Abstract
Indole-3-acetic acid (IAA) is a fundamental phytohormone with the ability to control many aspects of plant growth and development. Pseudomonas sp. strain UW4 is a rhizospheric plant growth-promoting bacterium that produces and secretes IAA. While several putative IAA biosynthetic genes have been reported in this bacterium, the pathways leading to the production of IAA in strain UW4 are unclear. Here, the presence of the indole-3-acetamide (IAM) and indole-3-acetaldoxime/indole-3-acetonitrile (IAOx/IAN) pathways of IAA biosynthesis is described, and the specific role of two of the enzymes (nitrilase and nitrile hydratase) that mediate these pathways is assessed. The genes encoding these two enzymes were expressed in Escherichia coli, and the enzymes were isolated and characterized. Substrate-feeding assays indicate that the nitrilase produces both IAM and IAA from the IAN substrate, while the nitrile hydratase only produces IAM. The two nitrile-hydrolyzing enzymes have very different temperature and pH optimums. Nitrilase prefers a temperature of 50°C and a pH of 6, while nitrile hydratase prefers 4°C and a pH of 7.5. Based on multiple sequence alignments and motif analyses, physicochemical properties and enzyme assays, it is concluded that the UW4 nitrilase has an aromatic substrate specificity. The nitrile hydratase is identified as an iron-type metalloenzyme that does not require the help of a P47K activator protein to be active. These data are interpreted in terms of a preliminary model for the biosynthesis of IAA in this bacterium.
INTRODUCTION
The phytohormone indole-3-acetic acid (IAA) is a key modulator of plant growth and development. This compound impacts nearly every aspect of plant development, including cell division, elongation, fruit development, initiation of roots, leaves and flowers, cambial growth, vascular development, and senescence (1–4). Moreover, IAA acts as a signal within the plant, directing both physiological and tropic responses to a range of environmental stimuli (1, 2, 4–7).
Bacterial IAA is multifunctional: it not only has a physiological effect on plants and a role as a communication signal in plant-microbe interactions but also serves a purpose in the bacteria that produce it. For example, IAA has been reported to enhance bacterial adaptation to stress conditions, leading to improved survival and persistence in the environment (8). This phenomenon was demonstrated in Sinorhizobium meliloti cells treated with exogenous IAA or genetically modified to overproduce IAA. When these cells were exposed to various stress conditions such as acidity, osmotic shock, UV irradiation, and heat shock, more bacteria survived in both the IAA-treated and the IAA-overproducing strains than with wild-type or nontreated strains (9, 10). Exogenous IAA treatment also promoted intracellular trehalose accumulation, which can serve as a source of carbon and as an osmolyte that confers protection against freezing and desiccation (9, 10). The production of lipopolysaccharide, exopolysaccharide, and biofilm was also elevated in the IAA-overproducer compared to wild-type Sinorhizobium (9). These compounds protect the bacterial cells against the innate plant defense system and enable rhizobia to resist harsh environmental conditions (9). Bradyrhizobium japonicum also displays higher resistance to stresses when treated with exogenous IAA (11).
Some bacteria have the dual ability to both produce and metabolize IAA. This phytohormone can serve as an ideal food source, rich in carbon and nitrogen (12). Recently, bacterial chemotaxis toward IAA by the IAA degrader P. putida 1290 was reported (12). Bacteria that can chemotaxi toward IAA, degrade it, and utilize its components have a competitive advantage over other bacteria. Plants are biological sources of IAA, and perhaps bacteria can use plant-derived IAA as the attractant to actively colonize plants better than other bacteria (13). In Streptomyces species, IAA has been reported to stimulate spore germination, aerial mycelium formation, and antimicrobial activity against other bacteria (14, 15). Plant-associated Streptomyces spp. produce antibiotics, which are lethal to fungal and bacterial phytopathogens. Rhizospheric IAA derived from both plants and bacteria may act as a signal for soil streptomycetes to enhance their antibiotic production, thereby inhibiting the growth of other competing microbes, while simultaneously offering protection to plants from phytopathogens (15).
IAA is a signaling molecule for different cellular processes within bacterial cells. In Azospirillum brasilense Sp245, interfering with IAA biosynthesis affects the expression of ribosomal proteins, which are involved in protein synthesis. IAA also affects aerobic denitrification and adaptation to an anaerobic metabolism, cell respiration, and carbohydrate uptake (16). In the phytopathogen Agrobacterium tumefaciens C58, exogenous IAA leads to the upregulation of genes involved in amino acid synthesis and carbon metabolism (17). In both A. brasilense and Dickeya dadantii, exposure to exogenous IAA leads to increased expression of a type VI secretion system, which may be used to directly interact with plant cells. IAA is also required for the expression of effectors secreted by the type III secretion system in Erwinia chrysanthemi (16, 18). Bacterial effector proteins are injected into host cells via type III secretion systems to facilitate pathogenicity (19). Altogether, these studies show that IAA acts as a signaling molecule for a variety of different bacterial genes that play a role in both beneficial and deleterious plant-microbe interactions (16).
Over the past few decades, several pathways leading to the production of this phytohormone have been identified. Some of the enzymes that catalyze IAA biosynthesis have been characterized, with most of the focus being on the indole-3-pyruvic acid (IPA) and indole-3-acetamide (IAM) pathways (20). In some instances, the presence of multiple pathways operating within a single bacterium creates a robust IAA biosynthetic network. Some pathways intersect at a common intermediate, implying that that route may be the predominant one. The advantage of having a complex multiroute biosynthetic system protects the bacterium against the loss of any particular pathway, since IAA production can be fluxed through an alternate circuit (21).
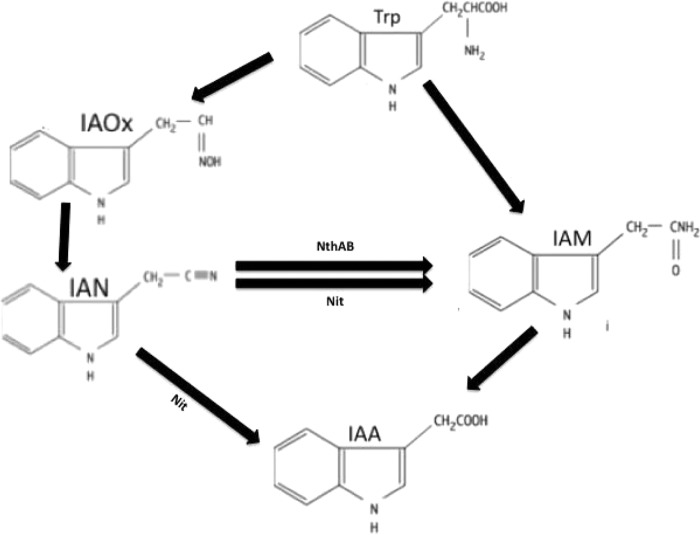
The genome of an IAA-producing, plant growth-promoting bacterium, Pseudomonas sp. UW4, has recently been completely sequenced (22). Several genes that encode enzymes potentially involved in the biosynthesis of IAA were identified (22). These genes appear to be part of at least two different IAA biosynthesis pathways. Thus, the present study is directed toward deciphering the IAA biosynthesis pathways operating in Pseudomonas sp. UW4 by isolating, purifying, and characterizing two of the implicated enzymes. Both the indole-3-acetaldoxime/indole-3-acetonitrile (IAOx/IAN) and the indole-3-acetamide (IAM) pathways are predicted to be operating in UW4. We hypothesize that in the former pathway, the tryptophan precursor is first converted to the IAOx intermediate by an unknown enzyme. Subsequently, a phenylacetyldoxime dehydratase converts the IAOx intermediate into the IAN intermediate. This IAN substrate is either directly converted into IAA by nitrilase (Nit) or is converted into IAM by either nitrilase (Nit) or nitrile hydratase (NthAB). The IAM intermediate would then be further converted into the final product IAA by an amidase (Fig. 1). The focus of the work reported here is the isolation, purification, and partial characterization of the Nit and NthAB enzymes.
FIG 1.
