Abstract
Cells use messenger RNAs (mRNAs) to ensure the accurate dissemination of genetic information encoded by DNA. Given that mRNAs largely direct the synthesis of a critical effector of cellular phenotype, i.e., proteins, tight regulation of both the quality and quantity of mRNA is a prerequisite for effective cellular homeostasis. Here, we review nonsense-mediated mRNA decay (NMD), which is the best-characterized posttranscriptional quality control mechanism that cells have evolved in their cytoplasm to ensure transcriptome fidelity. We use protein quality control as a conceptual framework to organize what is known about NMD, highlighting overarching similarities between these two polymer quality control pathways, where the protein quality control and NMD pathways intersect, and how protein quality control can suggest new avenues for research into mRNA quality control.
Keywords: posttranscriptional gene regulation, nonsense-mediated mRNA decay, translational repression, quality control, premature termination codon, posttranslational gene regulation, ubiquitin-proteasome system
INTRODUCTION
Cells use polymers to direct nearly all critical functions, including those that establish inheritance, environmental adaptation, and, for more evolved organisms, multicellularity. Genetic information encoded by the DNA polymer is transferred to mRNA polymers by transcription before ultimately being converted into polypeptides via translation. Owing to their vital functions, all three polymers must meet strict quality control standards. For both mRNA and protein, the cell has put in place multiple pathways to ensure fidelity. Efficiency and economy for these two quality control pathways are maximized bytightly coupling the recognition of defective products to their biosynthesis and/or stability (147, 157, 158). Among the detrimental features for proteins is a failure to attain the proper fold and thus an inability to properly function. Proteins may fail to properly fold for various reasons: mistaken amino acid incorporation or altered amino acid incorporation rate during protein synthesis, inability to locate a partner protein, or even changes that affect general cellular homeostasis. The critical function of mRNA is related to the primary sequence of its protein-coding region, and thus this is subject to inspection by the cell. In particular, creation of a premature termination codon (PTC) generally activates the nonsense-mediated mRNA decay (NMD) pathway. PTCs can be introduced as a result of genetic or somatic mutations, mistakes during transcription, or mistakes during splicing. For both proteins and mRNAs, the defective polymer must be first recognized, subsequently earmarked for degradation, and finally destroyed.
MECHANISMS
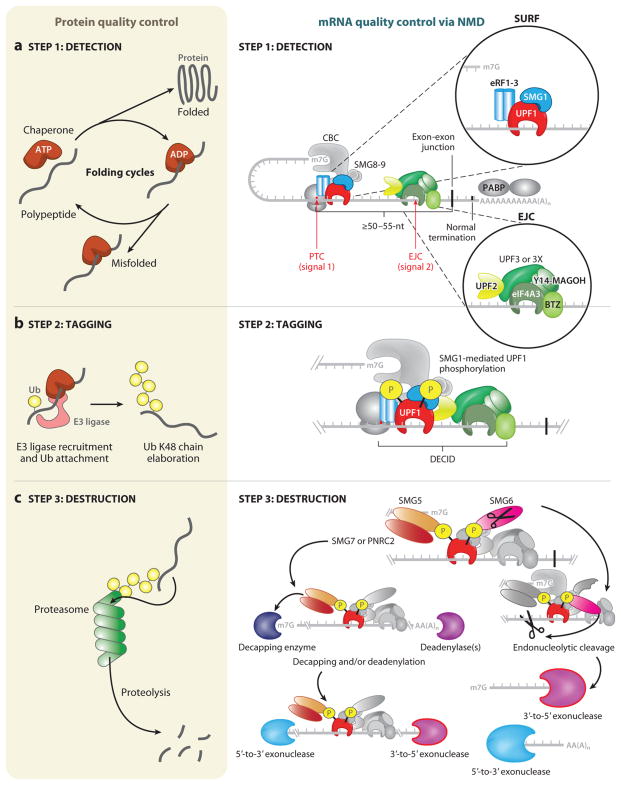
Although the methods utilized to achieve quality control are diverse, protein quality control and mRNA quality control as exemplified by NMD can be conceptually divided into three basic steps: detection, tagging, and destruction (Figure 1). Much is known about the molecular mechanism governing NMD activation, and this is the subject of recent extremely detailed reviews (20, 74, 75, 88, 114, 124, 128, 134, 146, 158, 162). We provide only a brief overview of the mechanism, focusing on mammalian cells, with the purpose of underscoring its similar design principles to protein quality control.
Figure 1.
Similar steps govern protein quality control and nonsense-mediated mRNA decay (NMD). A general overview of protein quality control (left) and mRNA quality control, as exemplified by NMD (right), can be separated into three distinct steps: detection, tagging, and destruction. (a) Detection for protein quality control proceeds through ATP-dependent cycles of chaperone binding, release, and rebinding to hydrophobic patches in unfolded and/or partially folded proteins, providing the opportunity to fold (left). A decision is made as to whether the client protein is terminally misfolded, and, if so, this leads to tagging. The detection step for NMD relies on two signals (right). The first signal is provided by the premature termination codon (PTC), which generally defines NMD substrates, and the proteins that associate with the translation termination complex that assembles at the PTC, including the terminating ribosome and the SURF complex (inset, upper right), which consists of SMG1, UPF1, eRF1, and eRF3. The second signal often derives from an exon-junction complex (EJC), which associates with mRNAs <24 nucleotides (nts) upstream of a splicing-generated exon-exon junction whether or not they are NMD substrates. Other signals, such as unusually long 3′ UTRs, also exist. The EJC (inset, lower right) is composed of four core components (eIF4A3, Y14, MAGOH, BTZ) and associated NMD factors UPF3 or UPF3X and UPF2. If a PTC is located ≥50–55 nts upstream of an exon-exon junction, the mRNA is recognized as aberrant and tagging proceeds. (b) (Left) Tagging for misfolded proteins is governed by the ubiquitylation system. In an ATP-dependent reaction, ubiquitin (Ub) is covalently transferred from an E1 enzyme to an E2 enzyme (not shown). Ub can be transferred in covalent linkage to E3 enzymes containing a HECT (homologous to the E6-AP carboxyl terminus) domain before transfer to the substrate protein, or Ub can be directly transferred from the E2 enzyme to substrates by E3 ligase enzymes containing a RING (really interesting new gene) domain. E3 ligases can mediate ubiquitin transfer to misfolded proteins by binding to molecular chaperones. Mono-Ub is elaborated into a Ub chain with linkages at lysine (K)48. This constitutes the tag that identifies the attached protein for destruction. (Right) For tagging during NMD to occur, all or some of the SURF complex joins the EJC, possibly while the terminating ribosome is still present, forming a decay-inducing complex (DECID). This configuration activates SMG1 to phosphorylate UPF1. Phosphorylated UPF1 signals the mRNA for destruction and has the added effect of inducing translational repression of the mRNA, which is a prerequisite for destruction. (c) (Left) Destruction of proteins is the job of the proteasome, a macromolecular proteolytic machine whose components include proteins that recognize Ub-chain tags and initiate feeding of the aberrant protein into the bore of the proteasome. Aberrant proteins are thereby degraded into short peptides. (Right) NMD-dependent destruction of mRNA relies on recruitment of SMG6 and/or SMG5-SMG7 or SMG5 -PNRC2 complexes via the phosphate tags on UPF1. SMG6 possesses its own endonucleolytic cleavage activity, cutting the mRNA target 5′ to the EJC. 3′-to-5′ exonucleases degrade the 5′-cleavage product. UPF1 helicase activity disassembles the RNP components bound to the 3′-cleavage product, and this is followed by 5′-to-3′ exonucleolytic degradation. The SMG5-SMG7 or SMG5-PNRC2 adaptor complexes, recruited via SMG5 to a UPF1-localized phosphate moiety, direct exonucleolytic degradation of the mRNA. These adaptors recruit decapping enzymes and/or deadenylation enzymes, and their activities are followed by 5′-to-3′ and 3′-to-5′ exonucleolytic decay, respectively. Proteins relevant to each step are shown in color. Abbreviation: CBC, cap-binding complex.
Step 1: Detection
In order to carry out their essential functions, proteins must be folded into the proper three-dimensional shape and further assume the proper quaternary structure. Terminally misfolded proteins are eliminated, but what distinguishes an unfolded protein from a nascent protein in the process of folding remains a key question in protein quality control. Mammalian cells contain a complex network of chaperones that promote folding (148), among them the heat shock protein Hsp70. Hsp70 uses cycles of ATP hydrolysis to repeatedly bind, protect, and release hydrophobic regions (normally located in the interior of proper folds) in client proteins, facilitating the opportunity to fold and preventing aggregation. Molecular chaperones bind nascent polypeptide chains as they are synthesized on the ribosome, ensuring a temporal window compatible with folding. In the case of proteins ultimately deemed to be misfolded, presumably at some point a decision is made that further folding cycles are of little utility toward achieving proper folding, and the protein is partitioned to degradation. Hsp70 can interact with ubiquitin E3 ligases (see Figure 1, Step 2) (3) that link detection to tagging. Thus, chaperone complexes make the key decision as to whether a protein is terminally misfolded and must be destroyed (28, 66).
In contrast to proteins, it is most often the protein-encoding potential of mRNAs that critically determines whether they fail to meet cytoplasmic quality control standards. Generation of a PTC can involve as little as a single nucleotide change. How such a subtle alteration can be detected as aberrant has led to several models and, for the sake of simplicity, we first discuss the best-documented model before layering on additional complexity. In all models, detection is intimately connected to translation termination that takes place during an initial pioneer round of translation (114). Newly synthesized cytoplasmic mRNAs are bound by the cap-binding protein heterodimer CBP80–CBP20, which constitutes their cap-binding complex (CBC), and if derived from intron-containing pre-mRNA, they are also bound by multiprotein assemblies called exon-junction complexes (EJCs), each deposited <20–24 nucleotides (nts) upstream of exon-exon junctions as a consequence of pre-mRNA splicing (100, 101). Translation termination, which involves eukaryotic release factor 1 (eRF1) and eRF3, provides the first signal necessary for activation of NMD. If assembly of eRF1–eRF3 at a termination codon occurs ≥50–55 nts upstream from an exon-exon junction, the footprint of the terminating ribosome is insufficient to physically remove the EJC, and signal two is engaged. Given that normal termination codons are usually located in the last exon, the presence of an EJC downstream of a termination codon generally constitutes an aberrant situation.
