Abstract
Since genome analysis did not allow unambiguous reconstruction of transport, catabolism, and substrate-specific regulation for several important carbohydrates in Phaeobacter inhibens DSM 17395, proteomic and metabolomic analyses of N-acetylglucosamine-, mannitol-, sucrose-, glucose-, and xylose-grown cells were carried out to close this knowledge gap. These carbohydrates can pass through the outer membrane via porins identified in the outer membrane fraction. For transport across the cytoplasmic membrane, carbohydrate-specific ABC transport systems were identified. Their coding genes mostly colocalize with the respective “catabolic” and “regulatory” genes. The degradation of N-acetylglucosamine proceeds via N-acetylglucosamine-6-phosphate and glucosamine-6-phosphate directly to fructose-6-phosphate; two of the three enzymes involved were newly predicted and identified. Mannitol is catabolized via fructose, sucrose via fructose and glucose, glucose via glucose-6-phosphate, and xylose via xylulose-5-phosphate. Of the 30 proteins predicted to be involved in uptake, regulation, and degradation, 28 were identified by proteomics and 19 were assigned to their respective functions for the first time. The peripheral degradation pathways feed into the Entner-Doudoroff (ED) pathway, which is connected to the lower branch of the Embden-Meyerhof-Parnas (EMP) pathway. The enzyme constituents of these pathways displayed higher abundances in P. inhibens DSM 17395 cells grown with any of the five carbohydrates tested than in succinate-grown cells. Conversely, gluconeogenesis is turned on during succinate utilization. While tricarboxylic acid (TCA) cycle proteins remained mainly unchanged, the abundance profiles of their metabolites reflected the differing growth rates achieved with the different substrates tested. Homologs of the 74 genes involved in the reconstructed catabolic pathways and central metabolism are present in various Roseobacter clade members.
INTRODUCTION
Carbohydrates are important constituents of the global carbon cycle. In marine surface waters they can account for about 21% of the dissolved organic carbon and reach concentrations of 7 to 33 μM (1). Although polysaccharides are dominant, monosaccharides have also been detected in the Atlantic, Pacific, and Antarctic Oceans at an average concentration of 4.3 μM (1). A prominent monosaccharide is N-acetylglucosamine, which is the monomer of chitin, the second most abundant polymer in the environment. Chitin is the key component of fungal cell walls, is present in the exoskeletons of worms, molluscs, and arthropods (2), and is estimated to account for about 10% of marine bacterial production (3). Another important source of N-acetylglucosamine is the peptidoglycan of the Gram-negative bacterial cell wall (4). Glucosamine (including deacetylated N-acetylglucosamine) has been detected as the most abundant amino sugar in marine surface waters (17 to 56 nM), with higher prevalences in the Atlantic and Pacific Oceans than in the Arctic Ocean (4). Glucose, on the other hand, is the most abundant neutral monosaccharide, with concentrations as high as 15 to 65 nM in the equatorial Pacific, where it accounts for 15 to 47% of bacterial production (5). Other abundant aldoses in seawater are galactose, mannose, and xylose (6). In addition to neutral monosaccharides and amino sugars, disaccharides and sugar alcohols are noteworthy carbohydrate species. One representative of the latter is mannitol, which acts as a “compatible solute” in algae (7). In general, the most abundant disaccharide in the environment is sucrose, due to its formation by photosynthesis in higher plants (8).
The carbohydrates in the marine water column are utilized predominantly by heterotrophic bacteria. A recent survey of transporter gene occurrence in coastal bacterioplankton communities revealed that Roseobacter clade members have the highest number of carbohydrate-associated transporter genes (9). This is in accord with the finding that roseobacters, as well as SAR11 affiliates, contribute significantly to glucose uptake in coastal North Sea waters (10). The striking dominance of the alphaproteobacterial Roseobacter clade among marine bacterioplankton members is paralleled by the wide range of metabolic capacities of its members (11). Due to this biogeographical prominence and ecophysiological relevance, more than 75 roseobacter genomes have been sequenced to date. Among them is that of Phaeobacter inhibens DSM 17395 (12), which was recently reclassified from Phaeobacter gallaeciensis DSM 17395 (13). Phaeobacter species are globally distributed in the marine system, e.g., in sediments of the Arctic Ocean (14), the Yellow Sea (15), and the Mediterranean Sea (16), and in surface water of the German Wadden Sea (17). Furthermore, they are constituents of marine biofilms (18) and are associated with scallops (19) and algae (12). The main physiological characteristics of P. inhibens DSM 17395 are the formation of the antibiotic tropodithietic acid (20) and its nutritional versatility (carbohydrates, organic acids, and all 20 proteinogenic amino acids [21]). Due to these characteristics, P. inhibens DSM 17395 was selected as a systems biology model for studying the molecular basis of the habitat success of heterotrophic roseobacters. An initial proteomic/metabolomic study with P. inhibens DSM 17395 demonstrated that its central metabolism was unaffected by the growth phase (22). Furthermore, the Entner-Doudoroff (ED) pathway was identified as the key route for glucose breakdown across roseobacters (22, 23). During growth with complex Marine Broth (MB) as a surrogate for the complex nutritional conditions typically resulting from the collapse of algal blooms, P. inhibens DSM 17395 simultaneously degraded various amino acids and other substrates (24). A subsequent time-resolved study revealed differing degrees of substrate preferences (high to low) during growth with Casamino Acids, containing 15 different detectable amino acids (25). Genomically unclear degradation pathways of nine amino acids could be elucidated by combined proteomics, enzymatics, and metabolomics and could be shown by comparative genomics to be archetypical for roseobacters (26). Besides amino acids, carbohydrates are common substrates for P. inhibens DSM 17395, but their catabolism has not been investigated so far. In the present study, the uptake and degradation of five representative carbohydrates were studied by combined proteomic and metabolomic analyses, and their dissemination across roseobacters and other marine heterotrophic bacteria was studied by comparative genomics.
MATERIALS AND METHODS
Cultivation.
Phaeobacter inhibens DSM 17395 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ; Braunschweig, Germany) and since then has been maintained in our laboratory.
The range of utilizable carbohydrates was assessed by incubating P. inhibens DSM 17395 in glass tubes filled with 5 ml defined mineral seawater medium (SWM) (22) supplemented with 1 of 18 different carbohydrates at 1 mg ml−1 (3 to 7 mM). Freshly grown precultures in rich (MB) medium were washed with substrate-free SWM prior to inoculation of carbohydrate-containing SWM. Test cultures were incubated on a rotary shaker at 28°C. Growth was monitored by measuring the optical density at 600 nm (OD600). Thirteen of the 18 carbohydrates investigated allowed growth (see Table S1 in the supplemental material).
For further analyses five growth substrates, representing different classes of carbohydrates, were selected: N-acetylglucosamine (5 mM), mannitol (5 mM), sucrose (2.5 mM), glucose (5 mM), and xylose (5 mM). Furthermore, succinate (20 mM) served as a reference. For substrate adaptation, P. inhibens DSM 17395 was grown with each substrate for 5 passages in Erlenmeyer flasks and was then stored in glycerol stocks, as described recently (26).
For each of the metabolomic and proteomic analyses, 12 replicate cell pellets (∼100 mg [wet weight]) were generated per substrate condition. Starting from a glycerol stock, 3 passages were conducted before six main cultures (250 ml) were inoculated with 2% [vol/vol] preculture (25). At half-maximal optical density (½ ODmax), 20 ml of each main culture was further incubated in 100-ml Erlenmeyer flasks (see Fig. S1 in the supplemental material), and the remainder was harvested as described previously (22), except for cell pellets allocated for metabolomics, which were washed with 3.7% (wt/vol) NaCl. The substrate-specific ½ ODmax values and corresponding protein contents were as follows: for succinate, an OD600 of 1.41 and 76.3 mg protein liter−1; for N-acetylglucosamine, an OD600 of 0.55 and 31.4 mg protein liter−1; for mannitol, an OD600 of 0.70 and 23.7 mg protein liter−1; for sucrose, an OD600 of 0.52 and 18.4 mg protein liter−1; for glucose, an OD600 of 0.48 and 25.2 mg protein liter−1; and for xylose, an OD600 of 0.46 and 18.3 mg protein liter−1. For sucrose- and succinate-grown cells, two additional main cultures were harvested for RNA isolation and subsequent reverse transcriptase PCR (RT-PCR). Cultures were harvested as described above. The resulting cell pellets were resuspended in 1.5 ml RNAprotect Bacteria reagent (Qiagen, Hilden, Germany) and were incubated for 10 min at room temperature prior to centrifugation and storage at −80°C.
All cultivation steps were purity controlled by microscopy (Axiostar; Zeiss AG, Göttingen, Germany) and plating on MB agar.
Proteomics. (i) Analysis of soluble proteome by 2D-DIGE and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) tandem mass spectrometry (MS-MS).
For 2-dimensional difference in gel electrophoresis (2D-DIGE) analysis, cell pellets of three biological replicates for each substrate condition were resuspended in lysis buffer {7 M urea, 2 M thiourea, 30 mM Tris-HCl, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS] [final pH 8.5]}, and cells were disrupted using the Plus One Sample Grinding kit (GE Healthcare, Munich, Germany) as described previously (27, 28). The protein content was determined by the method described by Bradford (29). Before electrophoresis, proteins were covalently labeled with CyDye DIGE Fluor dyes (GE Healthcare) essentially as reported previously (28). For each sample, 50 μg protein was labeled with 200 pmol of either Cy3 (for each of the five test states [carbohydrate-grown cells]), Cy5 (for the reference state [succinate-grown cells]), or Cy2 (for the internal standard [equal amounts of all test state samples and the reference state sample]). Then each labeled test state sample was mixed with the reference state sample and the internal standard, and the mixture was transferred to 24-cm-long, nonlinear (NL) immobilized pH gradient (IPG) strips with pH ranges from 3 to 5.6 and from 3 to 11 (GE Healthcare) for 1st-dimension separation. Isoelectric focusing, reduction, alkylation, and 2nd-dimension separation according to molecular mass by SDS-PAGE were carried out as described previously (22). After the completion of electrophoresis, 2-dimensional electrophoresis (2DE) gels were immediately scanned with a Typhoon 9400 scanner (GE Healthcare). Using DeCyder software (version 7.0; GE Healthcare), images were cropped and analyzed in two separate work packages according to the different pH gradients applied (pH 3 to 5.6 NL and pH 5 to 11 NL). The detection parameters were set as described previously (30). Those protein spots that changed in abundance by ≥|1.5|-fold, matched in at least 75% of the gels analyzed, and had P values of <0.05 by analysis of variance (ANOVA) and t test values of <10−4 were regarded as significantly regulated (31).
