Abstract
Members of Methanocellales are widespread in paddy field soils and play the key role in methane production. These methanogens feature largely in these organisms' adaptation to low H2 and syntrophic growth with anaerobic fatty acid oxidizers. The adaptive mechanisms, however, remain unknown. In the present study, we determined the transcripts of 21 genes involved in the key steps of methanogenesis and acetate assimilation of Methanocella conradii HZ254, a strain recently isolated from paddy field soil. M. conradii was grown in monoculture and syntrophically with Pelotomaculum thermopropionicum (a propionate syntroph) or Syntrophothermus lipocalidus (a butyrate syntroph). Comparison of the relative transcript abundances showed that three hydrogenase-encoding genes and all methanogenesis-related genes tested were upregulated in cocultures relative to monoculture. The genes encoding formylmethanofuran dehydrogenase (Fwd), heterodisulfide reductase (Hdr), and the membrane-bound energy-converting hydrogenase (Ech) were the most upregulated among the evaluated genes. The expression of the formate dehydrogenase (Fdh)-encoding gene also was significantly upregulated. In contrast, an acetate assimilation gene was downregulated in cocultures. The genes coding for Fwd, Hdr, and the D subunit of F420-nonreducing hydrogenase (Mvh) form a large predicted transcription unit; therefore, the Mvh/Hdr/Fwd complex, capable of mediating the electron bifurcation and connecting the first and last steps of methanogenesis, was predicted to be formed in M. conradii. We propose that Methanocella methanogens cope with low H2 and syntrophic growth by (i) stabilizing the Mvh/Hdr/Fwd complex and (ii) activating formate-dependent methanogenesis.
INTRODUCTION
The 16S rRNA gene of a new type of methanogen, Methanocellales, was discovered in 1998 from the surface of rice root (1). The 16S rRNA gene sequences were found phylogenetically branching off between Methanosarcinaceae and Methanomicrobiales. An enrichment culture at 50°C revealed that these organisms contained the methyl-coenzyme M (CoM) reductase-encoding genes, confirming their nature as methanogenic archaea (2, 3). They were found to be widespread in rice field soils and the environment, including desert soil (4–7). Due to the difficulty of cultivation, many molecular studies were conducted to characterize their ecological functions prior to isolation into pure culture. The application of stable isotope probing technology showed that these organisms outcompeted other methanogens under low-H2 conditions (8). This result suggested, for the first time, that these methanogens are intrinsically adaptive to low H2. Moreover, they were found to play the key role in CH4 production from root-derived material in the rice rhizosphere in situ, illustrating their ecological significance and niche specificity (9).
More evidence for the adaptation of Methanocellales to low H2 came from the investigations of syntrophic oxidation of short-chain fatty acids (10–14). In the investigations of syntrophic oxidations of acetate, propionate, and butyrate in Italian and Chinese rice soils, it was revealed that Methanocellales played the key role in H2 consumption for the syntrophic interactions (10–14). These ecological studies reconfirm that Methanocellales are adapted to low-H2 conditions. The mechanism, however, remains unknown.
H2 is the major electron donor for CH4 production and retains a central role in interspecies electron transfer for syntrophic fatty acid oxidation (15). Several studies have aimed to understand the mechanisms for the low-H2 adaptation of methanogens. A study on Methanothermobacter thermautotrophicus revealed that one of two methyl-coenzyme M reductases (MCRI) dominated under syntrophic conditions, while both MCRI and MCRII were expressed in pure culture (16). MCRI has a higher substrate affinity than MCRII; hence, it may play an important role in low-H2 adaptation. A study on Methanococcus maripaludis (17) showed that the genes coding for enzymes that reduce or oxidize the deazaflavin coenzyme F420 were significantly upregulated under H2 limitation. Similar results were observed in several other studies (16, 18–23). A recent study on M. maripaludis grown syntrophically with Desulfovibrio vulgaris revealed that most of the genes for methanogenesis were upregulated, while those for biosynthetic functions declined compared to levels in monoculture (24). In addition, this methanogen appears capable of activating the functions of paralogous genes to cope with the changing substrate availability (24). The transcript level of paralogs that were previously found upregulated with hydrogen limitation in monoculture (17) was significantly downregulated in the syntrophic coculture. Therefore, it appears that different paralogs are activated under pure low H2 and syntrophic growth conditions. Methanospirillum hungatei, on the other hand, showed that the gene expression levels of hydrogenase and formate dehydrogenase did not vary with culture conditions (25). Given that Methanocella are environmentally important, elucidating the mechanisms for their low-H2 adaptation is of great interest.
The first Methanocella strain, M. paludicola SANAE, was isolated from a Japanese rice field soil in 2007 (26). Thereafter, two more isolates, M. arvoryzae MRE50 and M. conradii HZ254, were obtained from Italian and Chinese rice field soils, respectively (27, 28). The whole-genome sequences of all three strains are now available (29–31). Of the three strains, M. conradii HZ254 shows the fastest growth, with a doubling time of 6.5 to 7.8 h under optimum conditions (27). In the present study, we constructed two cocultures by growing M. conradii syntrophically with Pelotomaculum thermopropionicum or Syntrophothermus lipocalidus. The transcript levels of key genes associated with methanogenesis and acetate assimilation in M. conradii were determined during the syntrophic growth compared to growth in monoculture. The genome of M. conradii lacks carbon monoxide dehydrogenase; hence, acetate is essential for growth (27). Therefore, the expression of acetate assimilation-related genes is related to the first step of M. conradii biosynthesis. Formate may serve as an alternative shuttle for the interspecies electron transfer in the syntrophic fatty acid oxidations (32, 33). Physiological tests showed no growth of M. conradii on formate alone (27), but the genes coding for formate dehydrogenase (Fdh) are present and were expressed under the experimental conditions. Therefore, we reevaluated the growth of M. conradii on formate in the presence of H2.
MATERIALS AND METHODS
Microorganisms and cultivation.
Methanocella conradii strain HZ254T was isolated in our laboratory and has been described previously (27). Pelotomaculum thermopropionicum strain SI (DSM13744) and Syntrophothermus lipocalidus strain TGB-C1 (DSM12680) were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany).
M. conradii monoculture was grown in 550-ml serum bottles containing 100 ml of minimal salts medium (with a 10% volume of inoculation) under H2:CO2 (80:20 [vol/vol]) at 1.7 atm and incubated for about 24 days to allow the full utilization of H2 in the headspace (Fig. 1A), as described previously (27). The production of CH4 and the consumption of H2 were determined. As a test of formate utilization, 4 mM H13COONa (99 atom%13C; Sigma-Aldrich) was added to a 125-ml serum bottle containing 50 ml medium. All other conditions were the same as those for H2-only incubation.
FIG 1.