Much is known about the molecular choreography that takes place during PTC detection. NMD is mediated by a core set of conserved up-frameshift proteins: UPF1, UPF2, UPF3 (also called UPF3a), and UPF3X (also called UPF3b) (75, 128, 143). UPF3 and UPF3X have partially redundant functions (see below). In multicellular organisms, NMD is aided by suppressors with morphological effects on genitalia proteins: SMG1, SMG5, SMG6, SMG7, SMG8, and SMG9 (83, 86, 132, 203). Among these proteins, UPF1 is the principal orchestrator of NMD. It is an ATP-dependent RNA helicase that initially interacts with mRNA cap-bound CBP80 and with eRF3 on the terminating ribosome. Remodeling of the pioneer translation initiation complex involves replacement of the CBC with eukaryotic translation initiation factor (eIF)4E, and this has been shown to render the remodeled mRNP an inefficient NMD target, in part because CBP80 is lost (see below) (22, 111, 153). UPF1 also associates with SMG1, a phosphatidylinositol 3-kinase-related serine/threonine kinase that forms the SURF (SMG1, UPF1, eRF1, and eRF3) complex (86). SMG1 activity in the SURF complex is silenced by SMG8 and SMG9 (202). Signal two of NMD is orchestrated by the EJC, whose core components include the RNA helicase eIF4A3, which serves as the EJC anchor, as well as the Y14–MAGOH heterodimer, which inhibits eIF4A3 ATPase activity (4, 130), and Barentsz (BTZ, or MLN51, or CASC3), which enhances eIF4A3 helicase activity (131). These core components are decorated with additional factors including RNA-binding protein S1 (RNPS1), Acinus, Sin3A-associated protein 18 (SAP18), Pinin, S6K1 Aly/REF-like substrate (SKAR), serine/arginine (SR)-related nuclear matrix protein 160 (SRm160), the DEAD-box protein UAP56, the mRNA export factor Aly/REF, and, importantly, UPF2, which is bound to the EJC via either UPF3 or UPF3X (9). The majority, but not every exon-exon junction, receives an EJC, and EJC composition is exceedingly complex and heterogeneous (52, 94, 154, 155, 164, 184). Other recently identified trans-effectors, such as the DEAH-box protein 34 (Dhx34) and neuroblastoma amplified gene (NAG), have been shown to function in human NMD, but their precise molecular roles remain to be determined (106).
Step 2: Tagging
Once misfolded proteins are identified, they are marked for destruction (27, 63). This mark takes the form of ubiquitylation, which is a covalent posttranslational modification. The C terminus of the 76–amino acid protein ubiquitin is covalently joined to the ε-amine of lysine residues in substrate proteins, forming an isopeptide bond. Ubiquitin addition proceeds via the ATP-dependent generation of a thioester bond between the C terminus of ubiquitin and a cysteine residue in an E1 enzyme. Ubiquitin is next transferred to one of more than twenty E2 ubiquitin-conjugating enzymes, which associate with E3 ubiquitin ligase enzymes. E3s transfer the ubiquitin moiety either directly to the substrate protein [in the case of RING (really interesting new gene) domain–containing E3s] or first to itself and then to the substrate [in the case of HECT (homologous to the E6-AP carboxyl terminus) domain–containing E3s]. Ubiquitin contains several internal lysine residues that can be targets of ubiquitylation, generating ubiquitin chains. Generally, Lys48-linked ubiquitin chains signal degradation by the protea-some.
Regarding NMD, once PTC-containing mRNPs are identified, their destruction also involves protein modifications and rearrangements. On substrates where both signal-one and signal-two requirements have been met, the complex of CBP80 and SMG1-UPF1 of the SURF complex associate with the UPF2-UPF3X (or UPF2-UPF3) complex located as part of the EJC, forming the decay-inducing complex (DECID) (76, 202). How are UPF1 and associated factors able to traverse the often considerable distance between the SURF complex and EJC? A recent model suggests that UPF1 may use its own helicase to “reel in” the mRNA that exists between the SURF complex and the EJC (160). UPF1 certainly associates with the 3′ UTRs of NMD targets in a way that is augmented by SURF and the presence of a 3′ UTR EJC (65, 73, 96, 209). In an incompletely understood but rate-limiting step, SMG1 kinase activity is stimulated (2, 82, 203), and UPF1 is phosphorylated at residues in its N- and C-terminal domains (86, 133). Phosphorylation of UPF1 triggers a critical step of translational repression that is required before the mRNP can be degraded (80). This step involves an interaction between phosphorylated UPF1 and eIF3 that is part of the 43S ribosomal complex at the initiation codon of an NMD target. This interaction inhibits 60S ribosomal subunit joining to form a translationally active 80S ribosome and thus further translation initiation events on the mRNP.
Step 3: Destruction
Proteins tagged with ubiquitin, i.e., those that are slated to be destroyed, are delivered to the proteasome for degradation (71, 174, 201). Collectively, the ubiquitin conjugation machinery and proteasome are referred to as the ubiquitin proteasome system (UPS). The 26S proteasome is composed of a barrel-shaped 20S core particle with two 19S regulatory cap complexes at either end. Polyubiquitylated substrates are recognized by one of the two 19S caps, either directly or by adaptor proteins, and fed into the interior of the core particle. The core particle is composed of seven different α subunits and seven different β subunits arranged into four stacks of rings. The outer two stacks, which abut the 19S cap, are composed of α subunits, and the inner two rings are composed of β subunits, of which β1, β2, and β5 are proteolytically active. It is here that terminally misfolded proteins that fail to meet quality control standards are recycled into their component amino acids.
During NMD, mRNAs decorated with phosphorylated UPF1 are similarly committed to destruction. Phosphorylated UPF1 recruits SMG6, which has endonucleolytic activity in its PIN (PilT N-terminal) domain (36, 72) and displaces UPF3 or UPF3X at the EJC (85). Irreversible endonucleolytic cleavage by SMG6 generates a 5′ cleavage product that includes the PTC and a 3′ cleavage product that contains the EJC and NMD components. The 5′ cleavage product is subject to 3′-to-5′ decay, possibly by the exosome (156). The 3′ cleavage product must meanwhile be stripped of its protein components in order to be accessible to nucleases, and this is the job of UPF1 (46). UPF1 activity is normally auto-inhibited by its own N- and C-terminal domains (43), but when UPF1 binds to UPF2, it undergoes a large conformational change that activates its helicase activity (15, 16). UPF1 helicase activity disassembles proteins bound to the 3′ cleavage product, recycling NMD factors and facilitating 5′-to-3′ exonucleolytic degradation by the exoribonuclease XRN1 (46), possibly consistent with cryoelectron microscopy structures showing UPF1 at the 3′ edge of the EJC (119). Phosphorylated UPF1 also recruits SMG5, an adaptor that binds either proline-rich nuclear receptor 2 (PNRC2) or SMG7 (24, 26, 83, 180), each of which in turn recruits activities that result in mRNA decapping followed by 5′-to-3′ degradation, deadenylation followed by 3′-to-5′ degradation, or both (49, 101, 180).
Because the events culminating in mRNA decay are preceded by a pioneer round of translation, destruction of the nascent peptide must be considered. Although not described for mammals, the UPS and NMD pathways likely converge in yeast (95, 173). The yeast UPF1 cysteine-histidine-rich domain interacts with an E2 enzyme and, together with UPF3, is sufficient to induce the autoubiquitylation of UPF1 in vitro, demonstrating the ability of UPF1 to participate in an E3-like reaction. This could potentially be a mechanism for rapid NMD-coupled degradation of the nascent peptide as a particularly effective means to downregulate the production of a truncated protein.
Post-mRNA Destruction Nonsense-Mediated mRNA Decay Events
After destruction of the mRNA, NMD factors must be recycled for further rounds of decay. As discussed, UPF1 helicase activity disassembles components of the NMD substrate for future use. UPF1 itself, however, must be returned to its basal hypophosphorylated state. This is accomplished by protein phosphatase 2A (PP2A), which is recruited by SMG5-SMG7 (23, 132). Dephosphorylation of UPF1 is critical because failure to recruit PP2A leads to an inhibition of NMD. Additionally, the ribosome must be recycled, but both the timing of this step with regard to mRNA destruction and the identity of the factors contributing to it are unknown (161).
Alternative Models for Nonsense-Mediated mRNA Decay Recognition
Although the EJC-dependent model for substrate recognition has been rigorously experimentally validated, exceptions exist. NMD is a highly conserved mechanism and the yeast Saccharomyces cerevisiae, which has few introns, can recognize aberrant termination on the basis of the distance between the terminating ribosome and the poly(A) tail (1), although there are exceptions to this faux 3′ UTR model (118), which has been difficult to prove (89). Mammalian NMD substrates have also been identified that bear unusually long 3′ UTRs that are devoid of an EJC. In this so-called “fail-safe” mechanism (115, 207), normal termination is, analogously to yeast, proposed to be prevented by an inefficient interaction between poly(A)-binding protein (PABP)C1 decorating the poly(A) tail and eRF3 at the PTC (37, 81, 165), although some data (10, 113, 115) indicate that efficient fail-safe NMD requires an EJC residing upstream of the 3′ UTR. In NMD substrates, an inefficient interaction between PABPC1 and eRF3 allows UPF1 to gain access to the mRNP. In support of this, manipulations that shorten the distance between the PTC and poly(A) tail by either deletion in yeast and mammalian cells (1, 59, 137, 165), by tethering PABPC1 within the 3′ UTR, or by formation of a secondary structure in mammalian cells (37) protect the transcript from NMD. Aunified model posits that NMD is a consequence of competition between 3′ UTR-bound factors that stimulate UPF1 recruitment to the terminating ribosome (such as the EJC) and factors that antagonize recruitment of UPF1 (such as PABPC1) (165). Importantly for models that rely on distance arguments, eIF4E-bound mRNAs are thought to circularize, and CBC-bound mRNAs may do likewise. This may explain why PTCs located near the 5′ end in certain substrates are less efficient at eliciting NMD (37, 136), although a role for translation reinitiation downstream of the PTC and upstream of an EJC offers an additional, if not alternative, explanation (125, 206).