For protein identification, separate replicate 2DE gels with 300-μg protein loads were run for each substrate condition and were stained with colloidal Coomassie brilliant blue (cCBB) after electrophoresis (30). Differentially abundant protein spots were excised with the EXQuest spot cutter (Bio-Rad, Munich, Germany). As described recently (24), excised protein spots were first washed and digested and were then spotted onto AnchorChip steel targets (Bruker Daltonik GmbH, Bremen, Germany) and subjected to mass spectrometric analysis with an UltrafleXtreme MALDI–two-stage TOF (TOF-TOF) mass spectrometer (Bruker Daltonik GmbH). Peptide mass fingerprint (PMF) searches and MS-MS searches were performed with a Mascot server (version 2.3; Matrix Science, London, United Kingdom) and the ProteinScape platform (version 3.0; Bruker Daltonik GmbH) as described previously (26).
(ii) Analysis of proteins from the cytoplasmic and outer membrane protein-enriched fractions by nano-liquid chromatography (nano-LC)-ESI-MS-MS.
Cytoplasmic and outer membrane proteins from two biological replicates per substrate condition were prepared via sequential solubilization with sarcosyl and SDS essentially as described recently (32). Outer membrane-enriched (2.5 μg) and cytoplasmic membrane-enriched (4.0 μg) protein fractions were separated according to molecular mass on 7-cm-long, 1-mm-thick 12.5% SDS-PAGE gels. Four slices per gel lane were cut into smaller pieces and were subjected to in-gel-digestion as described previously (32). The tryptic peptides generated were separated with an UltiMate 3000 RSLCnano system (Thermo Fisher Scientific, Germering, Germany) equipped with a trap column (PepMap C18 Nano-Trap column; pore size, 100 Å; bead size, 3 μm; inner diameter, 75 μm; length, 2 cm; Thermo Fisher Scientific) and a 15-cm analytical column (C18; pore size, 100 Å; bead size, 2 μm; inner diameter, 75 μm; length, 15 cm; Thermo Fisher Scientific). The following linear gradient of increasing acetonitrile concentrations was applied using solvent A (0.1% trifluoroacetic acid [TFA]) and solvent B (80% acetonitrile, 0.1% TFA): 0 to 12 min, 4% B; 12 to 120 min, 4 to 50% B; 120 to 130 min, 50 to 95% B; 130 to 135 min, 95% B. The eluent was continuously analyzed by an online coupled electrospray ionization (ESI) ion trap mass spectrometer (amaZon ETD; Bruker Daltonik GmbH) operated as described previously (24). Protein identification was performed with ProteinScape (version 3.1) on a Mascot server (version 2.3) against the translated genome sequence of P. inhibens DSM 17395 (12) by applying a target-decoy strategy and a false discovery rate of <1.0% (24).
(iii) Shotgun proteomics by nano-LC-ESI-MS-MS.
For shotgun proteomic analysis, cell pellets were resuspended in 200 μl lysis buffer (7 M urea, 2 M thiourea, 30 mM Tris-HCl [final pH 8.5]) and were disrupted by means of the Sample Grinding kit (GE Healthcare). Fifty micrograms of total protein was subjected to reduction and alkylation using 45 mM dithiothreitol (DTT) and 100 mM iodoacetamide, respectively, prior to tryptic digestion overnight (1 μg enzyme). Peptides were separated with an UltiMate 3000 RSLCnano system (Thermo Fisher Scientific) with a trap column setup (see above) on a 25-cm analytical column (C18; pore size, 100 Å; bead size, 2 μm; inner diameter, 75 μm; length, 25 cm; Thermo Fisher Scientific) by applying the following gradient: 0 to 12 min, 4% B; 12 to 255 min, 4 to 50% B; 255 to 270 min, 50 to 95% B; 270 to 275 min, 95% B. Protein identification was performed with ProteinScape on a Mascot server (see above for details). The peptide data of all three biological replicates were compiled, and only proteins identified with 2 or more peptides were accepted.
Metabolomic analyses.
Intracellular metabolites were extracted as described previously (22), except that for resuspension of the cell pellets, 1.5 ml ethanol containing 30 μl ribitol (0.2 mg ml−1) was used. Furthermore, after phase separation, 1 ml of the upper polar phase was transferred to a 2-ml reaction tube and was dried in a vacuum concentrator for gas chromatography (GC)-MS analysis. The two-step derivatization reaction was performed essentially as described previously (25).
Intracellular metabolites were analyzed by GC-MS as described previously (26) using a Leco Pegasus 4D GC×GC-TOF MS (Leco Instrumente, Mönchengladbach, Germany) operated in GC-TOF mode and equipped with an MPS 2 XL autosampler (Gerstel, Mülheim an der Ruhr, Germany). The solvent delay time was adjusted to 278 s, and the detector voltage was set to 1,750 V.
Additionally, the samples were analyzed using the JMS-T100GC AccuTOF GC (JEOL GmbH, Eching, Germany), equipped with an MPS 2 Twister autosampler (Gerstel). Tuning with perfluorokerosene (MasCom, Bremen, Germany) was carried out according to the manufacturer's instructions. Samples (1 μl) were injected in splitless mode into a programmed-temperature vaporizer (PTV) injector (Gerstel), equipped with a 71- by 1-mm CIS 4 glass liner filled with silanized glass wool (Gerstel). After an initial time of 0.02 min at 70°C, the temperature was increased at 12°C s−1 to 330°C, followed by holding at 330°C for 5 min. Gas chromatography was performed with a 6890N Agilent GC (Agilent Technologies, Waldbronn, Germany) equipped with a ZB-5MS column (length, 30 m; inner diameter, 0.25 mm; particle size, 25 μm; Phenomenex, Aschaffenburg, Germany). The helium flow was set to a constant 1.2 ml min−1. After 1 min at 70°C, the GC oven temperature was increased to 330°C at 10°C min−1, followed by an additional constant temperature period at 330°C for 3 min. The transfer line temperature was set to 250°C. The electron impact mode at 70 eV was used for ionization; the ion source temperature was set to 200°C, the detector voltage to 2,400 V, and the emission current to 300 μA. After 3.8 min (solvent delay time), full-scan mass spectra were collected from m/z 45 to 600 at 5 scans s−1 with MassCenter Main software (version 2.3.0.1; JEOL).
Data were analyzed as described recently (25, 26). The internal standard ribitol and the cell mass were used for normalization, and finally, data were centralized to the reference state succinate-grown cells.
RNA isolation and RT-PCR.
The primers (purchased from biomers.net, Ulm, Germany) designed for α-glucosidase (encoded by algA) (algA_F [5′-TGCTGTCGCATACCTCTGAC-3′] and algA_R [5′-GTATCAAGGCGAAAGCCATC-3′]) resulted in a theoretical PCR product of 304 bp. For RNA isolation, two biological replicates (100 mg each) of sucrose- and succinate-grown cell pellets were used (for harvest, see above). RNA was isolated as described previously (33) by using acidic phenol and PCI (phenol-chloroform-isoamyl alcohol, 25:24:1), followed by washing with ethanol. Then DNA was digested with DNase I (34). The completeness of the digestion was verified via PCR employing primers aglA_R and aglA_F.
For reverse transcription, the final mixture consisted of 2.5 μg RNA (isolated from sucrose- or succinate-grown cells), 1.0 μl deoxynucleoside triphosphates (dNTPs) (10 mM), 2.0 μl antisense primer (10 μM) (aglA_R), 4.0 μl RT buffer, 1.0 μl H Minus Moloney murine leukemia virus (M-MuLV) reverse transcriptase (200 U μl−1; MBI Fermentas, St. Leon-Roth, Germany), and PCR-H2O (to 20 μl). The reverse transcription took place at 42°C for 60 min.
Then a PCR was performed, the products of which were separated on a 1.5% agarose gel. For this purpose, 2 μl of aglA cDNA, RNA, or PCR-H2O (negative control), or 1 μl of P. inhibens DSM 17395 DNA (positive control), was used as the template. Furthermore, 0.4 μl of the forward and reverse primers, 10 μl 2× PCR master mix (Promega, Madison, WI, USA), and PCR-H2O (to 20 μl) were added. The PCR program for amplification of aglA was as follows: 94°C for 10 min; 39 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 2.5 min; 72°C for 5 min; and 4°C for 30 s.
BigDye sequencing (Applied Biosystems, Foster City, CA, USA) revealed the identities of the PCR products.
Genome-based metabolic reconstruction.
Carbohydrate degradation pathways were reconstructed based on the sequenced and annotated genome of P. inhibens DSM 17395 (12) and reanalysis using MetaCyc (35), KEGG (36), Artemis (version 10.0) (37), and BLASTP (38).
