Background: Zebrafish CNS oligodendrocytes up-regulate axon growth-promoting genes following axonal injury.
Results: Separate cis-regulatory elements mediate mpz expression in myelinating oligodendrocytes and its transcriptional induction by axonal damage.
Conclusion: Different mechanisms specify oligodendroglial mpz expression in the intact and injured CNS.
Significance: Mechanisms underlying the zebrafish repair response might be exploited to enhance axonal regeneration in the mammalian CNS.
Keywords: Axon, Myelin, Oligodendrocyte, Regenerative Medicine, Transcription Regulation, Protein Zero, Zebrafish
Abstract
Zebrafish CNS axons regenerate robustly following injury; it is thought that CNS oligodendrocytes contribute to this response by expressing growth-promoting molecules. We characterized the mpz gene, which encodes myelin protein zero and is up-regulated in oligodendroglia following axonal injury. The 2.5-kb mpz mRNA is expressed from a single TATA box promoter. Four independent Tg(mpz:egfp) transgenic zebrafish lines, in which GFP was expressed under the mpz promoter and 10 kb of genomic 5′-flanking sequence, showed transgene expression in CNS oligodendrocytes from larval development through adulthood. Following optic nerve crush injury, the mpz:egfp transgene was strongly up-regulated in oligodendrocytes along the regenerating retinotectal projection, mirroring up-regulation of endogenous mpz mRNA. GFP-expressing oligodendroglia were significantly more abundant in the regenerating optic pathway, resulting from both transgene induction in oligodendroglial precursors and the birth of new cells. Up-regulation of the mpz:egfp transgene was not dependent on axonal regeneration, suggesting that the primary signal may be axonal loss, debris, or microglial infiltration. Deletion experiments indicated that an oligodendroglial enhancer located in the region from −6 to −10 kb with respect to the mpz transcriptional start site is dissociable from the cis-regulatory element mediating the mpz transcriptional response to axonal injury, which is located between −1 and −4 kb. These data show that different mechanisms regulate expression of zebrafish mpz in myelinating oligodendrocytes and its induction following axonal injury. The underlying molecular events could potentially be exploited to enhance axonal repair following mammalian CNS injury. The transgenic lines and cis-regulatory constructs reported here will facilitate identification of the relevant signaling pathways.
Introduction
In contrast to mammals, CNS axons of teleosts show extensive capacity for regrowth after injury. For example, in the genetically manipulable zebrafish, retinal ganglion cell axons regenerate following an optic nerve crush lesion, with restoration of the retinotectal projection and recovery of vision. Furthermore, descending motor axons regenerate after spinal cord injury in adult zebrafish, allowing re-establishment of synaptic contacts and recovery of motor function (1). Understanding the molecular basis of these remarkable repair responses is an important objective; common human neurological diseases in which there is prominent axonal damage (including subcortical infarct, traumatic brain injury, and spinal cord compression) are associated with limited clinical recovery, which could be substantially improved by manipulations that enhance axonal regeneration.
Following axonal transection, adult zebrafish CNS neurons up-regulate genes that are expressed during larval axonogenesis but subsequently down-regulated, such as α-tubulin, GAP43 (2), and immunoglobulin superfamily cell surface proteins L1.1, L1.2, and NCAM (3). Neuronal transcriptional responses are thought to be essential for axonal regrowth; blocking up-regulation of L1.1 in brainstem neurons using a morpholino oligonucleotide prevented regeneration of reticulospinal axons following spinal cord injury (4). In addition to cell-autonomous events in zebrafish neurons that favor axonal regeneration, the extracellular environment of the zebrafish CNS is thought to provide a permissive setting for axon growth and guidance cues for anatomical restoration of fiber tracts. Unlike mammals, glial scar-derived growth-inhibitory chondroitin sulfates (5) and secreted axon-repellant semaphorins (6) were not increased in the injured zebrafish optic nerve. Furthermore, unlike mammalian CNS myelin, fish myelin and oligodendrocytes did not markedly inhibit axon growth (7); the zebrafish orthologue of the mammalian oligodendrocyte-derived axon growth inhibitor Nogo-A was found to lack a key inhibitory domain (8), which may partially account for these observations.
The expression of growth-promoting cell surface proteins in zebrafish oligodendrocytes following CNS injury is thought to stimulate axonal regeneration (6). Myelin protein zero (P0),4 expressed in Schwann cells in mammals, shows axonal growth-promoting properties in vitro (9), and the mpz gene encoding the zebrafish P0 orthologue is strongly up-regulated in oligodendrocytes in regenerating white matter tracts following CNS axonal injury (10). Similarly, contactin1a, implicated in axonal growth and myelination, is up-regulated in zebrafish optic nerve oligodendrocytes following crush injury (11).
The long term objective of this work is to understand how glial responses that promote axonal regeneration are regulated following injury, potentially permitting their manipulation in order to enhance CNS repair. As a first step toward this goal, we generated and analyzed novel transgenic zebrafish lines in which regulatory elements from the mpz gene drive expression of a reporter gene. Here, we demonstrate that oligodendrocytes in damaged CNS axon tracts show spatially restricted and highly reproducible responses to axonal loss, including temporally stereotyped changes in gene expression, morphology, and cell numbers. By comparing the responses of different mpz:egfp transgene constructs to axonal injury in stable transgenic zebrafish, we show that the enhancer element responsible for up-regulation of mpz during this response is located within the region −1 to −4 kb with respect to the transcriptional start site, whereas a separate enhancer located between −6 and −10 kb is necessary for basal transcription of mpz in developing and mature oligodendrocytes. These data suggest that the signaling pathways responsible for mpz expression during myelination and maintenance of CNS white matter are separable from those responding to axonal damage. The panel of transgenic animals we report here will be useful for further studies on myelination, oligodendroglial differentiation, and the role of oligodendrocytes in axonal regeneration.
EXPERIMENTAL PROCEDURES
Zebrafish
Experiments were carried out in accordance with National Institutes of Health guidelines for animal care and use and with approval from the University of Pittsburgh Institutional Animal Care and Use Committee. Adult stocks of WT strain AB zebrafish and transgenic lines were maintained at 28.5 °C and euthanized by deep tricaine anesthesia followed by exposure to ice-cold water. Embryos were raised in E3 buffer (5 mm NaCl, 0.17 mm KCl, 0.33 mm CaCl2, 0.33 mm MgSO4).
Transgene Constructs
Homologous arms derived from the zebrafish mpz gene were amplified from BAC zKp35B8 (a kind gift from Dr. R. Plasterk, Netherlands Institute for Developmental Biology) by PCR using the primers 5′-GGG GTA CCT GGA GTG AGT GAC ATA AAG-3′ and 5′-GGA ATT CTT TTA GCC TGA CCA GTT TAC-3′ (5′-arm for 10-kb construct), 5′-GGG GTA CCA ATA AGC GTA ATC TAG AGA C-3′ and 5′-GGA ATT CAG AAT TTG GCA TCT TTT AC-3′ (5′-arm for 6-kb construct), and 5′-GGA ATT CAG GAA GGA TTA CAG ACA AAC-3′ and 5′-GTA CCC ATG GAC AGC ATG ATC TCT CTC TG-3′ (3′-arm; mpz exon 1) and inserted into the EcoRI/NcoI sites (3′-arm) or KpnI/EcoRI sites (5′-arm) of pBS-I-SceI-egfp (12) to make a translational fusion such that the GFP ORF was in frame with exon 1 from the zebrafish mpz gene. The resulting plasmids (pBS-mpz[10/6kb]-5′/3′arms-egfp) were verified by restriction digest and DNA sequencing and then linearized by EcoRI digest. A fragment of the mpz gene encompassing the transcriptional start site, exon 1, and 6 or 10 kb of upstream flanking sequence was then captured from BAC zKp35B8 by gap repair recombination, using bacterial strain DY380 (13) as described in our previous work (12). Recombinants were identified by ampicillin resistance and verified by restriction digest and direct DNA sequencing. The resulting plasmids pBS-I-SceI-mpz[10kb]:egfp and pBS-I-SceI-mpz[6kb]:egfp were used to generate transgenic zebrafish. Deletion constructs were derived from pBS-I-SceI-mpz[10kb]:egfp by deleting a 2-kb EcoRI/PstI fragment (4 + 4-kb construct) or a 5-kb AgeI/MluI fragment (4 + 1-kb construct).
Transgenesis
A restriction digest containing the following components was prepared on ice: plasmid DNA (0.6 μg), injection dye (0.5% phenol red, 240 mm KCl, 40 mm Hepes, pH 7.4) (1 μl), 10× I-SceI buffer (100 mm Tris-HCl, 100 mm MgCl2, 10 mm dithiothreitol, pH 8.8) (0.5 μl), I-SceI (New England Biolabs) (1 μl) (5 units), double-distilled H2O to total 10 μl. Single cell embryos were microinjected with 0.5 nl of the restriction digest reaction, containing 30 pg of DNA, using a glass micropipette. Surviving embryos were raised to sexual maturity, and F0 chimeras that could pass the mpz:egfp transgene through the germ line were identified by GFP expression in their F1 progeny embryos. The genotype of Tg(mpz:egfp) lines was verified by PCR analysis of genomic DNA extracted from the tail fin as described previously (12) using primers 5′-AGG AAG GAT TAC AGA CAA AC-3′ and 5-CTT GTG GCC GTT TAC GTC GC-3′. Each line of transgenic zebrafish was expanded from a single F1 founder, and each F1 founder was derived from a single F0 germ line chimera. The data reported here are derived from F3–F9 generation zebrafish. All lines showed stable Mendelian inheritance of the transgene over multiple generations, indicating a single integration site.
Optic Nerve Crush Injury, Tracing, and Enucleation
Adult zebrafish were anesthetized in 0.016% tricaine and transferred to ice on a dissecting microscope. For optic nerve crush injury, the right eye was gently retracted to expose the optic nerve, which was crushed by applying pressure with Dumont number 5 forceps (Fine Science Tools, Foster City, CA). A translucent stripe across the optic nerve indicated successful production of a lesion. The same procedure was used for optic nerve tracing, except that the optic nerve was sectioned using dissecting scissors, and a small piece of Gelfoam (Pfizer, New York) soaked in 1 mg/10 μl biocytin (Sigma-Aldrich) dissolved in PBS was placed at the transection site for 4 h prior to perfusion. Enucleation was carried out similarly, except that the extraocular muscles and ophthalmic vessels were divided after sectioning the optic nerve, allowing removal of the eye.
