting samples of cold seep and hydrothermal vent sediments from the Guaymas Basin were recovered. These environments are known to contain important amounts of humic substances (39) that could impair subsequent molecular analyses (12, 23–25). Two different extraction methods and two primer sets targeting different hypervariable regions of the 16S rRNA gene for Bacteria, and two others for Archaea, were tested on each sample. Amplicons were sequenced using 454 pyrosequencing. Impacts of experimental designs were evaluated by comparing DNA extraction yields, OTU richness, and taxonomic diversity.
MATERIALS AND METHODS
Sample description and handling.
Sediment samples were collected from the Guaymas Basin by use of push cores operated by the manned submersible Nautile during the “BIG” cruise (June 2010). The target sites included the “Vasconcelos” site, in a cold seep area from the Sonora Margin (marker BIG18; 27°35.5770N, 111°28.9840W; 1,574-m depth; dive 1758-14 core CT8) (40, 41), and the “MegaMat” site, in a hydrothermal vent area from the Southern Trough (marker M27; 27°00.4451N, 111°24.5298W; 2,000-m depth; dive 1766-22 core CT3). At marker BIG18 (Vasconcelos site; cold seeps), the sediment surface was colonized by abundant populations of gray polychaetes and gastropods surrounding a large white microbial mat (40). At marker M27 (MegaMat site; hydrothermal vents), the sediment surface was covered with white microbial mats near a hydrothermal mound overgrown with Riftia clusters. After recovery on board, sediment cores were immediately transferred to a cold room (4°C). Cores were then subsampled aseptically in 2-cm-thick layers that were frozen at −80°C until required for molecular analysis.
Two different sections from the cold seep core (surface layer [0 to 2 centimeters below the seafloor {cmbsf}; called SC1] and bottom layer [10 to 12 cmbsf; called SC6]; dive 1758-14 core CT8) and the surface layer from the hydrothermal vent core (0 to 2 cmbsf; called SH1; dive 1766-22 core CT3) were studied.
DNA extraction methods.
We tested two of the most commonly used protocols to extract DNAs from deep marine sediment samples (42): the method proposed by Zhou et al. (12) (referred to here as “Zhou's method”) and a manufactured bead-beating kit. DNA extractions were carried out independently on each sample (SC1, SC6, and SH1), as follows. DNAs from three replicates of 2.5 g per sample were extracted using Zhou's method (12), with some modifications. Cycles of freeze-thawing in a high-salt extraction buffer were followed by chemical lysis at 65°C in the presence of sodium dodecyl sulfate (SDS), cetyltrimethylammonium bromide (CTAB), and proteinase K. Since lysis efficiencies vary between and within microbial groups (13, 28), the freeze-thawing step was repeated 10 times in order to increase the diversity of extracted lineages. Two-milliliter samples of this mix (total volume, 8 ml) were taken after 3, 5, 7, and 10 freeze-thaw cycles in order to protect DNAs released from easily disrupted cells and then were pooled before the chemical lysis step. This method allowed DNA extraction with minimal shearing, which might prevent the formation of chimeras (12). Moreover, CTAB has been used in previous studies to complex and remove contaminants from DNA (12).
In parallel, DNAs from four replicates of 0.6 g per sample were extracted by a bead-beating method. This method is based on a commercially available kit, FastDNA Spin for Soil (MP Biomedicals, Santa Ana, CA), that was modified following the method of Webster et al. (10). The FastDNA Spin kit is based on a mechanical lysis with ceramic and silica beads in a bead beater, which theoretically allows a more efficient lysis of all microorganisms, including bacterial spores, endospores, and Gram-positive bacteria (10, 11). Furthermore, this method uses silica columns to retain high-molecular-weight DNA while removing inhibitors, such as humic acids, salts, and proteins.
The quantities and purities of the extracted DNAs from all replicates were evaluated by optical density measurement using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE). Results were statistically compared via Student t tests and analysis of variance (ANOVA), using R software (R Development Core Team, 2011). DNA extract replicates were then pooled for each extraction method and purified using a Wizard DNA cleanup kit (Promega, Madison, WI) following the manufacturer's instructions. Purified DNA extracts were stored at −20°C until PCR amplification.
PCR amplification and pyrosequencing.
The 16S rRNA gene was amplified using four different primer sets: two targeting Bacteria and two others for Archaea (Table 1).
TABLE 1.
Sets of primers used for PCR amplifications and in silico PCR performances of the primer pairs
| Domain | Primer | Target region | Target positiona | Primer sequence (5′–3′) | Amplicon length (bp) | In silico performanceb (%) |
|---|---|---|---|---|---|---|
| Bacteria | Bact1F | V4 | 514 | GTGCCAGCAGCCGCGGTAATA | 275 | 87 |
| Bact1R | V4 | 789 | GCGTGGACTACCAGGGTATCT | |||
| Bact2F | V5–V6 | 778 | CAAACAGGATTAGATACCCTG | 304 | 80.5 | |
| Bact2R | V5–V6 | 1,082 | CGTTRCGGGACTTAACCCAACA | |||
| Archaea | Arch1F | V2–V3 | 155 | CGGRAAACTGGGGATAAT | 360 | 83.6 |
| Arch1R | V2–V3 | 515 | TRTTACCGCGGCGGCTGBCA | |||
| Arch2F | V5–V6 | 784 | GGATTAGATACCCSGGTAGTC | 313 | 64 | |
| Arch2R | V5–V6 | 1,097 | GYGGGTCTCGCTCGTTRCC |
According to Escherichia coli 16S rRNA gene numbering.
With two mismatches allowed compared to the SILVA databases (release 117), using TestPrime (107).
The primer sets for Bacteria targeted the V4 (Bact1F-Bact1R) and V5–V6 (Bact2F-Bact2R) hypervariable regions (Table 1), while the primer sets for Archaea targeted the V2–V3 (Arch1F-Arch1R) and V5–V6 (Arch2F-Arch2R) hypervariable regions (Table 1). To allow multiplexing, forward primers were fused to 6 different tags (10 nucleotides [nt]) and the 454 GS-FLX sequencing primer A (5′-CGTATCGCCTCCCTCGCGCCATCAG-Tag-PrimerF-3′). The reverse primers were fused to the 454 GS-FLX sequencing primer B (5′-CCTATCCCTGTGTGCCTTGGCAGTC-PrimerR-3′). Primer design and DNA amplifications were performed by Genoscreen (Lille, France). PCR amplifications were performed using the following reagents in a 25-μl reaction volume: 10× DNA polymerase buffer (Applied Biosystems), 1.5 mM MgCl2 (Applied Biosystems), a 0.24 mM concentration of each deoxynucleoside triphosphate (dNTP; Roche), 0.4 mM (each) primers, and 1 U of Taq DNA polymerase (Applied Biosystems). The PCR program involved an initial denaturation step at 95°C for 10 min, followed by 25 cycles for Bacteria and 40 cycles for Archaea, at 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 10 min.
Amplicons were purified, and their concentrations and sizes were analyzed using DNA 1000 chips on an Agilent 2100 bioanalyzer (Agilent Technologies, Waldbronn, Germany). Amplicon products were mixed at an equimolar ratio (108 molecules per microliter for each amplicon) and then sequenced on a 454 Life Sciences GS-FLX genome sequencer (Roche Diagnostics, Indianapolis, IN). Quantification, purification, and sequencing were performed by Genoscreen (Lille, France). Two replicates from the hydrothermal vent sample extracted via Zhou's method and two others for the kit method were amplified, sequenced, and analyzed independently to check for reproducibility.
Read filtering.
To minimize the effects of random sequencing errors (43, 44), pyrosequencing reads were filtered using the Mothur pipeline (45). We removed sequences (i) that were shorter than 200 bp, (ii) containing homopolymers longer than 8 bp, (iii) that aligned with the incorrect region of the 16S rRNA gene, (iv) identified as chimeras by the Uchime algorithm (45–47), and (v) affiliated with Archaea when Bacteria was targeted, and vice versa. Results were compared (Student t test and ANOVA) according to sample origin, targeted region, and extraction method, using R.
In order to minimize the impact of larger numbers of sequences per sample, all the following analyses were performed on the same number of sequences for the bacterial and archaeal regions.
OTU analysis.
Numbers of OTUs and beta diversity indexes (Yue and Clayton, Bray-Curtis, and Morisita-Horn indexes) were determined using the Mothur pipeline (45) at two different levels of dissimilarity (3% and 10%). Results were compared (Student t test and ANOVA) according to sample origin, targeted region, and extraction method, using R. OTU networks were constructed at 10% dissimilarity by using the QIIME pipeline (48) and Cytoscape software (49).
Taxonomic assignment.