For two reaction steps in the degradation of N-acetylglucosamine, no enzymes were predicted based on the genome of P. inhibens DSM 17395. To search for enzyme candidates, hidden Markov models (HMMs) (39) were used. Pretrained HMMs were taken from TIGRFAMs (http://www.jcvi.org/cgi-bin/tigrfams/index.cgi) if provided. Otherwise, an HMM was trained by collecting the sequence data of manually annotated enzymes for the required EC number obtained from the Braunschweig Enzyme Database (BRENDA) (40) and UniProt (41). The sequences collected were aligned by using Clustal Omega with standard settings (42, 43). After this, HMMBuild and finally HMMSearch were used to search candidates for the required enzymes in P. inhibens DSM 17395.
Comparative genomics.
The P. inhibens DSM 17395 genome, 27 other genomes from Roseobacter clade members, and genomes of 4 additional representatives of non-Roseobacter Rhodobacterales were collected from the National Center for Biotechnology Information (NCBI) proteome database (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria) (38). In addition, a selection of 30 other Proteobacteria and Bacteroidetes (Flavobacteria) was chosen in order to compare the genomes of marine heterotrophic bacteria lying outside the Rhodobacteraceae.
The predicted proteomes assembled in FASTA format were used to group orthologous/paralogous protein sequences across the genomes analyzed with the OrthoMCL algorithm, version 2.0.9 (44). The all-versus-all-BLASTP phase was carried out by BLAST, version 2.2.28+ (45). After the OrthoMCL software was run with standard parameters for bacteria (threshold for ortholog determination, an E value of ≤10−4 and an identity of ≥50%), the results were loaded into a relational database based on MySQL. Furthermore, the ortholog data were adjusted and supplemented by the “Genome Gene Best Homologs/Orthologous” service of the Integrated Microbial Genomes (IMG) database and comparative analysis system (46). Orthologous genes in the genomes of the 62 organisms tested were identified as bidirectional best BLAST hits with an E value below 10−4 and an identity higher than 50%. Since these criteria for the identification of orthologs were rather strict, and the same enzymatic activity can be performed by proteins with different sequences, an additional search for the presence of isofunctional enzymes was carried out in the EnzymeDetector website. The criteria for enzyme function prediction described in reference 47 were applied.
Information about additional gene loci in the P. inhibens DSM 17395 genome annotated with an identical EC number and an identical or better EnzymeDetector score was obtained from the EnzymeDetector website (47) and was integrated.
The information collected was analyzed with respect to the presence of orthologous genes, their genomic contexts, and their predicted enzyme functions. If two or more genes were in close proximity, the gene cluster was classified into six different categories of relatedness as reported previously (26). Predicted isofunctional enzymes that do not meet the strict criteria for orthologs are indicated by an additional color in Fig. 2 (see also Fig. S7 in the supplemental material).
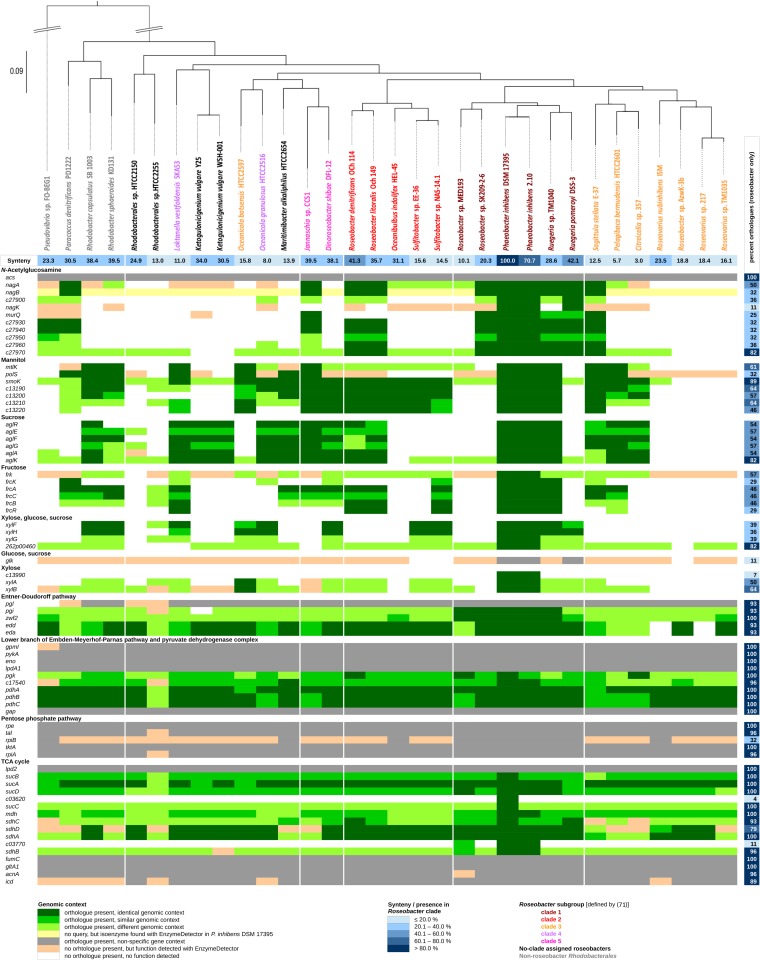
FIG 2.
Genome comparison of Roseobacter clade members and non-Roseobacter Rhodobacterales. Shown are genes of P. inhibens DSM 17395 related to the carbohydrate transporters and degradation pathways investigated (the names and functional assignments are those shown in Fig. 1 and 3), as well as to central metabolism, with their respective orthologs in the other species. Where applicable, the genomic contexts of the genes were investigated, leading to categorization, with P. inhibens DSM 17395 set as a reference. Table S7 in the supplemental material lists the P. inhibens DSM 17395 genes analyzed, together with their locus tags, EC numbers, and corresponding enzyme names. The synteny and phylogenetic tree are as previously published (26).
The synteny of the genomes compared and the rRNA tree representing the phylogenetic taxonomy of Rhodobacterales have been determined previously (26).
RESULTS AND DISCUSSION
Phaeobacter inhibens DSM 17395 is currently known to utilize 13 different carbohydrates as sole sources of carbon and energy for growth (see Table S1 in the supplemental material). Five of these carbohydrates, which are environmentally relevant and represent different chemical compound classes, were selected for this study: N-acetylglucosamine (amino sugar), mannitol (sugar alcohol), sucrose (disaccharide), glucose (hexose), and xylose (pentose). A combination of differential proteomics and metabolomics allowed the reconstruction of the uptake and catabolic routes for these five carbohydrates. The catabolic network, together with the genomic locations of the genes involved, is schematically drawn in Fig. 1, and dissemination across the Roseobacter clade is shown in Fig. 2.
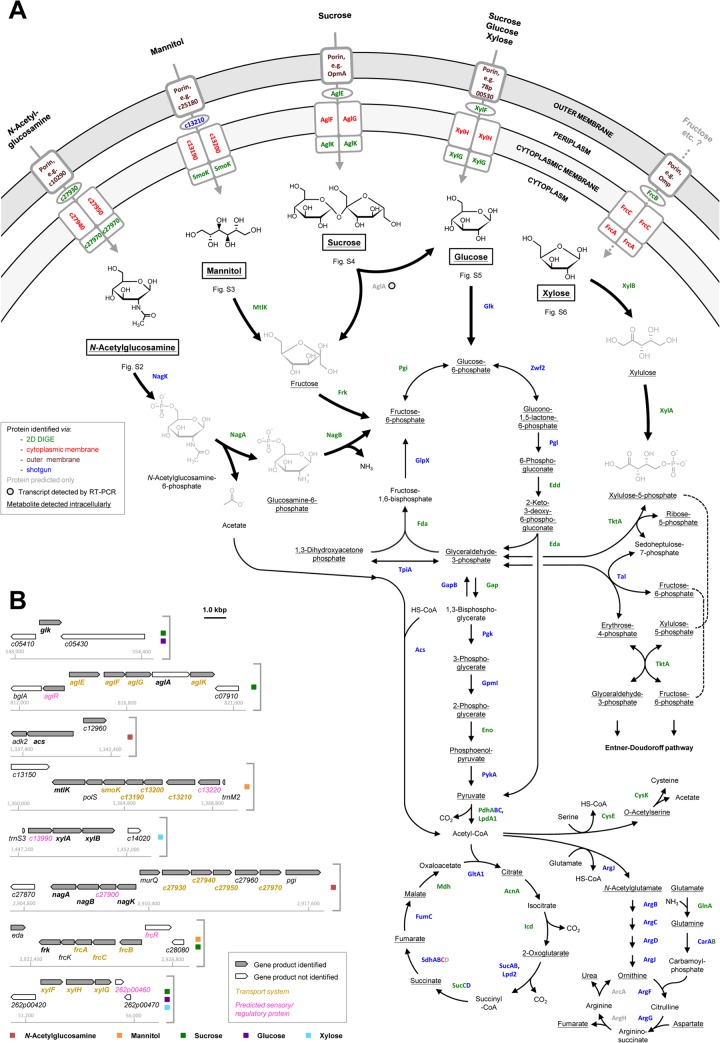
FIG 1.
(A) Reconstruction of transport and degradation pathways for five selected carbohydrates in P. inhibens DSM 17395. The names, predicted functions, and proteomic data of the proteins indicated are summarized in Fig. 3. The metabolites detected are detailed in Fig. 4. Additional information for each of the five carbohydrates is provided in the supplemental material. (B) Genomic locations and contexts of genes encoding the transport systems and degradation enzymes. Genes whose products appear in panel A are shown in boldface. Additional information on predicted regulatory proteins is provided in Table S6 in the supplemental material.
Growth physiology.