Probes and RNA in Situ Hybridization
Probe templates were generated by PCR amplification using primers to GFP (5′-CCA TGG TGA GCA AGG GCG AGG A-3 and 5′-CGA TGT TGT GGC GGA TCT TGA AG-3′) or mpz (5′-CGG GAT CCC TCT GGT CTC ATT CTC TTT T-3′ and 5′-CCG CTC GAG TAA CAA ACA TAT TTC CCA CA-3′), followed by ligation of the products into pBlueScript. For Northern blot, an mpz ORF probe template was made by PCR using primers 5′-ATG CTG TCC GTA CTG GCA CTG ACC-3′ and 5′-CAC TAC ACC AAT AAT GGA GCC CGT-3′ and ligated into pGEM-T. Sequence-verified plasmid clones were linearized, and digoxigenin-labeled antisense cRNA probes were synthesized by in vitro transcription, as described previously (12, 14). Whole mount RNA in situ hybridization (12), RNA in situ hybridization to brain cryosections (12), and double label in situ hybridization/immunohistochemistry (15) were carried out exactly as reported in our previous work.
Northern Blot
RNA was separated on a 1.2% agarose formaldehyde/MOPS gel and transferred to Nytran-N membrane (Schleicher & Schuell). Antisense cRNA probe was hybridized with the blot and detected using alkaline phosphatase-conjugated anti-DIG FAb and a light-emitting alkaline phosphatase substrate CDP-star (Roche Applied Science) as described previously (12).
RT-PCR and 5′-RACE
For RT-PCR, total RNA from whole adult zebrafish brains was subject to reverse transcription using SuperScript III (Invitrogen). PCR amplification of mpz was carried out using the mpz ORF primers listed above. 5′-RACE was carried out as described (16), using zebrafish mpz gene-specific reverse primer 5′-TGG AAT GGG CCT TTA TTA GCT GGA T-3′. Products were cloned in pGEM-T (Promega, Madison, WI) and sequenced. Multiple sequences were aligned and compared using AlignX.
Immunofluorescence and Antibodies
Adult zebrafish were perfused, and brains were postfixed in 4% paraformaldehyde, followed by cryoprotection in PBS-sucrose. 14-μm-thick cryosections were mounted on glass slides. For P0 immunolabeling, sections were treated with PBS-T (0.3% Triton X) for 1 h prior to blocking, as described previously (17). Sections were blocked with 10% goat serum in PBS for 2 h and then incubated overnight at 4 °C with primary antibody, diluted 1:5000 (GFP; catalog no. ab13970, Abcam, Cambridge, MA), 1:1000 (Sox10; a kind gift from Dr. Bruce Appel, University of Colorado) (18), 1:2000 (GFAP; catalog no. Z0334, Dako, Glostrup, Denmark), 1:500 (HuC; catalog no. A-21271, Invitrogen), 1:20 (7.4.C4, purified from hybridoma clone; catalog no. 92092321, HPA Culture Collections), 1:400 (TH; catalog no. MAB318, Millipore, Billerica, MA), or 1:500 (P0) (17) in PBS with 1% goat serum. Primary antibodies were detected using Alexa-488 (anti-chicken or anti-mouse)-, Alexa-405 (anti-mouse)-, or Alexa-555 (anti-rabbit)-conjugated secondary antibodies (Invitrogen) diluted 1:1000 in carrier buffer. Biocytin axonal tracing was detected using streptavidin conjugated to Alexa-555 or Alexa-488 (Invitrogen), diluted 1:1000 in carrier buffer.
Bromodeoxyuridine (BrdU) Labeling
Animals received 50 μl of 2.5 mg/ml BrdU (catalog no. B5002-1G, Sigma-Aldrich) in PBS by intraperitoneal injection prior to optic nerve crush injury and then every day for 7 days postinjury; brains were harvested for histological analysis at 7 and 14 days postinjury. Cryosections were prepared as above, depurinated with 2 n HCl at 37 °C for 30 min, and then incubated with 3% H2O2 in PBS at room temperature for 10 min and then 0.1% trypsin in PBS for 5 min at 37 °C, following which they were labeled with anti-BrdU primary antibody (1:200; Sigma-Aldrich) and a peroxidase-conjugated secondary antibody with ABC NovaRed (Vector Laboratories, Burlingame, CA).
Near-infrared Quantitative Immunofluorescence (nIR-qIF)
Sections were immunolabeled as described above, but secondary antibodies conjugated to infrared fluorophores, IRDye-800 and IRDye-680 (LI-COR, Lincoln, NE), were diluted 1:1000. Slides were scanned at high resolution using an Odyssey Infrared Imager (catalog no. 9120; LI-COR). Regions of interest were identified by anatomical landmarks in the P0 channel, and GFP or 4.C4 expression was quantified blindly in the other channel using Odyssey version 3.0 software. Validation controls were carried out to ensure that no saturated pixels were present and that regions of interest measured did not differ in area between treatment groups, sides, animals, or time points.
Microscopy, Image Acquisition, and Analysis
Bright field micrographs were captured using an Olympus BX-51 microscope and Olympus DP70 digital camera and software; whole mount images were photographed using an Olympus MVX10 stereoscope with SPOT RT3 camera (SPOT Imaging Solutions, Sterling Heights, MI), and confocal images were captured using an Olympus IX-71 confocal microscope with Fluoview software. All images in each series were acquired using identical microscope, camera, and laser settings, chosen to avoid pixel saturation. Where necessary for clarity in preparing figures, brightness and contrast adjustments were made equally to all parts of an image and equally to all panels in a multipanel set. For quantification of in situ hybridization signal, slides were processed in parallel using the same reagents to minimize variability in hybridization and chromogenic staining. Micrographs of labeled sections were analyzed using ImageJ (19), by quantifying the mean pixel grayscale value in anatomically defined regions of interest relative to background measurements in regions of the same sections that did not express mpz. Validation controls were carried out to ensure that no saturated pixels were present and that regions of interest measured did not differ in area between treatment groups, sides, or time points. For cell counting, labeled cells (EGFP-expressing cells in transgenic lines, cells labeled for mpz mRNA by in situ hybridization, BrdU-labeled cells, or Sox10-immunoreactive cells) within a region of interest encompassing the stratum opticum or optic tract were counted manually and corrected to the area of the ROI (measured using ImageJ) to allow analysis of data from sections at different levels.
Statistical Analysis
Data were shown to be normally distributed by d'Agostino's test and were analyzed using parametric statistics. Paired data (comparison between regenerating and control sides of an individual animal) were analyzed using two-tailed paired Student's t tests with Bonferroni correction for multiple comparisons. Multiple time points or transgenic lines were analyzed using one-way analysis of variance with Tukey's test for post hoc pairwise comparisons.
RESULTS
mpz Transcript and Promoter
In order to generate transgenic zebrafish lines to study oligodendroglial responses to axonal injury, we characterized the mpz gene, which is expressed in oligodendrocytes in zebrafish (20) and encodes myelin P0 (17). Up-regulation of mpz mRNA has previously been demonstrated following optic nerve or spinal cord injury in zebrafish (10), and the initial aim of this work was to clone cis-regulatory elements underlying this response. We recently showed that mpz is alternatively spliced to yield two different transcripts with divergent 3′-exons (17). In order to determine the size of the major mpz mRNA transcript expressed in the CNS, we carried out Northern blot analysis using a cRNA probe that is complementary to sequences shared between the known isoforms. We detected a single 2.5-kb mpz transcript in adult brain, which was not expressed in the gut or muscle (Fig. 1A). This is similar to the size of the mpz mRNA reported previously (10) and compatible with the predicted size of mpz transcript variant 1 (see below), which encodes the IP1-like isoform of P0 that we recently demonstrated to be most abundant in the CNS (17).
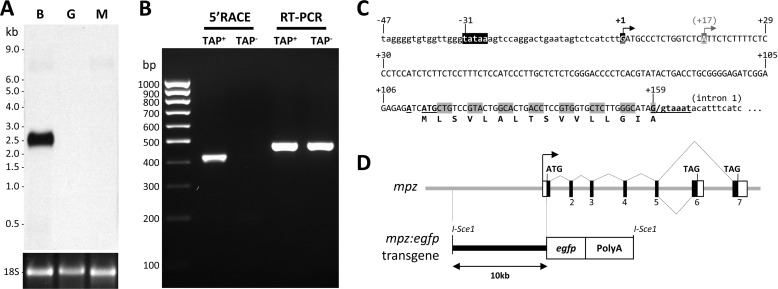
FIGURE 1.
mpz transcript and promoter. A, a Northern blot of total RNA (3 μg/lane) from brain (B), muscle (M), and gut (G) was probed using a cRNA probe to the open reading frame of mpz shared by all known splice variants. The ethidium bromide-stained 18 S rRNA loading control is shown below the blot, and molecular size markers are annotated to the left. B, 5′-RACE was used to map the transcriptional start site of mpz. The picture shows an ethidium bromide-stained agarose gel under UV illumination following electrophoretic separation of PCR products. 5′-RACE (lanes 2 and 3) and RT-PCR (lanes 4 and 5) were carried out using cDNA derived from total brain RNA that was either pretreated with tobacco acid pyrophosphatase (TAP+; lanes 2 and 4) or received no treatment (TAP−; lanes 3 and 5) prior to adapter ligation. C, the promoter region of mpz is shown. The arrows indicate the major and minor start sites at positions +1 and +17, respectively. The consensus TATA box at position −31 is highlighted in black. The open reading frame of exon 1 is indicated. The consensus translational initiation sequence at position +114 and splice donor consensus at position +159 are underlined and in boldface type. D, the diagram depicts the genomic organization and alternative splicing of mpz. Exons are numbered as previously reported (17). A schematic map of the 10-kb mpz:egfp construct is shown below the genomic map.
5′-RACE (Fig. 1B) was employed to map the 5′-end of the mpz mRNA transcript in adult zebrafish brain, using the tobacco acid pyrophosphatase (TAP) method and a gene-specific reverse primer complementary to sequence in exon 3. TAP− and RT-PCR controls were used as described previously (12, 14) to verify that the boundary between the RACE adapter and the cDNA sequence occurred at the mRNA cap site. 5′-RACE generated a single major ∼410-bp product, which was cloned; 12 randomly selected clones were sequenced. Comparison of 5′-RACE clone sequences with the genomic sequence demonstrated that mpz has a single major transcriptional start site, located 31 nucleotides 3′ to a consensus TATA box motif in the 5′ genomic flanking region (Fig. 1C). Two of the 12 RACE clones showed transcripts with 5′-ends located 17 nucleotides 3′ to the major start site, suggesting that there may be an additional minor start site. No alternative 5′-exons were identified, showing that there is a single promoter. The major transcriptional start site gives rise to a first exon that is 159 nt in length, predicting that the full mpz_TV1 mRNA is 2345 nt, compatible with the Northern blot data shown in Fig. 1A after the addition of the poly(A) tail to the transcript.