Alternative Nonsense-Mediated mRNA Decay Branches
Further complexity is added by the discovery that different mRNA substrates require different combinations of NMD trans-effectors. The classical branch of NMD requires the EJC and all of the UPF factors, whereas the fail-safe branch does not require that an EJC reside in the 3′ UTR; however, it does need an EJC situated upstream of the 3′ UTR (208). Through molecular tethering experiments, a UPF2-independent branch requiring the core EJC components, but not RNPS1, and a pathway requiring UPF2, UPF3X, and RNPS1, but not some of the core EJC components, have been described (52). Finally, a branch lacking any UPF3 or UPF3X requirements has been reported (18, 53). How these different dependences impact the molecular rearrangements that lead to NMD is unknown. However, it is clear that EJCs can manifest differences in their composition (52, 94, 154, 155, 164, 184).
Subcytoplasmic Sites of Quality Control
The cytoplasm of mammalian cells contains Hsp70 and other chaperones that help determine if their client proteins are properly folded (Figure 1, Step 1). In addition to nuclear proteasomes necessary for nuclear protein quality control (185), the protein degradation machinery is likewise cytoplasmically disposed. Thus, much of protein quality control takes place in what is topologically the cytoplasmic portion of the cell. An interesting exception is provided by proteins that traverse the secretory pathway and thus reside in what is topologically outside the cell. Sensing misfolded proteins is the job of endoplasmic reticulum (ER)-localized chaperones, some of which leverage glycan processing to decide when a protein has become terminally misfolded (167). These misfolded proteins must be ejected back into the cytoplasm, where they are ubiquitylated and degraded.
In contrast, the subcellular site where NMD takes place has been a matter of debate. Many biochemical fractionation studies have shown that NMD activity is nucleus-associated (5, 14, 21, 90, 108, 163, 177, 181, 207). This led to the hypothesis that the NMD of most mRNAs takes place during mRNA export and on mRNAs that have yet to be released into the cytoplasm, although some NMD substrates are degraded within the cytoplasm after release from an association with nuclei (123, 152). Alternatively, a nuclear surveillance pathway was invoked (12, 77, 195). Given that NMD is a process that is dependent on translation and that the consensus view does not support nuclear translation (29), such a nuclear surveillance model seems unlikely. Furthermore, recent evidence strongly supports that nucleus-associated NMD is not nucleoplasmic: Single-RNA fluorescent in situ hybridization (FISH) experiments that measured the NMD of PTC-containing mRNA in intact single cells after the induction of PTC-containing gene transcription have revealed that, for those substrates studied, NMD occurs not in the nucleoplasm but on the cytoplasmic face of the nuclear envelope soon after the mRNA is accessible to translation by cytoplasmic ribosomes so that the NMD target has a half-life of <1 min once in the cytoplasm (178). The remainder is degraded with a half-life that is comparable to the half-life of normal PTC-free mRNA. These results are consistent with early ensemble studies that demonstrate the biphasic decay of NMD targets (5, 21). Recent studies show that eIF4E-bound PTC-containing mRNAs may be subject to NMD (35, 68, 149); however, the fast degradation kinetics observed may imply that, for a particular transcript, the bulk of its degradation occurs before exchange of CBC for eIF4E. Although processing bodies (P-bodies) have been implicated as the cellular site of NMD, so have polysomes. P-bodies are cytoplasmic foci that are enriched in decapping enzymes and the 5′-to-3′ degradation machinery (39, 42, 45). Although some studies have shown localization of NMD components to P-bodies (26, 34, 180), depletion of Ge-1, an essential component of P-bodies, does not inhibit NMD (170). P-bodies may therefore simply be a by-product of extreme demand on the mRNA degradation machinery, manifested as visible mRNP aggregates rather than a cause of mRNA degradation (39). Evidence for polysome-associated NMD is derived from yeast (69), where decapped NMD substrates are detected in the polyribosome fraction. The finding that translational repression is required for NMD in mammals (80) also indicates that at least the first stages of NMD occur on polysomes (22).
BEYOND QUALITY CONTROL
Both the protein quality control pathway and NMD exert regulatory roles in the cell. These functions are generally an adjunct to their roles in eliminating misfolded proteins and transcripts that produce truncated proteins. Controlling protein and transcript levels after biosynthesis via the UPS and NMD pathways, respectively, offers the cell a rapid means of responding to acute insults. Here, we detail examples of these other functions and how they are exploited for the benefit of the cell.
Regulation of Normal Proteins or Transcripts
In addition to its job of disposing misfolded proteins, the UPS is involved in the regulation of properly folded proteins whose destruction yields a biological outcome. p53, the tumor-suppressor protein that integrates inputs from a diverse array of cellular damages, is a constitutive target of the UPS through its interaction with the mouse double-minute 2 homolog (MDM2) protein, which is an E3 ubiquitin ligase. In unstressed cells, p53 has a short half-life, on the order of 5–30 min, and its steady-state levels remain low because of ubiquitylation by MDM2 (122). During lethal insults to the cell, this interaction is disrupted, leading to rapid accumulation of p53 and ultimately growth arrest, senescence, and apoptosis. Posttranslational degradation of p53 by MDM2 allows for a rapid response, and provides a way by which nonlethal insults can be repaired.
As a second example, flux through the cholesterol biosynthesis pathway is regulated by adjusting HMG-CoA-reductase (HMGR) levels in the mammalian ER. HMGR is the rate-limiting enzyme in sterol biosynthesis. When levels of lanosterol, which is the first intermediate in sterol production, accumulate because flux is too high, lanosterol binds to HMGR and triggers an association with insulin-induced gene 1 (Insig1), another membrane-embedded protein. This complex then binds to glycoprotein 78 (gp78), a polytopic RING-finger E3 ligase via the Insig1 adaptor, and HMGR is degraded by the UPS (61, 168), functionally lowering cholesterol production.
NMD similarly regulates the quantity of physiological transcripts. Several studies using human cells have now shown that mRNAs encoding full-length, functional proteins are subject to NMD (120, 200). Evidence for conservation of this function comes from additional studies in Arabidopsis thaliana (67, 205), Drosophila melanogaster (145), Caenorhabditis elegans (142), and Saccharomyces cerevisiae (56, 60, 102). Many of the transcript lists produced in these studies must be interpreted with caution: Given that most studies involve ablation of an NMD protein (e.g., UPF1 or UPF2) and measuring the resultant changes in mRNA levels, not all of those mRNAs that are upregulated are direct targets of NMD. If, for example, a transcript encoding a transcription factor is a direct target (see below), downregulating this transcript will lead to additional indirect changes in the abundance of other transcripts. How can a quality control mechanism that presumably evolved to recognize aberrant transcription products recognize normal mRNAs? At least five classes of NMD-inducing features have been described: (a) an upstream open reading frame (uORF) in the 5′ UTR where the stop codon of the uORF is interpreted as a PTC relative to the main ORF; (b) alternative splicing (AS), in which the resulting shift in the translational reading frame generates a stop codon ≥50–55 nts upstream of an exon-exon junction; (c) abnormally long 3′ UTRs; (d ) a normal termination codon ≥50–55 nts upstream of an exon-exon junction (so that when translation terminates normally, the 3′ UTR EJC is not removed); and (e) UGA codons within certain selenoprotein-encoding mRNAs that are recognized as selenocysteine codons, depending on the cellular concentrations of selenium and the presence of a selenocysteine insertion sequence (SECIS) in the mRNA 3′ UTR (33, 123, 169). The magnitude of NMD-induced downregulation of these un-mutated transcripts is generally less than that of authentic PTC-bearing transcripts, leading to the idea that this is a method for fine-tuning rather than eliminating gene expression.
Alternatively spliced transcripts represent an interesting class of NMD targets because approximately 35% of AS events result in inclusion of an exon-bearing PTC (55, 103). AS-coupled NMD (AS-NMD), which is also called regulated unproductive splicing and translation (RUST), is likely a general strategy for fine-tuning the abundance of mRNA species that can be used to produce physiological amounts of protein without altering transcription levels (64, 103). Interestingly, among the AS-NMD targets are mRNAs that encode components of the splicing machinery itself, such as polypyrimidine tract-binding protein (PTB) and the SR protein SC35. When the levels of their encoded proteins become too high, this is thought to provide an autoregulatory mechanism for controlling the levels of the splicing machinery: High levels of these splicing factors direct inclusion of a termination codon into the mRNA from which each splicing factor is derived, triggering NMD and splicing downregulation (98, 117, 127, 151).
Regulation of Quality Control Itself
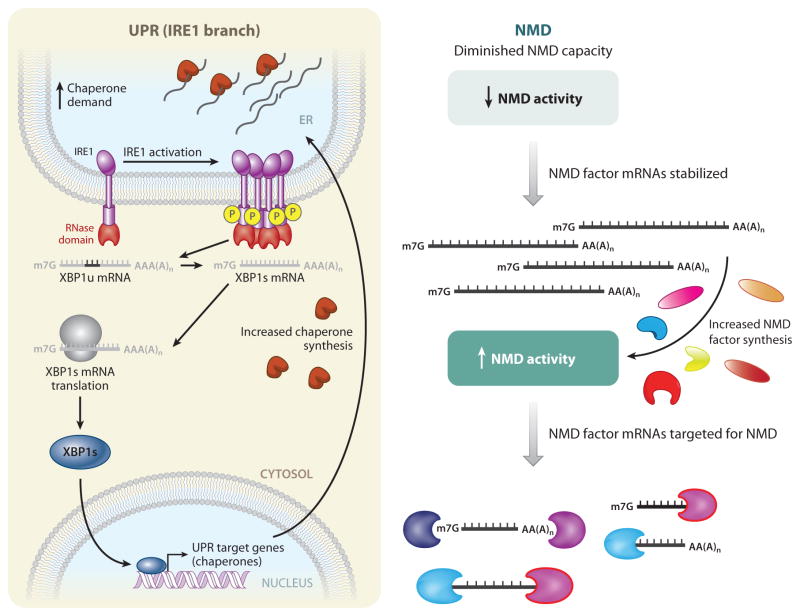
Protein quality control activity as a whole is not constant but is instead subject to regulation by the cell for several purposes. This is illustrated by a conserved form of feedback control called the unfolded protein response (UPR), which buffers the cell against deleterious effects that arise as a consequence of an increase in protein-folding load in the ER (187). When the chaperones involved in detection (Figure 1, Step 1) of unfolded proteins in the ER are overwhelmed by their client proteins, a suite of three sensors—inositol-requiring 1 (IRE1), activating transcription factor 6 (ATF6), and protein kinase R-like ER kinase (PERK)—are activated. The UPR takes a two-pronged attack to buffer against unfolded proteins. IRE1 mediates an unconventional post-transcriptional splicing event in the cytoplasm that ultimately results in production of the transcription factor X box–binding protein 1 spliced (XBP-1s) from the spliced mRNA (Figure 2), whereas ATF6 is cleaved by Site-1 protease (S1P) and Site-2 protease (S2P) to yield a fragment that translocates into the nucleus. In the nucleus, both XBP-1s and ATF6 activate a group of UPR target genes, among them the very chaperones and ER quality control components that are used to sense the increased protein load. This helps to match the folding capacity of the ER with an increase in demand. Activation of PERK results in eIF2α phosphorylation (see below), which decreases overall translation and thus protein-folding demand.