P. inhibens DSM 17395 was cultivated in defined mineral seawater medium containing one of the five selected carbohydrates (5 mM each, except for sucrose [2.5 mM]) or succinate (20 mM) (see Fig. S1 in the supplemental material). Since a higher concentration of succinate was applied, the highest maximal optical density (ODmax) of 2.4 was observed with this substrate. The sugars tested yielded ODmax values of 1.1 to 1.2, except for xylose (ODmax, 0.7), which, as a pentose, contains the fewest carbon atoms. The highest maximal specific growth rates (μmax) were observed with cultures growing with succinate (0.22 h−1) or N-acetylglucosamine (0.20 h−1). The optimization of P. inhibens DSM 17395 to fast growth with N-acetylglucosamine agrees with the fact that this compound represents one of the most abundant carbohydrates in marine habitats (4). Growth with glucose (μmax, 0.12 h−1) or xylose (μmax, 0.11 h−1) was about half as fast. With mannitol and sucrose, considerably lower μmax were achieved (0.04 and 0.02 h−1, respectively). The observed mean μmax value (0.10 h−1) is in the same range as that previously observed with a variety of amino acids (on average, 0.11 h−1) (26). A slightly higher μmax of 0.17 h−1 was reached by cultures of P. inhibens DSM 17395 growing with Casamino Acids (25). The fastest growth of P. inhibens DSM 17395 is currently known for cultures in complex Marine Broth (MB) containing peptone and yeast extract (μmax, 0.36 h−1) (24).
Passage through the outer membrane.
In Gram-negative bacteria, compounds have to traverse the outer membrane prior to entering the periplasm. Passage of smaller hydrophilic molecules (up to ∼550 Da) occurs via facilitated diffusion mediated by membrane channels, so-called porins (48). To identify these in P. inhibens DSM 17395, a recently established method for the isolation of outer membrane protein-enriched fractions (32) was applied. By this method, seven outer membrane proteins could be identified (Fig. 3). One of them, protein c06130, is a predicted OmpA-like porin, as known from Escherichia coli K-12 (49, 50), and was detected under all substrate conditions studied. In contrast, the outer membrane protein c10290 was specifically identified in N-acetylglucosamine-grown cells, whereas plasmid-encoded protein 78p00530 was found under the other substrate conditions. This may imply that some outer membrane proteins could serve substrate-specific functions.
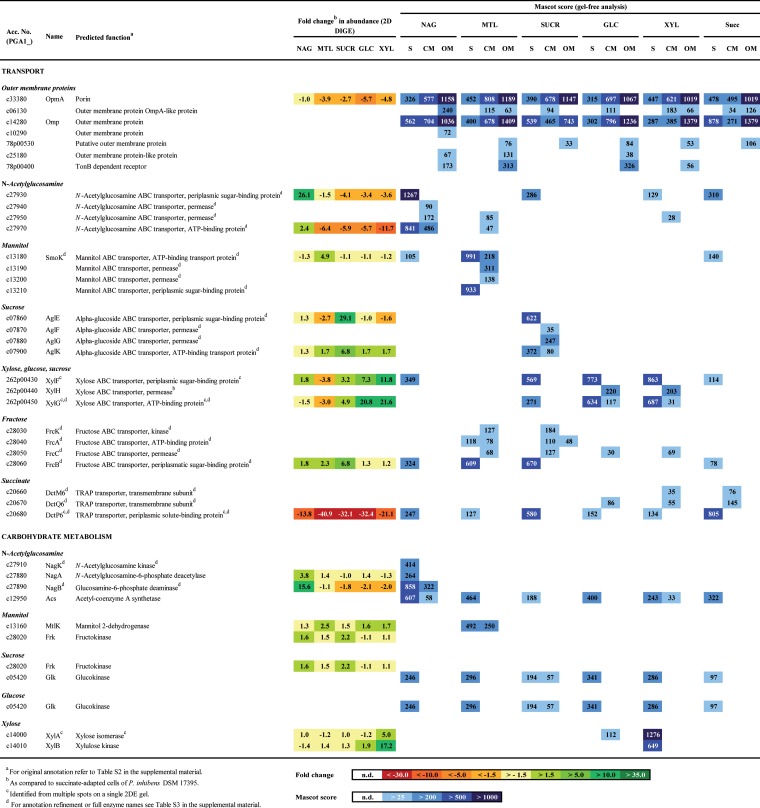
FIG 3.
Selected proteins of P. inhibens DSM 17395 identified from 2DE gels (2D DIGE), by shotgun analysis (S), and from fractions enriched in cytoplasmic membrane protein (CM) or outer membrane protein (OM) and resolved by SDS-PAGE (see also Fig. S8 in the supplemental material). Proteins involved in carbohydrate transport and degradation pathways or in central metabolism are shown. Abbreviations of substrates: NAG, N-acetylglucosamine; MTL, mannitol; SUCR, sucrose; GLC, glucose; XYL, xylose; Succ, succinate.
Carbohydrate transport and degradation pathways. (i) N-Acetylglucosamine degradation.
The amino sugar N-acetylglucosamine is apparently transported into the cytoplasm via a specifically formed ATP-binding cassette (ABC) transporter, c27930 to c27970 (Fig. 1A and 3), reannotated as the N-acetylglucosamine ABC transport system (see Table S3 in the supplemental material).
As in Xanthomonas campestris pv. campestris (51), N-acetylglucosamine in P. inhibens DSM 17395 is degraded to fructose-6-phosphate via three steps (see Fig. S2 in the supplemental material). Two steps could not be reconstructed based on the original genome annotation but could be resolved via differential proteomics and metabolomics. In the first step, intracellularly detected N-acetylglucosamine (Fig. 4) is phosphorylated to N-acetylglucosamine-6-phosphate. This reaction is generally known to be performed by N-acetylglucosamine kinase (NagK). A BLAST search (homology to an N-acetylglucosamine kinase from Roseobacter denitrificans OCh 114; E value, 2e−48) and an HMM search (E value, 6e−30), as well as specific identification in N-acetylglucosamine-grown cells of P. inhibens DSM 17395, suggest that protein c27910 functions as NagK. In the second step, N-acetylglucosamine-6-phosphate deacetylase (NagA) cleaves off acetate, forming the detected metabolite glucosamine-6-phosphate. Acetyl coenzyme A (acetyl-CoA) synthetase activates the acetate for funneling into the TCA cycle or into lipid or amino acid biosynthesis (Fig. 1A; see also Table S4 in the supplemental material). In the third step, glucosamine-6-phosphate is isomerized and deaminated to fructose-6-phosphate. This transformation is proposed to be performed by protein c27890, which was 15.6-fold more abundant in N-acetylglucosamine-grown cells than in succinate-grown cells and was reannotated as glucosamine-6-phosphate deaminase (NagB; EC 3.5.99.6). Although protein c27890 is also similar to glucosamine-fructose-6-phosphate aminotransferase (GlmS; EC 2.6.1.16), its substrate-specific formation supports its proposed role in glucosamine-6-phosphate degradation (Fig. 3). In accord with such an assumption is the general function of the deaminase in the catabolic direction (2, 51, 52), whereas the activity of the biosynthetic transaminase is known from E. coli to be irreversible (53).
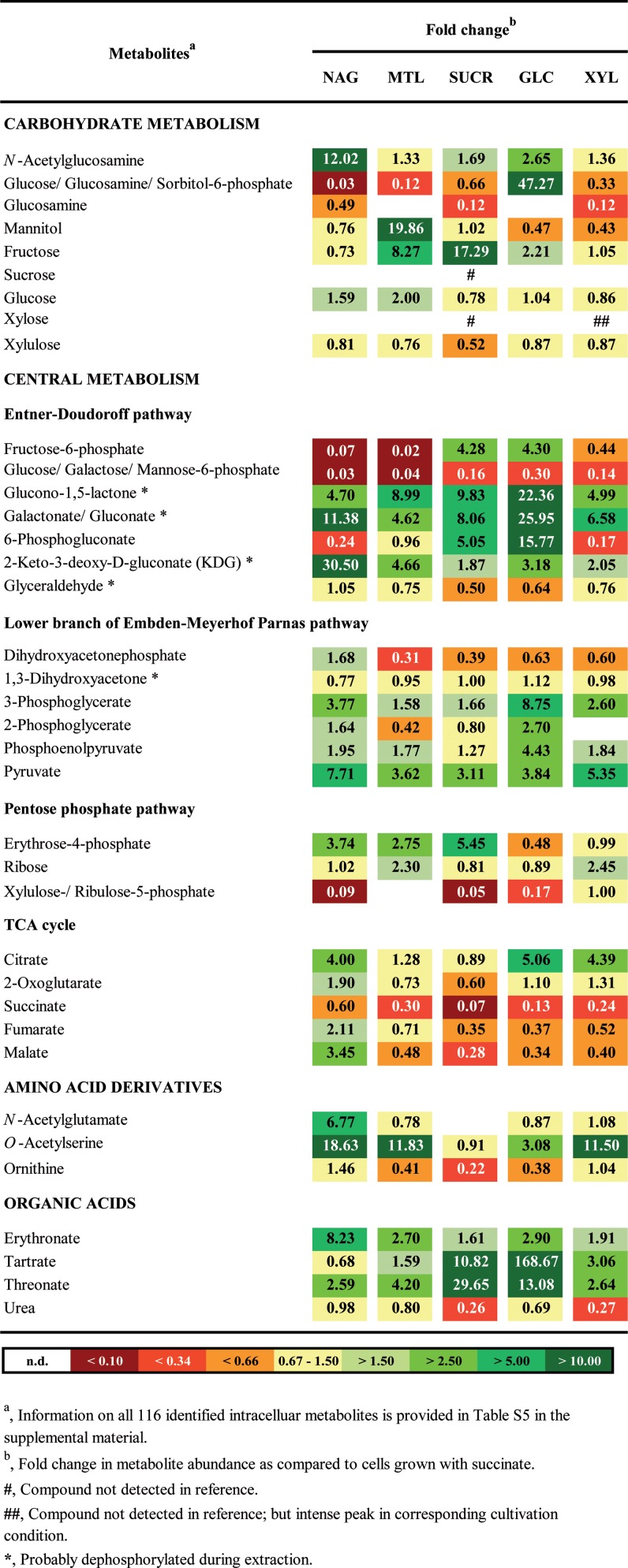
FIG 4.