Putative IAOx/IAN and IAM pathways operating in UW4. The IAN substrate is either directly converted into IAA by Nit or into IAM by Nit or NthAB. Only the two enzymes characterized in the present study are included in the figure.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli DH5α (Invitrogen/Life Technologies) was used as an initial cloning host, and E. coli BL21(DE3) (Novagen/Merck, KGaA) was used as the host for recombinant protein expression. This expression host is designed for expression from pET vectors and contains an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible T7 RNA polymerase (Novagen/Merck, KGaA). Pseudomonas sp. UW4 (22) was grown and maintained aerobically at 30°C in tryptic soy broth (Bacto) supplemented with 100 μg of ampicillin/ml. All E. coli strains were maintained aerobically at 37°C in Luria-Bertani broth (Fischer Bioreagents). When appropriate, 50 μg of kanamycin/ml and 0.1 mM IPTG were added. The plasmid, pET30b+, was used for the expression of Nit and NthAB. The plasmid constructs generated in the present study are described in detail in Table 1.
TABLE 1.
Expression plasmid constructs and primers used in this studya
| Plasmid or primer | Sequence (5′–3′) | Description |
|---|---|---|
| Plasmids | ||
| pETnit | pET30b(+) vector with the nit gene from UW4 cloned into the NdeI/XhoI site | |
| pETnthAB | pET30b(+) vector with the nthAB gene from UW4 cloned into the NdeI/XhoI site | |
| Primers | ||
| nit-F | TAATCATATGCCCAAATCAATCGTTGCGGC | nit (NdeI)-fwd |
| nit-R | ATTACTCGAGGGAAGTGAAGCGCACCCCC | nit (XhoI)-rev |
| nthA-F | TAATCATATGAGCGCCACTGTATCCCCAG | nthA (NdeI)-fwd |
| nthB-R | ATTACTCGAGTGCGGCCACCGTTTTC | nthB (XhoI)-rev |
Restriction sites within primers are underlined. Start and stop codons were not included in any of the primer sequences. All primers have the NdeI restriction site in the forward primer and XhoI in the reverse primer, preceded by a short 4-bp linker sequence to facilitate efficient digestion.
DNA extraction and expression plasmid construction.
Genomic DNA from Pseudomonas sp. UW4 was isolated using the Wizard Genomic DNA purification kit (Promega, catalog no. A1120). The primer sequences used to amplify UW4 nit, nthAB, and P47K genes are also given in Table 1. Both the forward and the reverse primer sequences were based on the fully sequenced UW4 genome (GenBank accession number CP003880.1). For the NthAB protein composed of α and β subunits, the forward primer was designed starting from the second codon of the α subunit, while the reverse primer was designed starting from the second last codon of the β subunit. The α-subunit, short intergenic region (42 bp), and β-subunit were amplified as a single fragment. The amplified fragments were subcloned between the NdeI and XhoI sites of pET30b+, resulting in the pETnit and pETnthAB constructs. Hot-start PCR was performed with KOD Hot Start DNA polymerase (Novagen, Mississauga, Ontario, Canada). The reaction mixture (50 μl) was set up on ice and included 5 μl of KOD hot start buffer (1×), 3 μl of 25 mM MgSO4 (final concentration, 1.5 mM), 5 μl of 2 M deoxynucleoside triphosphates (final concentration, 0.2 mM), 1.5 μl of forward primer (final concentration, 0.3 mM), 1.5 μl of reverse primer (final concentration, 0.3 mM), 100 ng of template genomic DNA, 1 μl of KOD hot start polymerase. and PCR-grade water up to the final volume of 50 μl. The PCR was performed in an Eppendorf MasterCycler gradient machine using the following amplification conditions for nit: 95°C for 2 min, 95°C for 20 s, 71°C for 10 s, 70°C for 15 s, 70°C for 5 min, and ending at 4°C. For nthAB, the following amplification conditions were used: 95°C for 2 min, 95°C for 20 s, 67°C for 10 s, 70°C for 23 s, 70°C for 5 min, and ending at 4°C.
Expression of recombinant proteins in E. coli.
The expression plasmids were initially transformed into E. coli DH5α to maintain recombinant plasmids without any background basal protein expression. For expression of the recombinant proteins, the plasmids were transformed into E. coli BL21(DE3). Cells transformed with pETnit or pETnthAB were grown in 500 ml of Luria-Bertani medium containing kanamycin (50 μg/ml) at 37°C with shaking for aeration, until the optical density at 600 nm (OD600) reached 0.5. At this point, the cultures were induced with 0.1 mM IPTG and incubated at 30°C (pETnit) or room temperature (pETnthAB) overnight. The following day, the cells were collected by centrifugation at 2,000 × g for 20 min at 4°C. The cell pellet was washed with 20 ml of 50 mM KH2PO4 (pH 7.5) with gentle vortexing, followed by centrifugation at 2,000 × g for 10 min at 4°C. This wash step was repeated twice more. The washed pellet was resuspended in 50 ml of binding buffer (20 mM NaH2PO4, 500 mM NaCl, 20 mM imidazole [pH 7.4]), and the cells were disrupted by sonication 30 times for 30 s each time with 10-s pauses between each round. The lysate was kept on ice at all times during sonication. After sonication, the lysate was centrifuged at 8,000 × g for 30 min at 4°C to separate the soluble and insoluble fractions. The soluble fraction was saved at 4°C for purification.
Purification of the recombinant proteins.
The recombinant proteins, which have a C-terminal His6 tag, were purified under native conditions using a His GraviTrap prepacked, single-use gravity-flow column containing precharged Ni Sepharose 6 Fast Flow. The protocol was followed according to the manufacturer's instructions (GE Healthcare, United Kingdom). The purified protein samples were analyzed by SDS-PAGE.
Enzyme assays.
The purified recombinant proteins were tested for the ability to convert a nitrile substrate into its corresponding acid or amide product. Specifically, Nit was tested for the ability to convert IAN directly into IAA, whereas NthAB was tested for the ability to convert IAN into IAM. The enzyme activity was generally assayed in a 1-ml reaction mixture containing 50 μg of purified enzyme (Nit or NthAB), 1 mM IAN substrate, and 50 mM KH2PO4 (pH 7.5) buffer. The reactions were stopped by adding 100 μl of 1 M HCl. Samples were analyzed by high-pressure liquid chromatography (HPLC).
Temperature and pH optima.
To determine the ideal temperature conditions for both Nit and NthAB activity, a 1-ml enzyme assay reaction mixture was set up as previously described. The tubes were incubated at various temperatures: 20 to 60°C for Nit or 4 to 60°C for NthAB. Reaction mixtures were allowed to proceed for 3 h. To determine the ideal pH for both Nit and NthAB, the following buffers were used: sodium acetate at pH 5 to 5.5, sodium citrate at pH 6 to 6.5, potassium phosphate at pH 7 to 7.5, Tris-HCl at pH 8 to 8.5, and glycine at pH 9. The 1-ml enzyme assay reaction mixture was set up in the corresponding pH buffers. The tubes were incubated at 30°C for Nit and at room temperature for NthAB for 3 h.
Controls.
To determine whether IAN and IAM are spontaneously converted to IAA, nonenzymatic controls were set up in 1-ml reactions. For pH controls, 1 mM IAN or IAM was added to buffers based on sodium acetate at pH 5, sodium citrate at pH 6, potassium phosphate at pH 7, Tris-HCl at pH 8, and glycine at pH 9. The tubes were incubated at 30°C for 3 h. For temperature controls, 1 mM IAN or IAM was added to KH2PO4 (pH 7. 5) buffer. The tubes were incubated at temperatures between 22 and 55°C for 3 h. No enzyme was added.
To determine whether the E. coli BL21(DE3) expression host produces an IAN-converting enzyme without expressing the recombinant Nit/NthAB proteins, 100-ml cultures of either nontransformed E. coli BL21(DE3) or E. coli BL21(DE3) transformed with empty pET30b(+) were grown at 37°C until they reached mid-log phase (OD600 of 0.4 to 0.8). The cultures were induced with 0.1 mM IPTG for 3 h. The cells were collected by centrifugation at 2,000 × g for 20 min at 4°C. The cell pellets were washed with 10 ml of 50 mM KH2PO4 (pH 7.5), followed by centrifugation at 2,000 × g for 10 min at 4°C. This wash step was repeated twice more. The washed pellet was resuspended in 10 ml of 50 mM KH2PO4 (pH 7.5) and sonicated 10 to 15 times for 10 s each time, with 10-s pauses between each round. After sonication, the lysate was centrifuged at 8,000 × g for 30 min at 4°C. The soluble fraction was assayed for activity. The reaction mixture (1-ml total volume) consisted of ∼50 μg of crude protein extract and 1 mM IAN in 50 mM KH2PO4 (pH 7.5) buffer. The reaction mixtures were incubated at 30°C for 3 h and analyzed by HPLC.