Effects of DNA extraction methods and targeted 16S rRNA gene regions were also evaluated using taxonomic assignment of sequences by use of BLAST algorithms. The use of international databases, such as the EMBL, DDBJ, GenBank, and RFAM databases, and their associated taxonomies did not allow assignment at a sufficient level of precision to allow pertinent comparisons between our samples. Indeed, taxonomies used in these databases are sometimes inaccurate or not updated, and levels of classification are often too high to be informative (e.g., “uncultured archaeon”). Therefore, we designed a reference database for the purpose of this study, consisting of 786 archaeal and 1,677 bacterial sequences from deep-sea marine sediments, collected from and affiliated according to reference publications (1, 3, 40, 41, 50–74; L. Durand, M. Roumagnac, V. Cueff-Gauchard, C. Jan, M. Guri, C. Tessier, M. Haond, P. Crassous, M. Zbinden, S. Arnaud-Haond, and M.-A. Cambon Bonavita, unpublished data). Sequences were downloaded from the NCBI website and aligned with MAFFT 6.903 (75). The consistency of taxonomic affiliations was checked by phylogenetic reconstructions. Phylogenetic trees were estimated using distance (neighbor joining) and maximum likelihood (ML) methods. Pairwise nucleotide sequence divergences were calculated using the Kimura 2-parameter model of substitution (76), and neighbor-joining trees (77) were reconstructed using MEGA 4.0.2 (78). The robustness of topologies was assessed by bootstrap procedures, using 1,000 replicates (79). ML analyses were performed using MPI-parallelized RAxML 7.2.8 (80), using a GTR model with among-site rate variation modeled by a discrete gamma approximation with four categories. GTRCAT approximation of models was used for ML bootstrapping (1,000 replicates). ML analyses were conducted on the CIPRES Science Gateway (81). When groups were recovered on the two phylogenetic trees, corresponding names provided in reference publications were considered valid and kept in our reference database. When names did not yet exist in the NCBI database, new taxonomic IDs were created. When groups appeared to be para- or polyphyletic, sequences were affiliated in our database to the first higher level of classification that appeared to be monophyletic. The full list of sequences and associated details (NCBI accession numbers, reference publications, associated affiliations, and affiliations used in our database) is available in Table S2 in the supplemental material. The custom database and its associated taxonomy were implemented in Koriblast software (Korilog, Questembert, France) and can be downloaded from an FTP server (http://ftp.genostar.com/ifremer/Bacteries_Archees_genostar.fasta). The pyrosequencing reads were searched against this reference database for taxonomic assignment (BLAST was performed in Koriblast, in which sequences are assigned to the first BLAST hit). Since errors in taxonomic assignments may be due to the restricted lengths of sequences, correct taxonomic assignments of short sequences obtained with the pyrosequencing primers used in this study were theoretically validated by a congruent BLAST search of our sequence database with sequences cut to the amplified fragment length against the original database, using Koriblast software.
Microbial diversities were then statistically analyzed using Primer-E software (82). Ordination by multidimensional scaling, derived from the Euclidean distance between samples, was used to visualize diversity (83).
Nucleotide sequence accession number.
The raw sequencing data have been submitted to the NCBI database under SRA accession number SRP038099 (details are given in Table S1 in the supplemental material).
RESULTS
Extraction yields and DNA quality.
The DNA quantities and qualities recovered varied according to samples and extraction methods (see Table S3 in the supplemental material).
Quantities of DNA obtained by Zhou's method were significantly different between samples (ANOVA; P < 0.0001), while the quantities obtained with the commercial kit seemed steadier. Differences between DNA quantities obtained using either Zhou's method or the kit method were significant only for samples from the cold seep surface (SC1) (ANOVA; P < 0.0001). The highest yields of DNA were recovered from cold seep surface samples extracted by Zhou's method.
DNA qualities revealed by the 260/280 and 260/230 spectrophotometric ratios showed significant differences between samples (ANOVA; P < 0.01), except for the 260/230 ratio obtained with the commercial kit (see Table S3 in the supplemental material). For 260/280 ratios, Zhou's method gave better results than the commercial kit for the cold seep surface samples (ANOVA; P < 0.001). However, the commercial kit performed better than Zhou's method with other samples (ANOVA; P < 0.03 for cold seep sample SC6 and hydrothermal vent sample SH1). For 260/230 ratios, Zhou's method yielded higher ratios than those with the commercial kit for all samples (ANOVA; P < 0.001).
Numbers of reads and sequence filtering.
Overall, no significant differences were observed between the numbers of reads obtained from the different extraction methods and from the different samples (see Table S4 in the supplemental material). However, the numbers of reads obtained for each amplified region were significantly different (ANOVA; P < 0.0001).
During the filtering step, 17.36% to 48.78% of reads were removed, depending on sample origin and amplified region (see Table S4 in the supplemental material). No significant difference was detected between proportions of remaining sequences obtained with Zhou's method and the kit extraction method. However, significant differences were observed between proportions of remaining sequences among the different samples and the different regions (ANOVA; P < 0.03).
To determine the filtering step when sequences were removed, we compared (i) the numbers of detected chimeras between samples and (ii) the numbers of sequences belonging to kingdoms that were not supposed to be amplified by our kingdom-specific primer sets.
Sequences identified as chimeras represented only a small part of the reads (see Table S4 in the supplemental material). No significant difference was observed between proportions of detected chimeras obtained with Zhou's method and the kit extraction method. However, the proportions of chimeras were globally significantly different between samples for the Arch2 region and between the amplified regions (ANOVA; P < 0.02). The largest proportion of chimeras was observed for the Arch2 region amplified from the hydrothermal vent samples (average of 4.38%).
Primer sets were designed to amplify specifically Bacteria or Archaea. However, with the exception of the Bact2 primer set, some sequences affiliated with Archaea were also detected with the Bact1 primer set and some sequences affiliated with Bacteria were also detected with the archaeal primer sets (see Table S4). No significant difference was pointed out between proportions of removed sequences due to a lack of specificity when DNAs were extracted using Zhou's method or the kit method. In contrast, proportions of removed sequences were significantly different between samples for the Arch1 and Arch2 regions (ANOVA; P < 0.001). Likewise, significant differences were identified depending on the amplified region (ANOVA; P < 0.01).
OTU analysis.
The number of OTUs was calculated for each sample at two different levels of dissimilarity, i.e., 3% and 10%, corresponding to the phylogenetic distances conventionally used to define species (3%) and families (10%) based on full-length 16S rRNA gene sequences (29) (Table 2). Whatever the amplified region, no significant difference was observed between the numbers of OTUs obtained when DNAs were extracted using Zhou's method or the kit method. However, the numbers of OTUs were globally significantly different between the sequenced regions for the archaeal data set (Student t test; P < 0.03). Indeed, fewer OTUs were obtained for the Arch2 region than for the Arch1 region for the cold seep SC6 sample (141 and 299, respectively, on average, at the 3% level and 26 and 125, respectively, on average, at the 10% level). For the hydrothermal vent sample, fewer OTUs were found for the Arch1 region than for the Arch2 region at 3% dissimilarity (183 and 224, respectively), whereas the opposite was true at 10% dissimilarity (95 and 78, respectively). For the bacterial data set, the numbers of OTUs at 10% dissimilarity were significantly higher for the Bact1 region (86, 131, and 124 for cold seep SC1, cold seep SC6, and hydrothermal vent SH1 samples, respectively) than for the Bact2 region (65, 92, and 100 for cold seep SC1, cold seep SC6, and hydrothermal vent SH1 samples, respectively) (Student t test; P < 0.02).
TABLE 2.
Numbers of OTUs for each sample, according to primer set and DNA extraction method, determined at two levels of dissimilarity (3% and 10%)
| Sample | Extraction methoda | No. of OTUs with primer set (3%) |
No. of OTUs with primer set (10%) |
||||||
|---|---|---|---|---|---|---|---|---|---|
|
Bacteria |
Archaea |
Bacteria |
Archaea |
||||||
| Bact1 | Bact2 | Arch1 | Arch2 | Bact1 | Bact2 | Arch1 | Arch2 | ||
| Dive 1758 CT8 SC1 (cold seep surface layer; gray mat) | Zhou | 171 | 182 | 202 | 165 | 85 | 67 | 113 | 32 |
| Kit | 185 | 172 | 175 | 113 | 87 | 62 | 76 | 26 | |
| Dive 1758 CT8 SC6 (cold seep bottom layer; gray mat) | Zhou | 268 | 278 | 296 | 136 | 134 | 96 | 132 | 21 |
| Kit | 243 | 286 | 302 | 145 | 128 | 88 | 118 | 31 | |
| Dive 1766 CT3 SH1 (hydrothermal vent surface layer; white mat) | Zhou (replicate 1) | 243 | 269 | 199 | 257 | 137 | 104 | 94 | 91 |
| Kit (replicate 1) | 208 | 240 | 181 | 222 | 110 | 98 | 89 | 78 | |
| Zhou (replicate 2) | 202 | 240 | 179 | 199 | 119 | 94 | 106 | 69 | |
| Kit (replicate 2) | 231 | 246 | 173 | 219 | 130 | 102 | 91 | 73 | |
Zhou, method of Zhou et al. (12).
Yue and Clayton (84), Bray-Curtis (85), and Morisita-Horn (86, 87) diversity indexes (see Table S5 in the supplemental material) and OTU networks (Fig. 1) were used to compare OTU compositions between samples, replicates, extraction methods, and amplified regions, at a level of dissimilarity of 10%.
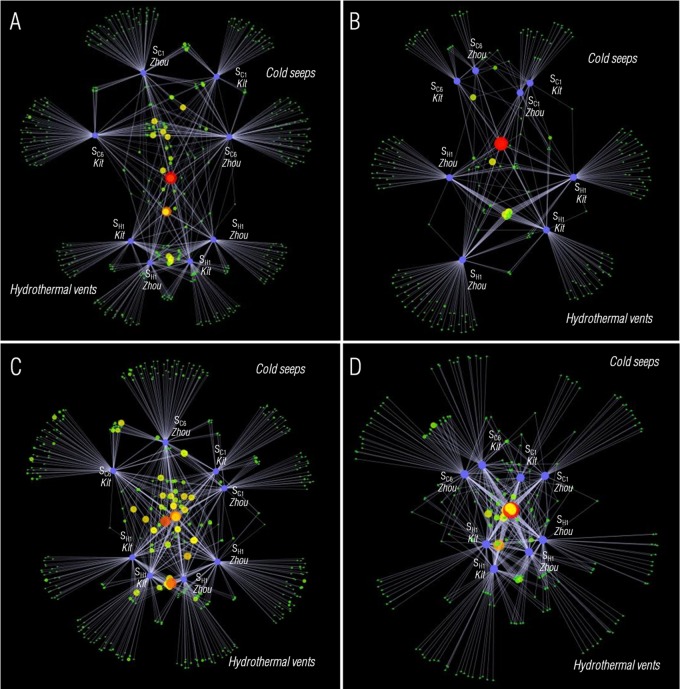
FIG 1.