Fold changes in abundances of selected intracellular metabolites of P. inhibens DSM 17395. For all intracellular metabolites of P. inhibens DSM 17395 identified in this study, see Table S5 in the supplemental material. Abbreviations of substrates: NAG, N-acetylglucosamine; MTL, mannitol; SUCR, sucrose; GLC, glucose; XYL, xylose.
The nagKBA genes are arranged in an operon-like structure on the chromosome of P. inhibens DSM 17395 (Fig. 1B), agreeing with their functions proposed above. In juxtaposition to nagKBA on the opposite strand are c27930 to c27970, encoding the above-mentioned ABC transporter. Between nagKBA and c27930 to c27970, murQ codes for N-acetylmuramate-6-phosphate etherase. MurQ (4.0-fold more abundant in N-acetylglucosamine-grown cells than in succinate-grown cells) converts N-acetylglucosamine-6-phosphate to N-acetylmuramate-6-phosphate; in their dephosphorylated forms, both are important components of the cell wall polymer murein.
(ii) N-Acetylglucosamine and anabolism.
In the absence of external N-acetylglucosamine, the murein building block UDP-N-acetylglucosamine has to be synthesized from fructose-6-phosphate by glucosamine-fructose-6-phosphate aminotransferase (GlmS), phosphoglucosamine mutase (GlmM), and the bifunctional enzyme GlmU, consisting of fused N-acetylglucosamine-1-phosphate uridyltransferase and glucosamine-1-phosphate acetyltransferase. As expected, these three enzymes were formed under all substrate conditions investigated except in N-acetylglucosamine-grown cells (see Table S4 in the supplemental material). Thus, P. inhibens DSM 17395 employs two different enzymes for the interconversion of glucosamine-6-phosphate and fructose-6-phosphate, i.e., GlmS for anabolic and NagB for catabolic purposes.
Degradation of N-acetylglucosamine yields acetyl-CoA via the catabolic pathway described above and via the lower part of the Embden-Meyerhof-Parnas (EMP) pathway. Acetyl-CoA can be used for the biosynthesis of, e.g., cysteine and arginine. The formation of cysteine involves acetyltransferase (CysE) and cysteine synthase (CysK) and proceeds via O-acetylserine. This intermediate possibly acts as a temporary acetyl sink and was more abundant in most carbohydrate-grown cells than in succinate-grown cells (Fig. 4). The first step of arginine biosynthesis is the ArgJ-catalyzed formation of N-acetylglutamate (specifically 6.8-fold more abundant in N-acetylglucosamine-grown cells than in succinate-grown cells) (Fig. 4). Except for the last enzyme (ArgH) of the biosynthetic sequence, all protein constituents (54) were identified (see Table S4 in the supplemental material). Proteins ArgFGH are also involved in the urea cycle employed by P. inhibens DSM 17395 to dispose of excess nitrogen as urea (24). Only slightly higher urea concentrations were detected in cells grown with the only nitrogen-containing substrate, N-acetylglucosamine, than in cells grown with the other four carbohydrates (Fig. 4). However, the possibility that the urea detected might also derive from cellular arginine decomposing during metabolic analysis should be considered.
(iii) Mannitol.
P. inhibens DSM 17395 imports mannitol presumably via a specifically formed ABC transporter, c13180 to c13210 (Fig. 1A and 3), here reannotated as a mannitol ABC transporter (see Table S3 in the supplemental material). In addition, a second ABC transporter, c28030 to c28060, was detected in mannitol- and sucrose-grown cells. The genes encoding this transporter colocalize with the frk gene, encoding fructokinase (Fig. 1B). Thus, one may speculate that the formation of this transporter is activated by fructose, which is formed during the degradation of mannitol and sucrose. BLAST analyses supported reannotation as a fructose ABC transporter (FrcBCAK), as known from Sinorhizobium meliloti (55).
Mannitol is probably converted to fructose by the 2.5-fold-upregulated (in mannitol-grown cells compared to succinate-grown cells) NADH-dependent mannitol 2-dehydrogenase (MtlK; EC 1.1.1.67) (see Fig. S3 in the supplemental material), as known from the related enzyme in Rhodobacter sphaeroides Si4 (56). Mannitol and fructose were detected in the intracellular metabolome. Fructose is then phosphorylated by fructokinase (Frk; EC 2.7.1.4), yielding fructose-6-phosphate.
The mtlK gene is located next to genes encoding the mannitol ABC transporter c13180 to c13210 (Fig. 1B), separated merely by polS (encoding sorbitol dehydrogenase), as in R. sphaeroides Si4 (57). Thus, the ABC transporter might also be active with other polyols, e.g., sorbitol. Sorbitol dehydrogenase (PolS) was identified exclusively by shotgun analysis of mannitol-grown cells, suggesting that the complete gene cluster is transcribed. Notably, P. inhibens DSM 17395 can also use sorbitol as a growth substrate (see Table S1 in the supplemental material).
(iv) Sucrose.
Differential proteomic data indicated sucrose uptake by the α-glucoside transport system (AglEFGK), as known from S. meliloti (58). The periplasmic binding protein (AglE) and the ATP-binding subunit (AglK) displayed the highest increases in abundance in sucrose-grown cells of P. inhibens DSM 17395 (Fig. 1A and 3). In addition to AglEFGK, two other ABC transporters are present in sucrose-grown cells: FrcBCAK (fructose) and XylFGH (xylose, fructose, and sucrose). Possibly, the formation of these two systems is coinduced by the sucrose degradation products fructose and glucose.
On the chromosome of P. inhibens DSM 17395, a predicted α-glucosidase (AglA) is encoded together with the genes of the sucrose transporter AglEFGK in an operon-like structure. Such an operon has been described previously for S. meliloti (58). Thus, one may assume that P. inhibens DSM 17395 also degrades sucrose to glucose and fructose by use of the α-glucosidase. While the AglA protein could not be identified in sucrose-grown cells, the aglA transcript was detected in sucrose-grown cells as well as in succinate-grown cells, indicating constitutive expression (see Fig. S4D in the supplemental material). The reaction products glucose and fructose were identified (Fig. 4) and could be further metabolized via phosphorylation by identified fructokinase (Frk) and glucokinase (Glk), respectively.
Notably, sucrose-grown cells contain a hypothetical protein, c11850 (92.1-fold upregulated in sucrose-grown cells compared to succinate-grown cells). It shows similarities to phage shock protein (PspA), which is generally associated with stress (59, 60). Growth with sucrose could result in a kind of growth-associated stress, since P. inhibens DSM 17395 displays the lowest μmax (0.02 h−1) with this substrate.
(v) Glucose.
Glucose is presumably taken up by the ABC transporter XylFGH (Fig. 1A and 3). Accordingly, the periplasmic binding protein (XylF) was previously found to be more abundant in cells grown with glucose than in cells grown in complex Marine Broth (24).
For degradation, glucose is initially phosphorylated by glucokinase (Glk; EC 2.7.1.2; identified by shotgun analysis) to glucose-6-phosphate (see Fig. S5 in the supplemental material). Further degradation proceeds via the ED pathway. Glucose and glucose-6-phosphate were detected in the intracellular metabolome. The glk gene has a solitary localization on the chromosome of P. inhibens DSM 17395 (Fig. 1B). The presence of its product, Glk, under all substrate conditions tested could point to constitutive expression (Fig. 3).
(vi) Xylose.
The uptake of xylose is assumed to involve the ABC transporter XylFGH, since the two soluble subunits (XylFG) displayed the greatest abundance increases in xylose-grown cells compared to succinate-grown cells (11.8-fold and 21.6-fold, respectively [Fig. 3]). Thus, the solute range of the XylFGH system in P. inhibens DSM 17395 may include xylose as well as glucose and sucrose (see above).
The degradation of xylose is initiated via isomerization to xylulose. As is known from enterobacteria (61, 62) and the marine bacterium Rhodopirellula baltica (63), this isomerization is performed by xylose isomerase (XylA), which was 5.0-fold more abundant in xylose-grown cells than in succinate-grown cells (see Fig. S6 in the supplemental material). Xylulose is then phosphorylated to xylulose-5-phosphate by xylulose kinase (XylB; 17.2-fold more abundant in xylose-grown cells than in succinate-grown cells). Both metabolites were detected in the intracellular fraction. Following the entry of xylulose-5-phosphate into the pentose phosphate pathway, fructose-6-phosphate is formed, which is further metabolized via the ED pathway.
In contrast to the pattern in E. coli K-12 (62), the genes for xylose degradation and transport do not colocalize in P. inhibens DSM 17395. Rather, xylAB form an operon-like structure on the chromosome, whereas xylFGH do so on the 262-kbp plasmid (Fig. 1B).
Regulation. (i) Transporters for carbohydrates and succinate.
Differential subcellular proteomics combined with genomic analysis in the present study revealed that P. inhibens DSM 17395 does not employ phosphotransferase systems (PTS) to import carbohydrates, as do E. coli and other bacteria (64). Rather, P. inhibens DSM 17395 uses ABC transporters. While these are less energy efficient than PTS, they are highly efficient at very low external substrate concentrations (up to a 104-fold concentration difference) (65), such as those encountered in the mostly oligotrophic environments of roseobacters. The substrate specificity of ABC transporter formation could be classified into three levels of stringency. The ABC transporters for N-acetylglucosamine, mannitol, and sucrose were formed with high substrate specificity, agreeing with the tight genomic association of their coding genes with the genes for the respective catabolic enzymes and proposed regulators (Fig. 1B; see also Table S6 in the supplemental material). In contrast, the XylFHG transporter, which is the only ABC transporter separately encoded on the 262-kbp plasmid, was identified in xylose-, glucose-, and sucrose-grown cells. Despite the colocalization of its genes with that for fructokinase (Frk) and the gene encoding the sensory/regulatory protein FrcR, the fructose transporter (FrcBCAK) was identified in mannitol- and sucrose-grown cells. Possibly, fructose as an intermediate of mannitol and sucrose degradation could serve as an inducer, as shown previously for S. meliloti (55).