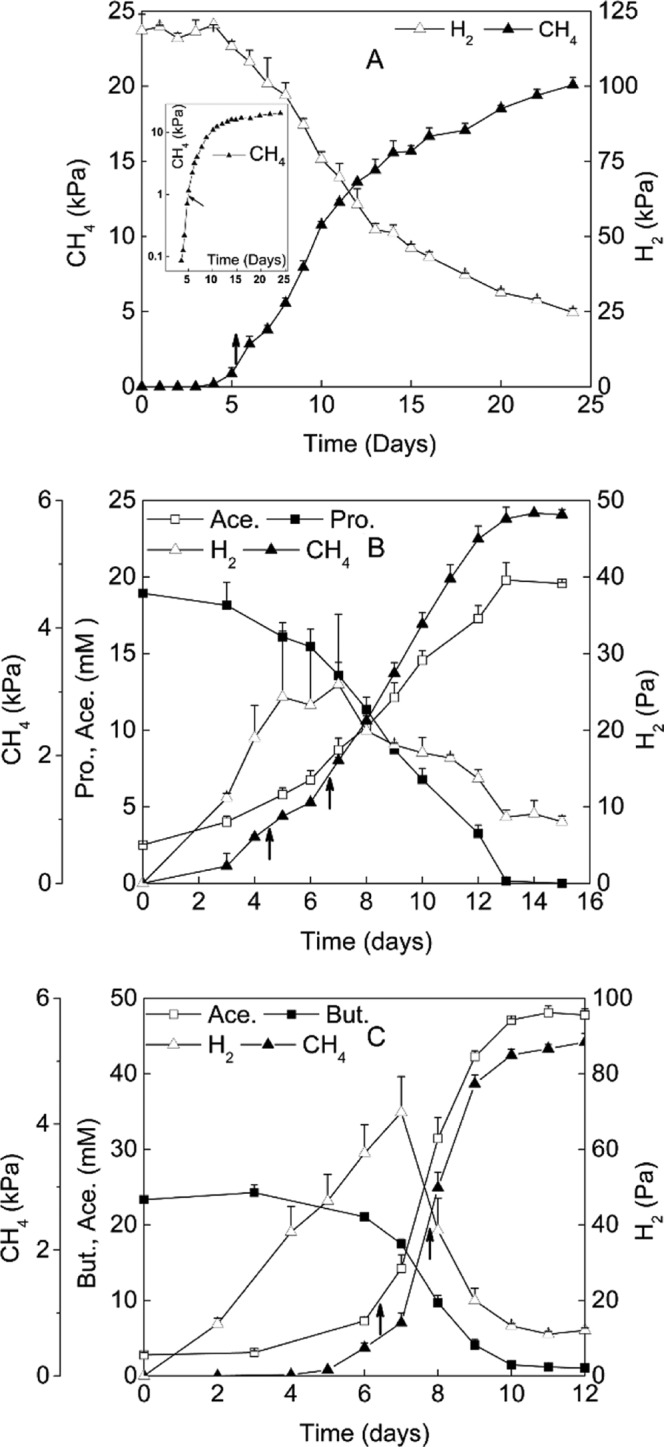
Time courses of substrates and products of M. conradii monoculture (A) and syntrophic cocultures with P. thermopropionicum (B) or S. lipocalidus (C). The inset in panel A shows log10(CH4) values. Ace., acetate; Pro., propionate; But., butyrate. Arrows indicated the time points for sampling. The values of gas samples (CH4 and H2) are means from three replicates, and the values of fatty acids (acetate, propionate, and butyrate) are means from two replicates. Error bars indicate standard deviations.
Syntrophic cocultures were obtained by adding an ∼5% volume of P. thermopropionicum or S. lipocalidus to a vigorously growing culture of M. conradii, flushing the headspace with N2, and finally adjusting the headspace with N2:CO2 (80:20 [vol/vol]) to a final pressure of 1.7 atm. The substrate sodium propionate or sodium butyrate was added to a final concentration of 20 mM. The production of CH4 and acetate and the consumption of propionate and butyrate were monitored to verify growth. After 14 and four successive transfers were conducted (each time with a 10% volume of inoculation) for propionate and butyrate cocultures, respectively, 550-ml serum bottles containing 100 ml of minimal salts medium plus 25 mM butyrate or 20 mM propionate were inoculated and incubated for cell collection. Cultivation was carried out at 55°C in the dark without agitation.
The cell density of HZ254 at the early stage of logarithmic growth was low, and it was technically problematic to harvest a sufficient amount of cells for RNA extraction. Therefore, cells were harvested only once for M. conradii monoculture at the middle point of logarithmic growth, which was about 4 to 5 days after inoculation (Fig. 1A, arrows). For two syntrophic cocultures, the time window of logarithmic growth was sufficient for collecting the cells twice at the early and mid-logarithmic phases, respectively (sampling was carried out about 5 and 7 days and 6 and 8 days after inoculation for syntrophic propionate and butyrate cocultures, respectively) (Fig. 1B and C, arrows). This allowed us to track the possible changes in gene expression during logarithmic growth. For all sampling points, three replicates were carried out, except for MP2, which had only two replicates. The cell suspensions were centrifuged at 16,000 × g at 4°C for 10 min (Avanti J-26XP; Beckman Coulter, USA), and the pellets were stored immediately at −80°C until RNA extraction.
Chemical analyses.
Gasses were sampled with a pressure-lock syringe (VICI, USA), and the concentrations of methane and hydrogen were determined using a gas chromatograph (GC; 7890A; Agilent, USA) equipped with an 80/100-mesh Porapak Q column (Supelco; Sigma-Aldrich, USA) and a thermal conductivity detector. Nitrogen was used as a carrier gas and as a reference for thermal conductivity detection (TCD). The column and the detector temperatures were 45°C and 250°C, respectively. The concentrations of propionate, butyrate, and acetate in the medium were measured with a high-performance liquid chromatograph (1200; Agilent, USA) equipped with a 4.6-mm-inner-diameter, 250-mm (5-μm) Zorbax SB-Aq C18 column (Agilent, USA) and a UV detector. The column was operated at 25°C, and 20 mM NaH2PO4 in 0.5% acetonitrile (pH adjusted to 2.0 with H3PO4) was used as a carrier at a flow rate of 0.8 ml min−1. The liquid samples were filtrated with a polyethersulfone (PES) membrane filter (0.22-μm pore size; Anpel, Shanghai, China) before the measurements.
For the measurements of 13CH4 and 13CO2, 0.5 ml of gas sample was taken from the headspace with a pressure-lock syringe (VICI, USA) and diluted into 15-ml serum tubes containing pure N2. Stable 13C/12C isotope ratios of CH4 and CO2 were determined using a GC-combustion-isotope ratio mass spectrometry (GC-C-IRMS) system (Thermo Finnigan, Germany) by following procedures described previously (34).
Nucleic acid extraction and purification and cDNA synthesis.
For each sample collected as described above, RNA was extracted using TRIzol reagent (Invitrogen) as described previously (35), with modifications. Cell pellets with 1.2 ml of TRIzol reagent were homogenized at 5.0 m s−1 for 20 s in a FastPrep-24 (MP Biomedicals, USA). Tubes were incubated at 25°C for 3 min and 250 μl chloroform was added, followed by 15 s of shaking by hand. After incubation at 25°C for 10 min, the suspension was centrifuged at 16,000 × g at 4°C for 15 min. The supernatant (750 to 800 μl) was transferred to a new tube, and 800 μl of 2-propanol was added, mixed, and incubated at 25°C for 15 min. The mixtures were centrifuged at 16,000 × g for 15 min at 4°C. The supernatants were discarded and the pellets were washed with 1,000 μl of 70% ethanol. The pellets were air dried, and the RNA samples were finally dissolved in 50 μl RNase-free water. DNA digestion was performed by incubation at 37°C for 1 h in a total volume of 50 μl containing 41 μl of RNA extracts, 5 μl of RQ1 RNase-free DNase, 10× reaction buffer, 3 μl of RQ1 RNase-free DNase, and 1 μl of recombinant RNasin RNase inhibitor (Promega, Madison, WI, USA). RNA was further purified using an RNeasy minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The quality and purity of RNA were checked by 1% agarose gel electrophoresis and NanoDrop-2000 spectrophotometry (NanoDrop Technologies, Wilmington, DE). Both indicated a good quality of RNA extracts. The ImProm-II reverse transcription system (Promega, Madison, WI, USA) was used to synthesize the complete cDNA according to the manufacturer's instructions. To verify the absence of DNA, a control reaction was performed with nuclease-free water instead of reverse transcriptase.
For the quantification of the 16S rRNA gene and transcripts, DNA and RNA were coextracted by using an AllPrep DNA/RNA minikit (Qiagen, Germany) according to the manufacturer's manual. DNA in coextracts was directly used for quantification. For RNA quantification, the coextracts were subjected to DNA digestion and purification, and then RNA was converted to cDNA as described above.
Primer design, qPCR, and data analysis.