Network analysis of microbial communities according to the sample origin and DNA extraction method used. OTUs were defined at a 10% dissimilarity. Sample nodes are denoted by purple nodes, while other nodes represent individual OTUs. OTUs with the largest numbers of sequences are represented by large red dots (1,046 sequences for Bact1, 3,560 for Bact2, 3,438 for Arch1, and 4,992 for Arch2), while OTUs including few sequences are represented by small green dots (1 sequence) and intermediates. Shared OTUs are connected by lines. (A) Arch1 region; (B) Arch2 region; (C) Bact1 region; (D) Bact2 region.
The beta diversity indexes obtained showed that OTU compositions between samples were not similar, though some of them were very close (see Table S5). Regarding the Bacteria, the highest similarity indexes were obtained between samples from the same origin. The two extraction methods did not seem to affect the similarity indexes. Furthermore, community structures obtained with the Bact2 primer set appeared to be more similar between all samples than those obtained with the Bact1 primer set (Student t test; P < 0.01 for the three indexes). Regarding the Archaea, the highest similarity indexes were obtained between the samples from the same origin in the case of the Arch2 region. However, community structures appeared to be different depending on the extraction method used for the hydrothermal vent sample in the case of the Arch1 region (Student t test; P < 0.001 for the three indexes).
On OTU networks, both OTUs and samples are represented by nodes. In Fig. 1, samples are represented by purple dots, while other nodes represent OTUs. OTUs with the largest numbers of sequences are represented by large red dots (1,046 sequences for Bact1, 3,560 for Bact2, 3,438 for Arch1, and 4,992 for Arch2), while OTUs including few sequences are represented by small green dots (1 sequence) and intermediates. When a sequence is affiliated with an OTU in a sample, the OTU is linked by a line to the corresponding sample node. The OTU networks show that OTUs represented by many sequences were shared between all samples (large red dots in Fig. 1), while nonshared OTUs contained only a few sequences (small green dots in Fig. 1). The more scattered Bact1 network shows that OTUs were more similar between samples when the Bact2 region was amplified.
Taxonomic affiliations.
After filtering of the reads, the taxonomic affiliations of the remaining sequences highlighted different community structures according to sampling sites, targeted regions, and extraction methods.
Similarities between samples are represented graphically for Bacteria (Fig. 2) and Archaea (Fig. 3), using Euclidean distances, and proportions of the different microbial groups are reported in Tables 3 and 4. If no bias existed between extraction methods and the primer set used, we would expect only three different microbial community structures, for the three physicochemically contrasting samples used.
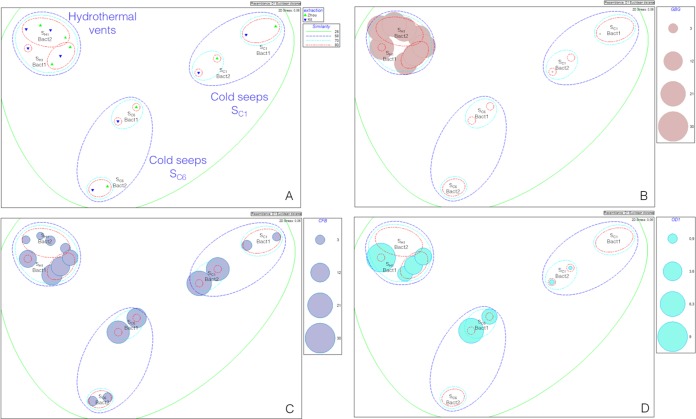
FIG 2.
Ordination by multidimensional scaling (MDS), derived from the Euclidean distances between samples, according to DNA extraction method and the primer set used for Bacteria. Dotted lines indicate percent similarities of the community compositions between grouped samples (green, 25%; blue, 58%; turquoise, 70%; red, 80%). (B to D) Sizes of colored circles are proportional to microbial group relative abundances in each sample (Table 3) (for the GBG [B], CFB [C], and OD1 [D] groups).
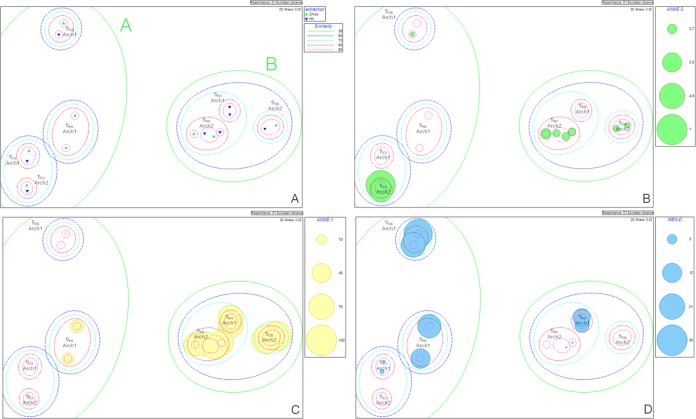
FIG 3.
Ordination by MDS, derived from the Euclidean distances between samples, according to DNA extraction method and the primer set used for Archaea. Dotted lines indicate percent similarities of the community compositions between grouped samples (green, 35%; blue, 60%; turquoise, 70%; red, 80%; pink, 90%). (B to D) Sizes of colored circles are proportional to microbial group relative abundances in each sample (Table 4) (for the ANME-3 [B], ANME-1 [C], and MBG-D [D] groups).
TABLE 3.
Taxonomic affiliations of pyrosequencing reads after filteringa
| Taxonomic group | % Contribution |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dive 1758 CT8 SC1 (cold seep surface layer; gray mat) |
Dive 1758 CT8 SC6 (cold seep bottom layer; gray mat) |
Dive 1766 CT3 SH1 (hydrothermal vent surface layer; white mat) |
||||||||||||||
| Zhou |
Kit |
Zhou |
Kit |
Zhou replicate 1 |
Kit replicate 1 |
Zhou vreplicate 2 |
Kit replicate 2 |
|||||||||
| Bact1 | Bact2 | Bact1 | Bact2 | Bact1 | Bact2 | Bact1 | Bact2 | Bact1 | Bact2 | Bact1 | Bact2 | Bact1 | Bact2 | Bact1 | Bact2 | |
| Acidobacteria | 0.13 | 0.72 | 0.07 | 0.59 | 1.12 | 0.72 | 0.26 | 0.79 | 2.83 | 3.42 | 3.95 | 2.96 | 2.30 | 3.29 | 3.36 | 3.55 |
| Actinobacteria | 3.75 | 2.57 | 5.53 | 5.13 | 0.53 | 1.84 | 1.45 | 3.16 | 0.33 | 0.86 | 0.07 | 0.26 | 0.46 | 0.72 | 0.86 | 1.18 |
| Caldithrix | 0.39 | 0.99 | 0.99 | 2.56 | 0.07 | 2.50 | 0.07 | 4.47 | 0.79 | 4.67 | 0.99 | 5.40 | 0.07 | 4.01 | ||
| CFB | 18.03 | 2.37 | 20.66 | 3.42 | 12.96 | 3.09 | 17.30 | 3.16 | 14.61 | 3.36 | 10.26 | 3.16 | 9.55 | 1.84 | 12.24 | 2.17 |
| Chloroflexi | 0.46 | 0.20 | 0.46 | 3.55 | 3.16 | 4.34 | 3.55 | 2.96 | 1.58 | 1.32 | 1.51 | 2.04 | 1.65 | 2.63 | 1.84 | |
| Firmicutes | 1.51 | 1.51 | 0.59 | 0.92 | 4.01 | 1.71 | 1.78 | 1.45 | 0.66 | 3.88 | 0.99 | 3.55 | 3.29 | 4.41 | 1.97 | 2.83 |