Transgenic Zebrafish Lines Expressing GFP under the Control of mpz Regulatory Elements
We next generated a construct in which expression of GFP was under transcriptional control of the mpz promoter and 10 kb of genomic sequence 5′ to the transcriptional start site (Fig. 1D). Transient assays in larval zebrafish showed that this construct expressed GFP in the developing CNS. We established four independent stable transgenic lines harboring the mpz:egfp transgene (Fig. 2, A and B); all four lines showed similar expression patterns, although lines Pt408 and Pt409 showed more robust expression and were used for subsequent studies.
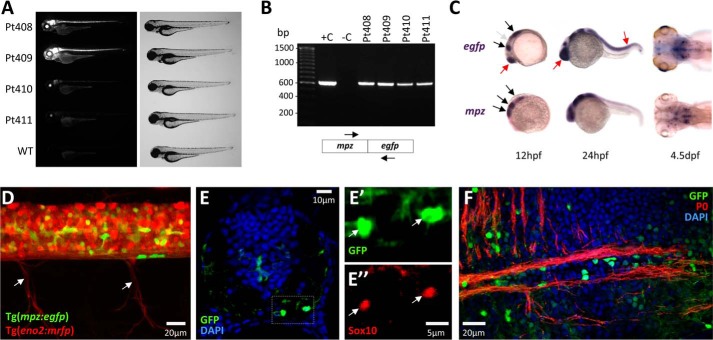
FIGURE 2.
Generation of Tg(mpz[10kb]:egfp) zebrafish and developmental expression of the transgene. A, epifluorescence micrograph (GFP channel; left) and light micrograph (right) of zebrafish from four independent stable Tg(mpz[10kb]:egfp) transgenic lines are shown at 96 h postfertilization compared with WT zebrafish. B, PCR genotyping of the transgenic lines shown in A. The picture shows PCR products separated by electrophoresis in an agarose gel containing ethidium bromide, under UV illumination. Lane 1, molecular marker; lane 2, positive control, WT zebrafish genomic DNA spiked with mpz:egfp transgene plasmid; lane 3, negative control, WT zebrafish genomic DNA; lanes 4–7, genomic DNA derived from each of the four stable transgenic lines. The positions of PCR primer sites in the transgene are shown below the gel image. C, whole mount RNA in situ hybridization using cRNA probes to egfp (top panels) or mpz (bottom panels) was employed to compare the early developmental expression patterns of the endogenous mpz gene and the mpz:egfp transgene. Hybridizing probe was revealed using a chromogenic reaction with a blue/purple product. The black arrows show expression domains common to mpz and mpz:egfp; the gray arrow shows an mpz expression domain that is not replicated by the transgene; red arrows show ectopic expression of the transgene. D, expression of GFP in spinal cord oligodendrocytes of a live double transgenic Tg(mpz:egfp);Tg(eno2:mrfp) zebrafish at 7 dpf. A lateral confocal z-plane projection of the intact spinal cord and exiting ventral nerve roots (arrows) is shown; GFP expression is shown in green, and mRFP expression is shown in red. E, a transverse section is shown of the spinal cord of a Tg(mpz:egfp) zebrafish at 7 dpf. Single confocal planes of the boxed area are shown in E′ and E″ at high magnification (green, GFP; red, Sox10; blue, DAPI). F, a longitudinal section is shown of the ventral hindbrain of a Tg(mpz:egfp) zebrafish at 10 dpf, illustrating brightly GFP-expressing oligodendrocytes adjacent to myelinated axons (green, GFP; red, P0; blue, DAPI).
In order to evaluate how closely developmental expression of the mpz:egfp transgene mirrored expression of the endogenous mpz transcript, we carried out RNA in situ hybridization using probes to mpz or egfp (Fig. 2C). Expression of mpz mRNA has been characterized previously in developing zebrafish (10, 20, 21). Similar to the endogenous mpz gene, the mpz:egfp transgene was expressed from 12 h postfertilization in the rostral neural tube; a rhombomeric pattern was visible between 24 and 48 h postfertilization. More robust expression then became evident in a small number of ventral hindbrain cells after 72 h postfertilization. These cells have previously been identified as the first myelinating oligodendrocytes in the zebrafish CNS (20). By 4–5 days postfertilization (dpf), endogenous mpz and the mpz:egfp transgene showed similar expression patterns in the developing hindbrain.
We crossed the Tg(mpz:egfp) line with a Tg(eno2:mrfp) line we reported previously, in which neurons are labeled with red fluorescent protein (22). Confocal imaging of the spinal cord in intact Tg(mpz:egfp);Tg(eno2:mrfp) larvae showed no overlap between GFP and mRFP expression at any developmental point. By 7 dpf, bright GFP expression was observed in cells located ventrally (Fig. 2, D and E) and dorso-laterally, adjacent to the mRFP-labeled axons of developing spinal cord white matter tracts. Double labeling studies in tissue sections using an antibody to Sox10 (18), a commonly used marker for oligodendroglial lineage cells in zebrafish (23–25), identified these GFP-expressing cells as oligodendrocytes (Fig. 2E). The hindbrain has previously been shown to contain discrete well defined myelinated tracts expressing P0 by 7 dpf (17). By 10 dpf, numerous GFP-expressing oligodendrocytes were located adjacent to and sent processes into myelinated fiber bundles (Fig. 2F).
In adult Tg(mpz:egfp) animals, GFP expression was evident in the brain, cranial nerves, and peripheral nervous system. GFP fluorescence was most prominent in the hindbrain (Fig. 3A), correlating with the location of the most densely myelinated axons. GFP was expressed in a regional distribution similar to that of the endogenous mpz transcript in tissue sections, with the strongest expression being evident in the brainstem and optic nerves (Fig. 3, B and C). Within the optic tectum, mpz:egfp transgene expression was localized to cells within white matter laminae, including the stratum opticum, stratum album centrale, tectobulbar tract, and tectal commissure (Fig. 3D). GFP-expressing cells were seen in close proximity to and sent numerous processes into bundles of P0-expressing myelinated fibers (Fig. 3E), although we did not observe co-localization between GFP and P0. We previously showed that P0 is only detected in myelin (17), and this may account for the lack of co-localization with cytoplasmic GFP. However, the identity of GFP-expressing cells as oligodendrocytes was established unequivocally by the following observations: (i) GFP-expressing cells in many parts of the brain showed typical oligodendroglial morphology (Fig. 3F); (ii) GFP-expressing cells also expressed Sox10 (Fig. 3G); (iii) there was no overlap between GFP expression and a number of different neural markers (Fig. 3, H and J) or GFAP; (iv) a double label in situ hybridization/immunohistochemistry method we developed previously (15) showed that cells expressing GFP also expressed the endogenous mpz mRNA transcript (Fig. 3, K and L).
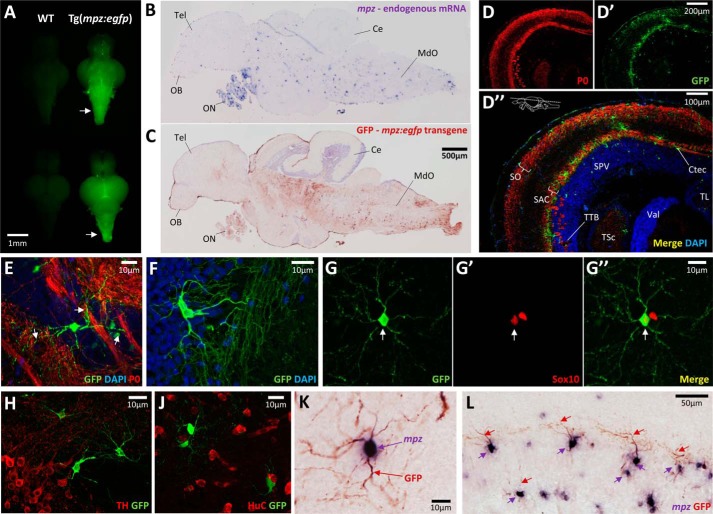
FIGURE 3.
Expression of the 10-kb mpz:egfp transgene in oligodendrocytes of the adult brain. A, GFP epifluorescence photomicrographs are shown of freshly dissected whole brains (top pair, ventral view; bottom pair, dorsal view) from WT (left) and Tg(mpz:egfp) (right) adult zebrafish. The arrows indicate bright GFP fluorescence in the brainstem of the Tg(mpz:egfp) brain. B and C, adjacent parasagittal sections are shown from an adult Tg(mpz:egfp) zebrafish brain. In B, the section was labeled by RNA in situ hybridization using a cRNA probe to the endogenous mpz mRNA (blue/purple chromogenic product). In C, the section was labeled by immunohistochemistry using an antibody to GFP (red chromogenic product). OB, olfactory bulb; Tel, telencephalon; ON, optic nerve; Ce, cerebellum; MdO, medulla oblongata. The scale bar for both panels is shown in C. D, confocal z-plane projections are shown of the optic tectum from a Tg(mpz:egfp) zebrafish, labeled for myelin P0 (red, D) and GFP (green, D′). D″ shows the two channels overlaid, with DAPI nuclear labeling; the plane of the section is indicated. SO, stratum opticum; SAC, stratum album centrale; SPV, stratum periventriculare; TTB, tetctobulbar tract; Ctec, tectal commissure; TSc, torus semicircularis; TL, torus longitudinalis; Val, valvula cerebelli. E, confocal z-plane projection of a single GFP-expressing oligodendrocyte in the midbrain of a Tg(mpz:egfp) zebrafish, sending processes (arrows) into bundles of myelinated axons (green, GFP; red, myelin P0; blue, DAPI). F, confocal z-plane projection of a single GFP-expressing oligodendrocyte in the tectobulbar tract of a Tg(mpz:egfp) zebrafish, shown to illustrate its morphology (green, GFP; blue, DAPI). G, confocal micrograph of a single GFP-expressing oligodendrocyte (G, green) in the hindbrain of a Tg(mpz:egfp) zebrafish, co-labeled with Sox10 (G′, red; arrow). The merged image and scale bar are shown in G″. H and J, confocal images of sections from a of a Tg(mpz:egfp) zebrafish, labeled for GFP (green) and neuronal markers (red; H, tyrosine hydroxylase; J, HuC), showing the absence of co-localization. K and L, double label RNA in situ hybridization (cRNA probe to mpz; blue/purple) and immunohistochemistry (antibody to GFP; red) showing that cells expressing GFP from the mpz:egfp transgene (seen best in processes; red arrows) also expressed the endogenous mpz mRNA (seen best in cell body; purple arrows). K, a single cell is shown at high magnification to illustrate the labeling method. L, the stratum album centrale of the optic tectum is shown to illustrate the high proportion of mpz-expressing cells that also expressed the mpz:egfp transgene.