Figure 2.
Regulation of protein quality control and nonsense-mediated mRNA decay (NMD) provides buffering capacity. Protein quality control (left), as exemplified by the IRE1-mediated branch of the unfolded protein response (UPR), and mRNA quality control, as exemplified by NMD (right), are both governed by regulatory loops that provide buffering capacity. (Left) For IRE1-mediated UPR activation, increased demand on the protein-folding load of the endoplasmic reticulum (ER) leads to an accumulation of unfolded and/or partially folded proteins and an increased demand for more folding chaperones. IRE1, a membrane-embedded sensor of unfolded proteins, is activated either by direct binding to unfolded proteins or by titration of chaperones (which prevent IRE1 oligomerization) away from IRE1. IRE1 subsequently oligomerizes and autophosphorylates. The RNase domain in activated IRE1 (red ) then mediates an unconventional cytoplasmic splicing reaction to generate spliced XBP1 (XBP1s) mRNA from unspliced (XBP1u) mRNA. XBP1s mRNA is translated into XBP1s protein, which is a transcriptional activator that upregulates a suite of UPR target genes. Among these genes are chaperones, leading to increased chaperone synthesis so as to match the protein-folding capacity of the ER with cellular demand. (Right) Some NMD factors (UPF1, UPF2, UPF3X, SMG1, SMG5, SMG6, and SMG7) are themselves normally targets of NMD by virtue of the presence of an unusually long 3′ UTR, a uORF, or both. When genetic insults diminish NMD capacity, NMD activity is decreased, and the mRNAs encoding the NMD factors are stabilized. These stabilized mRNAs direct protein synthesis in an effort to restore normal NMD activity.
A second example of regulated protein quality control concerns the degradation step (Figure 1, Step 3). Nucleated cells exploit protein degradation as a warning to alert the immune system of infection. Short peptide snippets that derive from proteasomal degradation products are displayed on the cell surface in a process called antigen presentation (186). One of the responses to infection includes the production of interferons, soluble cytokines that mediate changes in gene expression through the Janus kinase-signal transducers and activators of transcription ( JAK-STAT) signaling pathway. Among the interferon-induced genes are three that encode alternative proteasomal subunits (β1i, β2i, β5i). These subunits assemble with the remaining proteasomal α and β subunits to form immunoproteasomes. Formation of immunoproteasomes increases the total proteolytic capacity of the UPS and improves the quantity and quality of the peptides generated for antigen presentation (41, 140).
That normal nondefective genes are subject to NMD implies that the cell may modulate NMD activity like it modulates activity of the UPS, possibly exerting control over the levels of hundreds of transcripts at once. This concept is illustrated by several developmental processes. Portions of mammalian myogenesis are modeled by the differentiation of cultured mouse C2C12 myoblasts into multinucleated myotubes. During differentiation, the activity of another related mRNA degradation pathway, Staufen-mediated mRNA decay (SMD), is upregulated (91). UPF1 is essential for SMD and UPF1 binding to STAU1 is mutually exclusive with UPF1 binding to UPF2 (53). During the transition from myoblasts to myotubes, the level of STAU1 decreases less than the level of UPF2 decreases. Together, these observations imply that during myogenesis the activity of SMD is increased and the activity of NMD decreases, predictions that were met by assaying the expression of both endogenous and reporter SMD and NMD genes during myoblast differentiation. Why does this occur? Myogenin mRNA, which encodes a transcription factor that coordinates the myogenic program, is an NMD target bearing a uORF in its 5′ UTR. In contrast, the antimyogenic paired-box 3 (PAX3) transcription factor is an SMD target, illustrating how modulation of mRNA decay can achieve a physiological output. Additionally, the levels of UPF3X are increased during myogenesis, and targets of a UPF2-independent NMD pathway are suppressed. A similar competition between NMD and SMD occurs during adipogenesis for the purpose of regulating pro- and antiadipogenic factor abundance (25).
NMD activity is also modulated by production of microRNAs (miRNAs) that target UPF1 during neural development (11). Highly expressed in the brain, miR-128 is transcribed from two independent loci that are processed to yield the identical miRNA. This miRNA targets and represses expression of both UPF1 and the EJC constituent BTZ, downmodulating NMD activity, much like during myogenesis. miR-128 expression increases dramatically in the mouse embryonic brain between days E9.5 and E14.5, and high levels are maintained during adulthood. Transfection of miR-128 into murine neural stem cells normally devoid of it revealed upregulation of hundreds of genes, many of which both bear the structural features that would render them direct NMD targets and encode products that are necessary for neural differentiation. Thus, it appears that through upregulation of a single miRNA that targets the NMD apparatus, neural progenitors are able to coordinate upregulation of many genes that direct differentiation, and this upregulation is at least partially the result of mRNA stabilization. These findings fit nicely with other studies showing the importance of NMD, in particular the study concerning the UPF3-dependent pathway that when defective results in human intellectual disabilities (99, 176). Recently, deletions in the UPF2 gene were also identified in patients with forms of intellectual disability, and UPF3, SMG6, eIF4A3, and RNPS1 gene copy numbers were also aberrant in some of these patients (126).
These reports underscore the importance to gene expression of output from the NMD pathway during development. As such, activity of the NMD pathway, like transcription (44), should be buffered against insults that could inappropriately change its output. This is indeed the case and was first shown for the UPF3-dependent branch of NMD (17). Depletion of UPF3X protein augments the level of UPF3 protein, which partially compensates for the loss of UPF3X function. The authors propose that UPF3X normally outcompetes UPF3 for binding to UPF2, but when UPF3X is absent, UPF3 binds to UPF2, is posttranslationally stabilized, and partially restores NMD activity. In support of this, cells from patients with UPF3X loss (see above) have elevated levels of UPF3, and higher levels of functionally compensating UPF3 result in less severe intellectual disability.
Additional NMD components are also subject to feedback control. For example, the mRNAs encoding seven core NMD factors (UPF1, UPF2, UPF3B, SMG1, SMG5, SMG6, and SMG7) have unusually long 3′ UTRs, and the mRNAs of five NMD factors (UPF2, SMG1, SMG5, SMG6, and SMG7) have a uORF. Thus, in an elegant solution to buffer the capacity of the NMD pathway, the main trans-effectors of NMD themselves are derived from NMD targets (70, 204) (Figure 2). Huang et al. (70) found that various NMD factors are subject to feedback control via different NMD branches, and that different cell types have different abilities to buffer NMD via these pathways. Furthermore, mouse embryonic stem cells activate the UPF3X-dependent buffering mechanism during their differentiation into embryoid bodies.
Does NMD activity change as a result of acute insults to the cell? The results of studies investigating the stresses that cells encounter within the tumor microenvironment (hypoxia, generation of reactive oxygen species, nutrient deprivation) indicate that NMD is downmodulated in response to stress (51, 188, 189). The commonality among these stresses is that they all lead to phosphorylation of eIF2α, a subunit of eIF2 that delivers initiator Met-tRNAMet to the translational apparatus. Although the bulk of cellular translation is thus attenuated, it appears that this is not the cause for a decline in NMD activity, as select NMD-sensitive transcripts are still subject to ongoing translation in the face of eIF2α phosphorylation. The cause of NMD downmodulation must be investigated further, but the vital outcome is that transcripts encoding transcription factors involved in the integrated stress response (ATF-4, ATF-3, CHOP) are upregulated. These transcription factors ultimately direct a gene expression program needed to maintain cellular homeostasis and promote tumor growth. In particular, stabilization of ATF-4 mRNA, which is an NMD target, leads to induction of autophagy as a cellular method of coping with stress (194).
Viral Manipulation: Probes Designed by the Molecular Arms Race
Mammalian and viral genomes are in a constant state of flux, evolving mechanisms to prevent infection (host) and to overcome these preventive barriers (pathogen). Such zero-sum games or Red Queen genetic conflicts (“Now, here, you see, it takes all the running you can do, to keep in the same place. If you want to get somewhere else, you must run at least twice as fast as that!”- Red Queen, Lewis Carroll) (183) mean that viruses must constantly deploy new tactics to overcome the immune system.
When viewed through this lens, viruses represent powerful probes with which to discern the inner workings of the cell, and studying the effects of viruses on protein quality control has led to great insights. Antigen processing and presentation are vital tactics that nucleated cells use to report infection status to patrolling T lymphocytes. In this context, the cell makes use of the protein quality control system, specifically the UPS, to generate peptides (derived from both self-proteins and viral proteins) to be displayed on surface glycoprotein complexes called major histocompatibility complex (MHC) molecules (which are composed of an MHC heavy chain, the β2-microglobulin accessory protein, and an antigenic peptide) (186). In an effort to evade detection, viruses hijack the UPS for their own purposes. Human cytomegalovirus (HCMV) encodes two immunoevasins, US2 and US11, which bind to MHC heavy-chain proteins during the course of MHC molecule biosynthesis in the ER, dislocating them from the ER lumen and funneling them into the ubiquitin-mediated ER-associated degradation (ERAD) pathway for cytosolic destruction. Because they make use of host quality control processes, US2 and US11 represent useful probes to determine the composition of cellular complexes that normally mediate disposal of misfolded proteins from the ER lumen (104, 105, 107). Other examples are the Rubulavirus V proteins, which direct assembly of an E3 ligase complex containing UV-damaged DNA-binding protein 1 (DDB1) and cullin 4A (Cul4A). This complex functions to destroy the STAT proteins that normally coordinate the host antiviral interferon response (179).