| GBG (Guaymas bacterial group) | 0.07 | 0.07 | 17.38 | 17.30 | 20.00 | 19.74 | 18.89 | 20.61 | 16.25 | 21.97 | ||||||
| Gemmatimonadetes | 0.07 | 0.39 | 0.33 | 0.20 | 0.72 | 0.13 | 1.12 | 0.13 | 1.84 | 2.43 | 0.07 | 1.51 | 0.07 | 2.37 | ||
| JS-1 | 6.51 | 1.97 | 7.30 | 5.33 | 0.07 | 0.20 | 0.39 | 0.07 | 0.20 | 0.07 | 0.07 | |||||
| Mollicutes | 0.07 | 0.13 | 0.46 | 0.20 | 0.33 | 0.26 | 0.13 | 0.07 | 0.53 | 0.07 | 1.71 | 1.32 | ||||
| NC-10 | 0.07 | 0.13 | 0.66 | 0.59 | 0.66 | 0.39 | 0.53 | 0.26 | 0.20 | 0.39 | 0.13 | 0.46 | 0.33 | 0.20 | ||
| OD1 | 0.13 | 0.26 | 2.43 | 6.32 | 2.30 | 8.42 | 2.96 | 3.36 | ||||||||
| OP3 | 0.07 | 0.20 | 0.13 | 0.79 | 0.07 | 0.39 | 0.07 | 0.86 | 0.26 | 0.99 | 0.07 | 0.66 | 1.18 | |||
| OP8 | 0.07 | 0.33 | 0.07 | 0.39 | 0.20 | 0.33 | 0.99 | 0.59 | 0.20 | 0.72 | 0.07 | 0.79 | 0.46 | 0.59 | ||
| OP9 | 0.07 | 0.46 | 0.07 | 0.39 | 0.72 | 0.53 | 0.53 | |||||||||
| Planctomycetes | 0.39 | 0.39 | 0.20 | 0.26 | 0.92 | 0.39 | 2.17 | 0.20 | 2.11 | 0.46 | 0.33 | 0.53 | 2.44 | 0.99 | 1.97 | 0.20 |
| SAR406 | 0.13 | 0.33 | 0.33 | 1.05 | 0.46 | 1.31 | 0.07 | 0.07 | 0.13 | 0.07 | 0.20 | 0.13 | ||||
| Spirochaetes | 0.72 | 1.64 | 0.53 | 0.59 | 0.46 | 1.05 | 3.03 | 1.84 | 0.20 | 1.78 | 0.13 | 0.86 | 0.46 | 2.04 | 1.32 | 1.84 |
| Thermodesulfobacteria | 0.13 | 0.07 | 0.07 | 0.39 | 0.39 | 0.33 | 0.26 | 0.46 | 0.07 | 0.07 | ||||||
| Thermotogales | 0.07 | 4.08 | 0.33 | 0.07 | 0.07 | 0.39 | 0.13 | 0.13 | 0.13 | |||||||
| Verrucomicrobia | 2.04 | 0.72 | 0.86 | 0.92 | 0.33 | 0.79 | 1.91 | 0.53 | 0.92 | 1.25 | 0.79 | 0.72 | 1.25 | 1.51 | 0.53 | 1.58 |
| WS3/HG2 | 0.86 | 0.33 | 0.79 | 0.07 | 2.76 | 1.58 | 2.24 | 2.17 | 0.07 | 1.32 | 1.78 | 0.79 | 0.13 | 0.66 | 0.33 | 0.46 |
| Proteobacteria | ||||||||||||||||
| Alphaproteobacteria | ||||||||||||||||
| Rhodobacterales | 0.39 | 0.07 | 0.07 | 0.13 | 0.20 | 0.07 | 0.20 | 0.13 | 0.13 | 0.07 | 0.13 | |||||
| Other Alphaproteobacteria | 0.20 | 0.26 | 0.20 | 0.39 | 0.33 | 0.39 | 0.13 | 0.13 | 0.26 | 0.39 | 0.26 | 0.07 | 0.13 | 0.07 | 0.26 | |
| Deltaproteobacteria | ||||||||||||||||
| Myxococcales | 0.20 | 0.07 | 0.13 | 0.13 | 0.20 | 0.33 | 0.53 | 0.39 | 0.39 | 0.13 | 0.66 | 0.26 | 0.33 | |||
| Desulfuromonadales | 2.43 | 2.04 | 2.57 | 2.96 | 0.79 | 1.05 | 3.29 | 0.46 | 0.20 | 0.33 | 0.53 | 0.59 | 0.26 | 0.72 | 0.99 | 0.46 |
| Desulfobacterales | ||||||||||||||||
| Desulfobulbaceae | ||||||||||||||||
| SEEP-SRB3 | 2.50 | 3.22 | 4.67 | 5.20 | 1.05 | 1.18 | 1.51 | 0.59 | 0.99 | 1.58 | 1.12 | 0.72 | 0.20 | 1.05 | 1.25 | 2.11 |
| SEEP-SRB4 | 8.22 | 4.54 | 4.47 | 3.88 | 1.18 | 0.59 | 0.39 | 0.39 | 0.26 | 0.79 | 0.07 | 0.26 | 0.26 | 0.66 | 0.20 | 0.79 |
| Other | 11.64 | 10.00 | 5.92 | 7.11 | 0.86 | 1.25 | 2.24 | 0.92 | 2.76 | 1.91 | 0.46 | 1.51 | 1.32 | 1.45 | 0.79 | 1.12 |
| Desulfobacteraceae | ||||||||||||||||
| SEEP-SRB1a | 3.42 | 4.21 | 5.66 | 5.46 | 0.59 | 1.05 | 0.46 | 0.72 | 0.13 | 0.39 | 0.33 | 0.33 | 0.39 | 0.13 | 0.13 | |
| SEEP-SRB1b | 0.20 | 0.07 | 0.72 | 2.11 | 25.31 | 2.63 | 24.87 | 0.07 | 0.13 | 0.26 | 0.07 | 0.07 | 0.20 | 0.26 | ||
| SEEP-SRB1c | 0.07 | 1.25 | 0.07 | 1.25 | 0.59 | 0.07 | 1.38 | 0.07 | 0.20 | 0.39 | ||||||
| SEEP-SRB1d | 0.07 | 0.13 | 0.13 | 2.10 | 1.64 | 1.12 | 0.59 | 0.20 | 0.79 | 0.72 | ||||||
| SEEP-SRB1e | 0.26 | 0.26 | 0.39 | 0.46 | 0.26 | 0.59 | 0.39 | 0.07 | 1.18 | 0.26 | 0.26 | 0.07 | 1.71 | 0.13 | ||
| SEEP-SRB1f | 0.07 | 0.07 | 0.13 | 0.46 | ||||||||||||
| Other | 0.20 | 3.75 | 0.66 | 3.68 | 1.71 | 1.71 | 0.92 | 1.05 | 1.71 | 7.96 | 4.93 | 9.54 | 4.48 | 8.03 | 3.55 | 7.37 |
| Other Desulfobacterales | 3.75 | 1.45 | 3.09 | 1.25 | 9.93 | 5.33 | 3.22 | 4.41 | 2.04 | 2.37 | 2.89 | 2.11 | 2.83 | 2.44 | 3.68 | 2.30 |
| SEEP-SRB2 | 0.07 | 0.07 | 0.59 | 1.45 | 3.29 | 4.80 | 5.13 | 15.34 | 14.41 | 19.34 | 15.66 | 13.56 | 17.71 | 14.41 | 15.79 | |
| Other Deltaproteobacteria | 0.59 | 0.72 | 0.20 | 0.92 | 2.24 | 1.97 | 2.70 | 1.78 | 3.82 | 2.43 | 2.50 | 2.89 | 2.63 | 1.91 | 3.82 | 1.64 |
| Gammaproteobacteria | ||||||||||||||||
| Beggiatoa | 1.18 | 0.26 | 0.26 | 0.33 | 0.07 | 0.26 | 2.83 | 3.82 | 5.33 | 4.28 | 5.20 | 4.81 | 6.45 | 4.80 | ||
| CGS (clam gill symbiont group) | 1.58 | 1.78 | 1.18 | 1.78 | 1.51 | 1.31 | 0.79 | 1.78 | 0.66 | 0.13 | 0.86 | 0.46 | 0.99 | 0.33 | 1.05 | 0.46 |
| MHGSII (Maorithyas hadalis gill symbiont group) | 0.99 | 3.03 | 1.12 | 3.88 | 2.70 | 3.68 | 1.78 | 3.62 | 0.20 | 0.13 | 0.07 | 0.20 | 0.92 | 0.26 | 0.07 | 0.20 |
| Methylococcales | 0.53 | 0.20 | 2.57 | 0.20 | 4.41 | 0.26 | 3.29 | 0.20 | 0.26 | 0.53 | 0.13 | 0.07 | ||||
| Other Gammaproteobacteria | 2.24 | 4.47 | 8.55 | 3.09 | 12.76 | 10.19 | 13.03 | 10.20 | 5.07 | 4.67 | 1.84 | 3.62 | 3.62 | 3.09 | 3.29 | 3.55 |
| Epsilonproteobacteria | ||||||||||||||||
| Campylobacterales | 3.62 | 2.11 | 4.80 | 4.01 | 2.50 | 0.66 | 0.66 | 0.99 | 1.18 | 0.92 | 1.12 | 0.46 | 1.91 | 0.46 | 1.71 | 0.39 |
| Other Epsilonproteobacteria | 26.45 | 41.84 | 21.32 | 36.18 | 7.76 | 5.92 | 2.89 | 2.89 | 3.09 | 7.11 | 2.43 | 5.72 | 7.37 | 3.36 | 4.54 | 6.25 |
| Zetaproteobacteria | 0.13 | 0.79 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.13 | 0.13 | 0.07 | |||||
| Other Bacteria | 1.78 | 2.70 | 2.11 | 1.45 | 6.51 | 7.17 | 4.87 | 8.03 | 7.90 | 3.55 | 3.62 | 3.75 | 5.40 | 3.55 | 4.80 | 3.55 |
Results are given for Bacteria, according to sample, DNA extraction method, and primer set used. Empty cells indicate that no reads were affiliated with the corresponding microbial group.
TABLE 4.