Together, these data show that the mpz:egfp transgene is expressed in oligodendrocytes from larval development through adulthood. Consequently, the 10 kb of 5′ genomic sequence flanking the mpz transcriptional start site contains cis-regulatory elements necessary to direct expression of GFP in a pattern mimicking the endogenous mpz gene.
The 10-kb mpz:egfp Construct Contains an Axonal Injury Glial Response Element
We next asked whether the mpz:egfp transgene construct contains regulatory sequences from the endogenous mpz gene that are responsible for the previously reported induction of mpz mRNA following axonal injury (Fig. 4A) (10). Adult Tg(mpz:egfp) animals were subjected to unilateral optic nerve crush injury. Epifluorescence microscopy of whole dissected brains 2 weeks after the lesion showed an obvious and robust up-regulation of GFP in cells along the injured optic nerve, contralateral optic tract, and contralateral optic tectum (Fig. 4B). This was further illustrated by confocal microscopy of sections through the optic chiasm, showing an increase in both the number of GFP-expressing cells and the intensity of GFP expression, specifically along the path of the regenerating retinotectal projection (Fig. 4C). In order to quantify GFP up-regulation in the optic tract and stratum opticum, we developed a nIR-qIF method, using a high-resolution infrared scanner with a wide linear range. By labeling myelin P0 and GFP in separate nIR channels, we were able to draw regions of interest around the optic tract and stratum opticum, using the P0 signal to demarcate anatomical landmarks, thereby allowing blinded quantification of GFP expression in the other channel (Fig. 4, D and E; see “Experimental Procedures”). Using this method, we found a significant increase in GFP expression in both the optic tract and stratum opticum of the tectum by 5 days after optic nerve crush injury (Fig. 4F). The response peaked at 14–28 days postinjury, when GFP expression on the regenerating side was 3–4-fold higher than that on either the control side or sections from uninjured controls. This response started to wane by 56 days postinjury but still had not returned to baseline by 84 days postinjury. GFP expression in the regenerating optic pathway was restricted to cells also expressing Sox10 (Fig. 4J), confirming the identity of GFP+ cells as oligodendrocytes.
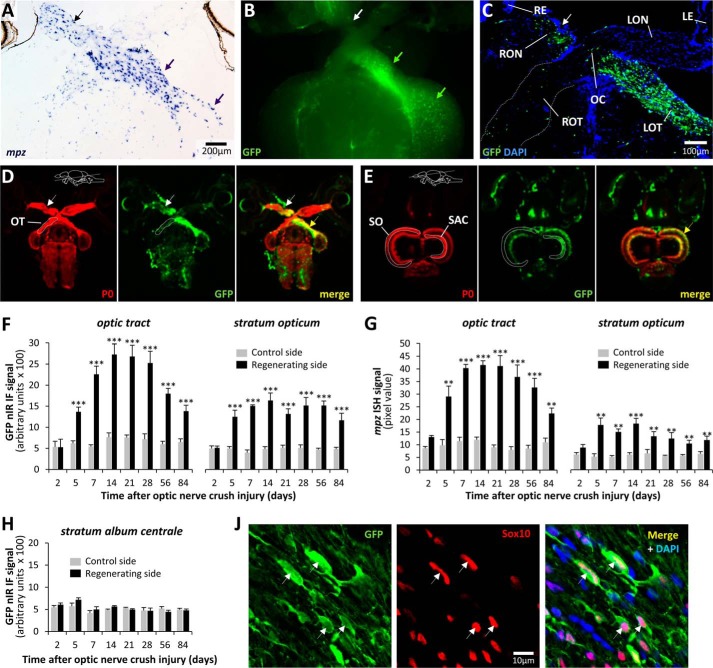
FIGURE 4.
The 10-kb mpz:egfp construct contains an oligodendroglial axonal injury response element. A, a section through the optic chiasm of a WT zebrafish is shown, 2 weeks after crush injury to the right optic nerve (black arrow; the convention followed throughout is that brain sections are shown as viewed from the ventral surface for consistency). The section was labeled by RNA in situ hybridization for endogenous mpz mRNA (blue/purple chromogenic product). Robust up-regulation of mpz is clearly seen in the right optic nerve, left optic tract, and left optic tectum (purple arrows). B, GFP epifluorescence (green) micrograph of the ventral surface of a freshly dissected Tg(mpz:egfp) brain, 2 weeks after right optic nerve crush injury (white arrow), showing robust GFP up-regulation in the right optic nerve, left optic tract, and left optic tectum (green arrows). C, confocal z-plane projection of a horizontal section through the optic chiasm of a Tg(mpz:egfp) brain, 2 weeks after right optic nerve crush injury (arrow), labeled for GFP (green) and DAPI (blue). RE, right eye; LE, left eye; RON, right optic nerve; LON, left optic nerve; OC, optic chiasm; ROT, right optic tract; LOT, left optic tract. D and E, high-resolution nIR-qIF scans of optic tracts (D) and optic tectum (E) from Tg(mpz:egfp) brain, 2 weeks after right optic nerve crush injury (arrow), labeled with antibodies to GFP (680-nm channel; pseudocolored green) and myelin P0 (800-nm channel; pseudocolored red). The planes of the sections and anatomical landmarks defining regions of interest are annotated in the P0 image for each set. The site of the optic nerve lesion is indicated with a white arrow, and up-regulation of the GFP transgene is shown with a yellow arrow. F, the nIRqIF GFP-immunoreactive signal from the optic tract and stratum opticum was quantified in Tg(mpz:egfp) brains, as shown in D and E, at time points between 2 and 84 days after optic nerve crush injury. For each time point, the regenerating and control sides of the same sections were analyzed in four animals. Graphs show mean and S.E. (error bars); ***, p < 0.0001, control versus regenerating side; paired t test with Bonferroni correction for multiple comparisons. G, the RNA in situ hybridization signal for mpz was quantified in the optic tract and stratum opticum of the same animals shown in F. For each time point, the regenerating and control sides of the same sections were analyzed. Graphs show mean and S.E. (error bars); ***, p < 0.0001; **, p < 0.001, control versus regenerating side; paired Student's t test with Bonferroni correction for multiple comparisons. H, the nIR-qIF GFP-immunoreactive signal from the stratum album centrale (see E) was quantified in the same sections from which data were acquired for F. Graphs show mean and S.E. J, a single confocal plane of the regenerating left optic tract, 2 weeks after optic nerve crush injury, is shown, labeled for indirect immunofluorescence with antibodies to GFP (green; left) and Sox10 (red; center), with a nuclear counterstain (DAPI; blue channel in merged image). Examples of cells showing co-localization of GFP and Sox10 are shown (arrows).
In order to compare the time course of mpz:egfp up-regulation following axonal injury with expression of the endogenous mpz gene, we quantified the RNA in situ hybridization signal for the mpz transcript in the same brain regions following optic nerve crush injury. The earliest statistically significant increase in mpz expression relative to the control side was found at 5 days postinjury (Fig. 4G). The response peaked at 14–28 days postinjury, with a 3–4-fold induction in mpz expression, following which there was a gradual decrease in expression that did not return to baseline by 84 days postinjury. Changes in expression of mpz along the optic tracts and stratum opticum after optic nerve crush injury thus showed a time course and magnitude remarkably similar to the changes in GFP expression we detected in Tg(mpz:egfp) animals, suggesting that the 10-kb transgene construct contains the cis-regulatory sequences mediating up-regulation of the endogenous gene following axonal injury.
We next determined whether changes in transgene expression were restricted to the regenerating visual pathway or were part of a more generalized response to injury by quantifying GFP expression in an adjacent tectal white matter lamina, the stratum album centrale, which does not receive a direct projection from the optic nerve. Unlike the robust changes in GFP expression that we observed in the stratum opticum, no changes in GFP expression were detected in the stratum album centrale at any time point following optic nerve injury (Fig. 4H). These data show that the transgene was induced by signals specific to the damaged retinotectal pathway.