In contrast to the carbohydrates, succinate is assumed to be taken up by P. inhibens DSM 17395 via the tripartite ATP-independent periplasmic (TRAP) transporter (DctM6P6Q6 [Fig. 3]). This system consists of two transmembrane proteins (DctMQ) and a periplasmic solute-binding protein (DctP) and was demonstrated in Rhodobacter capsulatus as a high-affinity transport system for succinate and other C4 dicarboxylates (66). Since TRAP transporters are ATP independent, the transport of succinate into the cells is less energy-consuming than the uptake of the carbohydrates studied.
(ii) Peripheral degradation pathways.
Most enzymes involved in channeling individual carbohydrates into the central metabolism are formed with high substrate specificity (Fig. 3). Six predicted regulatory/sensory proteins could serve as possible mediators of the underlying tight control of gene expression, since they are encoded in juxtaposition with the “catabolic” genes (Fig. 1B; see also Table S6 in the supplemental material). Four of these proteins were detected in the present study, and three of these are related to the LacI family of transcriptional regulators. The first, c07850 (reannotated as AglR), is more abundant in sucrose-grown cells than in succinate-grown cells and might regulate the expression of aglEFGK and aglA, as proposed previously for S. meliloti (58). The second, c13220, was specifically identified in mannitol-grown cells and probably controls the transcription of mtlK and the genes for the mannitol ABC transporter. The third, c13990, was specifically detected in xylose-grown cells and possibly regulates xylAB expression. The fourth probable regulatory protein identified is the GntR family-type transcriptional regulator c27900, which was identified exclusively in N-acetylglucosamine-grown cells and might control the expression of genes for the uptake and degradation of this amino sugar.
(iii) Central metabolism.
Following the formation of fructose-6-phosphate or glucose-6-phosphate, further metabolism proceeds via the ED pathway, connecting into the lower branch of the EMP pathway (see Fig. S9 in the supplemental material) (22, 23). Accordingly, the protein constituents of both pathways identified are abundant in cells adapted to any of the five carbohydrates tested, compared to succinate-adapted cells (see Table S4 in the supplemental material). This general profile was also observed with the respective metabolites identified (Fig. 4).
In succinate-utilizing cells, carbohydrates have to be generated via gluconeogenesis (see Fig. S12 in the supplemental material), involving pyruvate carboxylase (Pyc), phosphoenolpyruvate carboxykinase (PckA), glyceraldehyde-3-phosphate dehydrogenase 2 (GapB), and fructose-bisphosphate aldolase class 1 (Fda). As expected, these enzymes are mostly more abundant in succinate-adapted than in carbohydrate-adapted cells.
In contrast, proteins of the pentose phosphate pathway (see Fig. S10 in the supplemental material), pyruvate dehydrogenase complex, and TCA cycle (see Fig. S11 in the supplemental material) display mainly unchanged abundances across all six substrate conditions analyzed. This was also observed previously, when P. inhibens DSM 17395 was grown with nine different amino acids (26) and was analyzed for growth phase response (22). Notably, P. inhibens DSM 17395 does not possess a phosphogluconate dehydrogenase generating ribulose-5-phosphate from 6-phosphogluconate, indicating that the oxidative branch of the pentose phosphate pathway is not active.
Formation of C4 carboxylates.
Notably, P. inhibens DSM 17395 produces large amounts of threonate, erythronate, and tartrate during growth with carbohydrates (Fig. 4). It is not clear at present how these C4 carboxylates are formed and why. One may speculate that they are formed from erythrose-4-phosphate, an intermediate of the pentose phosphate pathway, which could be oxidized to 4-phospho-erythronate by an erythrose-4-phosphate dehydrogenase-like enzyme. In E. coli K-12 (67), this enzyme is involved in pyridoxine and pyridoxal-5′-phosphate biosynthesis. Notably, glycerol-aldehyde-3-phosphate dehydrogenase (Gap) from P. inhibens DSM 17395 shows high similarity to erythrose-4-phosphate dehydrogenase of E. coli K-12 (E value, 1e−108). Thus, abundantly formed Gap (see Table S4 in the supplemental material) may transform erythrose-4-phosphate in a side reaction. Dephosphorylation of the reaction product 4-phospho-erythronate would yield erythronate and its isomer threonate. In plants it was observed that threonate could be oxidized to tartrate (68). However, except for the possible involvement of Gap, no enzymes for the formation of erythronate, threonate, and tartrate could be delineated from the genome or the proteomic data.
Entner-Doudoroff pathway.
This study demonstrated that P. inhibens DSM 17395 uses the ED pathway (see Fig. S9 in the supplemental material) not only for the degradation of glucose (22, 23) but also for other carbohydrates. With the ED pathway, P. inhibens DSM 17395 gains, per mol of glucose-6-phosphate, 1 mol each of ATP, NADH, and NADPH instead of 2 mol of ATP and 2 mol of NADH, which are obtained if the EMP pathway is employed. To put it simply, 1 mol NADPH is formed instead of 1 mol each of ATP and NADH. NADPH is important for anabolic reactions, such as amino acid and fatty acid biosynthesis. It has been suggested that in a metabolic model of E. coli, most of the NADPH required is formed by the oxidative pentose phosphate pathway (69). Because the latter is not operative in P. inhibens DSM 17395, the ED pathway is supposed to take over its role in NADPH production. The advantage of using the ED pathway is that NADPH is generated directly without the need for transhydrogenases. In P. inhibens DSM 17395, the NAD(P) transhydrogenase (PntAB) is located in the cytoplasmic membrane (see Table S4 in the supplemental material), and the subunits have 5 and 10 transmembrane helices, respectively. Membrane-localized transhydrogenases use the proton electrochemical gradient to convert NADH and NADP+ to NAD+ and NADPH and thereby use energy indirectly (70). Considering the energy expenditure of transhydrogenases, the balance for the ED pathway approximates that of the EMP pathway.
Comparative genomics.
A total of 74 genes involved in the reconstructed carbohydrate catabolic network (Fig. 1 and 3) and central metabolism (see Table S4 in the supplemental material) were analyzed for their dissemination across Rhodobacterales (Fig. 2) and other heterotrophic members of the marine bacterioplankton (see Fig. S7 in the supplemental material). The query genes of P. inhibens DSM 17395 are compiled in Table S7 in the supplemental material, and those of the other Rhodobacterales selected for the comparative analysis have been reported recently (26).
Homologous enzymes of the central metabolism are widespread (average dissemination, 93% of genomes) across roseobacters. For example, three genes of the pyruvate dehydrogenase complex (pdhABC) are present in all genomes investigated and are mostly arranged in a genomic context identical to that in P. inhibens DSM 17395. The two key enzymes of the ED pathway (Edd and Eda) are encoded in 93% of the roseobacter genomes investigated (except for Roseovarius nubinhibens ISM and Roseovarius sp. strain 217, both from clade 2), corroborating our earlier observation on the general role of the ED pathway among roseobacters (22, 23).
In comparison to the central metabolism, homologous genes for carbohydrate transport, peripheral degradation, and regulation were less widespread among the roseobacter genomes analyzed (present in 48%). In particular, Roseobacter clade 3 members possess only a few homologs, while in the closely related but geographically distant strain P. inhibens 2.10, all genes not only are present but occur in the identical genomic context. In the case of N-acetylglucosamine, genes for the complete transporter are present in 11 genomes, mainly from clade 1, but also from clades 2, 3, and 5 and from non-Roseobacter Rhodobacterales. The reannotated N-acetylglucosamine kinase (nagK) could be found only in P. inhibens 2.10 and Ruegeria pomeroyi DSS-3, two other clade 1 members. However, in 41% of the genomes investigated, a nonhomologous enzyme replaces P. inhibens NagK. Genes involved in the uptake and initial conversion of sucrose and mannitol, as well as in their respective regulation, are present in more than half of the roseobacter genomes investigated. The only exception is the existence of sorbitol dehydrogenase, the coding gene of which (polS) is present in only 32% of the roseobacter genomes investigated. The genes coding for the fructose ABC transporter (frcABC) are confined to 46% of the roseobacter genomes, whereas the corresponding regulator (frcR) and a kinase (frcK) are present in only 29% of the genomes. The dissemination of the glk gene, encoding glucokinase, is restricted to P. inhibens 2.10 and Ruegeria pomeroyi DSS-3. Although the xylose degradation enzymes XylAB are encoded in about half of the genomes investigated, the corresponding regulator c13990 is confined to the two P. inhibens species analyzed.
Other heterotrophic members of the marine bacterioplankton, such as “Candidatus Pelagibacter” spp., Cytophaga spp., Flavobacterium spp., Gramella spp., or Polaribacter spp., possess considerably fewer homologs of the query genes: 28% of the genes involved in central metabolism and only 5% of the genes involved in carbohydrate transport, peripheral degradation, and regulation (see Fig. S7 in the supplemental material). Most of the functionalities unaccounted for in the central metabolism (except for the ED pathway) could be assigned in these bacteria based on the detection of nonorthologous genes by EnzymeDetector. Notably, however, such nonorthologous functions were mostly not found for the peripheral degradation reactions studied and the ED pathway in the selected non-Roseobacter heterotrophic bacteria (see Fig. S7 in the supplemental material). Taking our findings together, the P. inhibens DSM 17395 genes investigated here are present to various degrees in the different Roseobacter clades, as recently reported for genes of P. inhibens DSM 17395 involved in amino acid degradation (26). They could reflect an evolutionary shaping of roseobacter genomes directed at metabolic streamlining to optimize the habitat functioning and niche differentiation of this important group of marine bacteria.