HPLC.
HPLC analysis was performed using a Waters Alliance 2695 HPLC separation system (Mississauga, Ontario, Canada), which includes a Waters 2996 photodiode array detector. The system was connected to a PC with Empower 2 software (Waters) for data collection and processing. A Sunfire C18 column (50 by 4.6 mm [inner diameter], 2.5 μm; Waters, Ireland) was connected with a Security Guard C18 guard column (4 by 3.0 mm [inner diameter]; Phenomenex, Torrance, CA). Gradient HPLC was performed at room temperature using a mobile phase containing water-acetic acid (1% [vol/vol]) (A) and acetonitrile-acetic acid (1% [vol/vol]) (B). Starting with 80% A, the gradient began at 2 min and reached 60% A at 15 min. The flow rate was 1 ml/min. A 100-μl sample injection volume was used, and the eluent was monitored at 280 nm. The retention times observed were 2.3 min for indole-3-acetamide (IAM), 5.2 min for indole-3-acetic acid (IAA), 7.5 min for indole-3-propionic acid (IPA), and 8.4 min for indole-3-acetonitrile (IAN), respectively.
Multiple sequence alignments and phylogenetic analyses.
Multiple sequence alignments were constructed for Nit, NthAB, and a putative P47K activator protein using MEGA 5.10 software (23). Nucleotide sequences were translated into amino acid sequences using the web-based software from EMBL-EBI (http://www.ebi.ac.uk/Tools/emboss/transeq/). Protein BLAST searches were performed against the Nit, NthAB, and P47K proteins of Pseudomonas sp. UW4 to screen for similar amino acid sequences in the database. Only nonredundant coding sequence hits that shared at least 50% identity with the UW4 proteins were included in the alignments. Each alignment was manually refined, and the regions that could not be aligned reliably were removed. ML trees were constructed using MEGA 5.10 software. The Nit alignment utilized a WAG substitution model with a gamma distribution and invariant sites (WAG+G+I) (23). The NthAB α and β subunits both utilized a WAG+G model for maximum-likelihood (ML) analysis, whereas the P47K activator protein utilized a JTT+G model (23). Neighbor-joining (NJ) and maximum-parsimony (MP) trees were also constructed. One thousand bootstrap replicates were performed to assess the confidence for each clade of the tree.
RESULTS AND DISCUSSION
Nitrilase.
Nitrilase enzymes are part of a large superfamily, classified into 13 branches, of which only branch I (EC 3.5.5.1) convert nitriles to their corresponding carboxylic acids, releasing ammonia in the process (24). This branch of nitrilase enzymes has been implicated in IAA biosynthesis. However, their role in the biosynthesis of IAA has been neglected. Kobayashi et al. (25) characterized an indole-3-acetonitrile-specific nitrilase from Alcaligenes faecalis JM3. Howden et al. (26) also identified an arylacetonitrilase in the plant pathogen P. syringae B728a that was capable of hydrolyzing IAN to IAA.
In the present study, a nitrilase enzyme capable of producing both IAA and IAM has been identified in the plant growth-promoting bacterium Pseudomonas sp. UW4. Previous characterizations of nitrilases from various bacteria reveal diverse substrate specificities and the ability of a single nitrilase to act upon multiple nitriles (i.e., aromatic, aliphatic, arylacetonitriles) (27–32). Therefore, it is possible that IAN is neither the only nor the preferred substrate that UW4 nitrilase can act upon.
Nitrilase temperature and pH optima.
Optimal activity for the UW4 nitrilase was observed at 50°C and a pH of 6 (Fig. 2 and 3). These data agree well with other reported pseudomonad nitrilases, which have temperature optima from 40 to 55°C (27, 28, 33–35). The UW4 enzyme retained >50% activity at 55°C; however, <20% activity was retained at 60°C. The pH optima of the UW4 nitrilase is slightly lower than that of other pseudomonad nitrilases, which display maximum activity at pHs between 7 and 9 (27, 28, 33, 34). Rather, it is similar to that of R. rhodochrous K22 (pH 5.5) and Arthrobacter nitroguajacolicus ZJUTB06-99 (pH 6.5) (36, 37). However, the UW4 enzyme retained >80% activity at pH 7 and, in this respect, is comparable to other pseudomonad nitrilases. Plant roots can cause fluctuations in rhizospheric pH, which in turn can influence the activity of the enzymes produced by rhizospheric bacteria (38). The enzyme systems in these bacteria evolve to operate within these pH ranges (39). Pseudomonas sp. UW4 is a rhizosphere inhabitant that produces a cytoplasmic nitrilase most active at pH 6 to 7, conditions resembling those of the rhizosphere.
FIG 2.
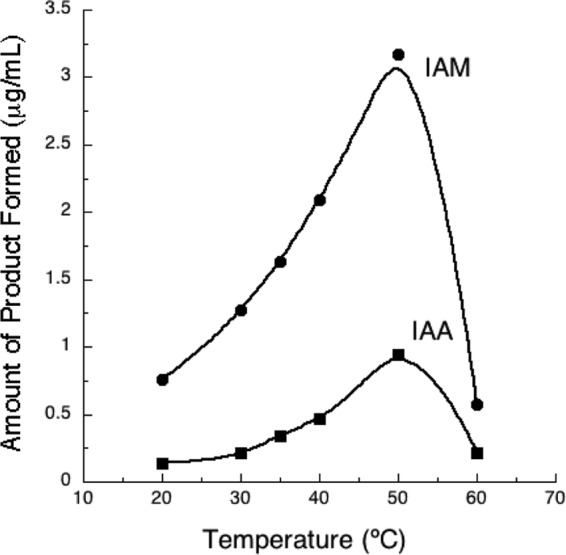
Temperature rate profile of recombinant Nit from Pseudomonas sp. UW4, expressed in E. coli BL21(DE3).
FIG 3.
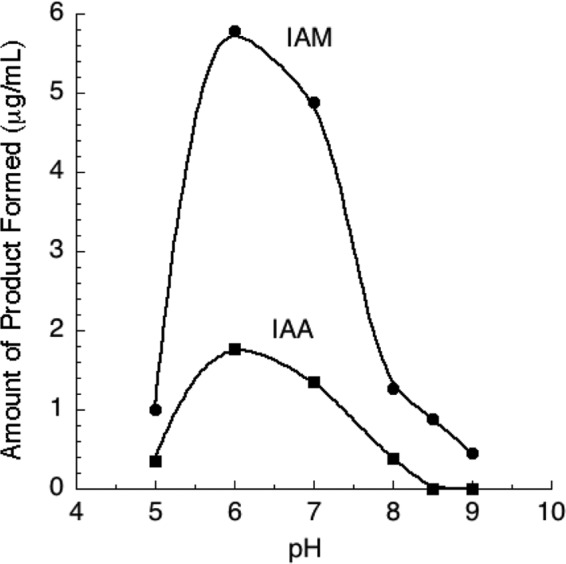
pH rate profile of recombinant Nit from Pseudomonas sp. UW4, expressed in E. coli BL21(DE3).
Amide production by nitrilase.
The conversion of IAN by the UW4 nitrilase resulted in the formation of IAM, as well as IAA. Significantly more of the amide product than carboxylic acid was produced. This activity does not conform to the commonly accepted acid-producing nitrilase mechanism. Nevertheless, other reports indicate that some nitrilases can produce modest amounts of amides (28, 40–43). Several studies suggest that the formation of acid relative to amide is dependent on both the electron-withdrawing substituents and the configuration at the α-carbon of the reactant (44, 45). Increasing the electronegativity of the α-substituent favors amide formation (44). The substrate IAN has a nonelectronegative methyl group at the α-position. In addition, smaller substituents near the catalytically active cysteine residue lead to the formation of larger amounts of amide (45). For example, substituting the tryptophan directly upstream of the catalytic cysteine for alanine in the nitrilase of Alcaligenes faecalis ATCC 8750, causes more amide production (45). The UW4 nitrilase contains a valine directly upstream of the catalytic cysteine. This residue is comparable in size to alanine and similarly has a hydrophobic side chain. The residue directly downstream of the catalytic cysteine is also important since its side chain points into the substrate-binding pocket of nitrilase (45). In the UW4 nitrilase, a large tryptophan residue occupies this position. This residue is conserved in most biochemically characterized nitrilases (45). It is therefore difficult to draw conclusions in regard to the preferential production of IAM by UW4 nitrilase based on electronic or steric factors. Additional experiments with various mutations in close proximity to the catalytic center and assays with different electron-withdrawing or -donating substrates may offer more insight into the capacity for amide production by this enzyme.