Oligodendroglial Morphology, Abundance, and Transgene Expression following Axonal Injury
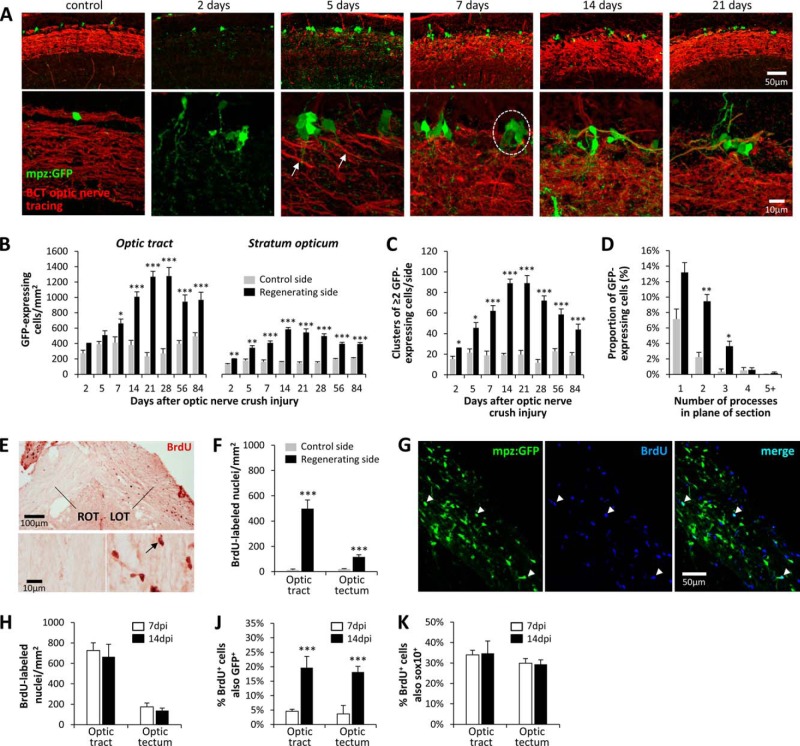
The unique availability of a cytoplasmic oligodendroglial marker that is induced by axonal injury allowed us to examine the number, distribution, morphology, and origin of oligodendrocytes in the control and regenerating visual pathways. This was most readily accomplished in the stratum opticum of the tectum, where GFP-expressing oligodendrocytes are less densely arranged than in the optic tract. At baseline, the cell bodies of tectal oligodendrocytes were situated at the superficial surface of the stratum opticum (Fig. 5A). The cells showed mostly unipolar morphology, with a single highly branched process passing into the underlying fiber tract. Single cells were uniformly spaced along the tract. By 2 days after optic nerve injury, there was a modest increase in the number of GFP-expressing cells in the stratum opticum, and clusters of multiple GFP-expressing cells in immediate proximity to one another started to emerge (Fig. 5A). We quantified both the number of GFP-expressing cells and the number of GFP cell clusters (two or more GFP-expressing cells in immediate contact; Fig. 5, B and C) over time following optic nerve crush injury. Both total cells and clusters of cells became more numerous between 2 and 14 days postinjury, both peaking at 14–28 days postinjury. The number of GFP-expressing cells and clusters then started to decline but had not reached baseline by 84 days postinjury. Examination of individual GFP-expressing cells suggested that their morphology had become more complex during the regeneration process. In order to determine whether this was the case, we counted the number of processes clearly emerging from the cell body within the plane of the section, on the control and regenerating sides of the brain. By 7 days postinjury, GFP-expressing cells on the regenerating side showed more processes than cells on the control side (Fig. 5D). Together, these data show that the increased GFP immunoreactivity observed in the visual pathway of Tg(mpz:egfp) zebrafish following optic nerve injury is attributable to an increase in the number of GFP-expressing cells in addition to changes in cellular morphology and increased GFP expression levels.
FIGURE 5.
Increased abundance of GFP-expressing oligodendrocytes in the regenerating optic pathway of Tg(mpz:egfp) zebrafish. A, low (top row) and high (bottom row) magnification confocal z-plane projections are shown of the stratum opticum in a control Tg(mpz:egfp) animal and at time points between 2 and 21 days following optic nerve crush injury. Animals underwent optic nerve tracing using biocytin prior to perfusion; sections are labeled for GFP (green) and biocytin (red). The arrows show the earliest regenerating axons reaching the optic tectum at 5 days postinjury. A cluster of GFP-expressing oligodendrocytes is circled in the 7 days postinjury panel. B, GFP-expressing cells were quantified in the optic tract and stratum opticum at time points between 2 and 84 days after optic nerve crush injury, on each side of the same sections. ***, p < 0.0001; **, p < 0.001; *, p < 0.01, control versus regenerating side; paired t test with Bonferroni correction for multiple comparisons. C, clusters of ≥2 GFP-expressing cells in direct contact with one another were quantified in the stratum opticum at time points between 2 and 84 days after optic nerve crush injury, on each side of the same sections. ***, p < 0.0001; *, p < 0.01, control versus regenerating side; paired t test with Bonferroni correction for multiple comparisons. D, the number of GFP-immunoreactive processes per cell was quantified in >100 GFP-expressing oligodendrocytes from the control and regenerating sides of the stratum opticum in four Tg(mpz:egfp) zebrafish, 7 days after optic nerve crush injury. The graphs show the mean percentage of cells on each side with 1, 2, 3, 4, or ≥5 processes seen leaving the cell body in the plane of the section. Error bars, S.E.; **, p < 0.001; *, p < 0.01, control versus regenerating side; paired t test with Bonferroni correction for multiple comparisons. E–K, BrdU was administered to Tg(mpz:egfp) zebrafish prior to optic nerve crush injury and then daily for 7 days. Histological sections were prepared at 7 or 14 days postinjury. E, a section encompassing the optic chiasm at 7 days postinjury was labeled with an antibody to BrdU (red chromogenic product). LOT, left optic tract; ROT, right optic tract. The bottom panels show the right and left optic tracts at high magnification; a BrdU-labeled cell nucleus is indicated (arrow). F, BrdU-labeled nuclei were quantified in the optic tract and optic tectum of five zebrafish on the control and regenerating sides at 7 days postinjury. ***, p < 0.0001 control versus regenerating side; paired t test with Bonferroni correction for multiple comparisons. G, single confocal planes showing the regenerating optic tract at 7 days postinjury were labeled for GFP (left, green) and BrdU (center, blue). BrdU+/GFP+ oligodendrocytes that were born after the crush injury are indicated with arrows. H, the density of BrdU+ cells in the regenerating optic tract and optic tectum was quantified at 7 and 14 days postinjury. J, the proportion of BrdU-labeled cells in the regenerating optic tract that also showed expression of the mpz:egfp transgene at 7 and 14 days postinjury was quantified. ***, p < 0.0001, two-tailed t test. J, the proportion of BrdU-labeled cells in the regenerating optic tract that also showed Sox10 immunoreactivity at 7 and 14 days postinjury was quantified. Graphs show mean and S.E.
Birth and Differentiation of New Oligodendrocytes following Axonal Injury
BrdU labeling was employed to address whether the increased abundance of GFP-expressing oligodendrocytes following axonal injury was attributable to cell division and birth of new cells. BrdU was administered prior to optic nerve injury and daily thereafter to label newly born cells until 7 days postinjury; tissue was harvested for analysis at 7 days postinjury or 14 days postinjury. By 7 days postinjury, there were highly significant increases in the abundance of BrdU-labeled cells in the regenerating optic tract (∼40-fold) and optic tectum (∼6-fold; Fig. 5, E and F) compared with the control side. At 7 days postinjury, occasional GFP+ oligodendrocytes were also BrdU+ (Fig. 5G), suggesting that birth of new cells may contribute to the increased abundance of GFP+ cells in the regenerating visual pathway. The total number of BrdU-labeled cells in the regenerating visual pathway did not change significantly between 7 and 14 days postinjury (Fig. 5H), and ∼30% of the BrdU-labeled cells at both time points were immunoreactive for the oligodendroglial lineage marker Sox10 (Fig. 5K). However, whereas only 5% of BrdU+ cells expressed the mpz:egfp transgene at 7 days postinjury, ∼20% of BrdU+ cells were also GFP+ by 14 days postinjury (Fig. 5H). These data suggest that axonal injury provoked birth of new Sox10+ oligodendroglial precursors, some of which subsequently differentiated into mpz-expressing oligodendrocytes. However, only ∼10% of GFP+ oligodendrocytes at 14 days postinjury were BrdU+, suggesting that the increased abundance of mpz:egfp transgene-expressing cells following axonal injury was mainly attributable to differentiation of progenitor cells or transcriptional induction in quiescent oligodendrocytes born prior to the injury.
Transcriptional Induction of mpz Is Not Dependent on Axonal Regrowth
We next sought to clarify the nature of the signal that drives the oligodendroglial response to axonal injury. Enucleation, which prevented retinotectal axonal regrowth, was employed to dissociate the contributions of axonal degeneration and axonal recontact to the observed response. The oligodendroglial response to enucleation was very similar to the response observed after crush injury. A 2–3-fold increase in the number of GFP-expressing oligodendrocytes was observed in both the optic tract and stratum opticum, peaking at 14–28 days following enucleation (Fig. 6, A and B). GFP-expressing oligodendrocyte clusters also appeared following enucleation, similar to the response seen after optic nerve crush injury (Fig. 6C). Up-regulation of the mpz:egfp transgene persisted to at least 56 days postinjury. Together, these data show that induction of the mpz:egfp transgene in oligodendrocytes following axonal injury was not dependent on axonal recontact.
FIGURE 6.
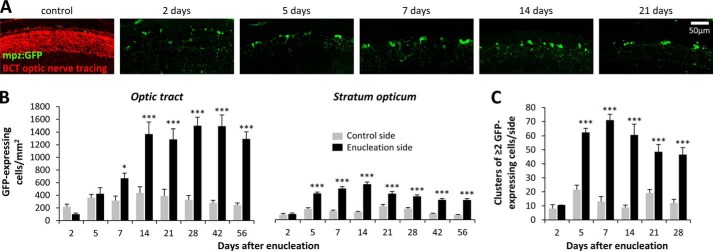
Axonal regeneration is not necessary for the oligodendroglial injury response. A, Tg(mpz:egfp) zebrafish underwent right-sided enucleation. At time points between 2 and 21 days after surgery, optic nerve tracing was carried out, and then histological sections were prepared and labeled for GFP (green) and biocytin (BCT, red). Confocal z-plane projections of the left stratum opticum (lesioned retinotectal pathway) are shown. B, GFP-expressing cells were quantified in the optic tract and stratum opticum on each side at time points between 2 and 56 days postenucleation. Graphs show mean and S.E. (error bars); ***, p < 0.0001; *, p < 0.01, control versus lesion side; paired t test with Bonferroni correction for multiple comparisons. C, clusters of ≥2 GFP-expressing cells in direct contact with one another were quantified on each side of the tectum at time points between 2 and 28 days postenucleation. The graph shows mean and S.E.; ***, p < 0.0001, control versus lesion side; paired t test with Bonferroni correction for multiple comparisons.