Although far less well-studied in this respect, NMD-mediated quality control represents yet another barrier that viruses must overcome. Cricket paralysis virus (CrPV) is a picorna-like virus whose host range includes field crickets and also D. melanogaster cell lines, where EJCs are dispensable for NMD and long 3′ UTRs promote decay. The genome encodes a dicistronic mRNA composed of two nonoverlapping ORFs separated by an intergenic region. How does such an mRNA escape quality control when its translation termination at the end of the first ORF would make the second ORF appear as an abnormally long 3′ UTR? CrPV has elegantly solved this problem by providing the host translational apparatus access to its ORFs through two independent upstream internal ribosome entry sites (IRESs). Assembly of 80S ribosomes by the CrPV IRES appears to proceed in the absence of any eIF (32), making the transcript resistant to UPF1-mediated translational repression, for which eIF3 is a prerequisite, and thus NMD. In this context, the CrPV IRES was used as a tool to demonstrate the requirement for eIF3 in UPF1-mediated translational repression in mammalian cells, although whether this occurs in insect cells remains to be determined (80).
Pre-mRNA splicing poses a significant problem for viruses such as human immunodeficiency virus-1 (HIV-1), which generates >30 unique mRNAs from a single transcript by alternative splicing. Uncontrolled deposition of the EJC in mammalian cells during the splicing of HIV-1 pre-mRNA would be expected to render many of these mRNAs targets of NMD. HIV-1 therefore exerts tight control over splicing, which proceeds in 5′-to-3′ order—intron excision depends on the complete removal of all upstream introns—and fails to occur downstream of the 3′ most-active termination codon (8). A by-product of this strategy is the generation of an abnormally long 3′ UTR for many transcripts, and how this is dealt with by HIV-1 is unknown (50).
A similar problem is faced by the avian Rous sarcoma virus (RSV), whose RNA is potentially targeted for NMD in the absence of splicing in avian cells (192). For the bulk of translation initiation events, only the first gene, which encodes the capsid protein gag, is translated, giving rise to a massive <7,000-nt 3′ UTR. If a specific sequence element in the 3′ UTR, called the RSV stability element (RSE), is deleted, the transcript half-life is shortened in a way that requires UPF1 and translation. The RSE contains significant secondary structure as well as a single-stranded region that may base pair with an element upstream of the poly(A) tail (191, 199). Such fold-back constructs that shorten the physical distance between the termination codon and poly(A) tail have proven resistance to NMD (37), and this may be the method whereby RSV mRNAs escape detection by NMD. Alternatively (but not mutually exclusively), the RSE may recruit unknown factors that directly inhibit NMD (199), or it may prevent UPF1 accumulation on the 3′ UTR (96).
There is also evidence that some viral proteins directly target the NMD apparatus, downregulating NMD activity in infected cells. Human T-lymphotropic virus type 1 (HTLV-1) encodes the protein Tax, which binds to the integration 6 (INT6, also called p48) subunit of eIF3 (30). The authors of a recent study (121) observed a direct interaction between Tax and UPF1, and formation of this complex excludes INT6. Tax binding to UPF1 increases the amount of UPF1 phosphorylation and enhances localization of UPF1 to P-bodies. It was hypothesized that tax inhibits NMD by two mechanisms: preventing the eIF3-dependent UPF1-mediated translational block as well as preventing dephosphorylation of UPF1, thus promoting UPF1 sequestration in P-bodies, where it cannot be recycled and participate in further rounds of NMD. Combined, these inhibitory mechanisms stabilize both viral transcripts and cellular NMD targets.
CLINICAL RELEVANCE
Because of their regulatory importance to the cell, a breakdown in the quality control apparatus or escape from detection of the polymers they inspect can lead to human pathologies. Methods for rectifying problems associated with protein quality control and NMD are currently being tested for clinical use.
Quality Control: A Double-Edged Sword
Protein quality control can exert protective effects in some cases and detrimental effects in others. It is increasingly clear that age-related pathologies result from a decreased capacity in the quality control systems to handle the protein-folding load (6). Although this breakdown in quality control illustrates its protective effect on cells, other pathologies result from an overly exuberant quality control system that degrades or sequesters partially misfolded proteins that could otherwise have some function and thus lessen the severity of disease.
Overcoming the protein and mRNA quality control systems is a concept that has been explored. For example, the most common cause of cystic fibrosis is a deletion of phenylalanine (Phe)508 (LlF508) from the cystic fibrosis transmembrane conductance regulator (CFTR) protein, causing a trafficking defect that traps the CFTRLlF508 protein within the lumen of the ER as a folding intermediate. Depletion of the protein activator of HSP90 ATPase 1 (Aha-1), a co-chaperone involved in the CFTR folding cycle, is able to overcome the trafficking block imposed by the protein quality control system, leading to delivery of the CFTRLlF508 variant to the cell surface and restitution of 50% of wild-type halide conductance (190). Small-molecule modulation of the proteostasis network that controls protein folding and, as a consequence, recognition (Figure 1, Step 1) of proteins as misfolded holds promise in correcting many diseases (141).
NMD is similarly intertwined with the disease state in humans (7, 97). In fact, nearly 33% of all inherited and acquired diseases result from acquisition of a PTC (48). It is noteworthy that mammalian-cell NMD was first described in the context of β0-thalassemia, a condition that illustrates its protective effect (19, 112). Hemoglobin is composed of two α subunits and two β subunits. The stoichiometry of these subunits is tightly regulated by protein quality control because unpaired α-globin subunits are rapidly degraded. Heterozygotes with one wild-type allele and one allele bearing a PTC located within an NMD-competent region of the transcript manifest no disease, whereas subjects with two PTC-bearing, NMD-competent alleles show anemia. However, a dominantly inherited form of β-thalassemia occurs when a PTC is located in the last exon of β-globin mRNA. Such a configuration escapes NMD and generates a truncated, toxic protein that can only dimerize with α-globin (93). Given that the disease state is not the result of haploinsufficiency, NMD exerts a protective effect in the case of patients harboring one normal and one NMD-susceptible allele. The long list of other PTC-containing genes that are not targeted for NMD and thus produce disease-causing truncated proteins includes those encoding truncated breast and ovarian cancer susceptibility 1 (BRCA1) protein (40, 139, 172), p53 (13, 87, 110, 182, 196), and Wilm’s tumor protein 1 (WT1) (38, 92, 144).
Analogous to the situation for protein quality control, many diseases result from depletion of PTC-bearing transcripts that have the potential to encode partially functional proteins that would lessen the severity of disease. Again, CFTR provides an illustrative example. Approximately 5% to 10% of CFTR gene mutations result in NMD of product mRNA because they contain a PTC. For those PTCs that are in-frame with the normal ORF, aminoglycosides have been explored as potential read-through therapeutics. Aminoglycosides are potent bactericidal antibiotics that bind in the ribosomal A-site to 16S rRNA, distort the A-site, and cause a misreading of the genetic code so near-cognate aminoacylated tRNAs are introduced into the growing peptide chain at stop codons. Owing to their reduced affinity for eukaryotic (compared with prokaryotic) ribosomes (109), high concentrations of aminoglycosides, such as gentamicin, have been explored as a method of ameliorating the effects of PTC-induced disease in patients with cystic fibrosis. Administration of gentamicin to the nasal epithelium for 14 days results in small increases in CFTR-mediated chloride conductance as well as an increase in the appearance of CFTR at the cell surface (198). However, the minimum threshold of corrected CFTR that must reach the cell surface for amelioration of disease symptoms is unknown, and there are serious side effects of prolonged aminoglycoside use that include kidney toxicity and, for unknown reasons, damage to the cochlea. Because patients would presumably require toxic administration of aminoglycosides for prolonged benefits, there is considerable interest in finding small molecules that are able to promote PTC read-through without these side-effects. PTC124 (Ataluren) (193), a small molecule bearing no structural similarity to aminoglycosides, was identified using cell-based screens for compounds that promote UGA read-through. Although the molecular target of PTC124 is unknown, it is currently in phase III clinical studies for cystic fibrosis. The compound is well tolerated, is selective for premature termination read-through as opposed to normal termination read-through, and shows general promise for treating a variety of PTC-based diseases (138). However, this drug is not without controversy: PTC124 activity has been attributed to off-target effects on the firefly luciferase (Fluc)-based reporter assay used in its discovery and development (116). Recently, Amlexanox, an anti-inflammatory drug used to treat asthma and ulcers, was also identified as a compound that promotes read-through (54), but its mechanism of action remains unknown. Targeting the process of NMD itself with small molecules has also been explored with the compound NMDI-1 (34). NMDI-1 bears an indole-containing pharmacophore and disrupts the interaction between SMG5 and UPF1. Although of considerable utility to the research community as a small molecule probe for NMD function, what relevance, if any, such compounds have to the clinic remains to be determined, especially in light of the critical function that NMD plays during, for example, lymphocyte maturation (see below).
In an interesting twist on the modulation of NMD activity, Pastor et al. (135) found that it was possible to coax tumor cells into producing neo-antigens by administrating a small interfering RNA (siRNA) targeting SMG1 that is also conjugated to a tumor-specific aptamer. Targeted tumor-cell ablation of SMG1 induces the immune system to attack the tumor, presumably because it upregulates protein products that derive from NMD substrates to a level that is above the threshold set by central and peripheral immune tolerance (i.e., the mechanisms put in place by the immune system to avoid a response to self-antigens). It should be noted that absolute precision in targeting the siRNA to the tumor is key in this strategy: thymocytes in mice bearing a dominant-negative allele of UPF1 (an arginine-to-cysteine mutation at amino acid 843) are unable to transit through the double-negative (CD4− CD8−) stage of T-cell development because of defects in pre-T-cell receptor signaling, for which a productively rearranged TCR β-chain is a requirement (47, 171). That NMD (or the lack of NMD) can be involved in neo-antigen production also finds support in studies showing that microsatellite instability-high (MSI-High) colorectal cancer cells express mutated proteins when PTC-inducing frameshift mutations occur in areas of the transcript that render them immune to NMD (197). The protein products of these mRNAs have altered C termini that may make them vaccination targets, provided they are processed by the antigen presentation machinery.
OPEN QUESTIONS
Many outstanding questions remain in the NMD field and are inspired by what is known about protein quality control. How NMD is integrated with the systems governing co-translational secretory protein biosynthesis remains to be determined. Eukaryotic cells are, by definition, organized into membrane-enclosed compartments, each with their own unique constituents and functions. Proteins traverse one compartment to another by virtue of localization signals. In the case of the secretory pathway, this takes the form of N-terminal signal peptides (for type I membrane proteins and soluble proteins) that are bound by the signal recognition particle (SRP) during the course of biosynthesis, halting translation. This block in translation is relieved when SRP-mediated delivery of the nascent polypeptide-ribosome complex to the translocon occurs, after which cotranslational insertion of the target protein ensues (159). Because the ER lumen is devoid of proteasomes, this presents a barrier to protein quality control. Detection (Figure 1, Step 1) of misfolded proteins is topologically separated from destruction (Figure 1, Step 3), and cells have evolved mechanisms for the dislocation of misfolded proteins from the ER lumen to the cytosol, where ubiquitin-mediated destruction takes place (105).