It has been suggested that lower temperatures and increased pH promote the formation of the amide (44). Consistent with this, UW4 nitrilase produced more amide at pH 8 than at pH 5 and more amide at 30°C than at 60°C. Some microbial nitrilases have dual abilities and can be exploited for both classic acid production and “nitrile hydratase-like” amide production, UW4 nitrilase being one of these enzymes (46). At this point, it is unclear whether the observed propensity to synthesize more IAM compared to IAA, at 30°C compared to 60°C, and at pH 8 compared to pH 5, has any physiological significance in terms of how strain UW4 responds to environmental changes.
Nitrilase motifs.
Aromatic and aliphatic nitrilases can be characterized based on four groups of conserved protein motifs (47). In the work reported here, a motif analysis revealed that the UW4 nitrilase protein contains all four of the motifs reported for aromatic nitrilases (Table 2). The conserved Glu-Lys-Cys (E-K-C) catalytic triad was identified in all of the nitrilase sequences. In agreement with the expected positions for aromatic nitrilase motifs (47), the four motif patterns identified in the UW4 nitrilase occur at positions 40 to 50, 127 to 139, 165 to 178, and 204 to 212, respectively.
TABLE 2.
Aliphatic versus aromatic nitrilase motif patternsa
| Nitrilase type | Consensus motif | Pseudomonas sp. UW4 nitrilase motif |
|---|---|---|
| Aliphatic | [FL]-[ILV]-[AV]-F-P-E-[VT]-[FW]-[IL]-P-[GY]-Y-P-[WY] | |
| R-R-K-[LI]-[KRI]-[PA]-T-[HY]-[VAH]-E-R | ||
| C-W-E-H-[FLX]-[NQ]-[PT]-L | ||
| [VA]-A-X-[AV]-Q-[AI]-X-P-[VA]-X-[LF]-[SD] | ||
| Aromatic | [ALV]-[LV]-[FLM]-PE-[AS]-[FLV]-[LV]- [AGP]-[AG]-Y-P | V-V-M-P-E-A-L-L-GG-Y |
| [AGN]-[KR]-H-R-K-L-[MK]-P-T-[AGN]-X-E-R | A-K-H-R-K-L-M-P-TG-T-E-R | |
| C-W-E-N-[HY]-M-P-[LM]-[AL]-R-X-X-[ML]-Y | C-W-E-N-M-M-P-L-LR-T-A-M-Y | |
| A-X-E-G-R-C-[FW]-V-[LIV] | A-H-E-G-R-C-F-V-V |
Letters (designating specific amino acid residues) inside brackets represent variable sites. Boldface letters represent the catalytic triad (E-K-C). Pseudomonas sp. UW4 motifs agree with those reported for aromatic nitrilases by Sharma and Bhalla (47).
Physicochemical properties of nitrilase.
Aliphatic and aromatic nitrilases can also be differentiated based on their amino acid sequence and physiochemical properties (48). The physicochemical properties of the UW4 nitrilase were deduced from an in silico analysis of the amino acid sequence using the ProtParam subroutine of the Expert Protein Analysis System (ExPASy) from the proteomic server of the Swiss Institute of Bioinformatics, in order to predict aromaticity or aliphaticity. Several of the parameters (number of amino acids, molecular mass [Da], number of negatively charged residues, extinction coefficients [M−1 cm−1] at 280 nm, and overall average hydropathicity) listed in Table 3 are closer to the consensus values reported for aromatic nitrilases, supporting the prediction that the UW4 nitrilase has aromatic substrate specificity. The nitrilase of UW4 has an instability index of 32.37 and a high aliphatic index of 94.43, suggesting that it is a thermostable enzyme. It also has a relatively low grand average hydropathicity value of 0.099 and is considered a hydrophilic protein.
TABLE 3.
Physicochemical properties of aliphatic, aromatic, and Pseudomonas sp. UW4 nitrilasesa
| Parameter | Avg values |
||
|---|---|---|---|
| Aliphatic nitrilase | Aromatic nitrilase | Pseudomonas sp. UW4 nitrilase | |
| No. of amino acids | 352 | 310 | 307 |
| Molecular mass (Da) | 38,274 | 33,694 | 32,848 |
| Theoretical pI | 5.50 | 5.51 | 5.69 |
| No. of negatively charged residues | 42 | 36 | 34 |
| No. of positively charged residues | 30 | 29 | 28 |
| Extinction coefficient (M−1 cm−1) at 280 nm | 50,213 | 43,975 | 40,210 |
| Instability index | 41.2 | 38.5 | 32.37 |
| Aliphatic index | 89.4 | 89.8 | 94.43 |
| Overall hydropathicity | 0.10 | 0.01 | 0.099 |
Average consensus values are as reported by Sharma et al. (48).
Nitrilase sequence alignments.
In our study, a total of 115 nitrilase amino acid sequences from bacteria, fungi, and plants were aligned using MEGA 5.0 software. The four characteristic nitrilase protein sequence motifs (47) were present in all of the sequences analyzed. The catalytic triad (E-K-C) was conserved in all sequences in alignment positions Glu44 (E), Lys131 (K), and Cys165 (C) (Table 4). The regions around the catalytic triads were further examined for conservation, considering only sites with at least 90% consensus (Table 5).
TABLE 4.
Reported consensus sequences flanking the invariant catalytic triad residues of nitrilasesa
| Conserved catalytic triad of nitrilases | Consensus sequence | Nitrilase sequence alignment (this study) |
|---|---|---|
| E44 | f P E a f | m P E A l |
| K131 | h R K l * p T | H R K L M P T |
| C165 | l * C W E n ** p | a i C W E N * M P |
Reported consensus sequences flanking the invariant catalytic triad residues of nitrilases as reported by Pace and Brenner (24) and the consensus sequences flanking the catalytic triad from the nitrilase multiple sequence alignment in this study. Uppercase letters indicate ≥90% consensus, whereas lowercase letters represent ≥50% consensus. An asterisk (*) represents residues with <50% consensus. Boldface indicates residues of the catalytic triad (E-K-C).
TABLE 5.
Further analysis of regions flanking the conserved catalytic triad of the nitrilase multiple sequence alignmenta
| Conserved catalytic triad of nitrilases | Nitrilase sequence alignment (this study) |
|---|---|
| E44 | LVV* PEA * |
| LGGYPKG | |
| K131 | HRKLMPT ** ERLIWG |
| *GDGST | |
| C165 | CWEN * MPL |
| LR* AMY |
Further analysis of regions flanking the conserved catalytic triad of the nitrilase multiple sequence alignment in the present study. Underlined residues were 100% conserved over all of the nitrilase sequences, and residues that are not underlined have at least 90% consensus. An asterisk (*) indicates residues with <90% consensus. Boldface indicates residues of the catalytic triad (E-K-C).
Nitrile hydratase.
Nitrile hydratases (E C 4.2.1.84) catalyze the hydration of nitriles to the corresponding amides (49). These enzymes are heterodimers composed of α and β subunits and are classified as either ferric or cobalt nitrile hydratases, depending on the type of metal cofactor within the α-subunit active site (49). Nitrile hydratases have been exploited as industrial biocatalysts in the commercial synthesis of acrylamide, nicotinamide, and 5-cyanovaleramide (50, 51). However, their involvement in IAA biosynthesis, specifically in the conversion of IAN into IAM, has not been well studied. Among the nitrile hydratases that have been experimentally shown to convert IAN into IAM are those from Agrobacterium, Rhizobium, and Bradyrhizobium (52, 53).
Some nitrile hydratase genes are flanked by sequences encoding activator proteins, which may be important for expression of both Fe and Co type enzymes (49, 54). This is the case for the Fe-type nitrile hydratase of Rhodococcus sp. N771, N774 and Pseudomonas chlororaphis B23 (55). The UW4 iron-type nitrile hydratase does not require an activator protein for activity, although there is a putative P47K activator protein encoded directly downstream of the β-subunit (22). However, at this time the possibility that P47K is involved in iron trafficking or modulating the activity of the UW4 nitrile hydratase cannot be ruled out.