Microglial Infiltration of the Damaged Optic Pathway Precedes the Oligodendroglial Response
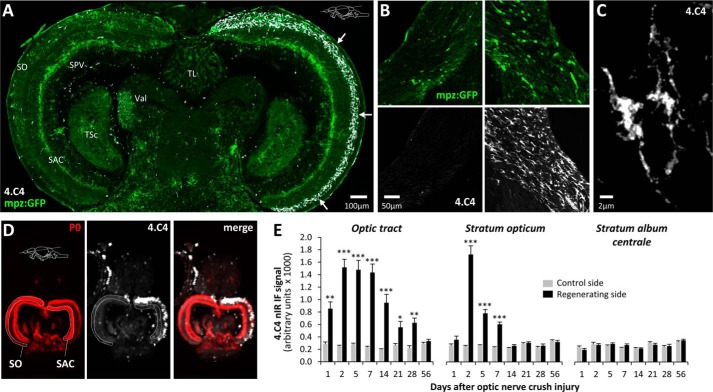
We next asked whether the neuroinflammatory response to axonal degeneration could underlie transgene induction following injury. Following optic nerve injury, robust microglial infiltration was evident in the stratum opticum (Fig. 7A) and the optic tract (Fig. 7, B and C). By employing a quantitative nIR immunofluorescence method to measure expression of the microglial marker 4.C4 (Fig. 7D), we compared the time course of the microglial response following crush injury with oligodendroglial GFP expression. The microglial response peaked at 2 days postinjury and then slowly resolved, reaching baseline by 14 days postinjury in the stratum opticum of the tectum and 56 days postinjury in the optic tract (Fig. 7E). This response was specific to the damaged retinotectal projection; there was no microglial infiltration in the stratum album centrale (Fig. 7E) or the contralateral stratum opticum. Microglial infiltration preceded the oligodendroglial response, which peaked at 14–28 days postinjury and persisted to 84 days postinjury (Figs. 4 (F and G) and 5B). It is conceivable that a microglial factor or an indirect microglia-dependent mechanism, such as removal of an axonal signal by phagocytosis, contributes to the oligodendroglial response. Reagents that prevent the microglial response to optic nerve injury in adult zebrafish would be necessary to evaluate this possibility directly.
FIGURE 7.
Microglial infiltration in the regenerating optic pathway precedes the oligodendroglial response. A, montage of confocal images showing a horizontal section of the midbrain region from an adult Tg(mpz:egfp) zebrafish 2 days after optic nerve crush injury, immunolabeled for GFP (green) and 4C.4 (microglia; white). The arrows show robust microglial infiltration limited to the stratum opticum contralateral to the optic nerve lesion. SO, stratum opticum; SAC, stratum album centrale; SPV, stratum periventriculare; TSc, torus semicircularis; TL, torus longitudinalis; Val, valvula cerebelli. B, single confocal planes of ventral optic tract (left column, control side; right column, regenerating side) from a Tg(mpz:egfp) zebrafish 5 days after optic nerve crush injury. Sections are labeled for GFP (green, top row) and 4.C4 (microglia; white, bottom row). The scale bar for all four panels is shown in the bottom left panel. C, high-magnification confocal z-plane projection showing the morphology of individual microglia in the regenerating optic tract. D, high-resolution nIR-qIF scan of the optic tectum from Tg(mpz:egfp) brain, 2 days after optic nerve crush injury, labeled with antibodies to 4.C4 (680-nm channel; pseudocolored white) and myelin P0 (800-nm channel; pseudocolored red). The plane of the sections and anatomical landmarks defining regions of interest are annotated in the P0 image. E, the nIR-qIF 4.C4-immunoreactive signal from the optic tract and stratum opticum was quantified in Tg(mpz:egfp) brains, as shown in D at time points between 2 and 56 days after optic nerve crush injury. For each time point, the regenerating and control sides of the same sections were analyzed in four animals. Graphs show mean and S.E. (error bars); ***, p < 0.0001; **, p < 0.001; *, p < 0.01, control versus regenerating side; paired Student's t test with Bonferroni correction for multiple comparisons.
Axonal Injury Response Element and Oligodendroglial Enhancer Are Separable
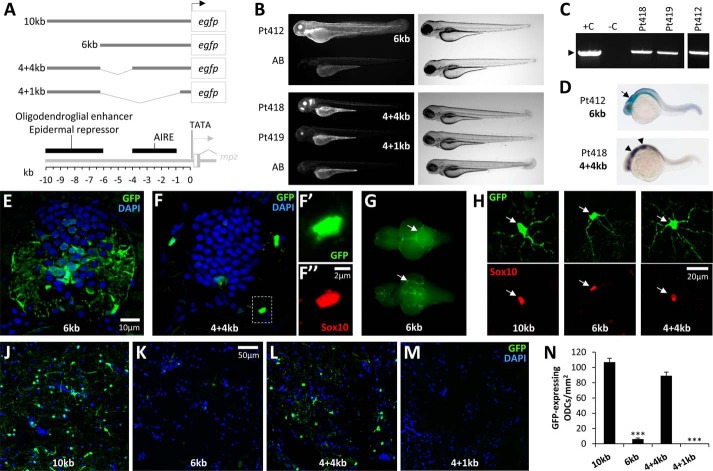
As a first step toward identifying cis-regulatory sequences underlying the mpz transcriptional response to axonal injury, we made a panel of mpz:egfp deletion constructs and tested the activity of each regulatory fragment in both transient assays and stable transgenic zebrafish (Figs. 8 and 9). First, we generated a 6-kb mpz:egfp construct lacking 4 kb of 5′ sequence contained in the 10-kb regulatory element (Fig. 8A). Larvae expressing the 6-kb construct either transiently or in a stable transgenic line (Fig. 8, B and C) showed weak CNS expression, but robust GFP fluorescence was visible throughout the epidermis and ocular lens. RNA in situ hybridization showed diffuse expression of the 6-kb mpz transgene at 24 h postfertilization that was dissimilar to the rhombomeric expression pattern of the endogenous mpz gene (Figs. 2C and 8D). In contrast to the 10-kb transgene, expression of the 6-kb transgene was not detected within oligodendrocytes of the developing spinal cord at 7 dpf (Figs. 2E and 8E). In adult Tg(mpz[6kb]:egfp) animals, the most robust GFP expression in the CNS was found in the meninges (Fig. 8G), although a few GFP-expressing, Sox10-immunoreactive oligodendrocytes were found scattered throughout the CNS under basal conditions (Fig. 8, H and K). Quantitative analysis showed that the density of GFP-expressing oligodendrocytes in the medulla oblongata of Tg(mpz[6kb]:egfp) animals was ∼20-fold less than in Tg(mpz[10kb]:egfp) brain (Fig. 8J). These data show that the region between −6 and −10 kb with respect to the transcriptional start site contains an oligodendroglial enhancer and additional signals that repress epidermal transcription of mpz.
FIGURE 8.
An oligodendroglial enhancer is located in the mpz upstream regulatory region. A, a panel of deletion mutants was constructed in order to localize the AIRE within the 10-kb mpz cis-regulatory construct shown in Figs. 2–7. The constructs shown schematically were tested in transient assays in zebrafish larvae and used to generate stable transgenic zebrafish lines. The map below summarizes the locations of key elements in the mpz regulatory region that were delineated in this study, including the TATA motif and transcription start site, AIRE, and upstream oligodendroglial enhancer/epidermal repressor sequences. B, epifluorescence micrographs (GFP channel; left panels) and light micrographs (right panels) showing stable Tg(mpz[6kb]:egfp) (Pt412; top panels), Tg(mpz[4+4kb]:egfp), and Tg(mpz[4+1kb]:egfp) (Pt418 and Pt419; bottom panels) zebrafish larvae at 72 h postfertilization. C, genotyping data confirming stable integration of the mpz:egfp transgenes in F3 generation zebrafish from lines Pt412, Pt418, and Pt419. Tail fin genomic DNA was subjected to PCR analysis as shown in Fig. 2B. Lane 1, positive control (+C), wild-type genomic DNA spiked with transgene plasmid; lane 2, negative control (−C), wild-type genomic DNA; lanes 3–5, genomic DNA from Pt418 (lane 3), Pt419 (lane 4), and Pt412 (lane 5, from a separate gel run in parallel). The expected PCR product is indicated by an arrowhead. D, whole mount RNA in situ hybridization using a cRNA probe to egfp and a chromogenic reaction with a blue/purple product was employed to show the early developmental expression of the 6-kb (top) and 4 + 4-kb (bottom) mpz constructs. E, transverse section of the spinal cord from a Tg(mpz[6kb]:egfp) zebrafish at 7 dpf labeled for GFP (green) and DAPI (blue). F, transverse section of the spinal cord from a Tg(mpz[4+4kb]:egfp) zebrafish at 7 dpf. Single confocal planes of the boxed area are shown in F′ and F″ at high magnification (green, GFP; red, Sox10; blue, DAPI). G, GFP epifluorescence photomicrographs are shown of a freshly dissected whole brain from a Tg(mpz[6kb]:egfp) zebrafish (dorsal view above, ventral view below). The arrows indicate bright GFP fluorescence on the meningeal surface of the brain. H, confocal micrographs of single GFP-expressing oligodendrocytes from the hindbrain of individual Tg(mpz:egfp) zebrafish lines, expressing a GFP reporter under different mpz regulatory fragments as indicated (see A). Top row, GFP (green); bottom row, Sox10 (red). The arrowheads show the position of the GFP-expressing cell bodies, confirming that the same cells were also Sox10-immunoreactive. The scale for all six panels is shown in the bottom right panel. J–M, fluorescence micrographs of the medulla oblongata of individual Tg(mpz:egfp) zebrafish lines, expressing a GFP reporter under different mpz regulatory fragments (see A). Sections are labeled for GFP (green) and DAPI (blue). The scale for all four panels is shown in the second panel. N, GFP-expressing oligodendrocytes were quantified in the medulla oblongata of Tg(mpz:egfp) zebrafish, expressing a GFP reporter under different mpz regulatory fragments as shown in A. Graphs show mean and S.E. (error bars). ***, p < 0.0001 compared with animals expressing the 10-kb mpz:egfp construct (one-way analysis of variance with Tukey's post hoc test).
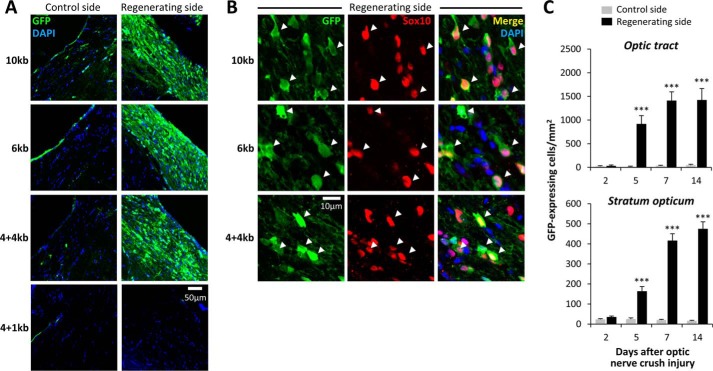
FIGURE 9.