Taxonomic affiliations of pyrosequencing reads after filteringa
| Taxonomic group | % Contribution |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dive 1758 CT8 SC1 (cold seep surface layer; gray mat) |
Dive 1758 CT8 SC6 (cold seep bottom layer; gray mat) |
Dive 1766 CT3 SH1 (hydrothermal vent surface layer; white mat) |
||||||||||||||
| Zhou |
Kit |
Zhou |
Kit |
Zhou (replicate 1) |
Kit (replicate 1) |
Zhou (replicate 2) |
Kit (replicate 2) |
|||||||||
| Arch1 | Arch2 | Arch1 | Arch2 | Arch1 | Arch2 | Arch1 | Arch2 | Arch1 | Arch2 | Arch1 | Arch2 | Arch1 | Arch2 | Arch1 | Arch2 | |
| AAG | 2.86 | 3.58 | 0.08 | 0.08 | ||||||||||||
| DHVE-6 | 8.34 | 0.40 | 7.78 | 0.08 | 25.50 | 24.72 | 0.08 | 0.95 | 1.59 | 0.87 | 0.64 | 1.43 | 0.71 | 1.51 | 1.51 | |
| DHVE-8 | 1.19 | 0.24 | 0.24 | 0.32 | 0.08 | 0.08 | 0.48 | 0.40 | 0.40 | 0.48 | 0.87 | 0.24 | ||||
| RC-V | 12.39 | 2.07 | 7.31 | 1.11 | 13.82 | 0.32 | 5.88 | 0.40 | 7.47 | 1.43 | 3.65 | 1.91 | 3.89 | 0.56 | 3.10 | 1.43 |
| Thermoprotei | ||||||||||||||||
| Desulfurococcales | 0.16 | 0.64 | 0.71 | 0.72 | ||||||||||||
| Thermoproteales | 0.08 | 0.79 | 0.32 | 0.08 | 0.08 | 0.16 | ||||||||||
| Korarchaeota | 0.08 | 0.16 | 0.16 | 0.24 | 0.08 | |||||||||||
| Thaumarchaeota | ||||||||||||||||
| MCG | 1.59 | 0.87 | 0.24 | 3.57 | 0.16 | 10.89 | 0.48 | 1.75 | 1.59 | 1.59 | 1.35 | 1.99 | 3.26 | 0.79 | 1.99 | |
| MBG-B | 0.95 | 3.18 | 1.35 | 1.19 | 3.26 | 2.86 | 8.19 | 3.89 | 1.51 | 1.51 | 0.64 | 0.56 | 1.19 | 0.71 | 0.79 | 0.24 |
| MG-I | 0.16 | 0.32 | 0.32 | 0.08 | 0.79 | 0.16 | 0.16 | 0.32 | 0.40 | 0.08 | 0.24 | |||||
| Other Thaumarchaeota | 0.08 | 0.16 | 0.08 | |||||||||||||
| Euryarchaeota | ||||||||||||||||
| Archaeoglobales | 0.32 | 0.56 | 0.48 | 0.08 | 0.40 | |||||||||||
| Halobacteriales | 0.08 | 0.08 | 0.08 | |||||||||||||
| Thermococcales | 0.08 | 0.40 | 1.27 | 0.24 | 2.62 | 1.43 | 0.08 | 1.99 | ||||||||
| MBG-D | 0.24 | 0.56 | 26.21 | 19.71 | 12.31 | 7.07 | 17.95 | 0.08 | 10.09 | 0.08 | ||||||
| Other Thermoplasmatales | 0.56 | 0.08 | 0.48 | 0.56 | 4.53 | 0.56 | 4.61 | 0.87 | 6.91 | 6.20 | 0.08 | |||||
| MBG-E | 0.08 | 0.08 | 0.40 | 0.32 | 0.08 | 0.08 | 0.08 | |||||||||
| SAGMEG | 0.32 | 0.08 | 0.08 | 0.16 | 0.48 | 0.16 | 0.16 | 0.08 | 0.24 | 0.08 | 0.16 | |||||
| GEG | 0.24 | 0.24 | 0.08 | 2.22 | 0.08 | 0.79 | 2.38 | 0.79 | 3.10 | 0.95 | 5.64 | 0.64 | 3.49 | |||
| ANME-1 Guaymas | 0.32 | 0.64 | 0.79 | 2.70 | 0.56 | 0.87 | 0.71 | 2.15 | ||||||||
| ANME-1 | 0.08 | 0.56 | 0.40 | 0.48 | 92.45 | 0.08 | 86.34 | 14.30 | 55.60 | 67.12 | 60.13 | 17.71 | 63.22 | 67.04 | 65.74 | |
| ANME-2ab | 17.87 | 24.70 | 23.27 | 23.75 | 3.34 | 0.71 | 2.70 | 1.43 | 0.08 | 0.32 | 0.08 | 0.08 | 0.64 | |||
| ANME-2c | 48.61 | 57.43 | 50.28 | 61.87 | 12.47 | 2.38 | 12.32 | 3.97 | 49.88 | 25.97 | 7.70 | 22.48 | 38.05 | 21.13 | 4.13 | 16.61 |
| ANME-2-Sonora | 0.71 | 1.11 | 0.16 | 0.08 | ||||||||||||
| ANME-2 | 0.08 | 1.59 | 0.79 | 4.21 | 0.08 | 0.24 | 4.05 | 1.19 | 0.48 | 0.24 | 0.24 | 0.72 | ||||
| ANME-3 | 6.59 | 3.81 | 0.24 | 0.16 | 0.32 | 0.87 | 0.40 | 0.48 | 0.48 | |||||||
| GoM-Arc-I | 0.08 | 0.24 | 0.32 | 0.08 | 1.11 | 0.32 | 0.48 | 0.16 | 0.95 | 0.08 | 1.59 | |||||
| Other Methanosarcinales | 1.19 | 2.94 | 0.40 | 2.78 | 0.08 | 0.08 | 0.32 | 0.95 | 0.40 | 1.43 | 0.08 | 0.24 | 0.40 | 1.27 | 0.32 | 0.48 |
| Methanomicrobiales | 0.24 | 0.08 | 0.32 | 0.08 | 0.08 | 0.08 | ||||||||||
| Methanocellales | 0.08 | 0.32 | 0.08 | |||||||||||||
| Methanobacteriales | 0.08 | 0.08 | 0.48 | 0.24 | 0.40 | |||||||||||
| Other Archaea | 5.40 | 0.16 | 5.48 | 0.08 | 4.53 | 4.53 | 0.32 | 2.22 | 2.30 | 1.67 | 0.95 | 2.62 | 1.19 | 1.27 | 2.38 | |
Results are given for Archaea, according to sample, DNA extraction method, and primer set used. Empty cells indicate that no reads were affiliated with the corresponding microbial group.
Regarding Bacteria, higher levels of dissimilarity were observed between samples from different origins. The three expected groups were observed as main clusters: the surface seep sediments, the deep-seep sediments, and the surface vent sediments (Fig. 2A). The differences observed between samples from different origins were mainly due to variations in some bacterial group proportions (Table 3). Similarity percentage analysis (SIMPER) (83) highlighted that the Epsilonproteobacteria (21.75% contribution) and SEEP-SRB1b (sulfate-reducing Bacteria) (71, 72, 88) (11.03% contribution) groups were mainly responsible for the differences between surface and deep-seep samples (Table 3). The difference between seep and vent samples was mainly due to the Guaymas bacterial group (GBG), also known as the HotSeep-1 cluster (63, 89) (Fig. 2B) (15.10% contribution), the Epsilonproteobacteria (11.40% contribution), and SEEP-SRB2 (11.01% contribution) (Table 3). This is congruent with previous molecular surveys of these ecosystems (41, 52, 90). Within these three main clusters, samples grouped according to the primer set used (Fig. 2A). SIMPER analysis highlighted that the difference in community structures observed between Bact1 and Bact2 amplifications was due mainly to variations in amplification of CFB (Bacteroidetes group) (Fig. 2C) (25.56%, 14.62%, and 15.11% contributions for surface seep samples, deep-seep samples, and vent samples, respectively). However, Epsilonproteobacteria from surface seep sediments (23.55% contribution) and SEEP-SRB1b from deep-seep sediments (27.65% contribution) were amplified more with the Bact2 primer set (SIMPER analysis). Likewise, the candidate division OD1 (69) was detected only with the Bact2 primer set (Fig. 2D), contributing 5.32% and 7.66% of the variation within the SC6 and SH1 samples, respectively (Fig. 2D and Table 3). Finally, we observed low levels of dissimilarity between extraction methods (Fig. 2A).
Regarding Archaea, differences generated by the targeted regions were much more conspicuous than for Bacteria (Fig. 3A). For both the Arch1 and Arch2 regions, clustering between samples of the same origin was partially masked by “region and extraction method effects” (Fig. 3A). Indeed, for deep-seep and vent samples, we observed an important effect of the targeted regions. For deep-seep samples, ANME-1 (archaeal anaerobic methanotroph) (91, 92) (Fig. 3C) (49.20% contribution), DHVE-6 (deep-sea hydrothermal vent euryarchaeota) (93) (13.84% contribution), and MBG-D (marine benthic group D) (94) (Fig. 3D) (12.68% contribution) were mainly responsible for this cleavage (0.28% and 89.40% ANME-1, 25.11% and 0.04% DHVE-6, and 22.96% and 0% MBG-D for Arch1 and Arch2, respectively, amplified from deep-seep samples) (Table 4). For vent samples, ANME-1 (Fig. 3C) (42.59% contribution), ANME-2c (20.03% contribution), and MBG-D (Fig. 3D) (14.81% contribution) were mainly responsible for this cleavage (13.01% and 59.41% ANME-1, 43.97% and 23.55% ANME-2c, and 15.13% and 0.04% MBG-D for Arch1 and Arch2, respectively, amplified from vent samples extracted by Zhou's method) (Table 4). Finally, the important difference between community structures obtained with the Arch1 and Arch2 primer sets and using Zhou's method on vent samples was not observed when the commercial kit was used (Fig. 3A).
DISCUSSION
In marine sediments, like the case in many other ecosystems, most microorganisms remain unculturable. Thus, the identification of the diverse microbial populations is based mainly on molecular characterization. Numerous studies investigating the microbial compositions of environments have focused on DNA profiling (3, 64), which requires efficient and unbiased DNA extraction procedures as well as the use of universal primers and relevant bioinformatic tools. These requirements become critical with the emergence and increasing use of high-throughput sequencing and analyses of complex environments harboring various contaminants and almost unknown microbial communities. In this study, we investigated the influence of DNA extraction methods and 16S rRNA gene regions targeted by four different primer sets through 454 pyrosequencing of three physicochemically contrasting samples from extreme marine environments (cold seeps and hydrothermal vents).
Impact of DNA extraction methods.
We showed that the two extraction methods led to various levels of DNA quantity and quality (see Table S3 in the supplemental material). Indeed, commercial kit extraction (FastDNA Spin for Soil), which used bead-beating lysis in reduced-size tubes and included a purification step, yielded steadier DNA quantities between samples than Zhou's method (12) (performed with modifications), which used chemical, enzymatic, and freeze-thaw lysis steps. The quantity of DNA extracted with the commercial kit could be limited by the maximum DNA concentrations allowed by the filters used in the filtration step. Variations between DNA quantities might also be dependent on sample features, such as presence of macrofauna or high-molecular-weight compounds, such as humic acids. The higher concentrations of DNA obtained with surface seep samples and Zhou's method could be explained by the coextraction of DNAs from the macrofauna living on the surface of this sediment sample (e.g., small gastropods and polychaetes). In contrast, the tube sizes used for the bead-beating extraction could have excluded these animals, whose DNA would then not be extracted. Interestingly, previous studies on soils highlighted that coextraction of eukaryotic DNAs or other polyvalent polymers among microbial DNAs enhanced the DNA extraction efficiency, minimizing adsorption and degradation of target DNA (21, 95). Thus, coextraction of eukaryotic DNAs by Zhou's method could eventually provide a benefice in case of low-microbial-biomass environments. Furthermore, DNA quality also seemed to be dependent on the extraction method used. While the commercial kit provided higher 260/280 ratios, Zhou's method led to higher 260/230 ratios. Therefore, the commercial kit may perform better in removing contaminant proteins, while Zhou's method may be more efficient at removing humic substances (96–98).