Nitrile hydratase temperature and pH optima.
In the present study, NthAB was found to exclusively convert IAN to IAM (Fig. 4 and 5). The maximum activity of the UW4 Fe-type nitrile hydratase was observed at 4°C and a pH 7.5. The pH optimum is narrow, since <20% activity was retained at pH 7 and <40% was retained at pH 8. These values are in agreement with what has been observed with other characterized nitrile hydratases, which display maximum activity at a pH between 7 and 8 (52, 56–58). The enzyme retained >50% of its activity at temperatures of 10 to 20°C; however, the activity decreased significantly as the temperature increased above 20°C. Since this is a heterodimeric enzyme, the elevated temperature may adversely affect enzyme stability, causing the two subunits to dissociate. Generally, the nitrile hydratases of mesophilic bacteria exhibit maximum activity at ambient temperatures between 20 and 40°C (59). Pseudomonas sp. UW4 naturally inhabits Canadian soils, where it is often exposed to cold temperatures. Although the optimum growth temperature of this bacterium is 25 to 30°C, it has the ability to grow at temperatures as low as 4°C so that it may be classified as a psychrotroph (60). Generally, the activity of most enzymes decreases at low temperatures; however, remarkably, UW4 nitrile hydratase activity increases as the temperature is lowered. This enzyme's ability to produce IAM at 4°C gives strain UW4 the potential to produce and secrete IAA into the plant rhizosphere in the spring, when Canadian soil temperatures are low (5 to 10°C) and new plantlets require IAA for growth.
FIG 4.
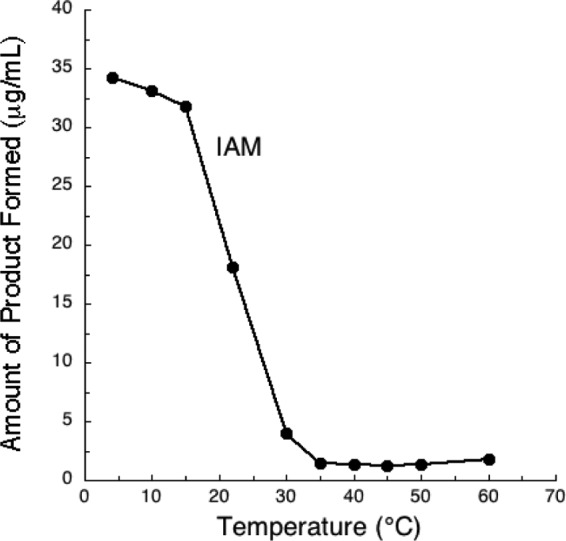
Temperature rate profile of recombinant NthAB from Pseudomonas sp. UW4, expressed in E. coli BL21(DE3).
FIG 5.
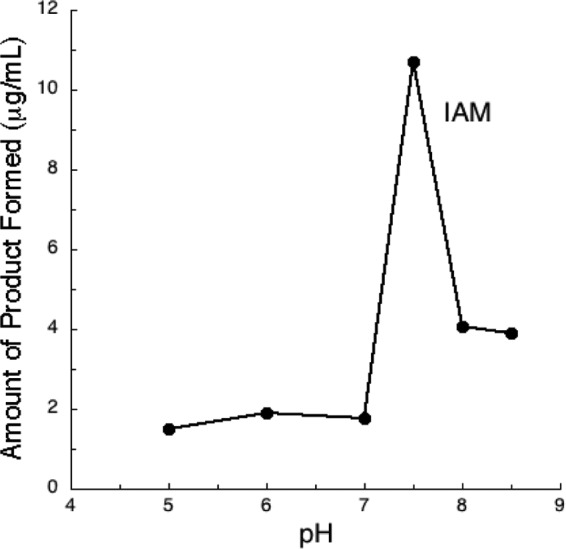
pH rate profile of recombinant NthAB from Pseudomonas sp. UW4, expressed in E. coli BL21(DE3).
Nitrile hydratase sequence alignments.
Multiple sequence alignments were constructed from protein BLAST hits against the UW4 nitrile hydratase α-subunit and β-subunit amino acid sequences. A total of 73 microbial nitrile hydratase α-subunit amino acid sequences and 66 β-subunit sequences were aligned. Nitrile hydratases have a highly conserved CXLCSC motif, where “X” corresponds to a T (threonine) residue for cobalt-type and an S (serine) residue for iron-type nitrile hydratases (61). Analysis of the multiple sequence alignment reveals that the metal binding motif was conserved in all α-subunit sequences at positions 98 to 103. Both cobalt and iron-type nitrile hydratases were included in the alignment. The UW4 nitrile hydratase contains an iron-type active site in the α-subunit. There is more conservation between the different β-subunits analyzed than between the α-subunits.
P47K putative activator multiple sequence alignment.
A multiple sequence alignment was also constructed from protein BLAST searches against the UW4 P47K putative activator protein amino acid sequence. Members of the nitrile hydratase activator proteins contain a highly conserved CXCC motif (49). Analysis of the multiple sequence alignment reveals this cysteine-rich motif nested within a highly conserved region NGCICCTLR[E/A]DLL encompassing positions 69 to 81 (Table 6). Studies suggest that this motif may be involved in metal trafficking for nitrile hydratase (49).
TABLE 6.
Conserved motifs in putative P47K activator protein sequence alignment in this studya
| Residue range | Motif sequence |
|---|---|
| 69–81 | NGCICCTLR[E/A]DLL |
| 8–18 | TVLSGFLG[S/A]GK |
| 93–115 | FDYlLIEStGIsEP[L/M]PvaETF[T/A]F |
| 360–371 | GDCRQE[L/I]vFIGQ |
| 34–47 | vAvIVNDMSe[I/V]NxD |
Uppercase letters indicate 100% consensus, whereas lowercase letters indicate at least 90% consensus.
The UW4 P47K protein is a member of the COG0523 proteins, which comprise cobalamin biosynthesis proteins, nitrile hydratase activators, and zinc-transport proteins (62). The COG0523 proteins belong to the G3E family, which either help incorporate a cofactor into the target protein's catalytic site or store/deliver a metal cofactor to a target metalloprotein (62). These proteins are characterized by the Walker A (GxxGxGK), Walker B (hhhExxG), and NKxD motifs (62). The two Walker motifs were identified and conserved in all of the sequences in our alignment, while the NKxD motif was only conserved in some sequences. The UW4 P47K sequence does not contain the NKxD motif. Further analysis of the multiple sequence alignment reveals several highly conserved regions (Table 6).
The UW4 P47K putative activator protein shares 58 to 98% sequence identity with proteins that have been annotated as 4-hydroxytetrahydrobiopterin dehydratases and 56 to 74% identity with cobalamin synthesis proteins in the NCBI database. Most bacteria produce enzymes that require cobalamin as a cofactor and thus have the ability to synthesize it. Cobalamin has a centrally chelated metal ion such as cobalt, which is delivered by ATP-dependent cobalt chelatases (63). Since the UW4 P47K is annotated as a cobalamin synthesis protein (22), the amino acid sequence was compared to those of known cobalt chelatases; CobN, CobS, and CobT from P. denitrificans, CbiK from Salmonella enterica serovar Typhimurium, and CbiX from B. megaterium. There is no significant sequence similarity with any of these proteins. Although the UW4 P47K protein is annotated as a “cobalamin biosynthesis protein” based on sequence similarity to other proteins annotated as such, it is not necessarily a true labeling of function. Haas et al. (62) reported that cobW genes are located within the cobalamin biosynthesis gene clusters and CobW proteins contain a histidine stretch of 4 to 15 residues. Neither of these criteria hold true for the P47K protein of UW4. On the other hand, UW4 P47K does share significant sequence identity with some experimentally confirmed Fe-type nitrile hydratase activators. For example, it shares 74% with P47K of Pseudomonas chlororaphis B23, 59% identity with the P44K activator of Rhodococcus erythropolis AJ270, and 46% identity with the P47K activator of Rhodococcus sp. strains N-771 and N-774 (49, 54, 55, 64). The UW4 P47K protein does not share any significant sequence identity with experimentally confirmed activators of Co-type nitrile hydratases (65, 66). It is reported that nitrile hydratase activators represent <1% of the COG0523 protein family (62). Moreover, these activators are clustered exclusively with the genes encoding the two subunits of the iron-type nitrile hydratase (62). The chromosomal location of the UW4 P47K gene (directly downstream of the β-subunit), the characteristic activator motifs and the sequence similarity with other characterized Fe-type nitrile hydratase activators suggest the possibility that UW4 P47K is such a protein. However, it remains to be experimentally determined whether this protein plays a role as a metallochaperone modulating NthAB activity in UW4.