Conclusion.
This study allowed the reconstruction of transport systems and degradation pathways of five environmentally relevant carbohydrates in the model roseobacter P. inhibens DSM 17395. One of the most abundant carbohydrates in the world's oceans is the chitin monomer N-acetylglucosamine. Correspondingly, among the five carbohydrates tested, the fastest growth of P. inhibens DSM 17395 was observed with this compound. Based on the original genome annotation, only two proteins (NagK and Acs) were associated with N-acetylglucosamine degradation. By means of proteogenomics, this study revealed two additional catabolic enzymes, a predicted regulatory protein, and a complete ABC transport system, all of which were formed exclusively in the presence of N-acetylglucosamine. These proteins are not restricted to P. inhibens DSM 17395 but are also predicted for most of the members of Roseobacter clade 1 whose genomes have been sequenced.
It is noteworthy that highly substrate specific formation of metabolic modules (regulators, uptake system, and catabolic enzymes) was observed for all five carbohydrates investigated, indicating a fine-tuned regulatory network similar to that observed recently for the degradation of nine selected amino acids in P. inhibens DSM 17395 (26). Taking these results together, such metabolic adaptability could be advantageous for coping with the naturally fluctuating availability of carbohydrates and other organic substrates in marine systems.
Considering that roseobacters contribute significantly to marine carbohydrate catabolism, the multiple new functional assignments of the present study could be beneficial for the metabolic interpretation of the steadily increasing marine meta-omics data.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to R. Gahl-Janssen (Oldenburg, Germany) for growth tests with carbohydrates and to C. Hinrichs (Oldenburg, Germany) and D. Thies (Bremen, Germany) for experimental support with 2D DIGE and 2DE. We thank K. Trautwein (Oldenburg, Germany), S. Koßmehl (Oldenburg, Germany), and K. Schmidt-Hohagen (Braunschweig, Germany) for expert discussions. We are indebted to F. Widdel for general support of proteomic work in our group.
This study was supported by the Deutsche Forschungsgemeinschaft (SFB TRR 51).
Footnotes
Published ahead of print 23 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00719-14.
REFERENCES
- 1.Pakulski JD, Benner R. 1994. Abundance and distribution of carbohydrates in the ocean. Limnol. Oceanogr. 39:930–940. 10.4319/lo.1994.39.4.0930 [DOI] [Google Scholar]
- 2.Yang C, Rodionov DA, Xiaoqing L, Laikova ON, Gelfand MS, Zagnitko OP, Romine MF, Obraztsova AY, Nealson KH, Osterman AL. 2006. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 281:29872–29885. 10.1074/jbc.M605052200 [DOI] [PubMed] [Google Scholar]
- 3.Kirchman DL, White J. 1999. Hydrolysis and mineralization of chitin in the Delaware estuary. Aquat. Microb. Ecol. 18:187–196. 10.3354/ame018187 [DOI] [Google Scholar]
- 4.Benner R, Kaiser K. 2003. Abundance of amino sugars and peptidoglycan in marine particulate and dissolved organic matter. Limnol. Oceanogr. 48:118–128. 10.4319/lo.2003.48.1.0118 [DOI] [Google Scholar]
- 5.Rich JH, Ducklow HW, Kirchman DL. 1996. Concentrations and uptake of neutral monosaccharides along 140°W in the equatorial Pacific: contribution of glucose to heterotrophic bacterial activity and the DOM flux. Limnol. Oceanogr. 41:595–604. 10.4319/lo.1996.41.4.0595 [DOI] [Google Scholar]
- 6.Skoog A, Benner R. 1997. Aldoses in various size fractions of marine organic matter: implications for carbon cycling. Limnol. Oceanogr. 42:1803–1813. 10.4319/lo.1997.42.8.1803 [DOI] [Google Scholar]
- 7.Iwamoto K, Shiraiwa Y. 2005. Salt-regulated mannitol metabolism in algae. Mar. Biotechnol. 7:407–415. 10.1007/s10126-005-0029-4 [DOI] [PubMed] [Google Scholar]
- 8.Reid SJ, Abratt VR. 2005. Sucrose utilisation in bacteria: genetic organization and regulation. Appl. Microbiol. Biotechnol. 67:312–321. 10.1007/s00253-004-1885-y [DOI] [PubMed] [Google Scholar]
- 9.Poretsky RS, Sun S, Mou X, Moran MA. 2010. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ. Microbiol. 12:616–627. 10.1111/j.1462-2920.2009.02102.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alonso C, Pernthaler J. 2006. Roseobacter and SAR11 dominate microbial glucose uptake in coastal North Sea waters. Environ. Microbiol. 8:2022–2030. 10.1111/j.1462-2920.2006.01082.x [DOI] [PubMed] [Google Scholar]
- 11.Wagner-Döbler I, Biebl H. 2006. Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60:255–280. 10.1146/annurev.micro.60.080805.142115 [DOI] [PubMed] [Google Scholar]
- 12.Thole S, Kalhöfer D, Voget S, Berger M, Engelhardt T, Liesegang H, Wollherr A, Kjelleberg S, Daniel R, Simon M, Thomas T, Brinkhoff T. 2012. Phaeobacter gallaeciensis genomes from globally opposite locations reveal high similarity of adaptation to surface life. ISME J. 6:2229–2244. 10.1038/ismej.2012.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buddruhs N, Pradella S, Göker M, Päuker O, Pukall R, Spröer C, Schumann P, Petersen J, Brinkhoff T. 2013. Molecular and phenotypic analyses reveal the non-identity of the Phaeobacter gallaeciensis type strain deposits CIP 105210T and DSM 17395. Int. J. Syst. Evol. Microbiol. 63:4340–4349. 10.1099/ijs.0.053900-0 [DOI] [PubMed] [Google Scholar]
- 14.Zhang D-C, Li H-R, Xin YH, Liu H-C, Chi Z-M, Zhou P-J, Yu Y. 2008. Phaeobacter arcticus sp. nov., a psychrophilic bacterium isolated from the Arctic. Int. J. Syst. Evol. Microbiol. 58:1384–1387. 10.1099/ijs.0.65708-0 [DOI] [PubMed] [Google Scholar]
- 15.Yoon J-H, Kang S-J, Lee S-Y, Oh T-K. 2007. Phaeobacter daeponensis sp. nov., isolated from a tidal flat of the Yellow Sea in Korea. Int. J. Syst. Evol. Microbiol. 57:856–861. 10.1099/ijs.0.64779-0 [DOI] [PubMed] [Google Scholar]
- 16.Gaboyer F, Tindall BJ, Ciobanu M-C, Duthoit F, Le Romancer M, Alain K. 2013. Phaeobacter leonis sp. nov., an alphaproteobacterium from Mediterranean Sea sediments. Int. J. Syst. Evol. Microbiol. 63:3301–3306. 10.1099/ijs.0.046128-0 [DOI] [PubMed] [Google Scholar]
- 17.Brinkhoff T, Bach G, Heidorn T, Liang L, Schlingloff A, Simon M. 2004. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 70:2560–2565. 10.1128/AEM.70.4.2560-2565.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandecandelaere I, Segaert E, Mollica A, Faimali M, Vandamme P. 2009. Phaeobacter caeruleus sp. nov., a blue-coloured, colony-forming bacterium isolated from a marine electroactive biofilm. Int. J. Syst. Evol. Microbiol. 59:1209–1214. 10.1099/ijs.0.002642-0 [DOI] [PubMed] [Google Scholar]
- 19.Ruiz-Ponte C, Cilia V, Lambert C, Nicolas JL. 1998. Roseobacter gallaeciensis sp. nov., a new marine bacterium isolated from rearings and collectors of the scallop Pecten maximus. Int. J. Syst. Evol. Microbiol. 48:537–542 [DOI] [PubMed] [Google Scholar]
- 20.Berger M, Neumann A, Schulz S, Simon M, Brinkhoff T. 2011. Tropodithietic acid production in Phaeobacter gallaeciensis is regulated by N-acyl homoserine lactone-mediated quorum sensing. J. Bacteriol. 193:6576–6585. 10.1128/JB.05818-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martens T, Heidorn T, Pukall R, Simon M, Tindall BJ, Brinkhoff T. 2006. Reclassification of Roseobacter gallaeciensis Ruiz-Ponte et al. 1998 as Phaeobacter gallaeciensis gen. nov., comb. nov., description of Phaeobacter inhibens sp. nov., reclassification of Ruegeria algicola (Lafay et al. 1995) Uchino et al. 1999 as Marinovum algicola gen. nov., comb. nov., and emended descriptions of the genera Roseobacter, Ruegeria and Leisingera. Int. J. Syst. Evol. Microbiol. 56:1293–1304. 10.1099/ijs.0.63724-0 [DOI] [PubMed] [Google Scholar]
- 22.Zech H, Thole S, Schreiber K, Kalhöfer D, Voget S, Brinkhoff T, Simon M, Schomburg D, Rabus R. 2009. Growth phase-dependent global protein and metabolite profiles of Phaeobacter gallaeciensis strain DSM 17395, a member of the marine Roseobacter-clade. Proteomics 9:3677–3697. 10.1002/pmic.200900120 [DOI] [PubMed] [Google Scholar]
- 23.Fürch T, Preusse M, Tomasch J, Zech H, Wagner-Döbler I, Rabus R, Wittmann C. 2009. Metabolic fluxes in the central carbon metabolism of Dinoroseobacter shibae and Phaeobacter gallaeciensis, two members of the marine Roseobacter clade. BMC Microbiol. 9:209. 10.1186/1471-2180-9-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zech H, Hensler M, Koßmehl S, Drüppel K, Wöhlbrand L, Trautwein K, Hulsch R, Maschmann U, Colby T, Schmidt J, Reinhardt R, Schmidt-Hohagen K, Schomburg D, Rabus R. 2013. Adaptation of Phaeobacter inhibens DSM 17395 to growth with complex nutrients. Proteomics 13:2851–2868. 10.1002/pmic.201200513 [DOI] [PubMed] [Google Scholar]
- 25.Zech H, Hensler M, Koßmehl S, Drüppel K, Wöhlbrand L, Trautwein K, Colby T, Schmidt J, Reinhardt R, Schmidt-Hohagen K, Schomburg D, Rabus R. 2013. Dynamics of amino acid utilization in Phaeobacter inhibens DSM 17395. Proteomics 13:2869–2885. 10.1002/pmic.201200560 [DOI] [PubMed] [Google Scholar]