Primers for the relative quantification of functional genes were designed with Primer Premier 6 (Premier, Canada) and synthesized by Life Technologies (Shanghai, China) (see Table S1 in the supplemental material). The optimal melting temperature (58°C) of each primer set was experimentally verified with genomic DNA from M. conradii as the template. Quantitative PCR (qPCR) was performed with the 7500 real-time PCR system (Applied Biosystems, USA). Each 25-μl reaction mixture contained 5 μl 25× diluted cDNA, 12.5 μl 2× SYBR green PCR mix (Applied Biosystems, USA), 0.5 μl of bovine serum albumin, and 0.2 μM each primer. Thermocycling conditions were 50°C for 2 min, 95°C for 10 min, 45 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 1 min, with fluorescence detection at the end of each extension step. Amplification was followed by a melting program consisting of 95°C for 1 min, 60°C for 1 min, and a stepwise temperature increase of 0.5°C per 10 s with fluorescence detection at each temperature transition. A single peak of the melting curve and the presence of only one band in the electrophoresis gel indicated that a single amplicon was generated from each primer pair. The optimal baseline and threshold cycle (CT) values were calculated by using the ABI 7500 real-time PCR system sequence detection software, version 1.3.1, according to the user's manual. Amplification efficiency for each individual reaction was calculated from the amplification profile of each sample (36). All transcript abundances were normalized to 16S rRNA levels of M. conradii to obtain the relative expression levels. It is known that rRNA constitutes the major part of total RNA (over 90%), and the ratio of 16S rRNA to total RNA is relatively constant (37). Therefore, we assumed that the ratio of a given mRNA target to 16S rRNA transcripts represented the relative expression level of that gene against the total RNA. Three biological replicates (two for MP2) and three technical replicates were performed for measurement.
For the quantification of 16S rRNA genes and transcripts, primer pair Ar364f/Ar915r (38) was used. Each 25-μl reaction mixture contained 12.5 μl 2× Go Taq qPCR Master mix (Promega, Madison, WI, USA) and 0.2 μM Ar364f/Ar915r, 0.5 μl of bovine serum albumin (10 mg/ml), and 5 μl of DNA (1/103 dilution) or cDNA (1/104 dilution). Thermocycling conditions were 95°C for 2 min, 45 cycles of 95°C for 30 s, 65°C for 30 s, 72°C for 1 min, and 82°C for 1 min, with fluorescence detection at the end of each extension step. Amplification was followed by a melting program consisting of 95°C for 1 min, 60°C for 1 min, and a stepwise temperature increase of 0.5°C per 10 s, with fluorescence detection at each temperature transition. The DNA standard and transcript standard were prepared as described earlier (39), excepted that we used the purified PCR product of a 16S rRNA clone amplified by T7/M13R primer pairs as the DNA standard. The concentration of DNA and transcript standard ranged from 1.0 × 102 to 1.0 × 108 copies μl−1. Each measurement was performed in three replicates. Since the genome of M. conradii contains two copies of 16S rRNA gene, the cell number of M. conradii per ml was estimated by dividing the total quantity of 16S rRNA gene per ml by 2.
The means ± standard deviations (SD) from all replicates were calculated, and the analysis of variance was performed to test significant differences between treatments using the DUNCAN test in the SAS program (SAS Institute, Cary, NC).
RESULTS
M. conradii monoculture showed a lag phase of 3 to 4 days before exponential growth (Fig. 1A). CH4 was stoichiometrically produced from H2 and CO2. The production of CH4 showed a lag phase of 3 to 4 days in propionate coculture and 6 days in butyrate coculture (Fig. 1B and C). H2 in the headspace reached a maximal partial pressure of 30 to 40 Pa in propionate and 80 Pa in butyrate cocultures, respectively. CH4 and acetate were produced stoichiometrically during the syntrophic oxidations of propionate and butyrate. The doubling time of M. conradii was estimated to be 7 to 8 h in monoculture based on CH4 production, consistent with the maximal growth rate under optimum conditions (27). The doubling time during syntrophic growth increased to 14 to 16 h in butyrate and 40 to 45 h in propionate cocultures, respectively.
RNA samples in monoculture were collected in the mid-exponential phase (Fig. 1A, arrows). RNA sampling in syntrophic cocultures was performed twice, in the early and mid-exponential phases (Fig. 1B and C, arrows). Quantification of rRNA transcripts indicated that the rRNA expression of M. conradii in syntrophic cultures decreased by a factor of 1.8 to 2.4 compared to that in monoculture (Table 1). Twenty-one genes involved in methanogenesis and acetate assimilation were selected for the relative expression analyses (see Table S1 in the supplemental material). Primers targeting each gene were designed according to the genome sequence. If a predicted transcription unit contains multiple genes, one gene was selected for the measurement (see Table S1).
TABLE 1.
Numbers of 16S rRNA transcripts per cell in M. conradii growing in monoculture and syntrophic cocultures with P. thermopropionicum or S. lipocalidus
| Culturea | Transcript no. per cellb | Significancec |
|---|---|---|
| M | 509.5 ± 87.6 | A |
| MB1 | 294.2 ± 54.7 | B |
| MB2 | 252.7 ± 94.3 | B |
| MP1 | 195.1 ± 106 | B |
| MP2 | 216.3 ± 35.1 | B |
M, monoculture; MB and MP, syntrophic butyrate and propionate cocultures, respectively; the number after the culture designation (1 or 2) indicates the early- and mid-exponential phase, respectively.
Values are means from four biological replicates (three for MP1 and MP2) for each sample, and means ± SD are given.
Different letters after the transcript number indicate significant differences at a level of P < 0.05.
Hydrogenases and methanogenesis.
M. conradii contains two types of membrane-bound hydrogenases, the energy-converting hydrogenase (Ech) and the methanophenazine-reducing hydrogenase (Vht), and two types of cytoplasmic hydrogenase, the F420-reducing hydrogenase (Frh) and the methyl viologen-reducing hydrogenase (Mvh). An Ech-like hydrogenase was predicted which was similar to Ech but lacking EchA, the subunit coding for proton transporter. Frh has two predicted transcription units, fhrA1G1B1 and fhrA2DG2B2, and a third single frhB3 gene. Mvh also has two predicted transcription units, mvhG1A1 and mvhG2A2, with the genes mvhD1 and mvhD2 located at other loci (see Fig. S1 in the supplemental material).
The transcript level of echE was significantly upregulated 2- to 3-fold in cocultures compared to that in pure culture (Table 2). The transcript level of vhtG did not change in response to coculture, except for an increase in propionate at the first sampling. The transcript level of frhA2 was upregulated 1.6- to 1.9-fold in coculture. The transcript abundance of frhA1 was approximately two orders of magnitude lower than that for frhA2 and showed no response to syntrophic growth. Similarly, the transcript level of mvhA2 increased 1.7- to 1.9-fold in cocultures, except in butyrate coculture, at the early sampling. The transcript abundance of mvhA1 was 20- to 33-fold lower than that of mvhA2 and showed no response to syntrophic growth.
TABLE 2.