The mpz AIRE is dissociable from the mpz upstream oligodendroglial enhancer. A, expression of GFP in the ventral optic tract of Tg(mpz:egfp) zebrafish. Each row of confocal micrographs shows a different transgenic line expressing GFP under transcriptional control of distinct fragments of the mpz regulatory region (indicated to the left; see Fig. 8A). The columns show control optic tract (left) and regenerating optic tract (right) at 14 days postinjury. All panels are labeled for GFP (green) and DAPI (blue). The scale bar for all eight panels is shown in the bottom right panel. B, single confocal planes through the regenerating optic tract are shown at high magnification for the same transgenic lines as shown in A. The scale bar for all nine panels is shown at the bottom left. Sections are labeled for GFP (green; first column) and Sox10 (red; second column). The third column shows the merged image with a nuclear counterstain (DAPI, blue). The arrowheads show the positions of GFP-expressing cells, confirming that they were also immunoreactive for Sox10. C, GFP-expressing cells in Tg(mpz[6kb]:egfp) zebrafish were quantified in the optic tract and stratum opticum at time points between 2 and 14 days after optic nerve crush injury, on each side of the same sections. The graphs show mean and S.E. (error bars). ***, p < 0.0001, control versus regenerating side; paired t test with Bonferroni correction for multiple comparisons.
Following optic nerve crush injury, robust up-regulation of GFP was apparent in the regenerating visual pathway of Tg(mpz[6kb]:egfp) animals (Fig. 9A). Double labeling confirmed that, similar to the 10-kb construct, the GFP-expressing cells also expressed Sox10, identifying them as oligodendrocytes (Fig. 9B). Transgene induction in the regenerating optic pathway in Tg(mpz[6kb]:egfp) showed kinetics and magnitude similar to those of Tg(mpz[10kb]:egfp) animals (Fig. 9C). At 14 days postinjury, there was no significant difference between the two genotypes in the density of GFP-expressing cells on the regenerating side, although the robust difference on the control side remained (Figs. 5B and 9C). These data show that an oligodendroglial axonal injury response element (AIRE) is located in the 6 kb of sequence upstream of the mpz promoter that is present in the mpz[6kb]:egfp construct. The AIRE is distinct from the upstream oligodendroglial enhancer that is missing from the 6-kb construct.
In order to further clarify the location of the AIRE, we generated two further internal deletion constructs that remove 2 or 5 kb of 5′ sequence from the 6-kb element while retaining the 4 kb of upstream sequence between −6 and −10 kb that contains the oligodendroglial enhancer and epidermal repressor (Fig. 8A). The expression pattern of the 4 + 4-kb construct was similar to the 10-kb construct, both in transiently transgenic larvae and in a stable transgenic line. Thus, the mpz[4 + 4kb]:egfp construct showed rhombomeric hindbrain expression during early development (Fig. 8D) and robust expression within Sox10-immunoreactive spinal cord oligodendrocytes by 7 dpf (Fig. 8F). Under basal conditions, numerous GFP-expressing Sox10-immunoreactive oligodendrocytes were visible throughout the CNS of adult Tg(mpz[4+4kb]:egfp) animals (Fig. 8, H and L), and there was no significant difference between Tg(mpz[4 + 4kb]:egfp) and Tg(mpz[10kb]:egfp) animals in the density of GFP-expressing oligodendrocytes in the medulla oblongata (Fig. 8N). Following optic nerve crush injury, there was robust up-regulation of GFP expression in the regenerating visual pathway of Tg(mpz[4+4kb]:egfp) animals (Fig. 9A), and the GFP-labeled cells co-labeled with Sox10, confirming their identity as oligodendrocytes (Fig. 9B). In contrast, the 4 + 1-kb construct did not show GFP expression in either transiently transgenic larvae or a stable transgenic line, except for weak GFP fluorescence in the ocular lens (Fig. 8B). In the adult CNS, GFP expression was not visible in Tg(mpz[4+1kb]:egfp) zebrafish, either under basal conditions or following optic nerve crush injury (Figs. 8 (M and N) and 9A). These data suggest that the AIRE is located in the region between −1 and −4 kb with respect to the promoter. This region may also contain additional enhancer elements necessary for activity of the mpz promoter.
Together these data show that the AIRE is dissociable from the upstream mpz enhancer necessary for transgene expression in oligodendrocytes in the uninjured CNS. Consequently, we conclude that different signaling mechanisms drive mpz expression in myelinating CNS oligodendrocytes and oligodendroglial mpz induction in response to axonal injury.
DISCUSSION
Transgenic zebrafish expressing GFP in oligodendrocytes have been generated previously, using cis-regulatory sequences derived from the genes encoding Olig2 (26), myelin basic protein (27), Sox10 (28), or Claudin K (29). However, up-regulation of an oligodendroglial reporter gene in response to axonal injury has not previously been demonstrated. Consequently, the novel Tg(mpz:egfp) zebrafish lines we generated allowed us to make a number of new observations concerning the oligodendroglial response to axonal injury.
Three factors contributed to the spatially restricted and temporally stereotyped increase in GFP expression that occurred along the retinotectal pathway of Tg(mpz:egfp) zebrafish following optic nerve crush injury.
First, oligodendroglial transcription of the mpz:egfp transgene was strongly up-regulated following axonal injury; this response was dependent on a cis-acting oligodendroglial axonal injury response element. Analysis of truncated transgene constructs in vivo showed that the AIRE is located between −1 and −4 kb with respect to the mpz transcriptional start site and is separable from an upstream enhancer necessary for GFP expression in myelinating oligodendrocytes in the adult brain, which is located between −6 and −10 kb. This is the first demonstration that cis-regulatory elements responsible for expression of a gene in myelinating CNS oligodendrocytes are dissociable from sequences underlying induction of the same gene by axonal injury. Our data strongly suggest that separate trans-acting factors drive mpz expression through these discrete cis-regulatory modules, compatible with distinct cellular signaling mechanisms being triggered by myelination and axonal damage. The specificity of transgene expression in both cases suggests that the underlying mechanisms are restricted to oligodendrocytes, and the pathway mediating up-regulation of mpz following axonal injury presumably also applies to mpz induction during differentiation of oligodendroglial precursors following axonal injury. Distinct cis-regulatory elements have previously been shown to control expression of genes in retinal ganglion cells during development versus retinotectal axonal regeneration (30). A 1.7-kb fragment of the α1-tubulin gene recapitulated the expression pattern of the endogenous gene during development and CNS repair in the adult (31). Different deletions in the regulatory region selectively abrogated transgene expression either during development (32) or following optic nerve injury (30). Similarly, 708 bp of genomic 5′ sequence from the fugu gap43 gene was sufficient to drive transgene expression in developing retinal ganglion cell neurons, but induction of transgene expression by optic nerve crush injury required additional sequences located further upstream (33, 34). These data suggest that there may be a common theme in which induction of gene expression following adult CNS injury occurs through mechanisms that differ from those dictating developmental expression of the same gene.
Second, oligodendrocytes expressing the GFP reporter gene became more numerous following axonal injury. Although the number of BrdU+ cells in the regenerating optic tract was elevated with respect to the control side, cells that had divided after optic nerve injury contributed relatively little to the substantial increase in GFP+ oligodendrocytes that occurred after the lesion. At 14 days postinjury, ∼10% of GFP+ oligodendrocytes in the regenerating visual pathway had undergone division following the injury. This is comparable with the 7.5% previously found in the regenerating goldfish visual pathway by culturing and immunolabeling cells after exposing the animal to BrdU (35). These data suggest that the major mechanism contributing to the observed increase in GFP+ oligodendrocytes was induction of transgene expression in cells that were born prior to the injury. Sox10+/Olig2+ cells that do not express myelin genes, such as mbp or mpz, are thought to represent oligodendroglial precursors in the adult zebrafish CNS (24). GFP+ cells were a subset of Sox10+ cells in the uninjured Tg(mpz:egfp) CNS, and it seems likely that the majority of GFP-expressing oligodendrocytes in the regenerating visual pathway were derived from resident precursor cells that underwent differentiation in response to axonal injury and thereby activated mpz gene expression without undergoing cell division. Compatible with this possibility, it has recently been demonstrated that oligodendroglial precursor cells in the mammalian brain usually differentiate without prior cell division (36). However, their differentiation frequently triggers division of adjacent cells to maintain the oligodendroglial precursor population, which may explain the origin of the Sox10+/BrdU+/GFP− cells we observed. In addition, evidence for migration of oligodendroglial precursors to the site of a CNS injury has been found previously in adult zebrafish (37); it is possible that oligodendroglial precursor cells also migrated into the damaged visual pathway and contributed to the enhanced abundance of GFP+ cells following their differentiation.
Third, oligodendroglial morphology in the stratum opticum of Tg(mpz:egfp) zebrafish became more complex following axonal injury. A previous study in goldfish, using Lucifer Yellow iontophoresis to label single cells, showed a series of stereotyped morphological changes in optic tract oligodendroglia following optic nerve injury (35). Loss of distal processes and myelin segments was observed following axonal injury, but typical oligodendroglial morphology was re-established during axon regrowth. At baseline, zebrafish stratum opticum oligodendrocytes differ morphologically from goldfish optic tract oligodendrocytes. Furthermore, the cytosolic GFP reporter that we employed labels oligodendroglial cell bodies and proximal processes but not myelin. Consequently, direct comparison of the findings from the two studies is difficult. However, the increased number of oligodendroglial processes that we observed during axonal regrowth in Tg(mpz:egfp) zebrafish may parallel the re-establishment of complex oligodendroglial morphology reported in the goldfish optic tract during axonal regeneration. It is possible that these changes in oligodendroglial structure promote axonal regrowth or the remyelination of regenerated axons.
The origin of the signals that mediate the oligodendroglial response to axonal injury is currently unknown. The strikingly restricted spatial distribution of increased oligodendroglial abundance and mpz up-regulation that we observed following optic nerve lesion suggests that the driving signal originates from within the damaged axonal tract and is not a generalized tissue response to the crush injury. There were marked similarities in the kinetics of mpz:egfp transgene expression following both crush injury and enucleation, suggesting that regenerating axons do not provide the primary signal for mpz up-regulation or differentiation of oligodendroglial precursors, although separate signals dependent on axonal contact are probably involved in remyelination. Robust microglial infiltration occurred in the damaged visual pathway before mpz up-regulation, raising the possibility that the oligodendroglial response was triggered by microglia. This could potentially occur directly through a secreted or contact factor or indirectly through removal of an axonal signal by phagocytosis. The development of methods that disrupt microglial activation in zebrafish will enable clarification of the role that neuroinflammation plays in this response. Other features shared by both optic nerve crush injury and enucleation include axonal loss and the presence of axonal debris. Either of these putative signals could also drive the initial oligodendroglial response to axonal degeneration, independent of microglial functions.