- 26.Drüppel K, Hensler M, Trautwein K, Koßmehl S, Wöhlbrand L, Schmidt-Hohagen K, Ulbrich M, Bergen N, Meier-Kolthoff JP, Göker M, Klenk HP, Schomburg D, Rabus R. 2014. Pathways and substrate-specific regulation of amino acid degradation in Phaeobacter inhibens DSM 17395 (archetype of the marine Roseobacter clade). Environ. Microbiol. 16:218–238. 10.1111/1462-2920.12276 [DOI] [PubMed] [Google Scholar]
- 27.Gade D, Thiermann J, Markowsky D, Rabus R. 2003. Evaluation of two-dimensional difference gel electrophoresis for protein profiling. J. Mol. Microbiol. Biotechnol. 5:240–251. 10.1159/000071076 [DOI] [PubMed] [Google Scholar]
- 28.Rabus R, Trautwein K. 2010. Proteogenomics to study the anaerobic degradation of aromatic compounds and hydrocarbons, p 4385–4405 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany [Google Scholar]
- 29.Bradford MM. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 30.Wöhlbrand L, Kallerhoff B, Lange D, Hufnagel P, Thiermann J, Reinhardt R, Rabus R. 2007. Functional proteomic view of metabolic regulation in “Aromatoleum aromaticum” strain EbN1. Proteomics 7:2222–2239. 10.1002/pmic.200600987 [DOI] [PubMed] [Google Scholar]
- 31.Zech H, Echtermeyer C, Wöhlbrand L, Blasius B, Rabus R. 2011. Biological versus technical variability in 2D DIGE experiments with environmental bacteria. Proteomics 11:3380–3389. 10.1002/pmic.201100071 [DOI] [PubMed] [Google Scholar]
- 32.Koßmehl S, Wöhlbrand L, Drüppel K, Feenders C, Blasius B, Rabus R. 2013. Subcellular protein localization (cell envelope) in Phaeobacter inhibens DSM 17395. Proteomics 13:2743–2760. 10.1002/pmic.201300112 [DOI] [PubMed] [Google Scholar]
- 33.Oelmüller U, Krüger N, Steinbüchel A, Friedrich CG. 1990. Isolation of prokaryotic RNA and detection of specific mRNA with biotinylated probes. J. Microbiol. Methods 11:73–81. 10.1016/0167-7012(90)90050-G [DOI] [Google Scholar]
- 34.Kühner S, Wöhlbrand L, Fritz I, Wruck W, Hultschig C, Hufnagel P, Kube M, Reinhardt R, Rabus R. 2005. Substrate-dependent regulation of anaerobic degradation pathways for toluene and ethylbenzene in a denitrifying bacterium, strain EbN1. J. Bacteriol. 187:1493–1503. 10.1128/JB.187.4.1493-1503.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Pujar A, Shearer AG, Travers M, Weerasinghe D, Zhang P, Karp PD. 2012. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 40:D742–D753. 10.1093/nar/gkr1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114. 10.1093/nar/gkr988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrel B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 38.NCBI Resource Coordinators. 2013. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 41:D8–D20. 10.1093/nar/gks1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson LS, Eddy SR, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. 10.1186/1471-2105-11-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schomburg I, Chang A, Placzek S, Söhngen C, Rother M, Lang M, Munaretto C, Ulas S, Stelzer M, Scheer M, Schomburg D. 2013. BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res. 41:D764–D772. 10.1093/nar/gks1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.UniProt Consortium. 2012. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40:D71–D75. 10.1093/nar/gkr981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38:W695–W699. 10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups of eukaryotic genomes. Genome Res. 13:2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 46.Markowitz VM, Chen I, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 40:D115–D122. 10.1093/nar/gkr1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quester S, Schomburg D. 2011. EnzymeDetector: an integrated enzyme function prediction tool and database. BMC Bioinformatics 12:376. 10.1186/1471-2105-12-376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benz R. 1988. Structure and function of porins from Gram-negative bacteria. Annu. Rev. Microbiol. 42:359–393. 10.1146/annurev.mi.42.100188.002043 [DOI] [PubMed] [Google Scholar]
- 49.Chen R, Schmidmayr W, Krämer C, Chen-Schmeisser U, Henning U. 1980. Primary structure of major outer membrane protein II* (ompA protein) of Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 77:4592–4596. 10.1073/pnas.77.8.4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugawara E, Nikaido H. 1994. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J. Biol. Chem. 269:17981–17987 [PubMed] [Google Scholar]
- 51.Boulanger A, Déjean G, Lautier M, Glories M, Zischek C, Arlat M, Lauber E. 2010. Identification and regulation of the N-acetylglucosamine utilization pathway of the plant pathogenic bacterium Xanthomonas campestris pv. campestris. J. Bacteriol. 192:1487–1497. 10.1128/JB.01418-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Comb DG, Roseman S. 1958. Glucosamine metabolism. IV. Glucosamine-6-phosphate deaminase. J. Biol. Chem. 232:807–828 [PubMed] [Google Scholar]
- 53.Badet B, Vermoote P, Haumont PY, Lederer F, Le Goffic F. 1987. Glucosamine synthetase from Escherichia coli: purification, properties, and glutamine-utilizing site location. Biochemistry 26:1940–1948. 10.1021/bi00381a023 [DOI] [PubMed] [Google Scholar]
- 54.Cunin R, Glansdorff N, Piérard A, Stalon V. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lambert A, Østerås M, Mandon K, Poggi M-C, Le Rudulier D. 2001. Fructose uptake in Sinorhizobium meliloti is mediated by a high-affinity ATP-binding cassette transport system. J. Bacteriol. 183:4709–4717. 10.1128/JB.183.16.4709-4717.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider K-H, Giffhorn F, Kaplan S. 1993. Cloning, nucleotide sequence and characterization of the mannitol dehydrogenase gene from Rhodobacter sphaeroides. J. Gen. Microbiol. 139:2475–2484. 10.1099/00221287-139-10-2475 [DOI] [PubMed] [Google Scholar]
- 57.Stein MA, Schäfer A, Giffhorn F. 1997. Cloning, nucleotide sequence, and overexpression of smoS, a component of a novel operon encoding an ABC transporter and polyol dehydrogenases of Rhodobacter sphaeroides Si4. J. Bacteriol. 179:6335–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willis L, Walker GC. 1999. A novel Sinorhizobium meliloti operon encodes an α-glucosidase and a periplasmic-binding-protein-dependent transport system for α-glucosides. J. Bacteriol. 181:4176–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darwin AJ. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621–628. 10.1111/j.1365-2958.2005.04694.x [DOI] [PubMed] [Google Scholar]
- 60.Model P, Jovanovic G, Dworkin J. 1997. The Escherichia coli phage-shock-protein (psp) operon. Mol. Microbiol. 24:255–261. 10.1046/j.1365-2958.1997.3481712.x [DOI] [PubMed] [Google Scholar]
- 61.Shamanna DK, Sanderson KE. 1979. Uptake and catabolism of d-xylose in Salmonella typhimurium LT2. J. Bacteriol. 139:64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song S, Park C. 1997. Organization and regulation of the d-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J. Bacteriol. 179:7025–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gade D, Gobom J, Rabus R. 2005. Proteomic analysis of carbohydrate catabolism and regulation in the marine bacterium Rhodopirellula baltica. Proteomics 5:3672–3683. 10.1002/pmic.200401200 [DOI] [PubMed] [Google Scholar]
- 64.Barabote RD, Saier MH., Jr 2005. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 69:608–634. 10.1128/MMBR.69.4.608-634.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Higgins CF. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67–113. 10.1146/annurev.cb.08.110192.000435 [DOI] [PubMed] [Google Scholar]
- 66.Forward JA, Behrendt MC, Wyborn NR, Cross R, Kelly DJ. 1997. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse Gram-negative bacteria. J. Bacteriol. 179:5482–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao G, Pease AJ, Bharani N, Winkler ME. 1995. Biochemical characterization of gapB-encoded erythrose 4-phosphate dehydrogenase of Escherichia coli K-12 and its possible role in pyridoxal 5′-phosphate biosynthesis. J. Bacteriol. 177:2804–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Helsper JP, Loewus FA. 1982. Metabolism of l-threonic acid in Rumex x acutus L. and Pelargonium crispum (L.) L'Hér. Plant Physiol. 69:1365–1368. 10.1104/pp.69.6.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riemer AS, Rex R, Schomburg D. 2013. A metabolite-centric view on flux distributions in genome-scale metabolic models. BMC Syst. Biol. 7:33. 10.1186/1752-0509-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jackson JB. 2003. Proton translocation by transhydrogenase. FEBS Lett. 545:18–21. 10.1016/S0014-5793(03)00388-0 [DOI] [PubMed] [Google Scholar]
- 71.Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, Howard EC, King E, Oakley CA, Reisch CR, Rinta-Kanto JM, Sharma S, Sun S, Varaljay V, Vila-Costa M, Westrich JR, Moran MA. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J. 4:784–798. 10.1038/ismej.2009.150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.