Expression of hydrogenase-encoding, methanogenesis-associated, and acetate assimilation-associated genes in M. conradii monoculture and syntrophic cocultures and fold change of expression levels in cocultures relative to monoculturea
| Locus tag and category | Gene name | Expression levelb (×10−3; means ±SD) |
Fold changec |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M | MB1 | MB2 | MP1 | MP2 | MB1 | MB2 | MP1 | MP2 | ||
| Hydrogenases | ||||||||||
| Mtc_0802 | echE | 0.17 ± 0.01 B | 0.54 ± 0.11 A | 0.52 ± 0.17 A | 0.55 ± 0.14 A | 0.38 ± 0.04 AB | 3.10* | 2.98* | 3.16* | 2.17 |
| Mtc_0470 | vhtG | 0.80 ± 0.06 B | 0.63 ± 0.11 B | 0.59 ± 0.32 B | 1.56 ± 0.19 A | 0.76 ± 0.38 B | 0.79 | 0.74 | 1.95* | 0.95 |
| Mtc_1001 | frhA1 | 0.05 ± 0.01 | 0.032 ± 0.004 | 0.049 ± 0.018 | 0.056 ± 0.023 | 0.046 ± 0.032 | 0.59 | 0.91 | 1.04 | 0.85 |
| Mtc_2127 | frhA2 | 1.55 ± 0.31 B | 2.50 ± 0.29 A | 2.90 ± 0.59 A | 2.50 ± 0.52 A | 3.05 ± 0.63 A | 1.61* | 1.87* | 1.61* | 1.96* |
| Mtc_2479 | mvhA1 | 0.045 ± 0.013 B | 0.061 ± 0.009 B | 0.082 ± 0.028 B | 0.13 ± 0.03 A | 0.077 ± 0.004 B | 1.36 | 1.83 | 2.97* | 1.72 |
| Mtc_0470 | mvhA2 | 1.51 ± 0.30 C | 1.77 ± 0.17 BC | 2.96 ± 0.85 A | 2.77 ± 0.46 A | 2.59 ± 0.33 AB | 1.17 | 1.96* | 1.83* | 1.72* |
| Methanogenesis pathway proteins | ||||||||||
| Mtc_0163 | fmdB | 0.0007 ± 0.0003 | 0.0011 ± 0.0003 | 0.0010 ± 0.0002 | 0.0011 ± 0.0004 | 0.0009 ± 0.0002 | 1.70 | 1.49 | 1.66 | 1.28 |
| Mtc_2470 | fwdB | 1.74 ± 0.33 B | 5.99 ± 1.62 A | 6.75 ± 1.07 A | 8.08 ± 0.79 A | 7.26 ± 0.57 A | 3.44* | 3.88* | 4.64* | 4.17* |
| Mtc_1592 | mtd | 1.72 ± 0.16 B | 1.87 ± 0.13 AB | 2.82 ± 0.92 AB | 3.16 ± 0.46 A | 3.11 ± 1.38 AB | 1.09 | 1.64 | 1.84* | 1.81 |
| Mtc_0135 | mer | 2.71 ± 0.78 B | 4.99 ± 0.56 A | 5.87 ± 1.21 A | 5.47 ± 0.91 A | 5.01 ± 1.54 A | 1.84* | 2.17* | 2.02* | 1.85* |
| Mtc_0943 | mtrH | 1.25 ± 0.06 B | 2.04 ± 0.11 A | 2.50 ± 0.11 A | 2.36 ± 0.52 A | 2.57 ± 0.61 A | 1.63* | 2.00* | 1.88* | 2.05* |
| Mtc_0908 | mcrA | 11.07 ± 0.45 C | 19.37 ± 2.43 B C | 30.24 ± 6.47 A | 25.97 ± 4.77 AB | 25.98 ± 7.43 AB | 1.75 | 2.73* | 2.35* | 2.35* |
| Mtc_1470 | hdrB1 | 0.0015 ± 0.0004 B | 0.0042 ± 0.0019 AB | 0.0040 ± 0.0023 AB | 0.0055 ± 0.0015 A | 0.0056 ± 0.0011 A | 2.85 | 2.75 | 3.78* | 3.81* |
| Mtc_2474 | hdrB2 | 0.64 ± 0.18 C | 2.36 ± 0.66 A | 2.33 ± 0.30 A | 2.30 ± 0.11 A | 1.55 ± 0.18 B | 3.69* | 3.64* | 3.60* | 2.42* |
| Mtc_0481 | hdrB3 | 0.095 ± 0.029 B | 0.28 ± 0.11 B | 0.30 ± 0.13 B | 0.59 ± 0.15 A | 0.61 ± 0.20 A | 2.96 | 3.19 | 6.18* | 6.36* |
| Mtc_2125 | fdhA | 0.44 ± 0.07 B | 1.35 ± 0.14 A | 2.18 ± 0.80 A | 2.26 ± 0.44 A | 2.28 ± 1.34 A | 3.10* | 4.99* | 5.18* | 5.22* |
| Acetate assimilation related proteins | ||||||||||
| Mtc_1904 | acs1 | 0.020 ± 0.005 A | 0.018 ± 0.003 A | 0.013 ± 0.001 AB | 0.015 ± 0.003 AB | 0.009 ± 0.003 B | 0.93 | 0.66 | 0.78 | 0.48* |
| Mtc_2228 | acs2 | 0.016 ± 0.010 A | 0.0021 ± 0.0002 B | 0.002 ± 0.001 B | 0.0015 ± 0.0004 B | 0.0016 ± 0.0002 B | 0.13* | 0.13* | 0.09* | 0.10* |
| Mtc_1504 | ppa | 0.12 ± 0.03 AB | 0.18 ± 0.01 A | 0.17 ± 0.06 A | 0.085 ± 0.042 B | 0.1201 ± 0.0003 AB | 1.55 | 1.43 | 0.72 | 1.02 |
| Mtc_0842 | porA-1 | 0.045 ± 0.013 B | 0.12 ± 0.03 A | 0.12 ± 0.04 A | 0.029 ± 0.01 B | 0.040 ± 0.003 B | 2.58* | 2.56* | 0.64 | 0.89 |
| Mtc_1765 | porA-2 | 0.043 ± 0.010 | 0.065 ± 0.006 | 0.065 ± 0.024 | 0.038 ± 0.011 | 0.052 ± 0.005 | 1.51 | 1.50 | 0.89 | 1.21 |
Results shown are expression levels (normalized as ratios of mRNA transcripts to 16S rRNA) of hydrogenase-encoding genes, methanogenesis-associated genes, and acetate assimilation-associated genes in M. conradii monoculture and syntrophic cocultures with P. thermopropionicum or S. lipocalidus, as well as fold changes of expression levels in M. conradii cocultures relative to monoculture. The gene names, locus tags, and primers for PCR amplification are given in Tables S1 and S2 in the supplemental material. M, monoculture; MB and MP, syntrophic butyrate and propionate cocultures, respectively. The numbers 1 and 2 indicate the early- and mid-exponential-growth phases.
Values are means from three biological replicates (two for MP2) and three technical replicates for each sample; means ± SD are given. Different letters indicate significant differences (P < 0.05).
For the fold change, values are the ratios of relative transcript abundance of target genes in cocultures compared to that in monoculture. An asterisk means significant difference (P < 0.05) between monoculture and cocultures.
The enzymes involved in methanogenesis of M. conradii include formylmethanofuran dehydrogenase (Fmd and Fwd, containing Mo and W, respectively), formylmethanofuran:tetrahydromethanopterin (H4MPT) formyltransferase (Ftr), methenyl-H4MPT cyclohydrolase (Mch), methylene-H4MPT dehydrogense (Mtd), methylene-H4MPT reductase (Mer), methyl-H4MPT:CoM methyltransferase (Mtr), methyl-coenzyme M reductase (Mcr), and CoB-CoM heterodisulfide reductase (Hdr) (see Fig. S1 in the supplemental material). Transcript analysis was conducted for all but Ftr and Mch enzymes.
The transcript abundance of fwdB was significantly upregulated 3.4- to 4.6-fold in cocultures (Table 2). The transcript level of fmdB (encoding the B subunit of Fmd) was four orders of magnitude lower than that of fwdB and showed no response to syntrophic growth. The transcript levels of mer, mtrH, and mcrA were moderately upregulated (1.6- to 2.3-fold) in cocultures, while mtd showed an increase only in propionate coculture at the first sampling. The gene hdrB has three homologous copies. hdrB2 is located in the large predicted transcription unit fwdGF-hdrC2B2A2-mvhD2-fwdBAC, and its expression was significantly upregulated 2.4- to 3.7-fold in cocultures (Table 2). hdrB1 is located in the predicted transcription unit of hdrA1B1C1, and hdrB3 is linked to the predicted transcription unit coding for Ech-like hydrogenase (see Fig. S1 in the supplemental material). The transcripts of hdrB1 and hdrB3 were also upregulated, but, significantly, only in propionate coculture. The transcript abundance of hdrB1 was two orders of magnitude lower than levels for hdrB2 and hdrB3 (Table 2).
The gene coding for the A subunit of F420-reducing formate dehydrogenase (fdhA) was transcribed to a level similar to that for hdrB2 and mtrH and was markedly upregulated (3- to 5-fold) in cocultures relative to monoculture (Table 2).
Acetate assimilation.