The influence of the extraction methods on pyrosequencing results was investigated. No significant difference in terms of numbers of reads was observed between DNA extraction protocols in the pyrosequencing raw data outputs (see Table S4 in the supplemental material). This result indicates that despite differences in DNA quality after extraction, the purification step led to similar amplification efficiencies. Likewise, after the filtering steps with the raw pyrosequencing data, no significant difference was observed among the numbers of reads obtained, regardless of the DNA extraction method used (see Table S4). Bead beating has previously been suspected to shear DNA due to mechanical lysis and thus may give rise to the formation of more chimeric PCR products than Zhou's protocol (12, 99). However, our results indicated that the two extraction methods did not appear to drastically affect the quality of the reads or chimera formation.
Extraction methods have previously been shown to bias community structure and microbial diversity results (10, 13, 16–18, 21, 99). In our study, the OTUs obtained when DNA was extracted with the commercial kit or by Zhou's method were globally similar for each given sample (Table 2 and Fig. 1; see Table S5 in the supplemental material), which indicated that the two extraction methods might lead to similar estimations of the global microbial diversity. Interestingly, the differences in DNA quantity observed in the surface seep sample did not appear to change the results of the analysis, confirming that the supplementary DNA extracted by Zhou's method likely corresponded to nonprokaryotic DNA. However, the extraction method used seemed to affect the results we obtained for the vent samples when the Arch1 primer set was used (Fig. 3 and Table 4; see Table S5). This could indicate that some groups, amplified only by the Arch1 primer set, could be extracted more efficiently with one extraction method, as previously noted (21, 99–101). Thus, the picture of the community structure may sometimes be strongly biased by the extraction step, and a combination of several extraction methods would be a good alternative to gain a more accurate picture of microbial diversity.
Impacts of primers and targeted regions.
Besides the DNA extraction step, the choice of PCR primers and 16S rRNA gene target regions is also critical for molecular analysis of microbial diversity (14, 15, 33, 38). Two primer sets for Bacteria and two others for Archaea, each targeting a different 16S rRNA gene region, were tested, and contrasting results were obtained.
The primer set choice affected the results in the early stages of our analysis. Even if an equimolar mix of samples was used for sequencing, significantly different numbers of reads were obtained depending on the primer set used (see Table S4), as previously noted in other projects (A. Dheilly, Biogenouest Platform, Rennes, France, personal communication). This variability may reflect differences in the sample titration in the equimolar mix (16) or may have been due to the fusion primer set used. Indeed, association of primers with tag sequences and pyrosequencing adaptors may reduce the hybridization efficiency on DNA capture beads, which is required for emulsion PCR and sequencing, and/or may be a disadvantage in amplification. Fusion primers must be designed carefully to minimize the formation of potential secondary structures. Each sample might also be labeled with two different tags and sequenced twice to reduce amplification bias due to differential binding of the fusion primers. We also observed contrasting impacts of read filtering according to the primer set used (see Table S4). Chimera detection and specificities of the primer sets for their respective kingdoms led to the removal of more or less reads depending on the primer set used (see Table S4), thus reducing the number of sequences available for the rest of our study. The largest number of chimeras obtained, for the V5–V6 regions (amplified with the Arch2 primer set) (see Table S4), might reveal that these regions could be more prone to splitting than the V2–V3 regions (amplified with the Arch1 primer set). Such variations between 16S rRNA gene regions have previously been highlighted in pyrosequencing analyses (102). The amount of low-quality data removed during the filtering step could also depend on samples and their associated microbial communities, as previously observed for chimera formation (102). Indeed, our results reflected some differences in terms of diversity among the samples (Tables 2 to 4 and Fig. 1 to 3; see Table S5). For example, we highlighted that the number of detected chimeras was dependent on the sample for the Arch2 region (see Table S4). The origin of these chimeras could be explained by the presence of some particular hydrothermal vent microbial lineages. Due to their nucleotide composition, 16S rRNA gene V2–V3 regions could not be amplified for these lineages with the Arch1 primer set and thus do not appear in our analyses. Their successful amplification using the Arch2 primer for the V5–V6 regions could have generated more chimeras.
Regarding the microbial diversity, important differences in community structure were observed depending on the primer set used (Tables 3 and 4 and Fig. 1 to 3; see Table S5 in the supplemental material). The use of a custom database allowed a sufficient level of precision for read assignment to detect these differences for some specific groups (e.g., SEEP-SRB1 or MBG-D), while the NCBI databases (EMBL, DDBJ, and GenBank databases) and associated taxonomies were inaccurate, and the levels of classification were often too high to be informative. These differences were more pronounced for Archaea than for Bacteria and were probably due to a disparity of matching efficiencies between primers for some microbial groups (Table 1), as previously observed in marine sediments (64). Thus, the Arch1 primer set appeared to amplify some archaeal groups which were less or not amplified with the Arch2 primer set (Tables 2 and 4), notably some ANME-2c, MBG-D, and DHVE-6 lineages (Table 4 and Fig. 3). Conversely, the Arch2 primer set appeared to amplify some archaeal groups which were less or not amplified with the Arch1 primer set (Tables 2 and 4), such as the ANME-1 lineage (Table 4 and Fig. 3). Likewise, the Bact1 primer set appeared to successfully amplify some bacterial groups (Tables 2 and 3), particularly CFB and candidate division OD1 bacteria, that were less or not amplified with the Bact2 primers, while the Bact2 primers more efficiently amplified Epsilonproteobacteria and the Deltaproteobacteria subgroup SEEP-SRB1b (Table 3 and Fig. 2). This differential amplification success led to more similar microbial communities between samples when the Bact2 primer set was used instead of the Bact1 primer set (Fig. 1; see Table S5). Differences in terms of numbers of OTUs at a 3% level of dissimilarity were observed between the two archaeal primer sets but were not detected at 10% dissimilarity, suggesting that the variation in the community structure was probably due to misdetection of some archaeal subgroups. This result might suggest a great variability of primer hybridization efficiency and amplification due to sequence mismatches of close microbial lineages (above 97% similarity).
Therefore, the choice of primer sets considerably affects the representation of the microbial community and, more particularly in our study, the archaeal community. This is important in microbial ecology because many of the discussions in the literature are based on the presence/absence of lineages to explain environmental data and microbial community distribution and function. The design of universal primers remains very delicate, especially since we do not know a priori the microbial compositions of samples. The situation is obviously more delicate for lineages presenting an intragroup heterogeneity. The use of different primer sets might be a good solution to minimize these biases. However, it would be better to totally eliminate the PCR amplification step that can generate many biases (e.g., by using de novo analyses or metagenomic studies).
Conclusions.
In conclusion, we found that DNA extraction methods and primer sets could have major effects on species richness estimations for our samples. This revealed that every type of sample, because of its own nature, requires a very careful drawing of community structure conclusions. Indeed, community structure, microbial diversity, and proportions of read numbers, sometimes used as proxies for microbial relative abundance (103), could be strongly biased in some cases, and researchers must be very careful in drawing conclusions about community structures. Differences in DNA quantities and qualities observed after extraction did not always reflect a larger degree of microbial diversity. Similarly, if fewer reads are obtained after pyrosequencing for some samples, it does not necessarily reflect less microbial diversity. As long as molecular studies are dependent on a PCR amplification step, the choice of a good primer set will be crucial for giving a nonbiased vision of the true composition of the analyzed sample. In our study, the use of either primer set led to a totally different representation of the microbial communities. Indeed, for deep-seep samples, we obtained a proportion of 92.45% ANME-1 with one set of primers but only 0.48% with the other set. In the same way, while we detected a proportion of 25.31% for the Deltaproteobacteria subgroup SEEP-SRB1b with one set of primers and 2.11% with the other set, we did not observe differences for the other Deltaproteobacteria lineages between the two sets of primers. Next, even if the two extraction methods gave the same results for one given sample, they could lead to drastic changes for another sample. While the two extraction methods resulted in totally different archaeal community compositions for the vent samples with one primer set, no significant impact was detected for all other samples, regardless of the amplified region. Thus, using Zhou's method, we detected 12.31% MBG-D, 14.30% ANME-1, and 49.88% ANME-2c, while using the commercial kit on the same sample, we observed 7.07% MBG-D, 67.12% ANME-1, and 7.70% ANME-2. Therefore, in addition to the combination of several extraction methods and primer sets, we recommend combining sequencing methods with complementary approaches to gain a more comprehensive picture of the communities under scrutiny. For example, real-time PCR with different primer sets appeared to be very useful for obtaining more details about microbial group quantities (40). In addition, since we do not really know the numbers of 16S rRNA gene copies within bacterial and archaeal genomes, which could drastically increase microbial diversity results (104, 105), a fluorescence in situ hybridization (FISH/catalyzed reporter deposition [CARD]-FISH) approach (90, 106) could allow for confirmation of previous results.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to the crews of the research vessel L'Atalante and the submersible Nautile of the “BIG” cruise and to the scientific team, in particular Françoise Lesongeur, Mathilde Le Roy, Laurent Toffin, and Nolwenn Callac, for their work on board. We thank Maxime Galan, Alexandra Dheilly, and Sophie Coudouel for very helpful scientific discussions.
This cruise was funded by Ifremer (France) and benefited from a work permit in Mexican waters (permit DAPA/2/281009/3803; 28 October 2009). This study was supported by Carnot Institute funding and by an Ifremer/Brittany region Ph.D. grant for P.C.
Footnotes
Published ahead of print 16 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00592-14.