Phylogenetic trees.
The UW4 nitrilase (Nit), nitrile hydratase α (NthA), and β (NthB) subunits and putative P47K activator protein amino acid sequences were used for phylogenetic analysis. Maximum-likelihood (ML), neighbor-joining (NJ), and maximum-parsimony (MP) trees were constructed to infer the phylogeny of these proteins. Only the phylogeny inferred using the ML method is shown in Fig. SA1 to SA4 in the supplemental material; however, bootstrap values from NJ and MP analyses are also included in the tree figures. The topology between the three different types of trees was nearly identical for each data set.
The nitrilase phylogenetic analysis includes microbial, yeast, and plant enzymes (see Fig. SA1 in the supplemental material). The pseudomonad nitrilases form a large well-supported monophyletic group, which includes the UW4 nitrilase (pink box). Interestingly, two P. putida strains did not group with the rest of the pseudomonad clade but instead form a separate monophyletic clade with Acidovorax avenae ATCC 19860 and Frateuria aurantia DSM 622, both plant-associated bacteria. An intriguing finding from the nitrilase phylogeny analysis, is that two bacterial nitrilases (Rhizobium etli and Bradyrhizobium japonicum USDA 110) cluster with the plant rather than bacterial nitrilases. They form a well-supported monophyletic group with the Arabidopsis, rice, and tobacco nitrilases. Both of these bacteria are nitrogen-fixing plant-symbionts that colonize plant roots and live inside root nodules. Since bacteria evolved before plants and these rhizobia form intimate relationships with their host plant, perhaps the plant nitrilase genes were initially acquired from these bacteria via a lateral gene transfer. The phylogenetic tree shows that nitrilases exist in different organisms, inhabiting diverse environments from soil to water. It has been suggested that both positive selection and purifying selection has taken place in nitrilase subfamilies and that groups of microbial nitrilases diverged functionally and associated with other metabolic enzymes, acquiring diverse biochemical activities (67).
The ML tree generated for the α-subunit of the nitrile hydratase (see Fig. SA2 in the supplemental material) clusters all of the pseudomonads (including NthA of UW4) into a well-supported monophyletic clade. With the exception of NthA of UW4, none of the Pseudomonas nitrile hydratases within this clade have been experimentally characterized. However, their conserved metal cofactor binding motif identifies them as iron-type nitrile hydratases. This pseudomonad clade is sister to a well-supported iron-type nitrile hydratase Burkholderia clade, with the exception of Burkholderia terrae BS001, Burkholderia sp. strain BT03 and Burkholderia phytofirmans PsJN. The three latter strains possess cobalt-type enzymes and form a separate clade that shares the most recent common ancestor with the cobalt-type enzymes of Rhodopseudomonas and Bradyrhizobium clades. Therefore, it appears that bacteria of the same genera (Burkholderia) cluster separately based on the type of metal cofactor they associate with. The rhodococci clade forms a polytomy, with several immediate descendants coming off the same node. It has been reported that strains of Rhodococcus erythropolis from different geographical areas possess highly variable nitrile hydratase enzymatic activities. Some R. erythropolis strains possess identical nitrile hydratase gene sequences, even though they occupy different habitats (68). However, the activities of these enzymes have been shown to be different, reflecting a possible functional diversification to accommodate their environment and the substrates available to them (68). The tree topology demonstrates a clear separation between the iron-type and the cobalt-type α-subunits (see Fig. SA2 in the supplemental material). All of the iron-type nitrile hydratase descendants cluster into a monophyletic clade in the upper half of the tree (shown in the red box), while the cobalt-type enzymes cluster into a separate monophyletic clade in the bottom half of the tree (shown in the blue box).
The ML tree generated for the β-subunit (see Fig. SA3 in the supplemental material) also clusters all of the pseudomonads (including NthB of UW4) into a well-supported monophyletic clade. This pseudomonad clade is sister to the Burkholderia clade, a similar tree topology as observed for the α-subunit. The rhodococci fall into a cluster in the upper half of the tree and form a polytomy. This cluster is not well supported by bootstrap analysis. The relatedness of these lineages cannot be well deciphered or all of the descendants are very closely related to one another, since their sequence similarity is very high (80 to 90%). The β-subunit tree, is separated into two clusters, one that makes up the upper half of the tree and the other makes up the bottom half. This tree topology is the same as the α-subunit tree; however, in the case of the β-subunits, the separation of the two clades is not correlated with the type of metal that the nitrile hydratase associates with. For example, the UW4 nitrile hydratase is an iron-type and the β-subunit clusters in the bottom half of the tree, along with other iron-type pseudomonad β-subunits. However, the Rhodococcus sp. strains 312, N-771, and AJ270 are also iron-type β-subunits, and these cluster in the clade comprising the upper half of the tree, which includes cobalt-type enzymes as well.
The ML tree of putative nitrile hydratase activator proteins (see Fig. SA4 in the supplemental material) contains multiple clusters of pseudomonad proteins dispersed throughout the tree (pink boxes). Cluster 1 includes proteins that have the NKXD motif. The remaining pseudomonads in the analysis do not have this motif. This is a conserved motif of the G3E family of P-loop GTPases (62), so perhaps these Pseudomonas proteins belong to this family, whereas the others belong to a different GTPase class and thus cluster separately. The UW4 P47K protein forms a clade with other Pseudomonas spp. in cluster 6. Certain protein motifs are conserved in some of the pseudomonads analyzed and not in others. One such motif occurs at amino acid residues 89 to 92, where some of the pseudomonads have a conserved KEGR sequence, while others have an RQQR sequence. Those with the KEGR sequence form cluster 1, while those with the RQQR sequence (this includes P47K of UW4) form cluster 6. Perhaps these motifs are signatures of different protein families and are associated with different catalytic functions or protein structures. Many of the proteins included in the tree are “hypothetical” proteins that have not been characterized but share sequence similarity with the P47K of UW4. Others are arbitrarily annotated as COG0523 family proteins but can have various biological functions (62). Therefore, the function of the UW4 P47K cannot be inferred from the tree topology.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by funds from the Natural Science and Engineering Research Council of Canada.
We appreciate the laboratory assistance provided by Yu Gu and the Ron Johnson Lab at the University of Guelph with the HPLC analyses. We also thank Kirsten Muller at the University of Waterloo for assistance with the phylogenetic analyses.
Footnotes
Published ahead of print 16 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00649-14.