M. conradii employs the AMP-forming acetyl-coenzyme A (CoA) synthetase (ACS) for acetate assimilation, similar to most obligate hydrogenotrophic methanogens (40). This organism lacks genes encoding carbon monoxide dehydrogenase for the acetyl-CoA biosynthesis from CO2 (30, 41). Two putative pathways for the conversion between acetyl-CoA and pyruvate involved the pyruvate-ferredoxin oxidoreductase (Por) and the pyruvate dehydrogenase (Pdh) complex; the latter presumably was expressed under oxic conditions (29). M. conradii contains two acs copies. The transcript level was comparable between the two genes in the monoculture (Table 2). The transcript level of acs2, however, was significantly downregulated 8- to 10-fold in cocultures, while acs1 remained unaffected. The transcript abundance of a gene coding for the membrane-bound inorganic pyrophosphatase also was analyzed and showed no significant change among cultures (Table 2). Two por copies showed similar transcript levels (Table 2) and did not change significantly in cocultures, except for an increase of porA-1 in butyrate coculture.
Growth on formate.
To determine whether the strain could grow on formate in the presence of H2, we performed a carbon isotope labeling experiment. [13C]formate (99% 13C) was added to a final concentration of 4 mM in pure culture under an H2:CO2 atmosphere (4:1; 170 kPa). All of the formate was consumed in 6 days in parallel with the consumption of H2 (Fig. 2A). Meanwhile, the atomic 13C ratio of CO2 increased rapidly in the first 4 to 5 days; thereafter, it leveled off at around 9% (Fig. 2B). These data indicated that formate dehydrogenase catalyzed the conversion of formate to CO2 as formate + F420→F420H2 + CO2. However, the rapid increase of 13CO2 could result from vigorous isotope exchange. The 13CO2 produced in this process was then converted to 13CH4. Nevertheless, in total, 38.5 μmol of 13CH4 was recovered in the headspace at the end of incubation. If one molecule of CH4 was produced from four molecules of formate according to the reaction stoichiometry (4 HCOOH→CH4 + 3 CO2 + 2 H2O), the carbon mass balance indicated that at least 77% of formate added was utilized for CH4 production. These results evidenced that Fdh was functionally active.
FIG 2.
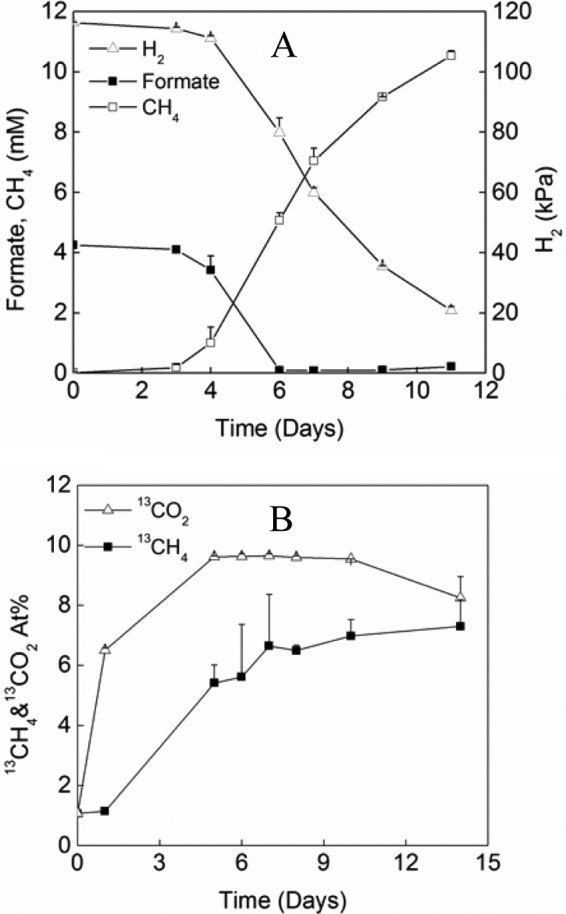
Growth of M. conradii in the presence of both H2 and formate. (A) Time courses of H2, CH4, and formate, where CH4 was expressed as mM equivalents (mMeq). mMeq is calculated according to C(mMeq) = n(CH4) × 103/V(liq) = PV(gas) × 103/RTV(liq), where n(CH4) is the amount of CH4 (mol), P is CH4 partial pressure (Pa), and V(liq) and V(gas) are the volume of the culture medium and headspace (liters), respectively. T is the cultivation temperature (328 K), and R = 8.314 J · K−1 · mol−1. (B) Atomic percent (At%) of 13CH4 and 13CO2 in M. conradii pure culture after addition of H13COONa. Values are means from three replicates. Error bars indicate standard deviations.
DISCUSSION
Environmental studies have demonstrated that Methanocellales methanogens are adapted to low H2 and syntrophic growth with fatty acid-oxidizing bacteria. To understand the adaptive mechanisms, we determined the transcripts of 21 genes for methanogenesis and acetate assimilation in M. conradii monoculture and syntrophic cocultures with P. thermopropionicum or S. lipocalidus. We found that the expression of three hydrogenase-encoding genes (ech, frh, and mvh) and six methanogenesis genes (fwd, mtd, mer, mtr, mcr, and hdr) were upregulated (summarized in Fig. 3), while the acetate assimilation gene (acs2) was downregulated in cocultures. In addition, the expression of the formate dehydrogenase-encoding gene (fdhA) was upregulated in cocultures. Apparently, M. conradii displayed a metabolic change, repressing energy-consuming biosynthesis while maintaining energy-generating methanogenesis during syntrophic growth. The methanogen possibly also activated the utilization of formate for the syntrophic interaction.
FIG 3.
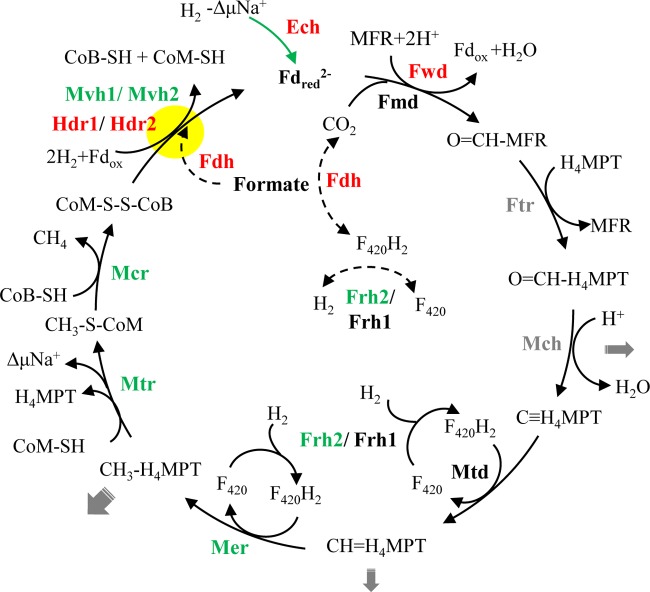
Conceptual model of the Wolfe cycle (modified from reference 46) for hydrogenotrophic methanogenesis in Methanocella conradii HZ254. Relative changes in transcript abundance during syntrophic growth are indicated by different colorations of the enzyme: red, >3-fold upregulation on average; green, significant but less than 3-fold upregulation; black, no significant change; gray, not analyzed in the present experiment. Gray arrows indicate the potential removal of intermediates for biosynthetic reactions (the thickness of the arrows reflects the quantitative importance), the green arrow illustrates the anaplerotic reaction catalyzed by Ech, dashed-line arrows indicate possible reactions deduced from this study, and the yellow closed circle highlights the electron-bifurcating reaction. Enzymes catalyzing each step include the following: Ech, energy-conserving hydrogenase; F420, coenzyme F420; Frh, F420-reducing hydrogenase; H4MPT, tetrahydromethanopterin; MFR, methanofuran; Mvh, F420-nonreducing hydrogenase; Fdh, formate dehydrogenase; Fwd, CHO-MFR dehydrogenase (W containing); Fmd, CHO-MFR dehydrogenase (Mo containing); Ftr, CHO-MFR:H4MPT formyltransferase; Mch, methenyl-H4MPT cyclohydrolase; Mtd, methylene-H4MPT dehydrogenase; Mer, methylene-H4MPT reductase; Mtr, methyl-H4MPT:CoM methyltransferase; Mcr, methyl-CoM reductase; Hdr, heterodisulfide reductase; Fdred/Fdox, reduced or oxidized ferredoxin; ΔμNa+, electrochemical sodium ion potential.