Secretory protein-encoding mRNAs are targets of NMD (5). As discussed, recent data indicate that NMD occurs rapidly after export on the cytoplasmic leaflet of the nuclear envelope (178). Does the SRP-mediated translational arrest of secretory protein-encoding transcripts occur while the mRNA is bound to CBC (i.e., during the pioneer round of translation)? If so, does the NMD of PTC-containing transcripts that encode secretory proteins occur in association with the ER? Are secreted proteins encoded by NMD targets in the class of proteins that are potentially degraded by the ubiquitin E3 ligase activity of UPF1? If UPF1 does indeed degrade a nascent peptide as it enters the ER (95, 161, 173), how is this conundrum resolved given that UPF1 is localized to the cytoplasm and the nascent peptide is partially localized to the lumen of the ER? Localization of particular mRNAs to ER subdomains has been described in plants, yeast, and flies (62). Are there specialized mRNA quality control subdomains, as has been hypothesized for ER protein quality control (61, 84)?
As indicated by its name, the field of NMD has focused on mRNAs. However, recent studies have generated catalogs of long noncoding RNAs (lncRNAs) (57) with demonstrable cellular functions (58). On the basis of bioinformatic analysis, the consensus view is that these lncRNAs have little protein-coding potential. Nevertheless, ribosomal footprinting studies have shown that a class of lncRNAs (sprcRNAs) is decorated with ribosomes (79). Given that lncRNAs can be cytoplasmic, polyadenylated, and spliced (129), at least some would likely be subject to a pioneer round of translation and NMD. Steady-state levels of one known noncoding RNA, Gas5, are subject to control by UPF1 (78, 175), and Gas5 lncRNA has long been known to associate with polyribosomes (166). How general is this control mechanism? Is this a posttranscriptional mechanism for maintaining a low basal steady-state level of certain lncRNAs?
Bortezomib (Velcade), a peptide-based inhibitor of the proteasome, is in clinical use for treating multiple myeloma patients. Administration of Bortezomib likely has many effects, but one of them is almost certainly a blockage in the clearance of misfolded proteins, leading to tonic UPR activation and apoptosis (31). Selectivity for cancer cells maybe due to their heightened metabolism and thus increased demand in protein folding. Could a small-molecule NMD inhibitor be used to likewise increase the protein-folding demand through production of aberrant proteins, and could such a molecule synergize its effects with Bortezomib? Experiments in C. elegans indicate that this may be possible (150). This idea would be subject to the same cautions outlined for NMD-I.
Are NMD factors subject to posttranslational modifications (i.e., phosphorylation, monoubiquitylation, methylation, and proteolytic processing)? UPF1 is phosphorylated as a consequence of the tagging step (Figure 1, Step 2) to promote mRNA destruction (Figure 1, Step 3), and this occurs for every cycle of NMD. However, do other posttranslational modifications exist that modulate the NMD activity of the cell as a whole? Such mechanisms would be a way for the cell to respond to changing environmental conditions by regulating whole swaths of the transcriptome with extreme temporal precision.
Although much is known about the mechanism of NMD, many mysteries still remain. Discovery of new NMD trans-effectors and what role they play in regulating NMD activity will likely be another important avenue of research. Taken as a whole, what is known about NMD is likely just the tip of the iceberg, and much interesting territory that is ripe for innovative research still remains.
SUMMARY POINTS.
Protein quality control and mRNA quality control, the latter as illustrated by NMD, share similar design principles.
Protein quality control and NMD are both used for destruction of aberrant targets, i.e., proteins and transcripts, respectively, and also for quantity control of normal/nonmutated targets.
Protein quality control and NMD activities are highly regulated by the cell.
Pathogen manipulation of protein quality control and NMD provides biological insight as well as molecular tools for dissecting these pathways.
Protein quality control and NMD can exert protective effects or detrimental effects with regard to human disease.
Manipulation of protein quality control and NMD holds great promise for correcting a variety of human diseases.
FUTURE ISSUES.
How secretory protein biosynthesis is coupled to NMD activity, which is largely restricted to newly synthesized mRNAs, remains an open question.
Whether, and if so, how, additional RNA species, such as lncRNAs, are regulated by NMD is unknown.
The possibility of synergism between small molecules targeting protein quality control and others targeting the NMD apparatus in cancer therapy is an important area for exploration.
Whether NMD activity can be acutely regulated on a cellular level by posttranslational modifications to the NMD apparatus itself (i.e., by phosphorylation, monoubiquitylation, methylation, proteolytic processing, etc.) awaits discovery.
Identification of new NMD trans-effectors will further elucidate the mechanism of NMD while also possibly providing important therapeutic targets.
Acknowledgments
We thank colleagues for providing manuscripts prior to their publication and apologize to those whose work we could not cite because of space limitations. We thank H.L. Ploegh, A.M. Van der Veen, and E.J. Klemm for help with figures. M.W.P. is an HHMI Postdoctoral Fellow of the Damon Runyon Cancer Research Foundation, grant number DRG-2119-12. Research on NMD in the Maquat lab is supported by grant number NIH R01 GM59614 to L.E.M.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–18. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 2.Arias-Palomo E, Yamashita A, Fernandez IS, Núñez-Ramirez R, Bamba Y, et al. The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev. 2011;25:153–64. doi: 10.1101/gad.606911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, et al. Identification of CHiP, a novel tetra-tricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–45. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballut L, Marchadier B, Baguet A, Tomasetto C, Séraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–69. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 5.Belgrader P, Cheng J, Zhou X, Stephenson LS, Maquat LE. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol Cell Biol. 1994;14:8219–28. doi: 10.1128/mcb.14.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Gedalya T, Cohen E. Quality control compartments coming of age. Traffic. 2012;13:635–42. doi: 10.1111/j.1600-0854.2012.01330.x. [DOI] [PubMed] [Google Scholar]
- 7.Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem J. 2010;430:365–77. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- 8.Bohne J, Wodrich H, Krausslich HG. Splicing of human immunodeficiency virus RNA is position-dependent suggesting sequential removal of introns from the 5′ end. Nucleic Acids Res. 2005;33:825–37. doi: 10.1093/nar/gki185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bono F, Gehring NH. Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol. 2011;8:24–29. doi: 10.4161/rna.8.1.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brocke KS, Neu-Yilik G, Gehring NH, Hentze MW, Kulozik AE. The human intronless melanocortin 4-receptor gene is NMD insensitive. Hum Mol Genet. 2002;11:331–35. doi: 10.1093/hmg/11.3.331. [DOI] [PubMed] [Google Scholar]
- 11.Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–10. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bühler M, Wilkinson MF, Mühlemann O. Intranuclear degradation of nonsense codon–containing mRNA. EMBO Rep. 2002;3:646–51. doi: 10.1093/embo-reports/kvf129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardinali M, Kratochvil FJ, Ensley JF, Robbins KC, Yeudall WA. Functional characterization in vivo of mutant p53 molecules derived from squamous cell carcinomas of the head and neck. Mol Carcinog. 1997;18:78–88. doi: 10.1002/(sici)1098-2744(199702)18:2<78::aid-mc3>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 14.Carter MS, Li S, Wilkinson MF. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–75. [PMC free article] [PubMed] [Google Scholar]
- 15.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, et al. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 17.Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, et al. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–53. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 18.Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–30. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang JC, Kan YW. beta 0 thalassemia, a nonsense mutation in man. Proc Natl Acad Sci USA. 1979;76:2886–89. doi: 10.1073/pnas.76.6.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 21.Cheng J, Maquat LE. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol. 1993;13:1892–902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–54. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu SY, Serin G, Ohara O, Maquat LE. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho H, Han S, Choe J, Park SG, Choi SS, Kim YK. SMG5-PNRC2 is functionally dominant compared with SMG5-SMG7 in mammalian nonsense-mediated mRNA decay. Nucleic Acids Res. 2013;41:1319–28. doi: 10.1093/nar/gks1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho H, Kim KM, Han S, Choe J, Park SG, et al. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell. 2012;46:495–506. doi: 10.1016/j.molcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell. 2009;33:75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 27.Ciechanover A. Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin-proteasome system, and onto human diseases and drug targeting (Nobel lecture) Angew Chem Int Ed Engl. 2005;44:5944–67. doi: 10.1002/anie.200501428. [DOI] [PubMed] [Google Scholar]
- 28.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 29.Dahlberg J, Lund E. Nuclear translation or nuclear peptidyl transferase? Nucleus. 2012;3:320–21. doi: 10.4161/nucl.20754. [DOI] [PubMed] [Google Scholar]
- 30.Desbois C, Rousset R, Bantignies F, Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I tax oncoprotein. Science. 1996;273:951–53. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- 31.Dong H, Chen L, Chen X, Gu H, Gao G, et al. Dysregulation of unfolded protein response partially underlies proapoptotic activity of bortezomib in multiple myeloma cells. Leuk Lymphoma. 2009;50:974–84. doi: 10.1080/10428190902895780. [DOI] [PubMed] [Google Scholar]
- 32.Doudna J, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey J, editors. Translational Control in Biology and Medicine. Cold Spring Harboy, NY: Cold Spring Harbor Lab. Press; 2007. pp. 129–54. [Google Scholar]
- 33.Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 34.Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, et al. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178:1145–60. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durand S, Lykke-Andersen J. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat Struct Mol Biol. 2013;20:702–9. doi: 10.1038/nsmb.2575. [DOI] [PubMed] [Google Scholar]
- 36.Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 37.Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Englert C, Vidal M, Maheswaran S, Ge Y, Ezzell RM, et al. Truncated WT1 mutants alter the subnuclear localization of the wild-type protein. Proc Natl Acad Sci USA. 1995;92:11960–64. doi: 10.1073/pnas.92.26.11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–81. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan S, Ma YX, Wang C, Yuan RQ, Meng Q, et al. Role of direct interaction in BRCA1 inhibition of estrogen receptor activity. Oncogene. 2001;20:77–87. doi: 10.1038/sj.onc.1204073. [DOI] [PubMed] [Google Scholar]