REFERENCES
- 1.Halliday KJ, Martínez-García JF, Josse EM. 2009. Integration of light and auxin signaling. Cold Spring Harb. Perspect. Biol. 1:6. 10.1101/cshperspect.a001586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grossmann K. 2010. Auxin herbicides: current status of mechanism and mode of action. Pest Manage. Sci. 66:113–120. 10.1002/ps.1860 [DOI] [PubMed] [Google Scholar]
- 3.McSteen P. 2010. Auxin and monocot development. Cold Spring Harb. Perspect. Biol. 2:3. 10.1101/cshperspect.a001479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P. 2011. Vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23:550–566. 10.1105/tpc.110.075267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uggla C, Moritz T, Sandberg G, Sundberg B. 1996. Auxin as a positional signal in pattern formation in plants. Proc. Natl. Acad. Sci. U. S. A. 93:9282–9286. 10.1073/pnas.93.17.9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao Y, Ferrer J, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, Cheng Y, Lim J, Zhao Y, Ballaré CL, Sandberg G, Noel JP, Chory J. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176. 10.1016/j.cell.2008.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casal JJ. 2013. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64:403–427. 10.1146/annurev-arplant-050312-120221 [DOI] [PubMed] [Google Scholar]
- 8.Bianco C, Imperlini E, Calogero R, Senatore B, Amoresano A, Carpentieri A, Pucci P, Defez R. 2006. Indole-3-acetic acid improves Escherichia coli's defenses to stress. Arch. Microbiol. 185:373–382. 10.1007/s00203-006-0103-y [DOI] [PubMed] [Google Scholar]
- 9.Bianco C, Imperlini E, Defez R. 2009. Legumes like more IAA. Plant Signal. Behav. 4:763–765. 10.4161/psb.4.8.9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imperlini E, Bianco C, Lonardo E, Camerini S, Cermola M, Moschetti G, Defez R. 2009. Effects of indole-3-acetic acid on Sinorhizobium meliloti survival and on symbiotic nitrogen fixation and stem dry weight production. Appl. Microbiol. Biotechnol. 83:727–738. 10.1007/s00253-009-1974-z [DOI] [PubMed] [Google Scholar]
- 11.Donati AJ, Lee H, Leveau JH, Chang W. 2013. Effects of indole-3-acetic acid on the transcriptional activities and stress tolerance of Bradyrhizobium japonicum. PLoS One 8:e76559. 10.1371/journal.pone.0076559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott JC, Greenhut IV, Leveau JH. 2013. Functional Characterization of the bacterial iac genes for degradation of the plant hormone indole-3-acetic acid. J. Chem. Ecol. 39:942–951. 10.1007/s10886-013-0324-x [DOI] [PubMed] [Google Scholar]
- 13.Monier J, Lindow S. 2005. Aggregates of resident bacteria facilitate survival of immigrant bacteria on leaf surfaces. Microb. Ecol. 49:343–352. 10.1007/s00248-004-0007-9 [DOI] [PubMed] [Google Scholar]
- 14.El-Shanshoury AR. 1991. Biosynthesis of indole-3-acetic acid in Streptomyces atroolivaceus and its changes during spore germination and mycelial growth. Microbios 67:159–164 [Google Scholar]
- 15.Matsukawa E, Nakagawa Y, Iimura Y, Hayakawa M. 2007. Stimulatory effect of indole-3-acetic acid on aerial mycelium formation and antibiotic production in Streptomyces spp. Actinomycetologica 21:32–39. 10.3209/saj.SAJ210105 [DOI] [Google Scholar]
- 16.Van Puyvelde S, Cloots L, Engelen K, Das F, Marchal K, Vanderleyden J, Spaepen S. 2011. Transcriptome analysis of the rhizosphere bacterium Azospirillum brasilense reveals an extensive auxin response. Microb. Ecol. 61:723–728. 10.1007/s00248-011-9819-6 [DOI] [PubMed] [Google Scholar]
- 17.Yuan Z, Haudecoeur E, Faure D, Kerr KF, Nester EW. 2008. Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole-3-acetic acid, and γ-amino butyric acid reveals signaling cross-talk and Agrobacterium–plant co-evolution. Cell. Microbiol. 10:2339–2354. 10.1111/j.1462-5822.2008.01215.x [DOI] [PubMed] [Google Scholar]
- 18.Yang S, Zhang Q, Guo J, Charkowski AO, Glick BR, Ibekwe AM, Cooksey DA, Yang C. 2007. Global effect of indole-3-acetic acid biosynthesis on multiple virulence factors of Erwinia chrysanthemi 3937. Appl. Environ. Microbiol. 73:1079–1088. 10.1128/AEM.01770-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alfano JR, Collmer A. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385–414. 10.1146/annurev.phyto.42.040103.110731 [DOI] [PubMed] [Google Scholar]
- 20.Duca D, Lorv J, Patten CL, Rose D, Glick BR. 2014. Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek. 10.1007/s10482-013-0095-y [DOI] [PubMed] [Google Scholar]
- 21.Lehmann T, Hoffmann M, Hentrich M, Pollmann S. 2010. Indole-3-acetamide-dependent auxin biosynthesis: a widely distributed way of indole-3-acetic acid production? Eur. J. Cell Biol. 89:895–905. 10.1016/j.ejcb.2010.06.021 [DOI] [PubMed] [Google Scholar]
- 22.Duan J, Jiang W, Cheng Z, Heikkila JJ, Glick BR. 2013. The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. PLoS One 8:e58640. 10.1371/journal.pone.0058640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum-parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pace HC, Brenner C. 2001. The nitrilase superfamily: classification, structure, and function. Genome Biol. 2:reviews0001.1-reviews0001.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi M, Izui H, Nagasawa T, Yamada H. 1993. Nitrilase in biosynthesis of the plant hormone indole-3-acetic acid from indole-3-acetonitrile: cloning of the Alcaligenes gene and site-directed mutagenesis of cysteine residues. Proc. Natl. Acad. Sci. U. S. A. 90:247–251. 10.1073/pnas.90.1.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howden AJM, Rico A, Mentlak T, Miguet L, Preston GM. 2009. Pseudomonas syringae pv. syringae B728a hydrolyzes indole-3-acetonitrile to the plant hormone indole-3-acetic acid. Mol. Plant Pathol. 10:857–865. 10.1111/j.1364-3703.2009.00595.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Tiwari MK, Moon H, Jeya M, Ramu T, Oh D, Kim I, Lee J. 2009. Identification and characterization of a novel nitrilase from Pseudomonas fluorescens Pf-5. Appl. Microbiol. Biotechnol. 83:273–283. 10.1007/s00253-009-1862-6 [DOI] [PubMed] [Google Scholar]
- 28.Kiziak C, Conradt D, Stolz A, Mattes R, Klein J. 2005. Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology 151:3639–3648. 10.1099/mic.0.28246-0 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Xu J, He Y, Ouyang L, Liu Y. 2011. Cloning and biochemical properties of a highly thermostable and enantioselective nitrilase from Alcaligenes sp. ECU0401 and its potential for (R)-(–)-mandelic acid production. Bioprocess Biosyst. Eng. 34:315–322. 10.1007/s00449-010-0473-z [DOI] [PubMed] [Google Scholar]
- 30.Nageshwar Y, Sheelu G, Shambhu RR, Muluka H, Mehdi N, Malik MS, Kamal A. 2011. Optimization of nitrilase production from Alcaligenes faecalis MTCC 10757 (IICT-A3): effect of inducers on substrate specificity. Bioprocess Biosyst. Eng. 34:515–523. 10.1007/s00449-010-0500-0 [DOI] [PubMed] [Google Scholar]
- 31.Prasad S, Misra A, Jangir VP, Awasthi A, Raj J, Bhalla TC. 2007. A propionitrile-induced nitrilase of Rhodococcus sp. NDB 1165 and its application in nicotinic acid synthesis. World J. Microbiol. Biotechnol. 23:345–353 [Google Scholar]
- 32.Wang H, Sun H, Wei D. 2013. Discovery and characterization of a highly efficient enantioselective mandelonitrile hydrolase from Burkholderia cenocepacia J2315 by phylogeny-based enzymatic substrate specificity prediction. BMC Biotechnol. 13:14. 10.1186/1472-6750-13-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Layh N, Parratt J, Willetts A. 1998. Characterization and partial purification of an enantioselective arylacetonitrilase from Pseudomonas fluorescens DSM 7155. J. Mol. Catal. B 5:467–474. 10.1016/S1381-1177(98)00075-7 [DOI] [Google Scholar]
- 34.Pawar SV, Meena VS, Kaushik S, Kamble A, Kumar S, Chisti Y, Banerjee U. 2012. Stereo-selective conversion of mandelonitrile to (R)-(–)-mandelic acid using immobilized cells of recombinant Escherichia coli. 3 Biotech. 2:319–326. 10.1007/s13205-012-0058-4 [DOI] [Google Scholar]