Among the genes upregulated, the expression of four genes, fwd, hdr, ech, and fdh, showed substantial increases (larger than 3-fold on average), while mer, mtr, mcr, mvh, and frh displayed moderate increases (Fig. 3). Since the transcript level of 16S rRNA in syntrophic cocultures decreased by a factor of about two compared to that of monoculture (Table 1), 2- to 3-fold increases of the relative expression levels of the genes described above indicated that M. conradii in cocultures maintained nearly the same absolute expression levels of methanogenesis genes as those in monoculture per cell, although their growth rates significantly decreased. Genes coding for Fwd, Hdr, and MvhD in M. conradii are linear in a same large predicted transcription unit (fwdGF-hdrC2B2A2-mvhD2-fwdCBAD). In hydrogenotrophic methanogens without cytochromes, MvhADG and HdrABC form a complex (42, 43) that reduces heterodisulfide CoM-S-S-CoB in conjunction with the reduction of oxidized ferredoxin via flavin-based electron bifurcation (43, 44). The reduced ferredoxin then is used to reduce CO2 to formylmethanofuran, the first step of methanogenesis. This coupling saves the Na+ motive force generated by Mtr that is available for ATP synthesis. Costa and colleagues (45) showed that in M. maripaludis, Hdr, Vhu, Fwd, and Fdh formed a supercomplex, proving that the first and last steps were physically connected. This pointed to a full cycle of the methanogenic pathway, now referred to as the Wolfe cycle (46). The presence of the large predicted transcription unit in M. conradii indicates that a complex consisting of Mvh, Hdr, and Fwd forms. The marked upregulation of hdr and fwd transcript levels indicates the response to energy limitation imposed by low H2 or syntrophic growth and suggests that M. conradii tends to stabilize the Fwd-Hdr interaction under these conditions. Given that electron bifurcation is crucial in the energy conservation of hydrogenotrophic methanogens, the increase of the related gene expression and, hence, the enzyme abundances will support an increased flux through this step.
The membrane-bound energy-converting hydrogenase is responsible for the reversible reduction of ferredoxin with H2, driven by a proton or sodium ion motive force. There are two types of energy-converting hydrogenases, namely, Eha and Ehb, in M. maripaludis. Ehb is responsible for anabolic ferredoxin synthesis (47, 48), while the function of Eha is probably related to anaplerotically replenishing the intermediates of the methanogenic pathway that is required to sustain the Wolfe cycle (46, 49). M. conradii has only one Ech hydrogenase (encoded by echABCDEF). Hence, its function is probably closer to that of methanogens with cytochromes like Methanosarcina barkeri, generating ferredoxin for both biosynthesis and methanogenesis (50). Since the growth rate of M. conradii was lower in the cocultures, it was unlikely that the upregulated Ech was due to the requirement of biosynthesis. Instead, the increased Ech very possibly functioned like Eha in M. maripaludis to generate ferredoxin for methanogenesis, stabilizing the Wolfe cycle (Fig. 3). The expression of other methanogenesis genes, including frh, mer, mtr, and mcr, was moderately upregulated during syntrophic growth. These enzymes all would be required for strengthening the central pathway of methanogenesis.
The formate dehydrogenase-encoding genes were actively transcribed and markedly upregulated in cocultures. Formate is needed for the biosynthesis of purine (51). However, it was unlikely that the significant upregulation of fdh in M. conradii cocultures was for biosynthesis, as the growth was slower. By using carbon isotope labeling, we showed that M. conradii could convert formate to CH4 in the presence of H2, indicating that formate dehydrogenase was functionally active for methanogenesis. The genomes of P. thermopropionicum and S. lipocalidus encode both hydrogenases and formate dehydrogenases (52–54). Thus, formate was produced possibly by the syntrophs and served as an alternative shuttle for interspecies electron transfer between syntrophic partners. Therefore, the upregulation of Fdh in methanogen could facilitate the use of formate for methanogenesis and growth. Similar upregulation of Fdh under H2 limitation or syntrophic conditions was detected in M. maripaludis (24). However, M. hungatei monoculture (on either formate or H2) and syntrophic coculture with Syntrophobacter fumaroxidans showed no difference in the transcript levels of hydrogenase- and formate dehydrogenase-encoding genes (25). Therefore, the function of Fdh in hydrogenotrophic methanogens deserves further investigation.
Among acetate assimilation genes, one gene (acs2), coding for the synthesis of acetyl-CoA from acetate, was significantly downregulated in cocultures, while the other genes, including acs1, remained unaffected. This result indicates that acs2 is more dynamic and functionally more important than acs1. Significant downregulation of acs genes also was observed in M. thermautotrophicus and M. maripaludis (18, 24). Apparently, the lower growth rate of M. conradii in cocultures was correlated with the downregulation of ACS that metabolized the first step of acetate assimilation. The expression of acs also might be influenced by the acetate concentration in the culture medium. Acetate was produced in the syntrophic cocultures to a much higher concentration (Fig. 1B and C) than the amount added in monoculture, which might exceed the Km of ACS and elevate the efficiency of ACS. Consequently, less ACS was needed, resulting in the downregulation of ACS.
We have shown that M. conradii exhibited substantial changes of gene expression associated with methanogenesis, acetate assimilation, and formate utilization during syntrophic growth. We assumed that the major factor causing the shift of gene expression was H2 limitation, but other factors could not be ruled out. Most obviously, the upregulation of fdh and, hence, very likely the activation of formate utilization have suggested that a more complicated effect than H2 limitation alone is present during syntrophic growth. Furthermore, it is also probable that the interspecies transfers of other metabolites happen that can influence gene expression. The transfer of alanine has been identified in the syntrophic coculture of M. maripaludis with D. vulgaris (24). Such kinds of metabolite transfers may diminish the pressure for biosynthesis in methanogens and the downregulation of genes like acs. As with the effect of H2 limitation alone, previous studies on M. thermoformicicum (55), M. thermautotrophicus (19, 20, 22), M. maripaludis (17), and M. jannaschii (21) indicated that mainly the genes (frh, mtd, and mer) coding for enzymes catalyzing the oxidation and reduction of coenzyme F420 showed a significant elevation of mRNA levels under H2 limitation. In comparison, the transcript levels of these genes were only moderately upregulated in the present study.
It has to be noted that batch cultivation was employed in the present study. The growth rate of M. conradii decreased in cocultures and, to a greater extent, in propionate than butyrate coculture. The difference between two syntrophic cocultures might be related to the thermodynamic conditions. The higher H2 partial pressure in butyrate coculture probably supported a greater growth rate for M. conradii compared to that in propionate coculture (56–58). The growth rate was believed to influence gene expression in both bacteria and archaea. The 16S rRNA expression per cell in M. conradii monoculture was 1.8 and 2.4 times higher than that in butyrate and propionate cocultures, respectively, consistent with the growth rate differences (Table 1).
The function of a few genes in M. conradii remains difficult to predict. For instance, we detected a high transcript level of vhtG (Table 2; also see Table S2 in the supplemental material), although there was no difference between coculture and monoculture. M. conradii lacks methanophenazine (30). Thus, the function of Vht remains unclear. We did not analyze the gene transcripts for the putative Ech-like hydrogenase. We found, however, that the gene hdrB3, which is located in the predicted transcription unit of ech-like genes (see Fig. S1), was significantly upregulated in cocultures (propionate coculture in particular) (Table 2). This implies that the Ech-like genes are transcribed. Its function, however, remains unknown. In addition, several enzymes, like Frh, Mvh, and Hdr, show multiple copies. While one copy was expressed usually to a relatively high level, others showed very low expression (orders of magnitude lower than the average). We found that the highly expressed genes responded to the growth conditions substantially, while the lower-expression copies did not show changes. Apparently, these results imply the functioning differences of homologous genes in M. conradii. The function and activity of multiple-copy genes deserve further investigations.