- 41.Ferrington DA, Gregerson DS. Immunoproteasomes: structure, function, and antigen presentation. Prog Mol Biol Transl Sci. 2012;109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fillman C, Lykke-Andersen J. RNA decapping inside and outside of processing bodies. Curr Opin Cell Biol. 2005;17:326–31. doi: 10.1016/j.ceb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Fiorini F, Boudvillain M, Le Hir H. Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res. 2013;41:2404–15. doi: 10.1093/nar/gks1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–93. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–15. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010;143:938–50. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frischmeyer-Guerrerio PA, Montgomery RA, Warren DS, Cooke SK, Lutz J, et al. Perturbation of thymocyte development in nonsense-mediated decay (NMD)-deficient mice. Proc Natl Acad Sci USA. 2011;108:10638–43. doi: 10.1073/pnas.1019352108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 49.Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14–3–3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–47. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 50.Gaglia MM, Glaunsinger BA. Viruses and the cellular RNA decay machinery. Wiley Interdiscip Rev RNA. 2010;1:47–59. doi: 10.1002/wrna.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 53.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez-Hilarion S, Beghyn T, Jia J, Debreuck N, Berte G, et al. Rescue of nonsense mutations by amlexanox in human cells. Orphanet J Rare Dis. 2012;7:58. doi: 10.1186/1750-1172-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green RE, Lewis BP, Hillman RT, Blanchette M, Lareau LF, et al. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19(Suppl 1):i118–21. doi: 10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- 56.Guan Q, Zheng W, Tang S, Liu X, Zinkel RA, et al. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2006;2:e203. doi: 10.1371/journal.pgen.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guttman M, Amit I, Garber M, French C, Lin MF, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–27. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hagan KW, Ruiz-Echevarria MJ, Quan Y, Peltz SW. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol. 1995;15:809–23. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–52. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 61.Hegde RS, Ploegh HL. Quality and quantity control at the endoplasmic reticulum. Curr Opin Cell Biol. 2010;22:437–46. doi: 10.1016/j.ceb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hermesh O, Jansen RP. Take the (RN)A-train: localization of mRNA to the endoplasmic reticulum. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbamcr.2013.01.013. pii:S0167-4889(13)00026–8. [DOI] [PubMed] [Google Scholar]
- 63.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle (Nobel lecture) Angew Chem Int Ed Engl. 2005;44:5932–43. doi: 10.1002/anie.200501724. [DOI] [PubMed] [Google Scholar]
- 64.Hillman RT, Green RE, Brenner SE. An unappreciated role for RNA surveillance. Genome Biol. 2004;5:R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hogg JR, Goff SP. Upf1 senses 3′ UTR length to potentiate mRNA decay. Cell. 2010;143:379–89. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hohfeld J, Cyr DM, Patterson C. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001;2:885–90. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hori K, Watanabe Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 2005;43:530–40. doi: 10.1111/j.1365-313X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- 68.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 69.Hu W, Petzold C, Coller J, Baker KE. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:244–47. doi: 10.1038/nsmb.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang L, Lou CH, Chan W, Shum EY, Shao A, et al. RNA homeostasis governed by cell type–specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–61. doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huber EM, Groll M. Inhibitors for the immuno- and constitutive proteasome: current and future trends in drug development. Angew Chem Int Ed Engl. 2012;51:8708–20. doi: 10.1002/anie.201201616. [DOI] [PubMed] [Google Scholar]
- 72.Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–17. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013 doi: 10.1101/gr.157354.113. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hwang J, Kim YK. When a ribosome encounters a premature termination codon. BMB Rep. 2013;46:9–16. doi: 10.5483/BMBRep.2013.46.1.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr Opin Genet Dev. 2011;21:422–30. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell. 2010;39:396–409. doi: 10.1016/j.molcel.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iborra FJ, Jackson DA, Cook PR. Coupled transcription and translation within nuclei of mammalian cells. Science. 2001;293:1139–42. doi: 10.1126/science.1061216. [DOI] [PubMed] [Google Scholar]
- 78.Ideue T, Sasaki YT, Hagiwara M, Hirose T. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev. 2007;21:1993–98. doi: 10.1101/gad.1557907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–27. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–47. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, et al. AAA+proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal. 2010;3:ra27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- 83.Jonas S, Weichenrieder O, Izaurralde E. An unusual arrangement of two 14-3-3-like domains in the SMG5-SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev. 2013;27:211–25. doi: 10.1101/gad.206672.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kamhi-Nesher S, Shenkman M, Tolchinsky S, Fromm SV, Ehrlich R, Lederkremer GZ. A novel quality control compartment derived from the endoplasmic reticulum. Mol Biol Cell. 2001;12:1711–23. doi: 10.1091/mbc.12.6.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, et al. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes Dev. 2010;24:2440–50. doi: 10.1101/gad.604610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–67. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kawasaki T, Tomita Y, Watanabe R, Tanikawa T, Kumanishi T, Sato S. mRNA and protein expression of p53 mutations in human bladder cancer cell lines. Cancer Lett. 1994;82:113–21. doi: 10.1016/0304-3835(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 88.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13:700–12. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kervestin S, Li C, Buckingham R, Jacobson A. Testing the faux-UTR model for NMD: analysis of Upf1p and Pab1p competition for binding to eRF3/Sup35p. Biochimie. 2012;94:1560–71. doi: 10.1016/j.biochi.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kessler O, Chasin LA. Effects of nonsense mutations on nuclear and cytoplasmic adenine phosphoribosyltransferase RNA. Mol Cell Biol. 1996;16:4426–35. doi: 10.1128/mcb.16.8.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–81. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.King-Underwood L, Pritchard-Jones K. Wilms’ tumor (WT1) gene mutations occur mainly in acute myeloid leukemia and may confer drug resistance. Blood. 1998;91:2961–68. [PubMed] [Google Scholar]
- 93.Kugler W, Enssle J, Hentze MW, Kulozik AE. Nuclear degradation of nonsense mutated β-globin mRNA: a post-transcriptional mechanism to protect heterozygotes from severe clinical manifestations of β-thalassemia? Nucleic Acids Res. 1995;23:413–18. doi: 10.1093/nar/23.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1015–22. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuroha K, Tatematsu T, Inada T. Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep. 2009;10:1265–71. doi: 10.1038/embor.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kurosaki T, Maquat LE. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc Natl Acad Sci USA. 2013;110(9):3357–62. doi: 10.1073/pnas.1219908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med. 2006;12:306–16. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 98.Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol. 2007;623:190–211. doi: 10.1007/978-0-387-77374-2_12. [DOI] [PubMed] [Google Scholar]
- 99.Laumonnier F, Shoubridge C, Antar C, Nguyen LS, Van Esch H, et al. Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol Psychiatry. 2010;15:767–76. doi: 10.1038/mp.2009.14. [DOI] [PubMed] [Google Scholar]
- 100.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–69. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–87. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 102.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–19. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–92. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–40. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 105.Lilley BN, Ploegh HL. Viral modulation of antigen presentation: manipulation of cellular targets in the ER and beyond. Immunol Rev. 2005;207:126–44. doi: 10.1111/j.0105-2896.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 106.Longman D, Plasterk RH, Johnstone IL, Cáceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21:1075–85. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Loureiro J, Lilley BN, Spooner E, Noriega V, Tortorella D, Ploegh HL. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature. 2006;441:894–97. doi: 10.1038/nature04830. [DOI] [PubMed] [Google Scholar]
- 108.Lozano F, Maertzdorf B, Pannell R, Milstein C. Low cytoplasmic mRNA levels of immunoglobulin κ light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J. 1994;13:4617–22. doi: 10.1002/j.1460-2075.1994.tb06783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lynch SR, Puglisi JD. Structural origins of aminoglycoside specificity for prokaryotic ribosomes. J Mol Biol. 2001;306:1037–58. doi: 10.1006/jmbi.2000.4420. [DOI] [PubMed] [Google Scholar]
- 110.Magnusson KP, Sandstrom M, Stahlberg M, Larsson M, Flygare J, et al. p53 splice acceptor site mutation and increased HsRAD51 protein expression in Bloom’s syndrome GM1492 fibroblasts. Gene. 2000;246:247–54. doi: 10.1016/s0378-1119(00)00068-8. [DOI] [PubMed] [Google Scholar]
- 111.Maquat LE, Hwang J, Sato H, Tang Y. CBP80-promoted mRNP rearrangements during the pioneer round of translation, nonsense-mediated mRNA decay, and thereafter. Cold Spring Harb Symp Quant Biol. 2010;75:127–34. doi: 10.1101/sqb.2010.75.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable β-globin mRNA in mRNA-deficient beta 0 thalassemia. Cell. 1981;27:543–53. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- 113.Maquat LE, Li X. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA. 2001;7:445–56. doi: 10.1017/s1355838201002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–74. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Matsuda D, Hosoda N, Kim YK, Maquat LE. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat Struct Mol Biol. 2007;14:974–79. doi: 10.1038/nsmb1297. [DOI] [PubMed] [Google Scholar]
- 116.McElroy SP, Nomura T, Torrie LS, Warbrick E, Gartner U, et al. A lack of premature termination codon read-through efficacy of PTC124 (Ataluren) in a diverse array of reporter assays. PLoS Biol. 2013;11:e1001593. doi: 10.1371/journal.pbio.1001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–93. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 118.Meaux S, van Hoof A, Baker KE. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol Cell. 2008;29:134–40. doi: 10.1016/j.molcel.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Melero R, Buchwald G, Castano R, Raabe M, Gil D, et al. The cryo-EM structure of the UPF-EJC complex shows UPF1 poised toward the RNA 3′ end. Nat Struct Mol Biol. 2012;19:498–505. S1–2. doi: 10.1038/nsmb.2287. [DOI] [PubMed] [Google Scholar]
- 120.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–78. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 121.Mocquet V, Neusiedler J, Rende F, Cluet D, Robin JP, et al. The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. J Virol. 2012;86:7530–43. doi: 10.1128/JVI.07021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001–8. [PubMed] [Google Scholar]
- 123.Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–39. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mühlemann O, Lykke-Andersen J. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol. 2010;7:28–32. doi: 10.4161/rna.7.1.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neu-Yilik G, Amthor B, Gehring NH, Bahri S, Paidassi H, et al. Mechanism of escape from nonsense-mediated mRNA decay of human β-globin transcripts with nonsense mutations in the first exon. RNA. 2011;17:843–54. doi: 10.1261/rna.2401811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nguyen LS, Kim HG, Rosenfeld JA, Shen Y, Gusella JF, et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22(9):1816–25. doi: 10.1093/hmg/ddt035. [DOI] [PubMed] [Google Scholar]