- 35.Banerjee A, Kaul P, Banerjee UC. 2006. Purification and characterization of an enantioselective arylacetonitrilase from Pseudomonas putida. Arch. Microbiol. 184:407–418. 10.1007/s00203-005-0061-9 [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi M, Yanaka N, Nagasawa T, Yamada H. 1990. Purification and characterization of a novel nitrilase of Rhodococcus rhodochrous K22 that acts on aliphatic nitriles. J. Bacteriol. 172:4807–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen M, Zheng Y, Shen Y. 2009. Isolation and characterization of a novel Arthrobacter nitroguajacolicus ZJUTB06-99, capable of converting acrylonitrile to acrylic acid. Process Bioche. 44:781–785. 10.1016/j.procbio.2009.03.006 [DOI] [Google Scholar]
- 38.McNear D., Jr 2013. The rhizosphere: roots, soil and everything in between. Nat. Education Knowledge 4(3):1 [Google Scholar]
- 39.Booth IR. 1985. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 49:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldlust A, Bohak Z. 1989. Induction, purification, and characterization of the nitrilase of Fusarium oxysporum f. sp. melonis. Biotechnol. Appl. Biochem. 11:581–601 [Google Scholar]
- 41.Stevenson D, Feng R, Dumas F, Groleau D, Mihoc A, Storer A. 1992. Mechanistic and structural studies on Rhodococcus ATCC 39484 nitrilase. Biotechnol. Appl. Biochem. 15:283–302 [PubMed] [Google Scholar]
- 42.Effenberger F, Osswald S. 2001. Enantioselective hydrolysis of (RS)-2-fluoroarylacetonitriles using nitrilase from Arabidopsis thaliana. Tetrahedron Asymmetry 12:279–285. 10.1016/S0957-4166(01)00034-9 [DOI] [Google Scholar]
- 43.Osswald S, Wajant H, Effenberger F. 2002. Characterization and synthetic applications of recombinant AtNIT1 from Arabidopsis thaliana. Eur. J. Biochem. 269:680–687. 10.1046/j.0014-2956.2001.02702.x [DOI] [PubMed] [Google Scholar]
- 44.Fernandes B, Mateo C, Kiziak C, Chmura A, Wacker J, van Rantwijk F, Stolz A, Sheldon RA. 2006. Nitrile hydratase activity of a recombinant nitrilase. Adv. Synth. Catal. 348:2597–2603. 10.1002/adsc.200600269 [DOI] [Google Scholar]
- 45.Kiziak C, Stolz A. 2009. Identification of amino acid residues responsible for the enantioselectivity and amide formation capacity of the arylacetonitrilase from Pseudomonas fluorescens EBC191. Appl. Environ. Microbiol. 75:5592–5599. 10.1128/AEM.00301-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banerjee A, Sharma R, Banerjee U. 2002. The nitrile-degrading enzymes: current status and future prospects. Appl. Microbiol. Biotechnol. 60:33–44. 10.1007/s00253-002-1062-0 [DOI] [PubMed] [Google Scholar]
- 47.Sharma N, Bhalla T. 2012. Motif design for nitrilases. J. Data Mining Genomics Proteomics 3(3):119 [Google Scholar]
- 48.Sharma N, Kushwaha R, Sodhi J, Bhalla T. 2009. In Silico analysis of amino acid sequences in relation to specificity and physiochemical properties of some microbial nitrilases. J. Proteomics Bioinform. 2:185–192. 10.4172/jpb.1000076 [DOI] [Google Scholar]
- 49.Lu J, Zheng Y, Yamagishi H, Odaka M, Tsujimura M, Maeda M, Endo I. 2003. Motif CXCC in nitrile hydratase activator is critical for NHase biogenesis in vivo. FEBS Lett. 553:391–396. 10.1016/S0014-5793(03)01070-6 [DOI] [PubMed] [Google Scholar]
- 50.Hann EC, Eisenberg A, Fager SK, Perkins NE, Gallagher FG, Cooper SM, Gavagan JE, Stieglitz B, Hennessey SM, DiCosimo R. 1999. 5-Cyanovaleramide production using immobilized Pseudomonas chlororaphis B23. Bioorg. Med. Chem. 7:2239–2245. 10.1016/S0968-0896(99)00157-1 [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi M, Shimizu S. 2000. Nitrile hydrolases. Curr. Opin. Chem. Biol. 4:95–102. 10.1016/S1367-5931(99)00058-7 [DOI] [PubMed] [Google Scholar]
- 52.Kobayashi M, Suzuki T, Fujita T, Masuda M, Shimizu S. 1995. Occurrence of enzymes involved in biosynthesis of indole-3-acetic acid from indole-3-acetonitrile in plant-associated bacteria, Agrobacterium and Rhizobium. Proc. Natl. Acad. Sci. U. S. A. 92:714–718. 10.1073/pnas.92.3.714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vega-Hernández MC, León-Barrios M, Pérez-Galdona R. 2002. Indole-3-acetic acid production from indole-3-acetonitrile in Bradyrhizobium. Soil Biol. Biochem. 34:665–668. 10.1016/S0038-0717(01)00229-2 [DOI] [Google Scholar]
- 54.Nojiri M, Yohda M, Odaka M, Matsushita Y, Tsujimura M, Yoshida T, Dohmae N, Takio K, Endo I. 1999. Functional expression of nitrile hydratase in Escherichia coli: requirement of a nitrile hydratase activator and post-translational modification of a ligand cysteine. J. Biochem. 125:696–704. 10.1093/oxfordjournals.jbchem.a022339 [DOI] [PubMed] [Google Scholar]
- 55.Hashimoto Y, Nishiyama M, Horinouchi S, Beppu T. 1994. Nitrile hydratase gene from Rhodococcus sp. N-774 requirement for its downstream region for efficient expression. Biosci. Biotechnol. Biochem. 58:1859–1865 [DOI] [PubMed] [Google Scholar]
- 56.Nagasawa T, Ryuno K, Yamada H. 1986. Nitrile hydratase of Brevibacterium R312: purification and characterization. Biochem. Biophys. Res. Commun. 139:1305–1312. 10.1016/S0006-291X(86)80320-5 [DOI] [PubMed] [Google Scholar]
- 57.Endo T, Watanabe I. 1989. Nitrile hydratase of Rhodococcus sp. N-774 purification and amino acid sequences. FEBS Lett. 243:61–64 [DOI] [PubMed] [Google Scholar]
- 58.Yamada H, Kobayashi M. 1996. Nitrile hydratase and its application to industrial production of acrylamide. Biosci. Biotechnol. Biochem. 60:1391–1400. 10.1271/bbb.60.1391 [DOI] [PubMed] [Google Scholar]
- 59.Prasad S, Raj J, Bhalla T. 2009. Purification of a hyperactive nitrile hydratase from resting cells of Rhodococcus rhodochrous PA-34. Indian J. Microbiol. 49:237–242. 10.1007/s12088-009-0033-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng Z, Woody OZ, Song J, Glick BR, McConkey BJ. 2009. Proteome reference map for the plant growth-promoting bacterium Pseudomonas putida UW4. Proteomics 9:4271–4274. 10.1002/pmic.200900142 [DOI] [PubMed] [Google Scholar]
- 61.Precigou S, Goulas P, Duran R. 2001. Rapid and specific identification of nitrile hydratase (NHase)-encoding genes in soil samples by polymerase chain reaction. FEMS Microbiol. Lett. 204:155–161. 10.1111/j.1574-6968.2001.tb10879.x [DOI] [PubMed] [Google Scholar]
- 62.Haas C, Rodionov D, Kropat J, Malasarn D, Merchant S, de Crécy-Lagard V. 2009. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10:470. 10.1186/1471-2164-10-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278:41148–41159. 10.1074/jbc.M305837200 [DOI] [PubMed] [Google Scholar]
- 64.Goda M, Hashimoto Y, Takase M, Herai S, Iwahara Y, Higashibata H, Kobayashi M. 2002. Isonitrile hydratase from Pseudomonas putida N19-2: cloning, sequencing, gene expression, and identification of its active amino acid residue. J. Biol. Chem. 277:45860–45865. 10.1074/jbc.M208571200 [DOI] [PubMed] [Google Scholar]
- 65.Zhou Z, Hashimoto Y, Kobayashi M. 2009. Self-subunit swapping chaperone needed for the maturation of multimeric metalloenzyme nitrile hydratase by a subunit exchange mechanism also carries out the oxidation of the metal ligand cysteine residues and insertion of cobalt. J. Biol. Chem. 284:14930–14938. 10.1074/jbc.M808464200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okamoto S, Van Petegem F, Patrauchan MA, Eltis LD. 2010. AnhE, a metallochaperone involved in the maturation of a cobalt-dependent nitrile hydratase. J. Biol. Chem. 285:25126–25133. 10.1074/jbc.M110.109223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Podar M, Eads JR, Richardson TH. 2005. Evolution of a microbial nitrilase gene family: a comparative and environmental genomics study. BMC Evol. Biol. 5:42. 10.1186/1471-2148-5-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brandão PF, Bull AT. 2003. Nitrile hydrolyzing activities of deep-sea and terrestrial mycolate actinomycetes. Antonie Van Leeuwenhoek 84:89–98. 10.1023/A:1025409818275 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.