In conclusion, by determining the gene expression of M. conradii grown in monoculture and syntrophically with P. thermopropionicum or S. lipocalidus, we illustrated that Methanocella methanogens used different strategies to cope with low-H2 and syntrophic growth conditions. First, they tend to strengthen the Wolfe cycle under syntrophic conditions. M. conradii is likely to form the Mvh/Hdr/Fwd complex. The significant upregulation of Hdr- and Fwd-encoding genes was in favor of maintaining this complex. In addition, Ech, which probably contributed to replenish the methanogenesis intermediates, was significantly upregulated during syntrophic growth. The expression of other genes within the Wolfe cycle was also moderately upregulated. All of these results point to a strategy of M. conradii to reinforce the Wolfe cycle during syntrophic growth. Second, M. conradii appeared to downregulate energy-demanding biosynthesis by repressing acetate assimilation. Third, M. conradii probably activated formate-dependent methanogenesis during syntrophic growth. In conclusion, we propose that maintaining the integration of the Wolfe cycle is crucial for the methanogen to cope with low-H2 and syntrophic growth conditions that prevail in nature. However, it remains unknown how methanogens initiate these metabolic shifts in response to environmental changes.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by the National Basic Research Program of China (2011CB100505) and the National Natural Science Foundation of China (41130527 and 40625003).
Footnotes
Published ahead of print 16 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01259-14.
REFERENCES
- 1.Groβkopf R, Stubner S, Liesack W. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fey A, Chin K, Conrad R. 2001. Thermophilic methanogens in rice field soil. Environ. Microbiol. 3:295–303. 10.1046/j.1462-2920.2001.00195.x [DOI] [PubMed] [Google Scholar]
- 3.Lueders T, Chin K, Conrad R, Friedrich M. 2001. Molecular analyses of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194–204. 10.1046/j.1462-2920.2001.00179.x [DOI] [PubMed] [Google Scholar]
- 4.Angel R, Claus P, Conrad R. 2012. Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J. 6:847–862. 10.1038/ismej.2011.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angel R, Matthies D, Conrad R. 2011. Activation of methanogenesis in arid biological soil crusts despite the presence of oxygen. PLoS One 6:e20453. 10.1371/journal.pone.0020453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad R, Erkel C, Liesack W. 2006. Rice Cluster I methanogens, an important group of Archaea producing greenhouse gas in soil. Curr. Opin. Biotech. 17:262–267. 10.1016/j.copbio.2006.04.002 [DOI] [PubMed] [Google Scholar]
- 7.Krüger M, Frenzel P, Kemnitz D, Conrad R. 2005. Activity, structure and dynamics of the methanogenic archaeal community in a flooded Italian rice field. FEMS Microbiol. Ecol. 51:323–331. 10.1016/j.femsec.2004.09.004 [DOI] [PubMed] [Google Scholar]
- 8.Lu YH, Lueders T, Friedrich MW, Conrad R. 2005. Detecting active methanogenic populations on rice roots using stable isotope probing. Environ. Microbiol. 7:326–336. 10.1111/j.1462-2920.2005.00697.x [DOI] [PubMed] [Google Scholar]
- 9.Lu YH, Conrad R. 2005. In situ stable isotope probing of methanogenic archaea in the rice rhizosphere. Science 309:1088–1090. 10.1126/science.1113435 [DOI] [PubMed] [Google Scholar]
- 10.Gan Y, Qiu Q, Liu P, Rui J, Lu Y. 2012. Syntrophic oxidation of propionate in rice field soil at 15 and 30°C under methanogenic conditions. Appl. Environ. Microbiol. 78:4923–4932. 10.1128/AEM.00688-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu FH, Conrad R. 2010. Thermoanaerobacteriaceae oxidize acetate in methanogenic rice field soil at 50°C. Environ. Microbiol. 12:2341–2354. 10.1111/j.1462-2920.2010.02289.x [DOI] [PubMed] [Google Scholar]
- 12.Liu P, Qiu Q, Lu Y. 2011. Syntrophomonadaceae-affiliated species as active butyrate-utilizing syntrophs in paddy field soil. Appl. Environ. Microbiol. 77:3884–3887. 10.1128/AEM.00190-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rui J, Qiu Q, Lu Y. 2011. Syntrophic acetate oxidation under thermophilic methanogenic condition in Chinese paddy field soil. FEMS Microbiol. Ecol. 77:264–273. 10.1111/j.1574-6941.2011.01104.x [DOI] [PubMed] [Google Scholar]
- 14.Lueders T, Pommerenke B, Friedrich MW. 2004. Stable-isotope probing of microorganisms thriving at thermodynamic limits: syntrophic propionate oxidation in flooded soil. Appl. Environ. Microbiol. 70:5778–5786. 10.1128/AEM.70.10.5778-5786.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJ, Schink B, Rohlin L, Gunsalus RP. 2008. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 1125:58–72. 10.1196/annals.1419.005 [DOI] [PubMed] [Google Scholar]
- 16.Luo H, Zhang H, Suzuki T, Hattori S, Kamagata Y. 2002. Differential expression of methanogenesis genes of Methanothermobacter thermoautotrophicus (formerly Methanobacterium thermoautotrophicum) in pure culture and in cocultures with fatty acid-oxidizing syntrophs. Appl. Environ. Microbiol. 68:1173–1179. 10.1128/AEM.68.3.1173-1179.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickson E, Haydock A, Moore B, Whitman W, Leigh J. 2007. Functionally distinct genes regulated by hydrogen limitation and growth rate in methanogenic Archaea. Proc. Natl. Acad. Sci. U. S. A. 104:8930–8934. 10.1073/pnas.0701157104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enoki M, Shinzato N, Sato H, Nakamura K, Kamagata Y. 2011. Comparative proteomic analysis of Methanothermobacter themautotrophicus ΔH in pure culture and in co-culture with a butyrate-oxidizing bacterium. PLoS One 6:e24309. 10.1371/journal.pone.0024309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato S, Kosaka T, Watanabe K. 2008. Comparative transcriptome analysis of responses of Methanothermobacter thermautotrophicus to different environmental stimuli. Environ. Microbiol. 10:893–905. 10.1111/j.1462-2920.2007.01508.x [DOI] [PubMed] [Google Scholar]
- 20.Morgan R, Pihl T, Nolling J, Reeve J. 1997. Hydrogen regulation of growth, growth yields, and methane gene transcription in Methanobacterium thermoautotrophicum deltaH. J. Bacteriol. 179:889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukhopadhyay B, Johnson EF, Wolfe RS. 2000. A novel pH2 control on the expression of flagella in the hyperthermophilic strictly hydrogenotrophic methanarchaeaon Methanococcus jannaschii. Proc. Natl. Acad. Sci. U. S. A. 97:11522–11527. 10.1073/pnas.97.21.11522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeve JN, Nolling J, Morgan RM, Smith DR. 1997. Methanogenesis: genes, genomes, and who's on first? J. Bacteriol. 179:5975–5986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinzato N, Enoki M, Sato H, Nakamura K, Matsui T, Kamagata Y. 2008. Specific DNA binding of a potential transcriptional regulator, inosine 5′-monophosphate dehydrogenase-related protein VII, to the promoter region of a methyl coenzyme M reductase I-encoding operon retrieved from Methanothermobacter thermautotrophicus strain ΔH. Appl. Environ. Microbiol. 74:6239–6247. 10.1128/AEM.02155-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker CB, Redding-Johanson AM, Baidoo EE, Rajeev L, He Z, Hendrickson EL, Joachimiak MP, Stolyar S, Arkin AP, Leigh JA, Zhou J, Keasling JD, Mukhopadhyay A, Stahl DA. 2012. Functional responses of methanogenic archaea to syntrophic growth. ISME J. 6:2045–2055. 10.1038/ismej.2012.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Worm P, Stams AJ, Cheng X, Plugge CM. 2011. Growth- and substrate-dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei. Microbiology 157:280–289. 10.1099/mic.0.043927-0 [DOI] [PubMed] [Google Scholar]