REFERENCES
- 1.Hugenholtz P, Goebel BM, Pace NR. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann RI, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rappé MS, Giovannoni SJ. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369–394. 10.1146/annurev.micro.57.030502.090759 [DOI] [PubMed] [Google Scholar]
- 4.Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541–555. 10.1016/j.mimet.2003.08.009 [DOI] [PubMed] [Google Scholar]
- 5.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.”. Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120. 10.1073/pnas.0605127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283–290. 10.1038/ismej.2007.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acosta-Martinez V, Dowd SE, Sun Y, Wester D, Allen V. 2010. Pyrosequencing analysis for characterization of soil bacterial populations as affected by an integrated livestock-cotton production system. Appl. Soil Ecol. 45:13–25. 10.1016/j.apsoil.2010.01.005 [DOI] [Google Scholar]
- 8.Buée M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F. 2009. 454 pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 184:449–456. 10.1111/j.1469-8137.2009.03003.x [DOI] [PubMed] [Google Scholar]
- 9.Lee OO, Wang Y, Yang J, Lafi FF, Al-Suwailem A, Qian P-Y. 2011. Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J. 5:650–664. 10.1038/ismej.2010.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webster G, Newberry CJ, Fry JC, Weightman AJ. 2003. Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA-based techniques: a cautionary tale. J. Microbiol. Methods 55:155–164. 10.1016/S0167-7012(03)00140-4 [DOI] [PubMed] [Google Scholar]
- 11.Roose-Amsaleg C, Garnier-Sillam E, Harry M. 2001. Extraction and purification of microbial DNA from soil and sediment samples. Appl. Soil Ecol. 18:47–60. 10.1016/S0929-1393(01)00149-4 [DOI] [Google Scholar]
- 12.Zhou J, Bruns M, Tiedje J. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maarit Niemi R, Heiskanen I, Wallenius K, Lindstrom K. 2001. Extraction and purification of DNA in rhizosphere soil samples for PCR-DGGE analysis of bacterial consortia. J. Microbiol. Methods 45:155–165. 10.1016/S0167-7012(01)00253-6 [DOI] [PubMed] [Google Scholar]
- 14.Sergeant MJ, Constantinidou C, Cogan T, Penn CW, Pallen MJ. 2012. High-throughput sequencing of 16S rRNA gene amplicons: effects of extraction procedure, primer length and annealing temperature. PLoS One 7:e38094. 10.1371/journal.pone.0038094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claesson MJ, O'Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O'Toole PW. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. 10.1371/journal.pone.0006669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vishnivetskaya TA, Layton AC, Lau MCY, Chauhan A, Cheng KR, Meyers AJ, Murphy JR, Rogers AW, Saarunya GS, Williams DE, Pfiffner SM, Biggerstaff JP, Stackhouse BT, Phelps TJ, Whyte L, Sayler GS, Onstott TC. 2014. Commercial DNA extraction kits impact observed microbial community composition in permafrost samples. FEMS Microbiol. Ecol. 87:217–230. 10.1111/1574-6941.12219 [DOI] [PubMed] [Google Scholar]
- 17.Morono Y, Terada T, Hoshino T, Inagaki F. 2014. Hot-alkaline DNA extraction method for deep-subseafloor archaeal communities. Appl. Environ. Microbiol. 80:1985–1994. 10.1128/AEM.04150-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazen TC, Rocha AM, Techtmann SM. 2013. Advances in monitoring environmental microbes. Curr. Opin. Biotechnol. 24:526–533. 10.1016/j.copbio.2012.10.020 [DOI] [PubMed] [Google Scholar]
- 19.Lloyd KG, MacGregor BJ, Teske A. 2010. Quantitative PCR methods for RNA and DNA in marine sediments: maximizing yield while overcoming inhibition. FEMS Microbiol. Ecol. 72:143–151. 10.1111/j.1574-6941.2009.00827.x [DOI] [PubMed] [Google Scholar]
- 20.Robe P, Nalin R, Capellano C, Vogel TM, Simonet P. 2003. Extraction of DNA from soil. Eur. J. Soil Biol. 39:183–190. 10.1016/S1164-5563(03)00033-5 [DOI] [Google Scholar]
- 21.Frostegård Å, Courtois S, Ramisse V, Clerc S, Bernillon D, Le Gall F, Jeannin P, Nesme X, Simonet P. 1999. Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65:5409–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon J, Soulas G, Catroux G. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354–2359. 10.1128/AEM.67.5.2354-2359.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tebbe CC, Vahjen W. 1993. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 59:2657–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juniper S, Cambon M-A, Lesongeur F, Barbier G. 2001. Extraction and purification of DNA from organic rich subsurface sediments (ODP Leg 169S). Mar. Geol. 174:241–247. 10.1016/S0025-3227(00)00153-5 [DOI] [Google Scholar]
- 25.Sørensen SJ, de Lipthay JR, Müller AK, Barkay T, Hansen LH, Rasmussen LD. 2002. Molecular methods for assessing and manipulating the diversity of microbial populations and processes, p 363–389 In Enzymes in the environment. Marcel Dekker Inc, New York, NY [Google Scholar]
- 26.Valentine DL. 2007. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 5:316–323. 10.1038/nrmicro1619 [DOI] [PubMed] [Google Scholar]
- 27.Kuske CR, Banton KL, Adorada DL, Stark PC, Hill KK, Jackson PJ. 1998. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl. Environ. Microbiol. 64:2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirk JL, Beaudette LA, Hart M, Moutoglis P, Klironomos JN, Lee H, Trevors JT. 2004. Methods of studying soil microbial diversity. J. Microbiol. Methods 58:169–188. 10.1016/j.mimet.2004.04.006 [DOI] [PubMed] [Google Scholar]
- 29.Schloss PD, Handelsman J. 2004. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68:686–691. 10.1128/MMBR.68.4.686-691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakravorty S, Helb D, Burday M, Connell N, Alland D. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69:330–339. 10.1016/j.mimet.2007.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms—proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U. S. A. 87:4576–4579. 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van de Peer Y, Chapelle S, DeWachter R. 1996. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 24:3381–3391. 10.1093/nar/24.17.3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim M, Morrison M, Yu Z. 2011. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J. Microbiol. Methods 84:81–87. 10.1016/j.mimet.2010.10.020 [DOI] [PubMed] [Google Scholar]
- 34.Schmalenberger A, Schwieger F, Tebbe CC. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557–3563. 10.1128/AEM.67.8.3557-3563.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu ZT, Morrison M. 2004. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:4800–4806. 10.1128/AEM.70.8.4800-4806.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stackebrandt E, Goebel B. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849. 10.1099/00207713-44-4-846 [DOI] [Google Scholar]
- 37.Galan M, Guivier E, Caraux G, Charbonnel N, Cosson J-F. 2010. A 454 multiplex sequencing method for rapid and reliable genotyping of highly polymorphic genes in large-scale studies. BMC Genomics 11:296. 10.1186/1471-2164-11-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vahjen W, Pieper R, Zentek J. 2010. Bar-coded pyrosequencing of 16S rRNA gene amplicons reveals changes in ileal porcine bacterial communities due to high dietary zinc intake. Appl. Environ. Microbiol. 76:6689–6691. 10.1128/AEM.03075-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simoneit BRT, Mazurek MA, Brenner S, Crisp PT, Kaplan IR. 1979. Organic geochemistry of recent sediments from Guaymas Basin, Gulf of California. Deep Sea Res. A 26:879–891. 10.1016/0198-0149(79)90102-X [DOI] [Google Scholar]
- 40.Vigneron A, Cruaud P, Pignet P, Caprais J-C, Cambon-Bonavita M-A, Godfroy A, Toffin L. 2013. Archaeal and anaerobic methane oxidizer communities in the Sonora Margin cold seeps, Guaymas Basin (Gulf of California). ISME J. 7:1595–1608. 10.1038/ismej.2013.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vigneron A, Cruaud P, Pignet P, Caprais JC, Gayet N, Cambon-Bonavita MA, Godfroy A, Toffin L. 14 November 2013. Bacterial communities and syntrophic associations involved in anaerobic oxidation of methane process of the Sonora Margin cold seeps, Guaymas Basin. Environ. Microbiol. 10.1111/1462-2920.12324 [DOI] [PubMed] [Google Scholar]
- 42.Alain K, Callac N, Ciobanu M-C, Reynaud Y, Duthoit F, Jebbar M. 2011. DNA extractions from deep subseafloor sediments: novel cryogenic-mill-based procedure and comparison to existing protocols. J. Microbiol. Methods 87:355–362. 10.1016/j.mimet.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 43.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12:118–123. 10.1111/j.1462-2920.2009.02051.x [DOI] [PubMed] [Google Scholar]
- 44.Huse S, Huber J, Morrison H, Sogin M, Welch D. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. 10.1186/gb-2007-8-7-r143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. 10.1371/journal.pone.0027310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K, Nealson KH, Horikoshi K. 2003. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69:7224–7235. 10.1128/AEM.69.12.7224-7235.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teske AP. 2006. Microbial communities of deep marine subsurface sediments: molecular and cultivation surveys. Geomicrobiol. J. 23:357–368. 10.1080/01490450600875613 [DOI] [Google Scholar]
- 52.Biddle JF, Cardman Z, Mendlovitz H, Albert DB, Lloyd KG, Boetius A, Teske A. 2012. Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J. 6:1018–1031. 10.1038/ismej.2011.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R, Fredricks HF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, Brenchley JE, Teske A, House CH, Hinrichs K-U. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U. S. A. 103:3846–3851. 10.1073/pnas.0600035103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cambon-Bonavita MA, Nadalig T, Roussel E, Delage E, Duperron S, Caprais JC, Boetius A, Sibuet M. 2009. Diversity and distribution of methane-oxidizing microbial communities associated with different faunal assemblages in a giant pockmark of the Gabon continental margin. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 56:2248–2258. 10.1016/j.dsr2.2009.04.007 [DOI] [Google Scholar]