- 127.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–18. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nicholson P, Mühlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem Soc Trans. 2010;38:1615–20. doi: 10.1042/BST0381615. [DOI] [PubMed] [Google Scholar]
- 129.Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM, et al. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res. 2012;4:127–50. [PMC free article] [PubMed] [Google Scholar]
- 130.Nielsen KH, Chamieh H, Andersen CB, Fredslund F, Hamborg K, et al. Mechanism of ATP turnover inhibition in the EJC. RNA. 2009;15:67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Noble CG, Song H. MLN51 stimulates the RNA-helicase activity of eIF4AIII. PLoS ONE. 2007;2:e303. doi: 10.1371/journal.pone.0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 133.Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–66. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Palacios IM. Nonsense-mediated mRNA decay: from mechanistic insights to impacts on human health. Brief Funct Genomics. 2013;12:25–36. doi: 10.1093/bfgp/els051. [DOI] [PubMed] [Google Scholar]
- 135.Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–30. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peixeiro I, Inácio Â, Barbosa C, Silva AL, Liebhaber SA, Romão L. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res. 2012;40:1160–73. doi: 10.1093/nar/gkr820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–54. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 138.Peltz SW, Morsy M, Welch EM, Jacobson A. Ataluren as an agent for therapeutic nonsense suppression. Annu Rev Med. 2013;64:407–25. doi: 10.1146/annurev-med-120611-144851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11:2805–14. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- 140.Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJ. The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J. 2010;432:585–94. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–91. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 142.Ramani AK, Nelson AC, Kapranov P, Bell I, Gingeras TR, Fraser AG. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol. 2009;10:R101. doi: 10.1186/gb-2009-10-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: What defines a substrate? Curr Opin Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 144.Reddy JC, Morris JC, Wang J, English MA, Haber DA, et al. WT1-mediated transcriptional activation is inhibited by dominant negative mutant proteins. J Biol Chem. 1995;270:10878–84. doi: 10.1074/jbc.270.18.10878. [DOI] [PubMed] [Google Scholar]
- 145.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–44. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Reznik B, Lykke-Andersen J. Regulated and quality-control mRNA turnover pathways in eukaryotes. Biochem Soc Trans. 2010;38:1506–10. doi: 10.1042/BST0381506. [DOI] [PubMed] [Google Scholar]
- 147.Rodrigo-Brenni MC, Hegde RS. Design principles of protein biosynthesis-coupled quality control. Dev Cell. 2012;23:896–907. doi: 10.1016/j.devcel.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 148.Roth DM, Balch WE. Modeling general proteostasis: proteome balance in health and disease. Curr Opin Cell Biol. 2011;23:126–34. doi: 10.1016/j.ceb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rufener SC, Mühlemann O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2013;20:710–17. doi: 10.1038/nsmb.2576. [DOI] [PubMed] [Google Scholar]
- 150.Sakaki K, Yoshina S, Shen X, Han J, DeSantis MR, et al. RNA surveillance is required for endoplasmic reticulum homeostasis. Proc Natl Acad Sci USA. 2012;109:8079–84. doi: 10.1073/pnas.1110589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28:4320–30. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sato H, Hosoda N, Maquat LE. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol Cell. 2008;29:255–62. doi: 10.1016/j.molcel.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 153.Sato H, Maquat LE. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin β. Genes Dev. 2009;23:2537–50. doi: 10.1101/gad.1817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Saulière J, Haque N, Harms S, Barbosa I, Blanchette M, Le Hir H. The exon junction complex differentially marks spliced junctions. Nat Struct Mol Biol. 2010;17:1269–71. doi: 10.1038/nsmb.1890. [DOI] [PubMed] [Google Scholar]
- 155.Saulière J, Murigneux V, Wang Z, Marquenet E, Barbosa I, et al. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19:1124–31. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- 156.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–10. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 157.Schmid M, Jensen TH. Transcription-associated quality control of mRNP. Biochim Biophys Acta. 2013;1829:158–68. doi: 10.1016/j.bbagrm.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 158.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–59. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shao S, Hegde RS. Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shigeoka T, Kato S, Kawaichi M, Ishida Y. Evidence that the Upf1-related molecular motor scans the 3′-UTR to ensure mRNA integrity. Nucleic Acids Res. 2012;40:6887–97. doi: 10.1093/nar/gks344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–81. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Simpson SB, Stoltzfus CM. Frameshift mutations in the v-src gene of avian sarcoma virus act in cis to specifically reduce v-src mRNA levels. Mol Cell Biol. 1994;14:1835–44. doi: 10.1128/mcb.14.3.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–64. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–90. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–40. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 169.Squires JE, Berry MJ. Eukaryotic selenoprotein synthesis: mechanistic insight incorporating new factors and new functions for old factors. Int Union Biochem Mol Biol Life. 2008;60:232–35. doi: 10.1002/iub.38. [DOI] [PubMed] [Google Scholar]
- 170.Stalder L, Mühlemann O. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA. 2009;15:1265–73. doi: 10.1261/rna.1672509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA. 1998;95:10009–14. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sylvain V, Lafarge S, Bignon YJ. Dominant-negative activity of a Brca1 truncation mutant: effects on proliferation, tumorigenicity in vivo, and chemosensitivity in a mouse ovarian cancer cell line. Int J Oncol. 2002;20:845–53. [PubMed] [Google Scholar]
- 173.Takahashi S, Araki Y, Ohya Y, Sakuno T, Hoshino S, et al. Upf1 potentially serves as a RING-related E3 ubiquitin ligase via its association with Upf3 in yeast. RNA. 2008;14:1950–58. doi: 10.1261/rna.536308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tanaka K, Mizushima T, Saeki Y. The proteasome: molecular machinery and pathophysiological roles. Biol Chem. 2012;393:217–34. doi: 10.1515/hsz-2011-0285. [DOI] [PubMed] [Google Scholar]
- 175.Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS ONE. 2013;8:e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–33. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, et al. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–94. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Trcek T, Sato H, Singer RH, Maquat LE. Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev. 2013;27(5):541–51. doi: 10.1101/gad.209635.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ulane CM, Kentsis A, Cruz CD, Parisien JP, Schneider KL, Horvath CM. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J Virol. 2005;79:10180–89. doi: 10.1128/JVI.79.16.10180-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–96. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 181.Urlaub G, Mitchell PJ, Ciudad CJ, Chasin LA. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989;9:2868–80. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Usuda J, Inomata M, Fukumoto H, Iwamoto Y, Suzuki T, et al. Restoration of p53 gene function in 12-O-tetradecanoylphorbor 13-acetate-resistant human leukemia K562/TPA cells. Int J Oncol. 2003;22:81–86. [PubMed] [Google Scholar]
- 183.Van Valen L. A new evolutionary law. Evol Theory. 1973;1:1–30. [Google Scholar]
- 184.Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the nonsense mediated decay pathway. Nucleic Acids Res. 2007;35:4542–51. doi: 10.1093/nar/gkm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.von Mikecz A. The nuclear ubiquitin-proteasome system. J Cell Sci. 2006;119:1977–84. doi: 10.1242/jcs.03008. [DOI] [PubMed] [Google Scholar]
- 186.Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8:607–18. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–86. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 188.Wang D, Wengrod J, Gardner LB. Overexpression of the c-myc oncogene inhibits nonsense-mediated RNA decay in B lymphocytes. J Biol Chem. 2011;286:40038–43. doi: 10.1074/jbc.M111.266361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang D, Zavadil J, Martin L, Parisi F, Friedman E, et al. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol. 2011;31:3670–80. doi: 10.1128/MCB.05704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, et al. Hsp90 cochaperone Aha1 down-regulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–15. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 191.Weil JE, Beemon KL. A 3′ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA. 2006;12:102–10. doi: 10.1261/rna.2129806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Weil JE, Hadjithomas M, Beemon KL. Structural characterization of the Rous sarcoma virus RNA stability element. J Virol. 2009;83:2119–29. doi: 10.1128/JVI.02113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 194.Wengrod J, Martin L, Wang D, Frischmeyer-Guerrerio P, Dietz HC, Gardner LB. Inhibition of nonsense-mediated RNA decay activates autophagy. Mol Cell Biol. 2013;33:2128–35. doi: 10.1128/MCB.00174-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wilkinson MF, Shyu AB. RNA surveillance by nuclear scanning? Nat Cell Biol. 2002;4:E144–47. doi: 10.1038/ncb0602-e144. [DOI] [PubMed] [Google Scholar]
- 196.Williams C, Norberg T, Ahmadian A, Ponten F, Bergh J, et al. Assessment of sequence-based p53 gene analysis in human breast cancer: messenger RNA in comparison with genomic DNA targets. Clin Chem. 1998;44:455–62. [PubMed] [Google Scholar]
- 197.Williams DS, Bird MJ, Jorissen RN, Yu YL, Walker F, et al. Nonsense mediated decay resistant mutations are a source of expressed mutant proteins in colon cancer cell lines with microsatellite instability. PLoS ONE. 2010;5:e16012. doi: 10.1371/journal.pone.0016012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wilschanski M, Yahav Y, Yaacov Y, Blau H, Bentur L, et al. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med. 2003;349:1433–41. doi: 10.1056/NEJMoa022170. [DOI] [PubMed] [Google Scholar]
- 199.Withers JB, Beemon KL. The structure and function of the rous sarcoma virus RNA stability element. J Cell Biochem. 2011;112:3085–92. doi: 10.1002/jcb.23272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wittmann J, Hol EM, Jäck HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:1272–87. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xie Y. Structure, assembly and homeostatic regulation of the 26S proteasome. J Mol Cell Biol. 2010;2:308–17. doi: 10.1093/jmcb/mjq030. [DOI] [PubMed] [Google Scholar]
- 202.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–28. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Mühlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–18. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 206.Zhang J, Maquat LE. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 1997;16:826–33. doi: 10.1093/emboj/16.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–83. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–15. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zünd D, Gruber AR, Zavolan M, Mühlemann O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol. 2013;20:936–43. doi: 10.1038/nsmb.2635. [DOI] [PubMed] [Google Scholar]