- 26.Sakai S, Imachi H, Sekiguchi Y, Ohashi A, Harada H, Kamagata Y. 2007. Isolation of key methanogens for global methane emission from rice paddy fields: a novel isolate affiliated with the clone cluster rice cluster I. Appl. Environ. Microbiol. 73:4326–4331. 10.1128/AEM.03008-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lü Z, Lu Y. 2012. Methanocella conradii sp. nov., a thermophilic, obligate hydrogenotrophic methanogen, isolated from Chinese rice field soil. PLoS One 7:e35279. 10.1371/journal.pone.0035279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai S, Conrad R, Liesack W, Imachi H. 2010. Methanocella arvoryzae sp. nov., a hydrogenotrophic methanogen isolated from rice field soil. Int. J. Syst. Evol. Microbiol. 60:2918–2923. 10.1099/ijs.0.020883-0 [DOI] [PubMed] [Google Scholar]
- 29.Erkel C, Kube M, Reinhardt R, Liesack W. 2006. Genome of Rice Cluster I archaea–the key methane producers in the rice rhizosphere. Science 313:370–372. 10.1126/science.1127062 [DOI] [PubMed] [Google Scholar]
- 30.Lü Z, Lu Y. 2012. Complete genome sequence of a thermophilic methanogen, Methanocella conradii HZ254, isolated from Chinese rice field soil. J. Bacteriol. 194:2398–2399. 10.1128/JB.00207-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakai S, Takaki Y, Shimamura S, Sekine M, Tajima T, Kosugi H, Ichikawa N, Tasumi E, Hiraki AT, Shimizu A, Kato Y, Nishiko R, Mori K, Fujita N, Imachi H, Takai K. 2011. Genome sequence of a mesophilic hydrogenotrophic methanogen Methanocella paludicola, the first cultivated representative of the order Methanocellales. PLoS One 6:e22898. 10.1371/journal.pone.0022898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stams AJM, Dong XZ. 1995. Role of formate and hydrogen in the degradation of propionate and butyrate by defined suspended cocultures of acetogenic and methanogenic bacteria. Antonie Van Leeuwenhoek 68:281–284. 10.1007/BF00874137 [DOI] [PubMed] [Google Scholar]
- 33.Thiele JH, Zeikus JG. 1988. Control of interspecies electron flow during anaerobic digestion: significance of formate transfer versus hydrogen transfer during syntrophic methanogenesis in flocs. Appl. Environ. Microbiol. 54:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan QA, Lu YH. 2009. Response of methanogenic archaeal community to nitrate addition in rice field soil. Environ. Microbiol. Rep. 1:362–369. 10.1111/j.1758-2229.2009.00065.x [DOI] [PubMed] [Google Scholar]
- 35.Culley D, Kovacikjr W, Brockman F, Zhang W. 2006. Optimization of RNA isolation from the archaebacterium Methanosarcina barkeri and validation for oligonucleotide microarray analysis. J. Microbiol. Methods 67:36–43 [DOI] [PubMed] [Google Scholar]
- 36.Peirson SN, Butler JN, Foster RG. 2003. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 31:e73. 10.1093/nar/gng073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hendrickson EL, Liu Y, Rosas-Sandoval G, Porat I, Söll D, Whitman WB, Leigh JA. 2008. Global responses of Methanococcus Maripaludis to specific nutrient limitations and growth rate. J. Bacteriol. 190:2198–2205. 10.1128/JB.01805-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kemnitz D, Kolb S, Conrad R. 2005. Phenotypic characterization of Rice Cluster III archaea without prior isolation by applying quantitative polymerase chain reaction to an enrichment culture. Environ. Microbiol. 7:553–565. 10.1111/j.1462-2920.2005.00723.x [DOI] [PubMed] [Google Scholar]
- 39.Ma K, Conrad R, Lu Y. 2012. Responses of methanogen mcrA genes and their transcripts to an alternate dry/wet cycle of paddy field soil. Appl. Environ. Microbiol. 78:445–454. 10.1128/AEM.06934-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oberlies G, Fuchs G, Thauer R. 1980. Acetate thiokinase and the assimilation of acetate in Methanobacterium thermoautotrophicum. Arch. Microbiol. 128:248–252. 10.1007/BF00406167 [DOI] [PubMed] [Google Scholar]
- 41.Shieh J, Whitman WB. 1988. Autotrophic acetyl coenzyme A biosynthesis in Methanococcus maripaludis. J. Bacteriol. 170:3072–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buckel W, Thauer RK. 2013. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827:94–113. 10.1016/j.bbabio.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 43.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6:579–591. 10.1038/nrmicro1931 [DOI] [PubMed] [Google Scholar]
- 44.Kaster AK, Moll J, Parey K, Thauer RK. 2011. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc. Natl. Acad. Sci. U. S. A. 108:2981–2986. 10.1073/pnas.1016761108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costa KC, Wong PM, Wang T, Lie TJ, Dodsworth JA, Swanson I, Burn JA, Hackett M, Leigh JA. 2010. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc. Natl. Acad. Sci. U. S. A. 107:11050–11055. 10.1073/pnas.1003653107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thauer RK. 2012. The Wolfe cycle comes full circle. Proc. Natl. Acad. Sci. U. S. A. 109:15084–15085. 10.1073/pnas.1213193109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Major TA, Liu YC, Whitman WB. 2010. Characterization of energy-conserving hydrogenase B in Methanococcus maripaludis. J. Bacteriol. 192:4022–4030. 10.1128/JB.01446-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porat I, Kim W, Hendrickson EL, Xia Q, Zhang Y, Wang T, Taub F, Moore BC, Anderson IJ, Hackett M. 2006. Disruption of the operon encoding Ehb hydrogenase limits anabolic CO2 assimilation in the archaeon Methanococcus maripaludis. J. Bacteriol. 188:1373–1380. 10.1128/JB.188.4.1373-1380.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lie TJ, Costa KC, Lupa B, Korpole S, Whitman WB, Leigh JA. 2012. Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc. Natl. Acad. Sci. U. S. A. 109:15473–15478. 10.1073/pnas.1208779109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meuer J, Kuettner HC, Zhang JK, Hedderich R, Metcalf WW. 2002. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. U. S. A. 99:5632–5637. 10.1073/pnas.072615499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White RH. 1997. Purine biosynthesis in the domain Archaea without folates or modified folates. J. Bacteriol. 179:3374–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Müller N, Worm P, Schink B, Stams AJM, Plugge CM. 2010. Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanisms. Environ. Microbiol. Rep. 2:489–499. 10.1111/j.1758-2229.2010.00147.x [DOI] [PubMed] [Google Scholar]
- 53.Sieber JR, McInerney MJ, Gunsalus RP. 2012. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu. Rev. Microbiol. 66:429–452. 10.1146/annurev-micro-090110-102844 [DOI] [PubMed] [Google Scholar]
- 54.Sieber JR, Sims DR, Han C, Kim E, Lykidis A, Lapidus AL, McDonnald E, Rohlin L, Culley DE, Gunsalus R, McInerney MJ. 2010. The genome of Syntrophomonas wolfei: new insights into syntrophic metabolism and biohydrogen production. Environ. Microbiol. 12:2289–2301. 10.1111/j.1462-2920.2010.02237.x [DOI] [PubMed] [Google Scholar]
- 55.Nolling J, Reeve JN. 1997. Growth- and substrate-dependent transcription of the formate dehydrogenase (fdhCAB) operon in Methanobacterium thermoformicicum Z-245. J. Bacteriol. 179:899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishii S, Kosaka T, Hori K, Hotta Y, Watanabe K. 2005. Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus. Appl. Environ. Microbiol. 71:7838–7845. 10.1128/AEM.71.12.7838-7845.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato S, Kosaka T, Watanabe K. 2009. Substrate-dependent transcriptomic shifts in Pelotomaculum thermopropionicum grown in syntrophic co-culture with Methanothermobacter thermautotrophicus. Microb. Biotechnol. 2:575–584. 10.1111/j.1751-7915.2009.00102.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schink B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61:262–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.