It is likely that enhanced mpz expression following axonal damage is a component of a wider transcriptional program that regulates multiple gene expression changes in oligodendrocytes. It will be of considerable interest to identify the molecular basis of this response, in particular to determine whether there is a master regulator driving transcriptional changes that promote repair, because manipulation of relevant regulatory mechanisms might be pursued as a means to enhance axonal regeneration and remyelination in the mammalian CNS. Identification of the AIRE should provide candidate trans-acting factors responsible for mpz gene induction following injury, leading to characterization of upstream signaling events mediating the oligodendroglial response to axonal injury. Isolation of GFP-expressing oligodendrocytes from the CNS of Tg(mpz:egfp) zebrafish may also allow detection of genes that are coordinately regulated following injury. This would further delineate molecular components of the oligodendroglial repair response and may allow identification of common regulatory mechanisms shared by participant genes.
Acknowledgments
We thank the following colleagues for generously sharing resources: Dr. Bruce Appel (University of Colorado; Sox10 antibody); Dr. Thomas Becker (University of Edinburgh; detailed experimental protocols for the optic nerve crush injury model); Dr. Neal Copeland (NCI, National Institutes of Health; DY30 cells); Dr. Ronald Plasterk (Netherlands Institute for Developmental Biology; BAC clone zKp35B8).
This work was supported, in whole or in part, by National Institutes of Health, NINDS, Grant NS058369. This work was also supported by the Ethel Vincent Charitable Trust.
- P0
- protein zero
- nIR
- near-infrared
- nIR-qIF
- near-infrared quantitative immunofluorescence
- TAP
- tobacco acid pyrophosphatase
- dpf
- days postfertilization
- AIRE
- axonal injury response element
- RACE
- rapid amplification of cDNA ends.
REFERENCES
- 1. Becker T., Wullimann M. F., Becker C. G., Bernhardt R. R., Schachner M. (1997) Axonal regrowth after spinal cord transection in adult zebrafish. J. Comp. Neurol. 377, 577–595 [DOI] [PubMed] [Google Scholar]
- 2. Bormann P., Zumsteg V. M., Roth L. W., Reinhard E. (1998) Target contact regulates GAP-43 and α-tubulin mRNA levels in regenerating retinal ganglion cells. J. Neurosci. Res. 52, 405–419 [DOI] [PubMed] [Google Scholar]
- 3. Bernhardt R. R., Tongiorgi E., Anzini P., Schachner M. (1996) Increased expression of specific recognition molecules by retinal ganglion cells and by optic pathway glia accompanies the successful regeneration of retinal axons in adult zebrafish. J. Comp. Neurol. 376, 253–264 [DOI] [PubMed] [Google Scholar]
- 4. Becker C. G., Lieberoth B. C., Morellini F., Feldner J., Becker T., Schachner M. (2004) L1.1 is involved in spinal cord regeneration in adult zebrafish. J. Neurosci. 24, 7837–7842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Becker C. G., Becker T. (2002) Repellent guidance of regenerating optic axons by chondroitin sulfate glycosaminoglycans in zebrafish. J. Neurosci. 22, 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Becker C. G., Becker T. (2008) Adult zebrafish as a model for successful central nervous system regeneration. Restor. Neurol. Neurosci. 26, 71–80 [PubMed] [Google Scholar]
- 7. Bastmeyer M., Beckmann M., Schwab M. E., Stuermer C. A. (1991) Growth of regenerating goldfish axons is inhibited by rat oligodendrocytes and CNS myelin but not but not by goldfish optic nerve tract oligodendrocytelike cells and fish CNS myelin. J. Neurosci. 11, 626–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diekmann H., Klinger M., Oertle T., Heinz D., Pogoda H. M., Schwab M. E., Stuermer C. A. (2005) Analysis of the reticulon gene family demonstrates the absence of the neurite growth inhibitor Nogo-A in fish. Mol. Biol. Evol. 22, 1635–1648 [DOI] [PubMed] [Google Scholar]
- 9. Yazaki T., Miura M., Asou H., Toya S., Uyemura K. (1994) Peripheral myelin P0 protein mediates neurite outgrowth of cortical neurons in vitro and axonal regeneration in vivo. Neurosci. Lett. 176, 13–16 [DOI] [PubMed] [Google Scholar]
- 10. Schweitzer J., Becker T., Becker C. G., Schachner M. (2003) Expression of protein zero is increased in lesioned axon pathways in the central nervous system of adult zebrafish. Glia 41, 301–317 [DOI] [PubMed] [Google Scholar]
- 11. Schweitzer J., Gimnopoulos D., Lieberoth B. C., Pogoda H. M., Feldner J., Ebert A., Schachner M., Becker T., Becker C. G. (2007) Contactin1a expression is associated with oligodendrocyte differentiation and axonal regeneration in the central nervous system of zebrafish. Mol. Cell Neurosci. 35, 194–207 [DOI] [PubMed] [Google Scholar]
- 12. Bai Q., Garver J. A., Hukriede N. A., Burton E. A. (2007) Generation of a transgenic zebrafish model of Tauopathy using a novel promoter element derived from the zebrafish eno2 gene. Nucleic Acids Res. 35, 6501–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee E. C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D. A., Court D. L., Jenkins N. A., Copeland N. G. (2001) A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65 [DOI] [PubMed] [Google Scholar]
- 14. Milanese C., Sager J. J., Bai Q., Farrell T. C., Cannon J. R., Greenamyre J. T., Burton E. A. (2012) Hypokinesia and reduced dopamine levels in zebrafish lacking β- and γ1-synucleins. J. Biol. Chem. 287, 2971–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bai Q., Burton E. A. (2009) Cis-acting elements responsible for dopaminergic neuron-specific expression of zebrafish slc6a3 (dopamine transporter) in vivo are located remote from the transcriptional start site. Neuroscience 164, 1138–1151 [DOI] [PubMed] [Google Scholar]
- 16. Bai Q., Mullett S. J., Garver J. A., Hinkle D. A., Burton E. A. (2006) Zebrafish DJ-1 is evolutionarily conserved and expressed in dopaminergic neurons. Brain Res. 1113, 33–44 [DOI] [PubMed] [Google Scholar]
- 17. Bai Q., Sun M., Stolz D. B., Burton E. A. (2011) Major isoform of zebrafish P0 is a 23.5 kDa myelin glycoprotein expressed in selected white matter tracts of the central nervous system. J. Comp. Neurol. 519, 1580–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park H. C., Boyce J., Shin J., Appel B. (2005) Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J. Neurosci. 25, 6836–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brösamle C., Halpern M. E. (2002) Characterization of myelination in the developing zebrafish. Glia 39, 47–57 [DOI] [PubMed] [Google Scholar]
- 21. Thisse C., Thisse B. (2004) Fast release clones: a high throughput expression analysis. ZFIN direct data submission [Google Scholar]
- 22. Bai Q., Wei X., Burton E. A. (2009) Expression of a 12-kb promoter element derived from the zebrafish enolase-2 gene in the zebrafish visual system. Neurosci. Lett. 449, 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park H. C., Mehta A., Richardson J. S., Appel B. (2002) olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev. Biol. 248, 356–368 [DOI] [PubMed] [Google Scholar]
- 24. März M., Schmidt R., Rastegar S., Strähle U. (2010) Expression of the transcription factor Olig2 in proliferating cells in the adult zebrafish telencephalon. Dev. Dyn. 239, 3336–3349 [DOI] [PubMed] [Google Scholar]
- 25. Chung A. Y., Kim P. S., Kim S., Kim E., Kim D., Jeong I., Kim H. K., Ryu J. H., Kim C. H., Choi J., Seo J. H., Park H. C. (2013) Generation of demyelination models by targeted ablation of oligodendrocytes in the zebrafish CNS. Mol. Cells 36, 82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shin J., Park H. C., Topczewska J. M., Mawdsley D. J., Appel B. (2003) Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 25, 7–14 [DOI] [PubMed] [Google Scholar]
- 27. Yoshida M., Macklin W. B. (2005) Oligodendrocyte development and myelination in GFP-transgenic zebrafish. J. Neurosci. Res. 81, 1–8 [DOI] [PubMed] [Google Scholar]
- 28. Carney T. J., Dutton K. A., Greenhill E., Delfino-Machín M., Dufourcq P., Blader P., Kelsh R. N. (2006) A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development 133, 4619–4630 [DOI] [PubMed] [Google Scholar]
- 29. Münzel E. J., Schaefer K., Obirei B., Kremmer E., Burton E. A., Kuscha V., Becker C. G., Brösamle C., Williams A., Becker T. (2012) Claudin K is specifically expressed in cells that form myelin during development of the nervous system and regeneration of the optic nerve in adult zebrafish. Glia 60, 253–270 [DOI] [PubMed] [Google Scholar]
- 30. Senut M. C., Gulati-Leekha A., Goldman D. (2004) An element in the α1-tubulin promoter is necessary for retinal expression during optic nerve regeneration but not after eye injury in the adult zebrafish. J. Neurosci. 24, 7663–7673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goldman D., Hankin M., Li Z., Dai X., Ding J. (2001) Transgenic zebrafish for studying nervous system development and regeneration. Transgenic Res. 10, 21–33 [DOI] [PubMed] [Google Scholar]
- 32. Goldman D., Ding J. (2000) Different regulatory elements are necessary for α1 tubulin induction during CNS development and regeneration. Neuroreport 11, 3859–3863 [DOI] [PubMed] [Google Scholar]
- 33. Udvadia A. J. (2008) 3.6 kb genomic sequence from Takifugu capable of promoting axon growth-associated gene expression in developing and regenerating zebrafish neurons. Gene Expr. Patterns 8, 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kusik B. W., Hammond D. R., Udvadia A. J. (2010) Transcriptional regulatory regions of gap43 needed in developing and regenerating retinal ganglion cells. Dev. Dyn. 239, 482–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ankerhold R., Stuermer C. A. (1999) Fate of oligodendrocytes during retinal axon degeneration and regeneration in the goldfish visual pathway. J. Neurobiol. 41, 572–584 [PubMed] [Google Scholar]
- 36. Hughes E. G., Kang S. H., Fukaya M., Bergles D. E. (2013) Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat. Neurosci. 16, 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. März M., Schmidt R., Rastegar S., Strähle U. (2011) Regenerative response following stab injury in the adult zebrafish telencephalon. Dev. Dyn. 240, 2221–2231 [DOI] [PubMed] [Google Scholar]