- 55.Dang H, Luan X-W, Chen R, Zhang X, Guo L, Klotz MG. 2010. Diversity, abundance and distribution of amoA-encoding archaea in deep-sea methane seep sediments of the Okhotsk Sea. FEMS Microbiol. Ecol. 72:370–385. 10.1111/j.1574-6941.2010.00870.x [DOI] [PubMed] [Google Scholar]
- 56.Dhillon A, Lever M, Lloyd KG, Albert DB, Sogin ML, Teske A. 2005. Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl. Environ. Microbiol. 71:4592–4601. 10.1128/AEM.71.8.4592-4601.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kubo K, Lloyd KG, Biddle FJ, Amann R, Teske A, Knittel K. 2012. Archaea of the miscellaneous crenarchaeotal group are abundant, diverse and widespread in marine sediments. ISME J. 6:1949–1965. 10.1038/ismej.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lloyd KG, Albert DB, Biddle JF, Chanton JP, Pizarro O, Teske A. 2010. Spatial structure and activity of sedimentary microbial communities underlying a Beggiatoa spp. mat in a Gulf of Mexico hydrocarbon seep. PLoS One 5:e8738. 10.1371/journal.pone.0008738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lloyd KG, Lapham L, Teske A. 2006. Anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl. Environ. Microbiol. 72:7218–7230. 10.1128/AEM.00886-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pachiadaki M, Kallionaki A, Dählmann A, De Lange G, Kormas K. 2011. Diversity and spatial distribution of prokaryotic communities along a sediment vertical profile of a deep-sea mud volcano. Microb. Ecol. 62:655–668. 10.1007/s00248-011-9855-2 [DOI] [PubMed] [Google Scholar]
- 61.Pachiadaki MG, Lykousis V, Stefanou EG, Kormas KA. 2010. Prokaryotic community structure and diversity in the sediments of an active submarine mud volcano (Kazan mud volcano, East Mediterranean Sea). FEMS Microbiol. Ecol. 72:429–444. 10.1111/j.1574-6941.2010.00857.x [DOI] [PubMed] [Google Scholar]
- 62.Sorensen KB, Teske A. 2006. Stratified communities of active archaea in deep marine subsurface sediments. Appl. Environ. Microbiol. 72:4596–4603. 10.1128/AEM.00562-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teske A, Hinrichs KU, Edgcomb V, Gomez AD, Kysela D, Sylva SP, Sogin ML, Jannasch HW. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994–2007. 10.1128/AEM.68.4.1994-2007.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teske A, Sorensen KB. 2008. Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J. 2:3–18. 10.1038/ismej.2007.90 [DOI] [PubMed] [Google Scholar]
- 65.Dhillon A, Teske A, Dillon J, Stahl DA, Sogin ML. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765–2772. 10.1128/AEM.69.5.2765-2772.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dojka MA, Hugenholtz P, Haack SK, Pace NR. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emerson D, Rentz JA, Lilburn TG, Davis RE, Aldrich H, Chan C, Moyer CL. 2007. A novel lineage of Proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS One 2:e667. 10.1371/journal.pone.0000667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MS, Strous M. 2009. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75:3656–3662. 10.1128/AEM.00067-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harris JK, Kelley ST, Pace NR. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845–849. 10.1128/AEM.70.2.845-849.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harrison BK, Zhang H, Berelson W, Orphan VJ. 2009. Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (Santa Barbara Basin, California). Appl. Environ. Microbiol. 75:1487–1499. 10.1128/AEM.01812-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Knittel K, Boetius A, Lemke A, Eilers H, Lochte K, Pfannkuche O. 2003. Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia margin, Oregon). Geomicrobiol. J. 20:269–294. 10.1080/01490450303896 [DOI] [Google Scholar]
- 72.Schreiber L, Holler T, Knittel K, Meyerdierks A, Amann R. 2010. Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade. Environ. Microbiol. 12:2327–2340. 10.1111/j.1462-2920.2010.02275.x [DOI] [PubMed] [Google Scholar]
- 73.Webster G, Parkes RJ, Cragg BA, Newberry CJ, Weightman AJ, Fry JC. 2006. Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol. Ecol. 58:65–85. 10.1111/j.1574-6941.2006.00147.x [DOI] [PubMed] [Google Scholar]
- 74.Webster G, Parkes RJ, Fry JC, Weightman AJ. 2004. Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl. Environ. Microbiol. 70:5708–5713. 10.1128/AEM.70.9.5708-5713.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katoh K, Kuma K-I, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- 77.Saitou N, Nei M. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 78.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 79.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- 80.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 81.Miller MA, Pfeiffer W, Schwartz T. 2010. The CIPRES science gateway, p 1–8 In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA. ACM Press, New York, NY [Google Scholar]
- 82.Clarke K, Warwick R. 2001. Changes in marine communities: an approach to statistical analysis and interpretation, p 144 Natural Environment Research Council, Plymouth, United Kingdom [Google Scholar]
- 83.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117–143. 10.1111/j.1442-9993.1993.tb00438.x [DOI] [Google Scholar]
- 84.Yue JC, Clayton MK. 2005. A similarity measure based on species proportions. Commun. Stat. Theory Methods 34:2123–2131. 10.1080/STA-200066418 [DOI] [Google Scholar]
- 85.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27:325–349. 10.2307/1942268 [DOI] [Google Scholar]
- 86.Horn HS. 1966. Measurement of ‘overlap' in comparative ecological studies. Annu. Rev. Ecol. Syst. 5:25–37 [Google Scholar]
- 87.Morisita M. 1959. Measuring of interspecific association and similarity between communities. Mem. Fac. Sci. Kyushu Univ. Series E 3:65–80 [Google Scholar]
- 88.Orcutt BN, Joye SB, Kleindienst S, Knittel K, Ramette A, Reitz A, Samarkin V, Treude T, Boetius A. 2010. Impact of natural oil and higher hydrocarbons on microbial diversity, distribution, and activity in Gulf of Mexico cold-seep sediments. Deep Sea Res. Part II Top. Stud. Oceanogr. 57:2008–2021. 10.1016/j.dsr2.2010.05.014 [DOI] [Google Scholar]
- 89.Kellermann MY, Wegener G, Elvert M, Yoshinaga MY, Lin Y-S, Holler T, Mollar XP, Knittel K, Hinrichs K-U. 2012. Autotrophy as a predominant mode of carbon fixation in anaerobic methane-oxidizing microbial communities. Proc. Natl. Acad. Sci. U. S. A. 109:19321–19326. 10.1073/pnas.1208795109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kleindienst S, Ramette A, Amann R, Knittel K. 2012. Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments. Environ. Microbiol. 14:2689–2710. 10.1111/j.1462-2920.2012.02832.x [DOI] [PubMed] [Google Scholar]
- 91.Hinrichs KU, Hayes JM, Sylva SP, Brewer PG, DeLong EF. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802–805. 10.1038/19751 [DOI] [PubMed] [Google Scholar]
- 92.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jorgensen BB, Witte U, Pfannkuche O. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626. 10.1038/35036572 [DOI] [PubMed] [Google Scholar]
- 93.Takai K, Horikoshi K. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach AL. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Volossiouk T, Robb EJ, Nazar RN. 1995. Direct DNA extraction for PCR-mediated assays of soil organisms. Appl. Environ. Microbiol. 61:3972–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Griffiths B, Dı′az-Raviña M, Ritz K, McNicol J, Ebblewhite N, Bååth E. 1997. Community DNA hybridisation and % G+C profiles of microbial communities from heavy metal polluted soils. FEMS Microbiol. Ecol. 24:103–112. 10.1111/j.1574-6941.1997.tb00427.x [DOI] [Google Scholar]
- 97.Logemann J, Schell J, Willmitzer L. 1987. Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163:16–20. 10.1016/0003-2697(87)90086-8 [DOI] [PubMed] [Google Scholar]
- 98.Schultz DJ, Craig R, Cox-Foster DL, Mumma RO, Medford JI. 1994. RNA isolation from recalcitrant plant tissue. Plant Mol. Biol. Report. 12:310–316. 10.1007/BF02669273 [DOI] [Google Scholar]
- 99.de Lipthay JR, Enzinger C, Johnsen K, Aamand J, Sørensen SJ. 2004. Impact of DNA extraction method on bacterial community composition measured by denaturing gradient gel electrophoresis. Soil Biol. Biochem. 36:1607–1614. 10.1016/j.soilbio.2004.03.011 [DOI] [Google Scholar]
- 100.Priemé A, Bonilla Sitaula JI, Klemedtsson ÅK, Bakken LR. 1996. Extraction of methane-oxidizing bacteria from soil particles. FEMS Microbiol. Ecol. 21:59–68. 10.1111/j.1574-6941.1996.tb00333.x [DOI] [Google Scholar]
- 101.Aakra Å, Hesselsøe M, Bakken L. 2000. Surface attachment of ammonia-oxidizing bacteria in soil. Microb. Ecol. 39:222–235 [DOI] [PubMed] [Google Scholar]
- 102.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504. 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jorgensen SL, Hannisdal B, Lanzén A, Baumberger T, Flesland K, Fonseca R, Øvreås L, Steen IH, Thorseth IH, Pedersen RB. 2012. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl. Acad. Sci. U. S. A. 109:E2846–E2855. 10.1073/pnas.1207574109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pei AY, Oberdorf WE, Nossa CW, Agarwal A, Chokshi P, Gerz EA, Jin Z, Lee P, Yang L, Poles M, Brown SM, Sotero S, DeSantis T, Brodie E, Nelson K, Pei Z. 2010. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl. Environ. Microbiol. 76:3886–3897. 10.1128/AEM.02953-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kormas KA. 2011. Interpreting diversity of proteobacteria based on 16S rRNA gene copy number, p 73–89 In Sezenna ML. (ed), Proteobacteria: phylogeny, metabolic diversity and ecological effects. Nova Publishers, Hauppauge, NY [Google Scholar]
- 106.Lloyd KG, May MK, Kevorkian R, Steen AD. 2013. Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor. Appl. Environ. Microbiol. 79:7790–7799. 10.1128/AEM.02090-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.